Abstract
Background
Accurate identification of Aspergillus species is a very important subject. Mass spectral fingerprinting using matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) is generally employed for the rapid identification of fungal isolates. However, the results are based on simple mass spectral pattern-matching, with no peak assignment and no taxonomic input. We propose here a ribosomal subunit protein (RSP) typing technique using MALDI-TOF MS for the identification and discrimination of Aspergillus species. The results are concluded to be phylogenetic in that they reflect the molecular evolution of housekeeping RSPs.
Results
The amino acid sequences of RSPs of genome-sequenced strains of Aspergillus species were first verified and compared to compile a reliable biomarker list for the identification of Aspergillus species. In this process, we revealed that many amino acid sequences of RSPs (about 10–60%, depending on strain) registered in the public protein databases needed to be corrected or newly added. The verified RSPs were allocated to RSP types based on their mass. Peak assignments of RSPs of each sample strain as observed by MALDI-TOF MS were then performed to set RSP type profiles, which were then further processed by means of cluster analysis. The resulting dendrogram based on RSP types showed a relatively good concordance with the tree based on β-tubulin gene sequences. RSP typing was able to further discriminate the strains belonging to Aspergillus section Fumigati.
Conclusions
The RSP typing method could be applied to identify Aspergillus species, even for species within section Fumigati. The discrimination power of RSP typing appears to be comparable to conventional β-tubulin gene analysis. This method would therefore be suitable for species identification and discrimination at the strain to species level. Because RSP typing can characterize the strains within section Fumigati, this method has potential as a powerful and reliable tool in the field of clinical microbiology.
Electronic supplementary material
The online version of this article (doi:10.1186/s12866-017-1009-3) contains supplementary material, which is available to authorized users.
Keywords: Aspergillus, Ribosomal subunit proteins, Matrix-assisted laser desorption ionization time-of-flight mass spectrometry
Background
Aspergillus is a saprophytic genus found in diverse environments [1]. Some species, typically A. fumigatus, are causative agents of aspergillosis [2]. Because the degree of virulence and susceptibility to antifungal agents are known to vary among species [3], accurate identification of Aspergillus species is a very important subject, especially in the field of clinical mycology.
The identification of fungal species has up to now been based on the morphological characteristics of colonies and filaments as observed by microscopy [4]. However, the morphology-based method suffers several drawbacks. It requires specialized skills and knowledge and is tedious and time-consuming work. Nevertheless, some strains lack obvious characteristic features under laboratory conditions. To achieve objective identification, molecular biological methods based on the DNA sequences of particular genes are increasingly being adopted. The internal transcribed spacer (ITS) regions between 18S rRNA, 5.8S rRNA, and 28S rRNA are regarded as “barcode regions” and are frequently used as biomarkers for species identification [5]. DNA sequences that code housekeeping proteins such as β-tubulin [6] and calmodulin [7] are also often used for detailed molecular studies. To improve the resolution of species discrimination, combinations of multi-genes have been attempted. For example, a combination of two genes (β-tubulin and calmodulin) [8] or four genes (β-tubulin, calmodulin, ITS and large-subunit rDNA, and RNA polymerase II) [9] has been used to characterize A. fumigatus strains. Multi-locus sequence typing (MLST), focusing on seven types of gene fragments, has also been applied to characterize A. fumigatus strains [10]. These DNA-based methods provide a more objective evaluation than the traditional morphological method.
In the field of medical microbiology, much attention has been paid recently to the mass spectrometric technique of matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI–TOF MS) as a tool for the rapid identification of fungal isolates [11]. MALDI–TOF MS has major clinical advantages, since it requires much smaller samples and the total process from sample preparation to data analysis is very rapid. This method is a type of mass spectral fingerprinting, for which mass spectral databases are commercially available from several mass spectrometer companies. The rapid identification and discrimination of clinical A. fumigatus isolates has been reported using this method [12, 13]. Several research groups have further attempted to discriminate Aspergillus isolates at the species and strain level [14–16]. However, Welker [17] has pointed out the following problems with this method in his review.
-
(i)
the general finding that the proteome is very dynamic in living cells and hence protein pattern expectedly could be subject to changes in response to growth conditions,
-
(ii)
doubts whether differences and similarities in mass spectral patterns are completely consistent with the established taxonomy,
-
(iii)
a lack of comprehensive databases covering all clinically relevant species.
