Abstract
The vacuolar ATPase (V‐ATPase; V1Vo‐ATPase) is a large multisubunit proton pump found in the endomembrane system of all eukaryotic cells where it acidifies the lumen of subcellular organelles including lysosomes, endosomes, the Golgi apparatus, and clathrin‐coated vesicles. V‐ATPase function is essential for pH and ion homeostasis, protein trafficking, endocytosis, mechanistic target of rapamycin (mTOR), and Notch signaling, as well as hormone secretion and neurotransmitter release. V‐ATPase can also be found in the plasma membrane of polarized animal cells where its proton pumping function is involved in bone remodeling, urine acidification, and sperm maturation. Aberrant (hypo or hyper) activity has been associated with numerous human diseases and the V‐ATPase has therefore been recognized as a potential drug target. Recent progress with moderate to high‐resolution structure determination by cryo electron microscopy and X‐ray crystallography together with sophisticated single‐molecule and biochemical experiments have provided a detailed picture of the structure and unique mode of regulation of the V‐ATPase. This review summarizes the recent advances, focusing on the structural and biophysical aspects of the field.
Keywords: vacuolar ATPase, V‐ATPase, V1Vo‐ATPase, rotary motor enzyme, rotary catalysis, protein structure, reversible disassembly, X‐ray crystallography, cryo electron microscopy, protein–protein interactions
Introduction
The vacuolar H+‐ATPase (V‐ATPase, V1Vo‐ATPase) is a ubiquitous multisubunit enzyme complex that acidifies subcellular compartments in all eukaryotic cells and the extracellular space in some specialized tissues of higher organisms.1, 2, 3, 4 V‐ATPase's proton pumping activity plays a vital role in numerous essential cellular processes such as pH and ion homeostasis, protein trafficking, autophagy, endocytosis, mTOR, and Notch signaling, as well as bone remodeling, urine acidification, hormone secretion, and neurotransmitter release.5, 6, 7, 8, 9 While complete loss of V‐ATPase function is embryonic lethal,10 aberrant (hypo‐ or hyper‐) activity is associated with a wide spectrum of human diseases including renal tubular acidosis,11 sensorineural deafness,12 osteoporosis,13 diabetes,8 microbial14, 15 and viral infection,16 male infertility,17 cancer,18 and AIDS.19 Because of its involvement in multiple widespread diseases, V‐ATPase is a potential drug target.20, 21, 22, 23 However, identifying and optimizing drugs that are designed to modulate V‐ATPase activity rather than causing complete inhibition will require a detailed understanding of the enzyme's structure and its catalytic and regulatory mechanisms.
V‐ATPase is made up of ∼14 unique subunits that are present in different stoichiometries, resulting in ∼30 polypeptides with a total relative molar mass of close to one million. The complex can be divided into two functional sectors, a water soluble ATP hydrolyzing catalytic V1 made of subunits A‐H, and a membrane embedded Vo proton channel composed of subunits a,d,e,f, and two to three “proteolipid” isoforms referred to as subunits c, (c′), and c″ [Fig. 1(A)]. While V‐ATPases from higher organisms can contain additional components such as Ac45 (ATP6AP1) or (pro)renin receptor (ATP6AP2), the core V‐ATPase complex is highly conserved from simple eukaryotes (such as baker's yeast) to higher animals, including human. Most of the subunits in higher organisms are expressed as multiple isoforms, some of which are organelle or tissue specific.24 However, in yeast only subunit a is expressed as two isoforms, with isoform Vph1p found in the vacuole and Stv1p in the Golgi.25 The well‐defined subunit composition, together with the ease of genetic manipulation, has made yeast a powerful model system for structural and mechanistic studies of intact V‐ATPase and its V1 and Vo sectors isolated from the native source. The composition of the V‐ATPase from the yeast Saccharomyces cerevisiae is A3B3(C)DE3FG3H for the V1 and a,c 8,c′,c″,d,e,f for the Vo [Fig. 1(A)]. Subunit f was only very recently found associated with yeast Vo,26 but since deletion of the corresponding gene did not produce the well characterized deletion phenotype,27 its designation as a V‐ATPase subunit requires further study.
Figure 1.
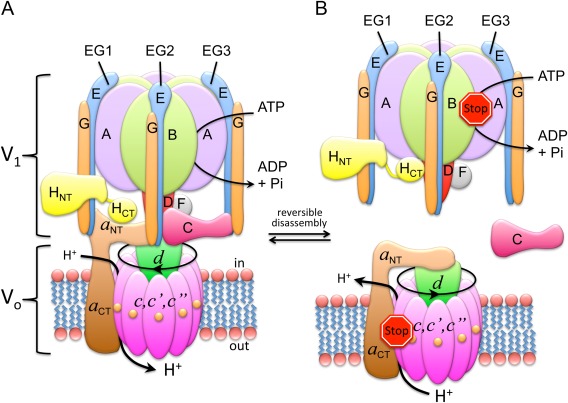
V‐ATPase subunit arrangement and regulation by reversible dissociation. (A) Subunit architecture of yeast V‐ATPase. (B) Regulation by reversible dissociation. In yeast, under conditions of starvation, V1‐ATPase disengages from the membrane bound Vo and the activity of both sectors is silenced. Activity silencing is accompanied by a large conformational change in V1's H subunit (HCT) and the N‐terminal domain of the Vo a subunit (a NT). Illustration adapted from Ref. 73.
V‐ATPase is a molecular motor enzyme, employing a rotary mechanism of energy coupling that is shared with the related F‐, A‐, and A/V‐type ATPases/ATP synthases.28, 29, 30 In the V‐ATPase, stepwise rotation of a central rotor domain made of V1 subunits DF and Vo subunits d and c couples the energy of ATP hydrolysis released from three catalytic sites on V1 to the uphill transport of protons across the lipid bilayer. However, unlike the related rotary motor ATPases, V‐ATPase activity in vivo is regulated via a unique mechanism referred to as “reversible dissociation” (or disassembly), a process that results in a transient pool of free cytosolic V1 and membrane integral Vo sectors that are functionally silenced [Fig. 1(B)]. V‐ATPase regulation by reversible dissociation was initially discovered in yeast31 and insect32 but more recent studies show that the process is conserved in higher animals including human.6, 33, 34, 35
This short review summarizes recent insights into the V‐ATPase's unique mode of regulation by reversible disassembly, focusing on the structural aspects of the process. Considering recent progress with structure determination of the autoinhibited V1 and Vo sectors from yeast, a model of the molecular mechanism of V‐ATPase disassembly is proposed. The review is concluded by a summary of some of the unresolved questions in the field.
V‐ATPase Structure
Throughout this review, V‐ATPase will refer to the eukaryotic enzyme unless stated otherwise. Where appropriate, the eukaryotic V‐ATPase from the yeast S. cerevisiae and its functional sectors will be designated by the prefix “Sc” as in ScV1Vo, ScV1, and ScVo. In analogy, the bacterial enzyme (from e.g. Enterococcus hirae) will be written as EhV1Vo, EhV1, and EhVo. By convention, subunits of the catalytic and membrane sectors will be in capital letters (ABCDEFGH) and lower case italics (acc'c“def”), respectively.
The structures of the holo V‐ATPase and its two separated sectors have been studied extensively using negative stain and cryo electron microscopy (cryoEM) together with single‐particle image averaging and three‐dimensional (3‐D) volume reconstruction. Early on, tagging of the enzyme with subunit specific probes such as monoclonal antibodies helped to elucidate the overall subunit arrangement and stoichiometry. At the same time, crystal structures of individual subunits or subunit complexes of yeast V‐ATPase (subunits H, C, and EG), and subcomplexes from related bacterial enzymes such as the sodium pumping V‐like ATPase from E. hirae (EhV1Vo) and the A‐ATPase from Thermus thermophilus (TtA1Ao) were becoming available. Fitting of the crystal structures into the low to medium‐resolution EM maps allowed building of partial and eventually complete pseudo atomic models of the V‐ATPase and its V1 and Vo sectors isolated from various sources including bovine brain,36, 37, 38 insect midgut,39 and yeast vacuoles.40, 41, 42, 43 The early models not only confirmed the V‐ATPase's structural similarity to the related F‐ATP synthases, which was already evident from primary sequence analysis of the catalytic and proteolipid subunits,44 but the EM models also revealed the increased complexity of the V‐ATPase compared to its ATP synthase counterparts. Recently, advances in electron detector technology have allowed determination of cryoEM maps of insect and yeast V‐ATPase at subnanometer resolution.45, 46 These maps revealed secondary structure elements in both the soluble V1 and, for the first time, in the membrane embedded Vo. Together with the available crystal structures, the density maps allowed building of pseudo atomic models of the complex in different catalytic states.