Furthermore, in the author’s opinion, the reported mass spectra of fungal samples sometimes show too few peaks when sample preparation is performed using the recommended protocol proposed by the manufacturers.
To overcome these problems relating to mass spectral fingerprinting, we have proposed a method using ribosomal proteins as biomarkers for microorganism analysis by MALDI–TOF MS [18–25]. Ribosomal proteins are typical housekeeping proteins and are abundantly present in microorganisms’ cells. Prokaryotic (bacterial) ribosomes consist of 57 kinds of ribosomal subunit proteins (RSPs), whereas eukaryotic ribosomes typically consist of 78 RSPs. The combination of subunit proteins and their structures are not influenced by culture conditions. Because most RSPs are basic proteins with higher proton affinity (i.e., easily producing [M + H]+ ions) and their masses are distributed in the range of ca. 4 - 30 kDa, RSPs can be easily observed in MALDI mass spectra [26]. We have reported that the identification of bacterial species and classification at the strain level can be accomplished based on the expressed mass types of RSPs [18–25]. The masses of RSPs used as biomarkers can be estimated from translational amino acid sequences of genome-sequenced strains, which can be obtained from public databases such as UniProt Knowledgebase (UniProtKB) [27]. Our proposed method is a form of molecular typing like MLST, based on bioinformatics. The biomarker RSPs are a complex of typical housekeeping proteins. Since the sequence variation of RSPs observed as the peak shift on the MALDI mass spectra results from molecular evolution, the results of identification and discrimination of microorganisms are assumed to phylogenetic ally. This is the crucial difference between our proposed RSP typing as “phylogenetic” method and the conventional mass spectral fingerprinting as “chemotaxonomic” method.
The aim of our project is to extend the RSP-based method to the identification of eukaryotic fungi. As the first step, we have investigated the actual state of information of RSPs of fungi registered in public protein databases through the characterization of ribosomal protein fractions extracted from genome-sequenced A. fumigatus strains as a model [28]. In our previous paper [28], we revealed that more than half of the amino acid sequences of RSPs registered in the public databases were incorrect, due chiefly to mis-annotation of exon/intron structures. We were able to successfully correct the sequence errors using a combination of in silico inspection by sequence homology analysis and MALDI–TOF MS measurements. Post-translational modifications such as acetylation and methylation could also be verified. In this way, the expressed masses of RSPs observed under 16,000 Da could finally be confirmed.
As the next step, this paper describes the results of comparable characterization of RSPs of eleven Aspergillus species to establish biomarker references for the reliable identification of Aspergillus species. First, verification and correction of the amino acid sequences of RSPs and confirmation of post-translational modifications common to all sample strains were performed to accurately determine the expressed mass, as described in our previous paper [28]. RSPs with appropriate intensity commonly observed in each strain were then selected as reliable biomarkers for the identification of Aspergillus species. The selected RSPs of each strain were categorized into “RSP types” based on their mass and used to construct a dendrogram. The resulting dendrogram was compared with that arrived at using the DNA-based method; the reliability of the species identification and discrimination of this method was then assessed.
Results and discussion
Characterization of RSPs of genome-sequenced Aspergillus strains
In our previous paper [28], the amino acid sequences of RSPs in A. fumigatus strains were verified to compile the reference mass list of expressed RSPs. We noted that more than half of the amino acid sequences in the public databases, such as UniProtKB [27] and the NCBI protein database [29] were incorrect. These errors could be corrected by a combination of in silico inspection using sequence homology analysis and verification of actual expressed masses of RSPs by MALDI–TOF MS measurements. In this study, by applying this strategy, the amino acid sequences of RSPs of ten genome-sequenced strains of Aspergillus species were further verified and compared to build a reliable biomarker list for the identification of Aspergillus species. The sample strains used in this study are summarized in Table 1. For RSPs with amino acid sequences not registered in the public databases, corresponding gene sequences were manually identified in the genome sequence by referring to gene sequences of A. fumigatus strains. The genome sequence of A. viridinutans is not yet published, but the authors have annotated the RSP sequences manually. Finally, 26 kinds of RSPs whose molecular weights were under 16,000 Da were selected that are common to those already verified for A. fumigatus strains [28]. Supporting Information Additional file 3: Figures SI-1 to SI-10 show the mass spectra of each genome-sequenced strain (except for A. fumigatus, for which the mass spectra were reported in ref. [28]). Additional file 1: Table SI-1 summarizes accession numbers in public protein databases of ribosomal protein biomarkers of genome sequenced strains used in this study. Here, the names of RSPs are adopted from the yeast nomenclature system [30] to prevent confusion in the RSP’s nomenclature. Supporting Information Additional file 2: Table SI-2 summarizes the data of the RSPs of ten genome-sequenced strains such as the accession number, post-translational modifications, corrected amino acid sequences, and corrected exon/intron structures (the corrected sequences of A. fumigatus A1163 and Af293 have been reported in ref [28]).