Overall architecture of holo V‐ATPase
The A and B subunits of V1 come together in an alternating fashion to form a “catalytic hexamer” (A3B3) with catalytic nucleotide binding sites located at three of the six AB interfaces47 [Fig. 2(A)]. Located within the hexamer and protruding ∼30 Å towards the membrane is part of the central rotor that is comprised by subunits D and F, which provide the functional link between V1 and Vo 48 [Fig. 2(B)]. Structural support between ATPase and proton channel is provided by the “peripheral stator”, which consists of three heterodimers formed by subunits E and G (“peripheral stalks” EG1‐3) [Fig. 2(C,D)], the single copy subunits H and C [Fig. 2(E,F)], and subunit a [Fig. 2(G); only the N‐terminal domain of subunit a (a NT) is shown]. The three EG heterodimers link the top of the catalytic hexamer to V1 subunits H and C and the membrane integral a subunit of the Vo [Fig. 1(A)]. The yeast EG heterodimer in complex with the head domain of subunit C has been crystallized in two conformations49 [Fig. 2(C,D)]. The structures show EG to be folded in an unusual right‐handed coiled‐coil (formed by the subunits' N‐termini) and a C‐terminal globular domain that provides the binding energy to the A3B3 hexamer. The H and C subunits are two domain polypeptides with N‐ and C‐terminal domains for subunit H (HNT and HCT)50 [Fig. 2(E)] and foot and head domains for subunit C (Cfoot and Chead)51 [Fig. 2(F)]. Subunit a of the Vo can be divided into a cytosolic N‐terminal (a NT) and a membrane‐integral C‐terminal domain (a CT). The a NT X‐ray crystal structure of the related A‐ATPase from M. ruber showed the protein to be folded in two domains designated as proximal and distal lobes (a NT(prox) and a NT(dist))52 [Fig. 2(G)]. a NT provides the scaffold to connect the peripheral stator components H, C, and EG1‐3 to the membrane.
Figure 2.
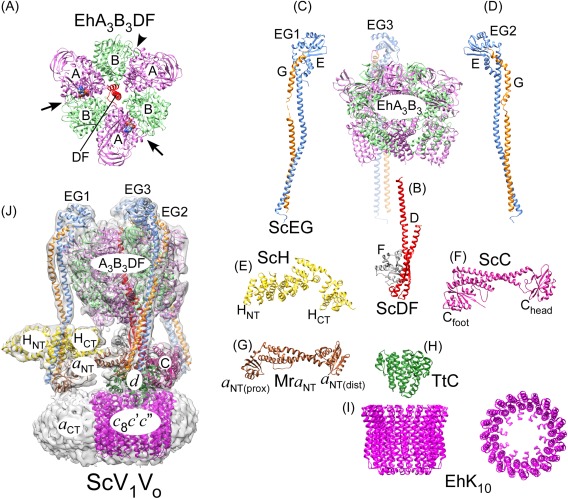
Overall subunit architecture of holo V‐ATPase. (A) Crystal structure of EhA3B3DF (3vr6; a side‐view of the structure is also shown on the right).47 The arrows indicate closed catalytic sites with AMPPNP bound and the arrowhead points to the open site. (B) Crystal structure of ScDF (4rnd).48 (C,D) Crystal structures of ScEGChead in two conformations (4dl0, 4efa; Chead not shown).49 (E) Crystal structure of ScH (1ho8).50 (F) Crystal structure of ScC (1u7l).51 (G) Crystal structure of the a NT homolog from M. ruber (Mra; 3rrk).52 (H) Crystal structure of the d homolog from T. thermophilus (TtC; 1r5z).103 (I) Crystal structure of the proteolipid ring from E. hirae (EhK10; 2bl2)53. (J) CryoEM map of ScV1Vo (emd‐6284)45 with fitted coordinate models of individual subunits and subcomplexes (3j9t).45 The overall model was generated from individual crystal structures of yeast subunits and homology models generated by threading yeast primary sequences into crystal structures of bacterial A‐ATPase subunits.
The ATPase is functionally coupled to the proton channel via binding of the central DF rotor to subunit d. A homolog of subunit d from T. thermophilus A‐ATPase (TtC) has been crystallized [Fig. 2(H)] and is an all α helical protein that links the central DF rotor to the cytoplasmic loops of the c subunits (“proteolipids”). The c subunits are arranged in a ring (referred to as proteolipid ring or c‐ring) as shown for the crystal structure of the related EhK10 53 [Fig. 2(I)]. V‐ATPase proteolipids contain four transmembrane α helices (TMH) except isoform c″, which has an additional α helix at its N‐terminus that is not essential for function.54 From sequence analysis it has been proposed that V‐ATPase four TMH proteolipids have originated by gene duplication from ancestral two TMH c subunits found in F‐ and A‐type motors.55 As of today, there is no high‐resolution crystal structure of eukaryotic holo V‐ATPase or of any of its bacterial or archaeal counterparts. The highest resolution information currently available for the intact enzyme has been obtained by cryoEM of the yeast V‐ATPase.45 Three‐dimensional reconstruction and classification of a large dataset of individual molecular projections allowed generation of maps of the enzyme in three distinct rotational states, which are distinguished by the three rotational positions of the DFd rotor within the catalytic A3B3 hexamer. The analysis showed “state 1” to be most populated, resulting in a map at 6.9 Å resolution [Fig. 2(J)]. To generate a pseudo atomic model of holo V‐ATPase [Fig. 2(J)], the individual crystal structures listed above [Fig. 2(A–I)] were placed into the map, and the individual subunits' structures were flexibly fit guided by the cryoEM map density. Note that there is no high‐resolution structure available for the membrane embedded C‐terminal domain of subunit a as part of holo V‐ATPase (but see below for the recent 3.9 Å cryoEM map of autoinhibited ScVo).
The V‐ATPase in animal cells
The comparison of the early low‐resolution EM studies of bovine brain V‐ATPase56 with images of the yeast enzyme57, 58 showed that the basic architecture of the V‐ATPase complex is conserved from lower eukaryotes to mammals. As mentioned above, in yeast only subunit a is expressed as two organelle specific isoforms, Vph1p and Stv1p.25 Although subunits c, c′, and c″ are sometimes called proteolipid isoforms, all three variants of the c subunits are essential for enzyme function in yeast59 (as of today, c′ has not been identified in higher animals). In the animal V‐ATPase, on the other hand, most of the subunits are expressed as multiple isoforms, some of which are organelle or tissue specific.24 Most notable are the four isoforms of subunit a with isoform a1 found in neurons, a 2 in endothelia, a 3 in osteoclast ruffled membrane and pancreatic β cells, and a 4 in the kidney. However, isoform distribution is not strictly defined, making it very challenging, if not impossible, to isolate and purify animal V‐ATPases with a homogeneous isoform composition.
V‐ATPase Mechanism
The V‐ATPase, like all members of the rotary ATPase family, contains two motors: a V1 motor that converts the chemical energy of ATP hydrolysis into mechanical energy (torque of central DF rotor), and a Vo proton turbine that converts mechanical energy into the potential energy store of an ion gradient28, 29, 30 [Fig. 3(A)]. Conformational changes powered by ATP hydrolysis at the three catalytic sites on the A3B3 hexamer drive rotation of the central DF rotor [Fig. 3(B)], which in turn, via its connection to subunit d, rotates the c‐ring past the membrane integral a CT for proton translocation across the lipid bilayer [Fig. 3(C)]. During catalysis, the three peripheral stalks (EG1‐3), in conjunction with subunits C, H, and a NT, function to resist the torque of rotation, thus keeping the A3B3 hexamer static against the proton channel to allow for the rotation to be productive [Fig. 3(A)].
Figure 3.