Table 1.
List of sample strains used in this study
| species | strain names a) |
|---|---|
| A. fumigatus | * IFM 53842 (= A1163), *IFM 54229 (= Af293), IFM 57323NT |
| N. fischeri | * IFM 57324T (= NRRL 181T) |
| A. lentulus | * IFM 54703T, IFM 47547, IFM 58399, IFM 60648, IFM 61392, IFM 62073, IFM 62096 |
| A. viridinutans | * IFM 47045T |
|
A. felis
(former A. viridinutans) |
IFM 59564, IFM 60053, IFM 62093 |
| A. pseudoviridinutans (former A. viridinutans) | IFM 55266, IFM 62075 |
| A. wyomingensis (former A. viridinutans) | IFM 62083 |
| A. udagawae | *IFM 46973T, IFM 46972, IFM 5058, IFM 51744, IFM 53868, IFM 61606, IFM 62070, IFM 62100 |
| A. clavatus | * IFM 60676NT (= NRRL 1NT) |
| A. niger | * CBS 513.88 |
| A. kawachii | * IFO 4308 |
| A. flavus | * IFM 60677 (= NRRL 3357) |
| A. oryzae | * IFM 59475 (=RIB 40) |
| A. nidulans | * IFM 60678 (= FGSC A4) |
a) * genome sequenced strain.
Table 2 shows the number and ratio of incorrect or not registered sequences among 26 RSPs for nine newly analyzed genome-sequenced strains (the result of A. viridinutans, annotated by the authors, was not added to this list). Depending on the species, about 10 - 60% RSPs needed to be corrected or newly added. The source of sequence errors in prokaryotic bacteria was chiefly due to misidentification of the start codon [20]. The main reason in Aspergillus fungi seemed to be due to misidentification of the exon/intron structure, resulting in incorrect CDS as well as an incorrect stop codon caused by frame shift. Because this type of error is unique to prokaryotes, a similar problem in the annotations of RSP genes might have occurred in other fungi.
Table 2.
The number of corrected RSPs and their ratio to analyzed RSPs
| Species | Strains | The number of the corrected RSPs | The ratio of the corrected RSPs to the analyzed RSPs (%) |
|---|---|---|---|
| N. fischeri | IFM 57324T (= NRRL 181T) | 11 | 42 |
| A. lentulus | IFM 54703T | 14 | 54 |
| A. udagawae | IFM 46973T | 15 | 58 |
| A. clavatus | IFM 60676NT (= NRRL 1NT) | 9 | 35 |
| A. niger | CBS 513.88 | 4 | 15 |
| A. kawachii | IFO 4308 | 11 | 42 |
| A. flavus | IFM 60677 (= NRRL 3357) | 7 | 27 |
| A. oryzae | IFM 59475 (=RIB 40) | 3 | 12 |
| A. nidulans | IFM 60678 (= FGSC A4) | 4 | 15 |
The post-translational modifications were then confirmed by referring to already-reported modifications in eukaryotic RSPs. Details of the assignments of each modification are described in the Supporting Information (Additional file 4: Figures SI-11 to SI-14). Acetylation (S16, S21, S24, S28, L31, and L35), methylation (L42), and two hydroxylations (S23) have been reported in several papers (see citations in ref. [28]) and have also observed in A. fumigatus [28]. A mass shift of these RSPs from the calculated sequence mass after taking into account N-terminal methionine loss was commonly observed in all sample strains (+42 Da for acetylation, +14 Da for methylation, and +32 Da for two hydroxylations). This result suggests these modifications to be evolutionarily-conserved modifications, at least in Aspergillus species. In addition, S27 showed a common +28 Da shift, suggesting two methylations. Although this modification has, to our knowledge, not been reported before, two methylations of S27 were concluded to be common modifications in Aspergillus species.