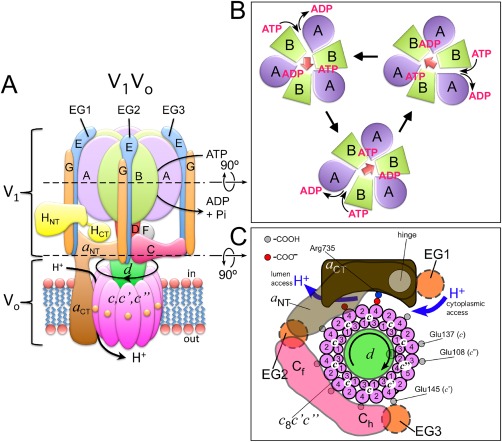
V‐ATPase mechanism. (A) V‐ATPase consists of two motors, a V1‐ATPase and a Vo proton channel. (B) ATP hydrolysis at three of the six AB interfaces drives counterclockwise rotation of the central DF rotor (A3B3 seen towards the cytosol). (C) Rotation of the central DF rotor drives rotation of the proteolipid ring past the essential arginine residue in a CT. Protons enter a cytoplasmic half‐channel and, after being carried 360º on lipid exposed glutamate residues on the c subunits, are released into the lumenal half‐channel.
From biochemical and biophysical work on related bacterial enzymes, we have some understanding of the molecular mechanism of ATP hydrolysis and the concomitant conformational changes on the A3B3 catalytic hexamer that drive rotation of the central DF rotor. For each ATP hydrolyzed, the central rotor rotates 120º counter‐clockwise (when looking towards the cytoplasm), driven by conformational changes that occur in the A3B3 hexamer upon ATP binding, hydrolysis, and product release. Interestingly, while in F‐ATP synthase each 120º rotation occurs in substeps of ∼80º (ATP binding/release) and 40º (ADP/Pi binding/release),60 no substeps have been observed for bacterial V‐like ATPase from, for example T. thermophilus 61 or E. hirae 62 as of today.
As previously mentioned, the central DF rotor, via its connection to subunit d, rotates the c‐ring past the membrane integral a CT to pump protons across the lipid bilayer. Each c subunit contains an essential glutamic acid residue mid‐membrane that, after getting protonated in an aqueous half channel accessible from the cytoplasmic side, is able to enter the lipid bilayer and following a close to 360º rotation of the c‐ring, delivers its proton to an aqueous half‐channel accessible from the lumen of the organelle or the outside of the cell [Fig. 3(C)]. A strictly conserved arginine residue in a CT located at the interface of a CT and the c‐ring (Arg735 in yeast63) serves to stabilize the essential glutamic acid residues of the c subunits while in the deprotonated state between lumen and cytoplasmic half channels. Besides the essential Arg735, site‐directed mutagenesis experiments in yeast V‐ATPase have uncovered several charged and polar residues in a CT that are essential for proton pumping.64 How these residues work together to guide the protons from the cytoplasmic to the lumen or cellular exterior, however, is not known, largely because of lack of high‐resolution structural information for the V‐ATPase's (or, for that matter, any of the rotary ATPases') membrane sector, as well as the dynamic nature of the process.
V‐ATPase Regulation
Breaking up…
V‐ATPase hydrolyzes up to ∼300 ATP·(sec−1)65 and as a major consumer of cellular energy1, its MgATPase activity must be tightly controlled. In eukaryotic organisms, the enzyme's proton pumping activity is regulated by reversible dissociation (or disassembly), a process resulting in free, water soluble cytoplasmic ATPase (V1) and membrane bound proton channel (Vo) sectors. In yeast, reversible dissociation is driven by environmental factors such as nutrient availability,31, 32 salinity,66 and extracellular pH.67 In higher animals, reversible disassembly or stimulated assembly (without prior disassembly) occurs in response to nutrients,33, 35 cell maturation,34 hormones,68 and growth factors.6 Upon enzyme dissociation, the activity of both sectors is silenced, resulting in a V1 that has no MgATPase activity69, 70, 71 and a Vo that does not catalyze passive proton translocation.72, 73 From studies in yeast and insect, we have some information as to the molecular mechanism of regulated disassembly. Upon initiating disassembly by, for example glucose withdrawal in yeast, the C subunit dissociates from the vacuolar membrane31 and is not part of either V1 or Vo when these complexes are purified subsequent to enzyme disassembly.40, 69, 70, 73 It is also known that inhibition of ATP hydrolysis by, for example mutation of residues involved in catalysis, nucleotide binding, or proton transport, or by binding of specific drugs that block rotation, results in “hyper‐assembly”,74, 75 which indicates that disassembly occurs while the enzyme is in a high‐energy conformation during rotary catalysis. Similar results were obtained from in vitro experiments conducted with detergent solubilized insect V‐ATPase in which MgATP, but not MgADP or the nonhydrolyzable MgAMPPNP, resulted in the formation of free V1 and Vo sectors.76 Thus, while some aspects of enzyme disassembly are known, there is still no clear understanding of the nature of the signal that initiates the process, though several possibilities have been proposed, including involvement of protein kinase A (either upstream or downstream of V‐ATPase), ATP to ADP ratio, and cytoplasmic pH to name only a few.75, 77, 78, 79 Recent structural models of autoinhibited ScV1 and ScVo summarized in the following paragraphs revealed some of the conformational changes that occur as a result of holo enzyme dissociation.
Structural features of autoinhibited ScV1
As mentioned above, membrane detached V1 isolated from yeast or insect midgut has no measurable MgATPase activity, though the complex can hydrolyze ATP in presence of Ca2+ (given the absence of magnesium ions).40, 69, 70 In yeast, deletion of subunit H allows purification of a V1ΔH complex that displays robust MgATPase activity.70, 71, 80 Interestingly, deletion of subunit H still allowed assembly of the resulting V1ΔH on the membrane, but the resulting complex has no measurable MgATPase or proton pumping activities.81 Biochemical work in yeast showed that it is HCT that is responsible for MgATPase inhibition.80, 82 However, even V1ΔH's ATPase activity is short lived because of trapping of the complex in the so‐called MgADP inhibited state.70, 71 The MgADP inhibited state is an off‐catalytic pathway conformation that is also found in other related rotary ATPase catalytic sectors.83 The role of subunit H in activity silencing has recently been illuminated in the 6.2 Å crystal structure of autoinhibited ScV1 [Fig. 4(A), right panel]. A comparison of the ScV1 crystal structure with the cryoEM models of holo V‐ATPase [Fig. 4(B)] revealed that autoinhibition is accompanied by a large conformational change of the C‐terminal domain of subunit H (HCT) [Fig. 4(A), left panel]. Upon release from the membrane, HCT moves and rotates 150º from its binding site on a NT in ScV1Vo to a position at the bottom of the A3B3 hexamer in autoinhibited ScV1 to contact the central rotor subunit D and the B subunit of the open catalytic site [Fig. 4(A)]. The structural and accompanying biochemical studies also showed that while V1ΔH contained only ∼0.4 moles/mol ADP, autoinhibited ScV1 had ∼1.3 ADP molecules per complex, consistent with ScV1 being in the MgADP inhibited state, a conformation likely stabilized by HCT's interaction with the central rotor and the open catalytic site. Curiously, the loop in HCT that was identified as being responsible for the subunit's ability to inhibit MgATPase activity is not present in higher animals including human71 and it remains to be seen whether mammalian subunit H is inhibitory, or whether autoinhibition of mammalian V1 is solely because of trapping of MgADP.
Figure 4.