Species identification using the RSP types
The amino acid sequences and the theoretical mass of RSPs thus determined mostly varied among species. This finding strongly suggests that species identification can be performed using RSPs as biomarkers. To make a reference table for the RSP typing, each RSP was classified into different types based on mass. For example, S29 has five types of different expressed mass, of which the peaks are distributed approximately from m/z 6570 to m/z 6650, as shown in Fig. 1 (the whole-range mass spectra are shown in Supporting Information Additional file 3: Figures SI-1 to SI-10). For S29, Type I was first allocated to A. fumigatus A1163 observed at m/z 6647, which was common with that of N. fischeri NRRL 181T, A. lentulus IFM 54703T, A. viridinutans IFM 47045T, and A. udagawae IFM 46973T. Interestingly, these species belong to Aspergillus section Fumigati. The mass of S29 of A. clavatus NRRL 1NT was different from Type I, so it was allocated to Type II. In the same manner, S29 of A. niger CBS 513.88 and A. kawachii IFO 4308 were allocated to Type III, that of A. flavus NRRL 3357 and A. oryzae RIB 40 to Type IV, and that of A. nidulans FGSC A4 to Type V. The type classification was conducted in the same way in other RSPs. Table 3 summarizes the mass and types of RSPs of each genome-sequenced strain, in which the post-translational modifications were taken into consideration as affecting the mass in this list.
Fig. 1.
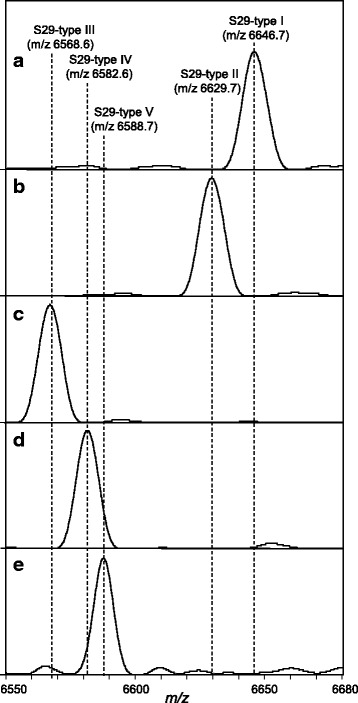
Expanded view of S29 (m/z 6550-6680) regions. a A. fumigatus A1163, b A. clavatus NRRL 1NT, (C) A. niger CBS 513.88, (D) A. flavus NRRL 3357, and (E) A. nidulans FGSC A4
Table 3.
The expressed masses and the types of ribosomal protein biomarkers of genome-sequenced strains used in this study
| RP name |
A. fumigatus
IFM 53842 |
A. fumigatus
IFM 54229 |
N. fischeri
IFM 57324T |
A. lentulus
IFM 54703T |
A. viridinutans
IFM 47045T |
A. udagawae
IFM 46973T |
| = A1163 | = Af293 | = NRRL 181T | ||||
| L40 | I (6002.3) | I | II (6016.3) | II | II | II |
| L39 | I (6151.2) | I | I | I | I | I |
| S29 | I (6646.7) | I | I | I | I | I |
| S30 | I (6789.1) | I | II (6807.1) | II | III (6793.1) | II |
| L29 | I (7456.6) | I | I | I | I | II (7470.6) |
| S28 | I (7710.0) | I | I | I | I | I |
| S27 | I (8765.3) | I | I | I | I | I |
| S31 | I (9134.9) | I | I | I | I | II (9120.9) |
| L38 | I (9153.8) | I | II (9169.8) | II | II | II |
| S21 | I (10038.1) | II (10052.2) | I | III (10035.1) | IV (10008.1) | V (10024.1) |
| L43 | I (10025.8) | I | I | I | I | I |
| L37 | I (10386.9) | I | I | II (10387.9) | III (10358.9) | II |
| L30 | I (11171.1) | I | I | I | I | I |
| L36 | I (11869.8) | I | II (11861.8) | III (11847.8) | II | IV (11875.8) |
| L42 | I (12028.3) | I | II (12009.2) | III (11979.2) | IV (11995.2) | V (11965.2) |
| L33 | I (12215.1) | I | I | I | II (12233.1) | III (12206.1) |
| L34 | I (13164.5) | I | I | I | I | I |
| S26 | I (13338.7) | I | I | I | I | I |
| L31 | I (13919.1) | I | II (13937.1) | II | II | III (13937.1) |
| L35 | I (14532.0) | I | II (14518.0) | II | a) | III (14548.0) |
| L32 | I (14836.6) | I | II (14822.6) | II | II | III (14880.6) |
| L26 | I (14979.4) | I | II (14982.5) | II | II | III (14954.4) |