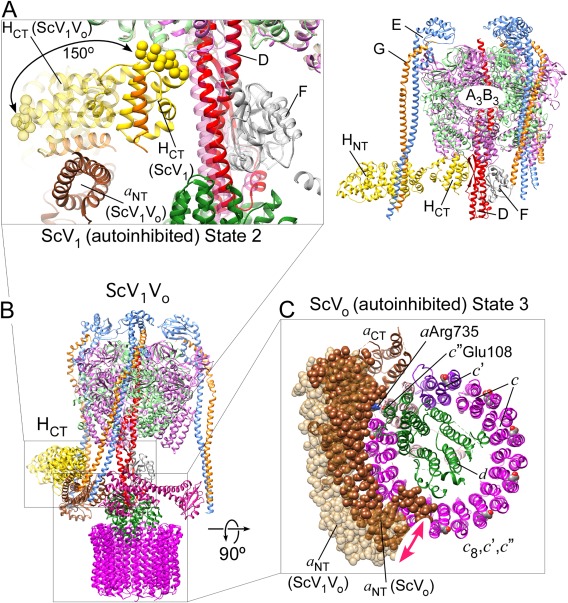
V‐ATPase conformational changes accompanying reversible disassembly. (A) Conformational changes in the auto‐inhibited ScV1‐ATPase (5d80).71 Upon holo enzyme dissociation, the C‐terminus of subunit H (HCT) is released from its binding site on a NT (in V1Vo) and rotates 150º to bind at the bottom of the A3B3 hexamer (in V1). HCT binds to the B subunit of the open catalytic site, thereby stabilizing inhibitory ADP in another site.71 The inhibitory loop is in spacefill and the α helix that binds a NT in V1Vo is in orange. (B) CryoEM model of holo ScV1Vo 45 (3j9u). (C) Conformational changes in the Vo proton channel (5tj5)26. Upon release of V1‐ATPase from the membrane, the N‐terminus of subunit a (a NT) moves from its ternary interface with Cfoot and EG2 to form a new binding interface with subunit d 26, 73, 88 and the c‐ring is positioned so that Glu108 of isoform c″ is in contact with the essential Arg735 in a CT.
Structural features of autoinhibited ScVo
Unlike F‐ATPase Fo membrane sector, which allows passive proton conductance when detached from F1,84, free Vo does not catalyze downhill translocation of protons along an electrochemical gradient.72, 73, 85 The autoinhibition of free Vo allows co‐existence of free V‐ATPase membrane sectors next to assembled holo enzyme complexes as has been observed in yeast and higher animals.75 Early negative stain EM images of bovine Vo showed an interaction of a NT with subunit d, an observation that lead to the proposal that blockage of passive proton transport is because of the interaction between rotor and stator. Subsequent studies in yeast confirmed the a NT–d interaction, however, selective removal of either a NT or subunit d did not relieve the inhibition,73, 86 suggesting that structural features of the c‐ring or a CT may be responsible for the process. Early genetic screens in yeast revealed the presence of multiple proteolipid isoforms referred to as subunit c, c′, and c″. As mentioned above, the c and c′ proteolipid isoforms have each four TMH while the c″ isoform has five. It was noted that while the essential glutamic acid residues in c and c′ are in TM helix 4 (Glu137 and Glu145, respectively), the corresponding residue in c″ is in helix 3 (Glu108).59 This led to the speculation that the resulting asymmetry in the spacing of the proton transporting glutamic acid residues at the periphery of the c‐ring may be the cause of autoinhibition as the large gap in carboxyls at the interface of c″ and c′ might not be able to move past a CT's Arg735, thereby preventing the reversal of the direction of rotation that would be required for passive proton transport.73 Recently, Mazhab‐Jafari et al. reported a cryoEM reconstruction of autoinhibited ScVo, allowing a first glimpse at the interactions between residues of the a CT stator and the c‐ring rotor within the membrane26 [Fig. 4(C)]. As first seen in the related F‐ATP synthase from Polytomella sp.,87 a CT contains two long α helices that cross the lipid bilayer at an angle of 30º with respect to the plane of the membrane and contact the periphery of the c‐ring mid‐membrane. In V‐ATPase, these two α helices contain some of the charged and polar residues identified in the site‐directed mutagenesis experiments mentioned above,64 including the essential Arg735. While the EM model of the autoinhibited ScVo showed a cavity on the cytoplasmic side likely to represent the aqueous access for protons to enter the membrane before they bind the glutamates on the proteolipids, no such cavity was identified on the lumenal side of the complex.26 The cryoEM model of autoinhibited yeast Vo also confirmed the earlier proposal in that Glu108 of c″ was seen to be in contact with Arg73526 [Fig. 4(C)]. However, whether the asymmetry in the glutamic acid spacing is the sole reason for Vo's autoinhibition remains to be seen.
… and making up
The comparison between ScV1 and holo ScV1Vo mentioned above also revealed that subunit H and MgADP facilitated autoinhibition halts ScV1 in state 2 based on the position of the central DF rotor71 [Fig. 4(A,B)]. The corresponding comparison of the ScVo sectors based on the position of the proteolipid ring or the d subunit showed that autoinhibited ScVo is arrested in state 3.26, 88 These observations lead to the proposal that the resulting mismatch between state 2 V1 and state 3 Vo conformations may serve to prevent unintended reassembly of the complex when its proton pumping activity is not required.88 Consequently, before reassembly can take place, the conformational mismatch must be overcome. Bringing V1 and Vo into matching rotational states can either happen by converting Vo from state 3 to state 2 or by converting V1 from state 2 to state 3. Since autoinhibited Vo is arrested in state 3 despite a transmembrane proton gradient, it is more likely that subunit H stabilized MgADP inhibition of V1 is relieved to allow hydrolysis of at least one ATP molecule with concomitant rotation of the central rotor from state 2 to state 3 to match Vo's rotational state. What could be the mechanism for relieving the ADP inhibition? From studies in yeast, it is known that assembly, be it the initial assembly following biosynthesis, or the re‐assembly following reversible disassembly, occurs in absence of turnover but requires presence of the “regulator of H+‐ATPases of vacuolar and endosomal membranes” (RAVE),89, 90 a heterotrimeric chaperone complex that has been shown to promote assembly by binding to the V1, the Vo, and the C subunit.91, 92 Curiously, for reasons that are currently not understood, the RAVE complex seems not to be required for the assembly of Stv1 containing V‐ATPase found in the Golgi.93 Besides the RAVE complex, several enzymes of the glycolytic pathway have been shown to be required for efficient V‐ATPase assembly, most notably aldolase94 and phosphofructokinase‐1.95 However, the mechanism by which RAVE and any of these enzymes promote the assembled state is not known.
The V1–Vo structural interface—a predetermined breaking point
Reversible disassembly requires breaking (and re‐forming) of several protein–protein interactions at the V1–Vo interface [Fig. 5(A)]. The affinities of many of these interactions have been determined with biophysical tools such as isothermal titration calorimetry and optical methods (surface plasmon resonance and biolayer interferometry) using recombinant subunits or subcomplexes of the yeast73, 88, 96, 97 and human98 V‐ATPase. From the interactions that need to be broken during disassembly of the yeast enzyme (EG3‐Chead; EG2‐aNT(dist); EG1‐a NT(prox); Cfoot‐a NT(dist); HCT‐a NT; DF‐d), all have affinities in the micromolar range except EG3‐Chead, which has a K d of ∼42 nM.96 It has been proposed that the junction of a NT with EG2 and Cfoot is stabilized through the high avidity of the multiple weak interactions involved and that breaking one of these weak interactions could destabilize the V1–Vo interface and initiate dissociation of the complex.97 A possible role in this process could be played by EG3. The SAXS solution and X‐ray crystal structures of the ScEGC42 and ScEGChead 49 subcomplexes suggested that EG3 may be under strain in holo V‐ATPase, and it has been proposed that this “spring‐loading” of EG3 that occurs upon assembly of the complex could play a role in the disassembly process by preventing re‐association of Cfoot with a NT(distal) once the interaction is broken at the initiation of the process [Fig. 5(B), blue arrow].
Figure 5.