| S24 | I (15226.6) | I | I | I | II (15212.6) | I |
| L27 | I (15682.6) | I | II (15654.5) | II | II | III (15667.5) |
| S23 | I (15802.5) | I | II (15786.5) | II | II | II |
| S16 | I(15883.4) | I | II (15853.4) | II | II | II |
| RP name |
A. clavatus
IFM 60676NT |
A. niger
CBS 513.88 |
A. kawachii
IFO 4308 |
A. flavus
NRRL 3357 |
A. oryzae
RIB 40 |
A. nidulans
FGSC A4 |
| = NRRL 1NT | ||||||
| L40 | II | II | II | III (6043.3) | III | IV (6057.3) |
| L39 | I | II(6123.2) | II | I | I | III (6123.2) |
| S29 | II (6629.7) | III | III (6568.6) | IV (6582.6) | IV | V (6588.7) |
| S30 | IV (6790.0) | V (6745.0) | VI (6729.0) | VII (6691.9) | VII | VIII (6761.1) |
| L29 | III (7433.6) | IV (7368.4) | IV | V (7414.5) | V | VI (7387.5) |
| S28 | I | II (7681.9) | II | III (7708.0) | III | IV (7754.0) |
| S27 | I | II (8735.3) | II | III (8792.4) | III | IV (8807.3) |
| S31 | II | III (9101.8) | III | IV (9090.8) | IV | V (9111.9) |
| L38 | III (9160.8) | IV (9073.7) | IV | V (9173.9) | V | VI (9128.8) |
| S21 | VI (9922.9) | VII (10027.1) | VIII (10041.2) | IX (10050.1) | IX | X (10064.1) |
| L43 | II (10011.8) | IV (9997.7) | IV | IV | IV | V (10011.8) |
| L37 | IV (10279.7) | V (10451.9) | VI (10435.9) | VII (10436.9) | VII | VIII (10371.8) |
| L30 | II (11169.1) | III (11210.1) | III | IV (11223.1) | IV | V (11259.2) |
| L36 | V (11850.8) | VI (11831.8) | VI | III | III | VII (11757.7) |
| L42 | VI (11921.1) | VII (12025.2) | VII | VIII (12029.2) | VIII | IX (11994.2) |
| L33 | II | IV (12250.1) | IV | V (12299.2) | V | VI (12216.1) |
| L34 | I | II (13147.4) | II | III (13178.5) | III | IV (13133.4) |
| S26 | I | II (13381.7) | II | II | II | IV (13349.7) |
| L31 | IV (13809.9) | V (13878.0) | V | VI (13981.2) | VI | VII (13847.9) |
| L35 | IV (14517.0) | V (14384.8) | VI (14411.9) | VII (14536.9) | VII | VIII (14458.0) |
| L32 | IV (14852.6) | V (14891.6) | V | VI (14794.5) | VI | VII (14838.5) |
| L26 | IV (14915.3) | V (14877.2) | VI (14891.2) | VII (14999.3) | VII | VIII (15084.6) |
| S24 | III (15130.5) | IV (15163.5) | IV | V (15157.5) | V | VI (15232.7) |
| L27 | IV (15636.5) | V (15610.4) | V | VI (15613.4) | VI | VII (15642.5) |
| S23 | II | III (15739.4) | III | IV (15752.4) | IV | V (15757.5) |
| S16 | III (15849.4) | IV (15881.4) | IV | V (15835.3) | V | VI (15852.3) |
a) The amino acid sequence of L35 in A. viridinutans IFM 47045T was not obtained from the draft genome sequences
The distribution of the RSP types shown in Table 3 was then processed using the unweighted pair group method with arithmetic mean (UPGMA) cluster analysis using a categorical coefficient. Fig. 2 compares the dendrogram based on the RSP types (Fig. 2a) with that based on the β-tubulin gene sequence (Fig. 2b). Among the Aspergillus species used in this study, A. fumigatus, N. fischeri, A. lentulus, A. viridinutans, and A. udagawae are known to be genetically related species, belonging to section Fumigati [31] (note that Neosartorya is a teleomorph of Aspergillus). Interestingly, these five species form a cluster in the dendrograms based on both the RSP type (Fig. 2a) and the β-tubulin gene sequence (Fig. 2b). In these sample strains, eight RSPs (L30, L34, L39, L43, S26, S27, S28, and S29) of the analyzed 26 RSPs (31%) matched completely.
Fig. 2.
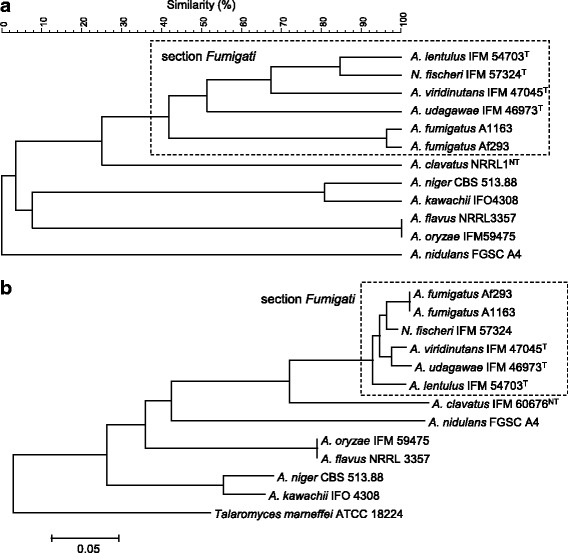
Dendrograms constructed based on RSP typing (a) and the DNA sequence of β-tubulin (b)
Aspergillus clavatus has been suggested to be closest to section Fumigati as shown in the dendrogram based on β-tubulin (Fig. 2b). RSP typing (Fig. 2a) also shows the closest position of A. clavatus next to section Fumigati. Five RSPs (S26, S27, S28, L34, and L39) of common eight RSPs in section Fumigati are also shared with A. clavatus.
The positions of A. niger, A. kawachii, A. flavus, A. oryzae, and A. nidulans are distinct from the cluster of section Fumigati. The RSP types of these species show little in common with section Fumigati species: only L39 of A. oryzae and A. flavus was common. Of these species, pairs of ‘A. niger and A. kawachii’ and ‘A. flavus and A. oryzae’ are known to be very closely interrelated: A. flavus and A. oryzae are assumed to be ecotypes of the same species because they have only 350 unique genes, even when comparing the total genome sequences [32]. In the types of 26 RSPs used in this study, 21 out of 26 RSPs agreed between A. niger and A. kawachii and also 100% of RSPs agreed between A. flavus and A. oryzae. The ratio of conformity between species thus appears to be reflected in the relatedness between species.
As discussed above, the dendrogram based on the RSP types shows relatively good concordance with the tree based on β-tubulin gene sequences (Fig. 2b) and with the tree arrived at by genomic analysis of Aspergillus fungi [1]. This result suggests that RSP typing can perform accurate species identification that reflects molecular evolution. Therefore, the dendrogram constructed by the RSP typing can be considered as a kind of phylogenetic tree.
The effectiveness of RSP typing for discriminating section Fumigati strains
Because sensitivity to antifungal agents differs between A. fumigatus and other species within section Fumigati, accurate discrimination of these strains is very important [31]. However, traditional morphological analysis does not always accurately locate species within section Fumigati, so misidentifications often occur. To overcome this problem, Yaguchi et al. have characterized the species within section Fumigati by molecular phylogenetic analysis using multiple genes [33]. On the other hand, as described in the previous section, RSP typing shows the potential of species discrimination within section Fumigati. To reveal the effectiveness of this method, the strains of A. fumigatus, A. lentulus, A. viridinutans, A. felis, A. pseudoviridinutans, A. wyomingensis, A. udagawae, and N. fischeri belonging to section Fumigati were characterized.
Table 4 shows the RSP types of the sample strains assigned using the RSP reference list (Table 3). In this section, 18 RSPs with clearly separated peaks observed for all strains were adopted from 26 RSPs. Some RSP peaks were not matched to the reference due to peak shift or not detected. These RSPs are designated as N (Not matched) in Table 4.
Table 4.