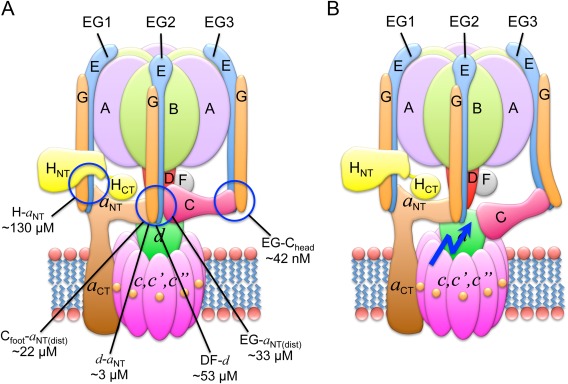
Interactions at the V1–Vo interface. (A) The affinities of the individual interactions have been measured using isothermal titration calorimetry of recombinant yeast V‐ATPase subunits.48, 96, 97 (B) Spring‐loading of EG3. It has been suggested that the tension in EG3 would prevent re‐forming of the EG2–a NT(distal)–Cfoot ternary interface after one of these interactions is broken to initiate disassembly.49
Mechanistic model for V‐ATPase dissociation
From recent structural and biochemical work, we now know what some of the conformational changes are that take place during regulated V‐ATPase disassembly. From the observations that autoinhibited Vo is arrested in rotational state 3, the same state that was the least populated in the cryoEM analysis of the holo enzyme,45 it has been speculated that disassembly occurs when V‐ATPase goes through this state during turnover.26 In our current working model [Fig. 6, Supporting Information Fig. S1], we therefore assume that disassembly starts when holo V‐ATPase is in the catalytic dwell in state 3 [Fig. 6(A)]. Once the (as yet unknown) signal for disassembly is received, the ternary interface EG2–a NT(distal)–Cfoot is broken, allowing a NT to bind to subunit d [Fig. 6(B)]. This in turn breaks the HCT–a NT interaction and frees up HCT. Since ScV1 is found to be arrested in state 2, DF has to rotate two 120º steps at the expense of the hydrolysis of two MgATP molecules [Fig. 6(C, D)]. As Vo remains in state 3, this would require uncoupling of the V1 central DF rotor from the d subunit. Once V1 is in state 2, MgADP inhibition sets in and A3B3DF presents a high‐affinity binding site for HCT [Fig. 6(E)]. Note that subunit C is released during this process, possibly because of the conformational changes in EG3 induced by the two steps of ATP hydrolysis. Since most of the weak interactions are now broken, the complex is destabilized to a point so that disassembly could occur [Fig. 6(F)]. It should be pointed out that the timing of the structural changes, a NT binding to d and HCT binding to A3B3DF [Fig. 6(G, H)] has not been firmly established. Furthermore, whether the final dissociation step [Fig. 6(F)] occurs right away or whether the weak affinity of some of the residual interactions keeps the V1 transiently attached to the membrane in vivo has been debated based on in vivo fluorescence and FRET imaging.99
Figure 6.
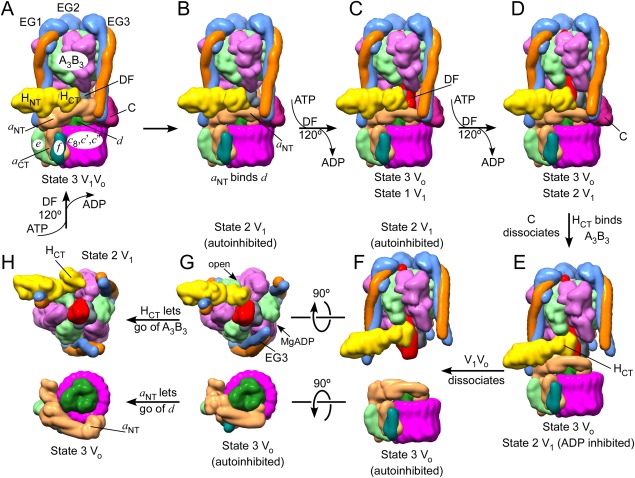
Model of the structural changes along the steps of holo V‐ATPase disassembly. (A) It has been proposed that disassembly is initiated in state 3 V1Vo. 26 (B) The ternary a NT–EG2–Cfoot interaction is destabilized, allowing a NT to bind subunit d. (C,D) Two uncoupled catalytic steps convert V1 into the state 2 conformation. (E) Subunit C dissociates and HCT rotates 150º to bind at the open catalytic site. V1 is now in state 2 and autoinhibited and Vo is in state 3, also autoinhibited. (F) V1Vo dissociates. (G) Views towards to bottom of autoinhibited V1, illustrating HCT's binding site on A3B3DF, and towards the top of Vo, showing the interaction of a NT with subunit d. (H) Reassembly must involve release of HCT and a NT from their autoinhibitory positions and one catalytic turnover of V1 to return both sectors into matching state 3 conformations. Note that the starting conformation of V1Vo and the order of the steps are not firmly established. See also Supporting Information Fig. S1 for an animation of the process.
Conclusions and the “To‐Do” List
While our understanding of the structure and regulatory mechanism of the vacuolar ATPase is better than ever before, more information leads to new questions and there is still much work to be done. Most pressing is the need for high‐resolution structural information for the active holo V‐ATPase and its autoinhibited V1 and Vo sectors, information that will be essential for our understanding of the proton‐pumping and regulatory mechanisms of the enzyme. However, V‐ATPase is a dynamic nano‐motor that races through many conformations during its catalytic and regulatory cycle, and structural information alone will not suffice to uncover V‐ATPase's many remaining secrets.
Currently, the highest resolution information available is for the autoinhibited Vo from yeast. However, whether the interactions between a CT and the c‐ring seen in Vo are the same as in state 3 holo V1Vo, and how the ring interacts with a CT's essential arginine in the remaining states 1 and 2 remains to be seen. Also unresolved is the question whether Vo autoinhibition is caused solely by the asymmetric spacing of the proton carrying glutamates in the c‐ring, or whether the conformational change in a NT upon enzyme disassembly is transmitted to a CT to help block the proton channel?
Static structures must be complemented with functional studies to resolve remaining mechanistic questions, including, how does the proteolipid ring interact with residues in a CT during rotation, and how do these interactions result in net transport of protons? Does the asymmetry in the angular steps of the proton carrying glutamates in the c‐ring serve primarily for autoinhibition of free Vo, or does the asymmetry also function to inhibit reverse rotation in holo V1Vo? What is the molecular signal for enzyme dissociation, and how does this signal cause breaking of protein–protein interactions at the V1–Vo interface, in particular the high‐affinity interaction between subunit C and peripheral stalk EG3? Where does subunit C go upon release from V1 and Vo, and does the binding and/or release of C involve post‐translational modifications? What is the order of the functional events (e.g. MgADP inhibition) and structural changes (movements of HCT and EG3 in V1 and a NT in Vo) that occur during enzyme dissociation? Is MgADP inhibition a prerequisite for enzyme disassembly? Conversely, what is the signal (or signals) that lead to reassembly, and what role do assembly chaperones such as the RAVE complex play in the process? Do the assembly chaperones merely serve to bring the components back together, or does the RAVE complex also function to overcome the conformational mismatch between state 2 V1 and state 3 Vo?
Furthermore, there is mounting evidence that Vo, possibly via its proteolipid ring, participates in cellular functions other than proton pumping such as membrane fusion and neuronal communication.100, 101, 102 However, the significance of these so‐called “noncanonical” functions of the free Vo are only beginning to emerge. Finally, aberrant V‐ATPase function is involved in many widespread human diseases. Therefore, the ultimate question is how we can apply our present knowledge and insights from future structural and functional studies to aid in the design of therapeutics to fight against human disease, with one attractive possibility being the targeting of reversible disassembly of the human V‐ATPase in an isoform and tissue specific manner.
Conflict of Interest
The authors declare that they have no conflicts of interest with the contents of this article.
Supporting information
Supporting Information Movie 1.
Footnotes
Specific activities of up to 18 µmoles·min–1·mg–1 have been reported for the enzyme from, for example yeast,65 corresponding to the hydrolysis of ∼300 ATP·sec–1.
References
- 1. Forgac M (2007) Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat Rev Mol Cell Biol 8:917–929. [DOI] [PubMed] [Google Scholar]
- 2. Kane PM (2006) The where, when, and how of organelle acidification by the yeast vacuolar H+‐ATPase. Microbiol Mol Biol Rev 70:177–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Marshansky V, Futai M (2008) The V‐type H+‐ATPase in vesicular trafficking: targeting, regulation and function. Curr Opin Cell Biol 20:415–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Graham LA, Flannery AR, Stevens TH (2003) Structure and assembly of the yeast V‐ATPase. J Bioenerg Biomembr 35:301–312. [DOI] [PubMed] [Google Scholar]
- 5. Zoncu R, Bar‐Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM (2011) mTORC1 senses lysosomal amino acids through an inside‐out mechanism that requires the vacuolar H(+)‐ATPase. Science 334:678–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Xu Y, Parmar A, Roux E, Balbis A, Dumas V, Chevalier S, Posner BI (2012) Epidermal growth factor‐induced vacuolar (H+)‐atpase assembly: a role in signaling via mTORC1 activation. J Biol Chem 287:26409–26422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yan Y, Denef N, Schupbach T (2009) The vacuolar proton pump, V‐ATPase, is required for notch signaling and endosomal trafficking in Drosophila. Dev Cell 17:387–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sun‐Wada GH, Toyomura T, Murata Y, Yamamoto A, Futai M, Wada Y (2006) The a3 isoform of V‐ATPase regulates insulin secretion from pancreatic beta‐cells. J Cell Sci 119:4531–4540. [DOI] [PubMed] [Google Scholar]
- 9. Vavassori S, Mayer A (2014) A new life for an old pump: V‐ATPase and neurotransmitter release. J Cell Biol 205:7–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Inoue H, Noumi T, Nagata M, Murakami H, Kanazawa H (1999) Targeted disruption of the gene encoding the proteolipid subunit of mouse vacuolar H(+)‐ATPase leads to early embryonic lethality. Biochim Biophys Acta 1413:130–138. [DOI] [PubMed] [Google Scholar]
- 11. Smith AN, Skaug J, Choate KA, Nayir A, Bakkaloglu A, Ozen S, Hulton SA, Sanjad SA, Al‐Sabban EA, Lifton RP, Scherer SW, Karet FE (2000) Mutations in ATP6N1B, encoding a new kidney vacuolar proton pump 116‐kD subunit, cause recessive distal renal tubular acidosis with preserved hearing. Nat Genet 26:71–75. [DOI] [PubMed] [Google Scholar]
- 12. Karet FE, Finberg KE, Nelson RD, Nayir A, Mocan H, Sanjad SA, Rodriguez‐Soriano J, Santos F, Cremers CW, Di Pietro A, Hoffbrand BI, Winiarski J, Bakkaloglu A, Ozen S, Dusunsel R, Goodyer P, Hulton SA, Wu DK, Skvorak AB, Morton CC, Cunningham MJ, Jha V, Lifton RP (1999) Mutations in the gene encoding B1 subunit of H+‐ATPase cause renal tubular acidosis with sensorineural deafness. Nat Genet 21:84–90. [DOI] [PubMed] [Google Scholar]
- 13. Thudium CS, Jensen VK, Karsdal MA, Henriksen K (2012) Disruption of the V‐ATPase functionality as a way to uncouple bone formation and resorption—a novel target for treatment of osteoporosis. Curr Prot Pept Sci 13:141–151. [DOI] [PubMed] [Google Scholar]
- 14. Wong D, Bach H, Sun J, Hmama Z, Av‐Gay Y (2011) Mycobacterium tuberculosis protein tyrosine phosphatase (PtpA) excludes host vacuolar‐H+‐ATPase to inhibit phagosome acidification. Proc Natl Acad Sci USA 108:19371–19376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xu L, Shen X, Bryan A, Banga S, Swanson MS, Luo ZQ (2010) Inhibition of host vacuolar H+‐ATPase activity by a Legionella pneumophila effector. PLoS Pathog 6:e1000822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lu X, Yu H, Liu SH, Brodsky FM, Peterlin BM (1998) Interactions between HIV1 Nef and vacuolar ATPase facilitate the internalization of CD4. Immunity 8:647–656. [DOI] [PubMed] [Google Scholar]
- 17. Brown D, Smith PJ, Breton S (1997) Role of V‐ATPase‐rich cells in acidification of the male reproductive tract. J Exp Biol 200:257–262. [DOI] [PubMed] [Google Scholar]
- 18. Sennoune SR, Bakunts K, Martinez GM, Chua‐Tuan JL, Kebir Y, Attaya MN, Martinez‐Zaguilan R (2004) Vacuolar H+‐ATPase in human breast cancer cells with distinct metastatic potential: distribution and functional activity. Am J Physiol Cell Physiol 286:C1443–C1452. [DOI] [PubMed] [Google Scholar]
- 19. Geyer M, Yu H, Mandic R, Linnemann T, Zheng YH, Fackler OT, Peterlin BM (2002) Subunit H of the V‐ATPase binds to the medium chain of adaptor protein complex 2 and connects Nef to the endocytic machinery. J Biol Chem 277:28521–28529. [DOI] [PubMed] [Google Scholar]
- 20. Kane PM (2012) Targeting reversible disassembly as a mechanism of controlling V‐ATPase activity. Curr Prot Pept Sci 13:117–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kartner N, Manolson MF (2014) Novel techniques in the development of osteoporosis drug therapy: the osteoclast ruffled‐border vacuolar H+‐ATPase as an emerging target. Expert Opin Drug Discov 9:505–522. [DOI] [PubMed] [Google Scholar]
- 22. Spugnini EP, Citro G, Fais S (2010) Proton pump inhibitors as anti vacuolar‐ATPases drugs: a novel anticancer strategy. J Exp Clin Cancer Res 29:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bowman EJ, Bowman BJ (2005) V‐ATPases as drug targets. J Bioenerg Biomembr 37:431–435. [DOI] [PubMed] [Google Scholar]
- 24. Toei M, Saum R, Forgac M (2010) Regulation and isoform function of the V‐ATPases. Biochemistry 49:4715–4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Manolson MF, Wu B, Proteau D, Taillon BE, Roberts BT, Hoyt MA, Jones EW (1994) STV1 gene encodes functional homologue of 95‐kDa yeast vacuolar H(+)‐ATPase subunit Vph1p. J Biol Chem 269:14064–14074. [PubMed] [Google Scholar]
- 26. Mazhab‐Jafari MT, Rohou A, Schmidt C, Bueler SA, Benlekbir S, Robinson CV, Rubinstein JL (2016) Atomic model for the membrane‐embedded VO motor of a eukaryotic V‐ATPase. Nature 539:118–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nelson H, Nelson N (1990) Disruption of genes encoding subunits of yeast vacuolar H(+)‐ATPase causes conditional lethality. Proc Natl Acad Sci USA 87:3503–3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wilkens S (2005) Rotary molecular motors. Adv Prot Chem 71:345–382. [DOI] [PubMed] [Google Scholar]
- 29. Muench SP, Trinick J, Harrison MA (2011) Structural divergence of the rotary ATPases. Q Rev Biophys 44:311–356. [DOI] [PubMed] [Google Scholar]
- 30. Futai M, Nakanishi‐Matsui M, Okamoto H, Sekiya M, Nakamoto RK (2012) Rotational catalysis in proton pumping ATPases: from E. coli F‐ATPase to mammalian V‐ATPase. Biochim Biophys Acta 1817:1711–1721. [DOI] [PubMed] [Google Scholar]
- 31. Kane PM (1995) Disassembly and reassembly of the yeast vacuolar H(+)‐ATPase in vivo . J Biol Chem 270:17025–17032. [PubMed] [Google Scholar]
- 32. Sumner JP, Dow JA, Earley FG, Klein U, Jager D, Wieczorek H (1995) Regulation of plasma membrane V‐ATPase activity by dissociation of peripheral subunits. J Biol Chem 270:5649–5653. [DOI] [PubMed] [Google Scholar]
- 33. Sautin YY, Lu M, Gaugler A, Zhang L, Gluck SL (2005) Phosphatidylinositol 3‐kinase‐mediated effects of glucose on vacuolar H+‐ATPase assembly, translocation, and acidification of intracellular compartments in renal epithelial cells. Mol Cell Biol 25:575–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Trombetta ES, Ebersold M, Garrett W, Pypaert M, Mellman I (2003) Activation of lysosomal function during dendritic cell maturation. Science 299:1400–1403. [DOI] [PubMed] [Google Scholar]
- 35. Stransky LA, Forgac M (2015) Amino acid availability modulates vacuolar H+‐ATPase assembly. J Biol Chem; 290:27360–27369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wilkens S, Forgac M (2001) Three‐dimensional structure of the vacuolar ATPase proton channel by electron microscopy. J Biol Chem 276:44064–44068. [DOI] [PubMed] [Google Scholar]
- 37. Wilkens S, Inoue T, Forgac M (2004) Three‐dimensional structure of the vacuolar ATPase. Localization of subunit H by difference imaging and chemical cross‐linking. J Biol Chem 279:41942–41949. [DOI] [PubMed] [Google Scholar]
- 38. Wilkens S, Zhang Z, Zheng Y (2005) A structural model of the vacuolar ATPase from transmission electron microscopy. Micron 36:109–126. [DOI] [PubMed] [Google Scholar]
- 39. Muench SP, Huss M, Song CF, Phillips C, Wieczorek H, Trinick J, Harrison MA (2009) Cryo‐electron microscopy of the vacuolar ATPase motor reveals its mechanical and regulatory complexity. J Mol Biol 386:989–999. [DOI] [PubMed] [Google Scholar]
- 40. Zhang Z, Charsky C, Kane PM, Wilkens S (2003) Yeast V1‐ATPase: affinity purification and structural features by electron microscopy. J Biol Chem 278:47299–47306. [DOI] [PubMed] [Google Scholar]
- 41. Zhang Z, Zheng Y, Mazon H, Milgrom E, Kitagawa N, Kish‐Trier E, Heck AJ, Kane PM, Wilkens S (2008) Structure of the yeast vacuolar ATPase. J Biol Chem 283:35983–35995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Diepholz M, Venzke D, Prinz S, Batisse C, Florchinger B, Rossle M, Svergun DI, Bottcher B, Fethiere J (2008) A different conformation for EGC stator subcomplex in solution and in the assembled yeast V‐ATPase: possible implications for regulatory disassembly. Structure 16:1789–1798. [DOI] [PubMed] [Google Scholar]
- 43. Benlekbir S, Bueler SA, Rubinstein JL (2012) Structure of the vacuolar‐type ATPase from Saccharomyces cerevisiae at 11‐A resolution. Nat Struct Mol Biol 19:1356–1362. [DOI] [PubMed] [Google Scholar]
- 44. Gogarten JP, Kibak H, Dittrich P, Taiz L, Bowman EJ, Bowman BJ, Manolson MF, Poole RJ, Date T, Oshima T, J Konishi, K Denda, M Yoshida (1989) Evolution of the vacuolar H+‐ATPase: implications for the origin of eukaryotes. Proc Natl Acad Sci USA 86:6661–6665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhao J, Benlekbir S, Rubinstein JL (2015) Electron cryomicroscopy observation of rotational states in a eukaryotic V‐ATPase. Nature 521:241–245. [DOI] [PubMed] [Google Scholar]
- 46. Rawson S, Phillips C, Huss M, Tiburcy F, Wieczorek H, Trinick J, Harrison MA, Muench SP (2015) Structure of the vacuolar H(+)‐ATPase rotary motor reveals new mechanistic insights. Structure 23:461–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Arai S, Saijo S, Suzuki K, Mizutani K, Kakinuma Y, Ishizuka‐Katsura Y, Ohsawa N, Terada T, Shirouzu M, Yokoyama S, Iwata S, Yamato I, Murata T (2013) Rotation mechanism of Enterococcus hirae V1‐ATPase based on asymmetric crystal structures. Nature 493:703–707. [DOI] [PubMed] [Google Scholar]
- 48. Balakrishna AM, Basak S, Manimekalai MS, Gruber G (2015) Crystal structure of subunits D and F in complex gives insight into energy transmission of the eukaryotic V‐ATPase from Saccharomyces cerevisiae . J Biol Chem 290:3183–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Oot RA, Huang LS, Berry EA, Wilkens S (2012) Crystal structure of the yeast vacuolar ATPase heterotrimeric EGC(head) peripheral stalk complex. Structure 20:1881–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sagermann M, Stevens TH, Matthews BW (2001) Crystal structure of the regulatory subunit H of the V‐type ATPase of Saccharomyces cerevisiae . Proc Natl Acad Sci USA 98:7134–7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Drory O, Frolow F, Nelson N (2004) Crystal structure of yeast V‐ATPase subunit C reveals its stator function. EMBO Rep 5:1148–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Srinivasan S, Vyas NK, Baker ML, Quiocho FA (2011) Crystal structure of the cytoplasmic N‐terminal domain of subunit I, a homolog of subunit a, of V‐ATPase. J Mol Biol 412:14–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Murata T, Yamato I, Kakinuma Y, Leslie AG, Walker JE (2005) Structure of the rotor of the V‐Type Na+‐ATPase from Enterococcus hirae . Science 308:654–659. [DOI] [PubMed] [Google Scholar]
- 54. Nishi T, Kawasaki‐Nishi S, Forgac M (2003) The first putative transmembrane segment of subunit c″ (Vma16p) of the yeast V‐ATPase is not necessary for function. J Biol Chem 278:5821–5827. [DOI] [PubMed] [Google Scholar]
- 55. Nelson H, Nelson N (1989) The progenitor of ATP synthases was closely related to the current vacuolar H+‐ATPase. FEBS Lett 247:147–153. [DOI] [PubMed] [Google Scholar]
- 56. Wilkens S, Vasilyeva E, Forgac M (1999) Structure of the vacuolar ATPase by electron microscopy. J Biol Chem 274:31804–31810. [DOI] [PubMed] [Google Scholar]
- 57. Venzke D, Domgall I, Kocher T, Fethiere J, Fischer S, Bottcher B (2005) Elucidation of the stator organization in the V‐ATPase of Neurospora crassa. J Mol Biol 349:659–669. [DOI] [PubMed] [Google Scholar]
- 58. Zhang Z, Inoue T, Forgac M, Wilkens S (2006) Localization of subunit C (Vma5p) in the yeast vacuolar ATPase by immuno electron microscopy. FEBS Lett 580:2006–2010. [DOI] [PubMed] [Google Scholar]
- 59. Hirata R, Graham LA, Takatsuki A, Stevens TH, Anraku Y (1997) VMA11 and VMA16 encode second and third proteolipid subunits of the Saccharomyces cerevisiae vacuolar membrane H+‐ATPase. J Biol Chem 272:4795–4803. [DOI] [PubMed] [Google Scholar]
- 60. Adachi K, Oiwa K, Nishizaka T, Furuike S, Noji H, Itoh H, Yoshida M, Kinosita K, Jr (2007) Coupling of rotation and catalysis in F(1)‐ATPase revealed by single‐molecule imaging and manipulation. Cell 130:309–321. [DOI] [PubMed] [Google Scholar]
- 61. Furuike S, Nakano M, Adachi K, Noji H, Kinosita K, Jr , Yokoyama K (2011) Resolving stepping rotation in Thermus thermophilus H(+)‐ATPase/synthase with an essentially drag‐free probe. Nat Commun 2:233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Minagawa Y, Ueno H, Hara M, Ishizuka‐Katsura Y, Ohsawa N, Terada T, Shirouzu M, Yokoyama S, Yamato I, Muneyuki E, Noji H, Murata T, Iino R (2013) Basic properties of rotary dynamics of the molecular motor Enterococcus hirae V1‐ATPase. J Biol Chem 288:32700–32707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kawasaki‐Nishi S, Nishi T, Forgac M (2001) Arg‐735 of the 100‐kDa subunit a of the yeast V‐ATPase is essential for proton translocation. Proc Natl Acad Sci USA 98:12397–12402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Toei M, Toei S, Forgac M (2011) Definition of membrane topology and identification of residues important for transport in subunit a of the vacuolar ATPase. J Biol Chem 286:35176–35186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Uchida E, Ohsumi Y, Anraku Y (1985) Purification and properties of H+‐translocating, Mg2+‐adenosine triphosphatase from vacuolar membranes of Saccharomyces cerevisiae . J Biol Chem 260:1090–1095. [PubMed] [Google Scholar]
- 66. Li SC, Diakov TT, Rizzo JM, Kane PM (2012) Vacuolar H+‐ATPase works in parallel with the HOG pathway to adapt Saccharomyces cerevisiae cells to osmotic stress. Eukaryot Cell 11:282–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Diakov TT, Kane PM (2010) Regulation of vacuolar proton‐translocating ATPase activity and assembly by extracellular pH. J Biol Chem 285:23771–23778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Voss M, Vitavska O, Walz B, Wieczorek H, Baumann O (2007) Stimulus‐induced phosphorylation of vacuolar H(+)‐ATPase by protein kinase A. J Biol Chem 282:33735–33742. [DOI] [PubMed] [Google Scholar]
- 69. Graf R, Harvey WR, Wieczorek H (1996) Purification and properties of a cytosolic V1‐ATPase. J Biol Chem 271:20908–20913. [DOI] [PubMed] [Google Scholar]
- 70. Parra KJ, Keenan KL, Kane PM (2000) The H subunit (Vma13p) of the yeast V‐ATPase inhibits the ATPase activity of cytosolic V1 complexes. J Biol Chem 275:21761–21767. [DOI] [PubMed] [Google Scholar]
- 71. Oot RA, Kane PM, Berry EA, Wilkens S (2016) Crystal structure of yeast V1‐ATPase in the autoinhibited state. Embo J 35:1694–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zhang J, Feng Y, Forgac M (1994) Proton conduction and bafilomycin binding by the V0 domain of the coated vesicle V‐ATPase. J Biol Chem 269:23518–23523. [PubMed] [Google Scholar]
- 73. Couoh‐Cardel S, Milgrom E, Wilkens S (2015) Affinity purification and structural features of the yeast vacuolar ATPase Vo membrane sector. J Biol Chem 290:27959–27971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. MacLeod KJ, Vasilyeva E, Merdek K, Vogel PD, Forgac M (1999) Photoaffinity labeling of wild‐type and mutant forms of the yeast V‐ATPase A subunit by 2‐azido‐[(32)P]ADP. J Biol Chem 274:32869–32874. [DOI] [PubMed] [Google Scholar]
- 75. Parra KJ, Kane PM (1998) Reversible association between the V1 and V0 domains of yeast vacuolar H+‐ATPase is an unconventional glucose‐induced effect. Mol Cell Biol 18:7064–7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Huss M, Wieczorek H (2007) Influence of ATP and ADP on dissociation of the V‐ATPase into its V(1) and V(O) complexes. FEBS Lett 581:5566–5572. [DOI] [PubMed] [Google Scholar]
- 77. Bond S, Forgac M (2008) The Ras/cAMP/protein kinase A pathway regulates glucose‐dependent assembly of the vacuolar (H+)‐ATPase in yeast. J Biol Chem 283:36513–36521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Dechant R, Binda M, Lee SS, Pelet S, Winderickx J, Peter M (2010) Cytosolic pH is a second messenger for glucose and regulates the PKA pathway through V‐ATPase. Embo J 29:2515–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Parra KJ, Chan CY, Chen J (2014) Saccharomyces cerevisiae vacuolar H+‐ATPase regulation by disassembly and reassembly: one structure and multiple signals. Eukaryot Cell 13:706–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Diab H, Ohira M, Liu M, Cobb E, Kane PM (2009) Subunit interactions and requirements for inhibition of the yeast V1‐ATPase. J Biol Chem 284:13316–13325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Ho MN, Hirata R, Umemoto N, Ohya Y, Takatsuki A, Stevens TH, Anraku Y (1993) VMA13 encodes a 54‐kDa vacuolar H(+)‐ATPase subunit required for activity but not assembly of the enzyme complex in Saccharomyces cerevisiae . J Biol Chem 8:18286–18292. [PubMed] [Google Scholar]
- 82. Liu M, Tarsio M, Charsky CM, Kane PM (2005) Structural and functional separation of the N‐ and C‐terminal domains of the yeast V‐ATPase subunit H. J Biol Chem 280:36978–36985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Nakano M, Imamura H, Toei M, Tamakoshi M, Yoshida M, Yokoyama K (2008) ATP hydrolysis and synthesis of a rotary motor V‐ATPase from Thermus thermophilus . J Biol Chem 283:20789–20796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Schneider E, Altendorf K (1985) All three subunits are required for the reconstitution of an active proton channel (F0) of Escherichia coli ATP synthase (F1F0). Embo J 4:515–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Couoh‐Cardel S, Milgrom E, Wilkens S (2015) Affinity purification and structural features of the yeast vacuolar ATPase Vo membrane sector. J Biol Chem 290:27959–27971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Qi J, Forgac M (2008) Function and subunit interactions of the N‐terminal domain of subunit a (Vph1p) of the yeast V‐ATPase. J Biol Chem 283:19274–19282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Allegretti M, Klusch N, Mills DJ, Vonck J, Kuhlbrandt W, Davies KM (2015) Horizontal membrane‐intrinsic alpha‐helices in the stator a‐subunit of an F‐type ATP synthase. Nature 521:237–240. [DOI] [PubMed] [Google Scholar]
- 88. Stam NJ, Wilkens S (2016) Structure of nanodisc reconstituted vacuolar ATPase proton channel: definition of the interaction of rotor and stator and implications for enzyme regulation by reversible dissociation. J Biol Chem 292:1749–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Seol JH, Shevchenko A, Shevchenko A, Deshaies RJ (2001) Skp1 forms multiple protein complexes, including RAVE, a regulator of V‐ATPase assembly. Nat Cell Biol 3:384–391. [DOI] [PubMed] [Google Scholar]
- 90. Smardon AM, Tarsio M, Kane PM (2002) The RAVE complex is essential for stable assembly of the yeast V‐ATPase. J Biol Chem 277:13831–13839. [DOI] [PubMed] [Google Scholar]
- 91. Smardon AM, Kane PM (2007) RAVE is essential for the efficient assembly of the C subunit with the vacuolar H(+)‐ATPase. J Biol Chem 282:26185–26194. [DOI] [PubMed] [Google Scholar]
- 92. Smardon AM, Nasab ND, Tarsio M, Diakov TT, Kane PM (2015) Molecular interactions and cellular itinerary of the yeast RAVE (regulator of the H+‐ATPase of vacuolar and endosomal membranes) complex. J Biol Chem 290:27511–27523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Smardon AM, Diab HI, Tarsio M, Diakov TT, Nasab ND, West RW, Kane PM (2014) The RAVE complex is an isoform‐specific V‐ATPase assembly factor in yeast. Mol Biol Cell 25:356–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Lu M, Ammar D, Ives H, Albrecht F, Gluck SL (2007) Physical interaction between aldolase and vacuolar H+‐ATPase is essential for the assembly and activity of the proton pump. J Biol Chem 282:24495–24503. [DOI] [PubMed] [Google Scholar]
- 95. Chan CY, Parra KJ (2014) Yeast phosphofructokinase‐1 subunit Pfk2p is necessary for pH homeostasis and glucose‐dependent vacuolar ATPase reassembly. J Biol Chem 289:19448–19457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Oot RA, Wilkens S (2010) Domain characterization and interaction of the yeast vacuolar ATPase subunit C with the peripheral stator stalk subunits E and G. J Biol Chem 285:24654–24664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Oot RA, Wilkens S (2012) Subunit interactions at the V1–Vo interface in yeast vacuolar ATPase. J Biol Chem 287:13396–13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Rahman S, Yamato I, Saijo S, Mizutani K, Ishizuka‐Katsura Y, Ohsawa N, Terada T, Shirouzu M, Yokoyama S, Iwata S, Murata T (2013) Biochemical and biophysical properties of interactions between subunits of the peripheral stalk region of human V‐ATPase. PLoS One 8:e55704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Tabke K, Albertmelcher A, Vitavska O, Huss M, Schmitz HP, Wieczorek H (2014) Reversible disassembly of the yeast V‐ATPase revisited under in vivo conditions. Biochem J 462:185–197. [DOI] [PubMed] [Google Scholar]
- 100. Peters C, Bayer MJ, Buhler S, Andersen JS, Mann M, Mayer A (2001) Trans‐complex formation by proteolipid channels in the terminal phase of membrane fusion. Nature 409:581–588. [DOI] [PubMed] [Google Scholar]
- 101. Morel N, Poea‐Guyon S (2015) The membrane domain of vacuolar H(+)ATPase: a crucial player in neurotransmitter exocytotic release. Cell Mol Life Sci 72:2561–2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Couoh‐Cardel S, Hsueh YC, Wilkens S, Movileanu L (2016) Yeast V‐ATPase proteolipid ring acts as a large‐conductance transmembrane protein pore. Sci Rep 6:24774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Iwata M, Imamura H, Stambouli E, Ikeda C, Tamakoshi M, Nagata K, Makyio H, Hankamer B, Barber J, Yoshida M, Yokoyama K, Iwata S (2004) Crystal structure of a central stalk subunit C and reversible association/dissociation of vacuole‐type ATPase. Proc Natl Acad Sci USA 101:59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Movie 1.