The RSP typing profile of the sample strains belonging to Aspergillus section Fumigati
| Sample strains a) | The types of biomarker RSPs | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L40 | L39 | S29 | S30 | S28 | S31 | L38 | L30 | L42 | L33 | L34 | S26 | L32 | L26 | S24 | L27 | S23 | S16 | |
| IFM 53842 (Afu) | I | I | I | I | I | I | I | I | I | I | I | I | I | I | I | I | I | I |
| IFM 54229 (Afu) | I | I | I | I | I | I | I | I | I | I | I | I | I | I | I | I | I | I |
| IFM 57323NT (Afu) | I | I | I | I | I | I | I | I | I | I | I | I | I | I | I | I | I | I |
| IFM 54703T (Al) | II | I | I | II | I | I | II | I | III | I | I | I | II | II | I | II | II | II |
| IFM 47457 (Al) | II | I | I | II | I | I | II | I | III | I | I | I | II | II | I | II | II | II |
| IFM 58399 (Al) | II | I | I | II | I | I | II | I | III | I | I | I | II | II | I | II | II | II |
| IFM 60648 (Al) | II | I | I | II | I | I | II | I | III | I | I | I | II | II | I | II | II | II |
| IFM 61392 (Al) | II | I | I | II | I | I | II | I | III | I | I | I | II | II | I | II | II | II |
| IFM 62073 (Al) | II | I | I | II | I | I | II | I | III | I | I | I | II | II | I | II | II | II |
| IFM 62096 (Al) | II | I | I | II | I | I | II | I | III | I | I | I | II | II | I | II | II | II |
| IFM 47045T (Av) | II | I | I | III | I | I | II | I | IV | II | I | I | II | II | II | II | II | II |
| IFM 55266 (Ap)b) | II | I | I | II | I | II | N | I | II | III | I | I | II | II | N | III | II | II |
| IFM 62075 (Ap) | II | I | I | II | I | II | II | I | II | III | I | I | II | II | I | III | II | II |
| IFM 59564 (Afe) b) | II | I | I | II | I | II | I | N | III | II | I | I | II | N | N | III | II | II |
| IFM 62093 (Afe) | II | I | I | II | I | II | I | N | III | II | I | I | II | N | N | III | II | II |
| IFM 60053 (Afe) | II | I | I | II | I | II | I | N | III | II | I | I | II | N | N | III | II | N |
| IFM 62083 (Aw) b) | II | I | N | II | I | II | N | I | N | II | I | I | II | N | I | I | II | II |
| IFM 46972T (Au) | II | I | I | II | I | II | II | I | V | III | I | N | III | III | I | III | II | II |
| IFM 46973 (Au) | II | I | I | II | I | II | II | I | V | III | I | I | III | III | I | III | II | II |
| IFM 5058 (Au) | II | I | I | II | I | II | II | I | V | III | I | I | III | III | I | III | II | II |
| IFM 51744 (Au) | II | I | I | II | I | II | II | I | V | III | I | N | III | III | N | III | II | N |
| IFM 53868 (Au) | II | I | I | II | I | N | II | I | V | III | I | I | III | III | I | III | II | II |
| IFM 61606 (Au) | II | I | I | II | I | II | II | I | V | III | I | N | III | III | I | III | II | II |
| IFM 62070 (Au) | II | I | I | II | I | N | II | I | V | III | I | I | III | III | I | III | II | II |
| IFM 62100 (Au) | II | I | I | II | I | II | II | I | V | III | I | I | III | III | I | III | II | II |
| IFM 57324T (Nf) | II | I | I | II | I | I | II | I | II | I | I | I | II | II | I | II | II | II |
a) Abbreviations: Afu; A. fumigatus, Afe; A. felis, Al; A. lentulus, Ap; A. pseudoviridinutans, Av; A. viridinutans, Au; A. udagawae, Aw; A. wyomingensis and Nf; N. fischeri
b) A. felis, A. pseudoviridinutans and A. wyomingensis are former A. viridinutans
The distribution of the RSP types within species tended to be consistent, and the variation was assumed to be small. This allowed the typing of sample strains to be conducted using the mass list of the genome-sequenced strain. Fig. 3 shows the result of UPGMA cluster analysis based on the RSP typing profile. In the dendrogram based on RSP typing, every species formed one general cluster.
Fig. 3.

The result of the UPGMA cluster analysis of the RSP typing of the strains belonging to Aspergillus section Fumigati. Abbreviations: Al; A. lentulus, Nf; N. fischeri, Av; A. viridinutans, Au; A. udagawae, Ap; A. pseudoviridinutans, Afe; A. felis, Aw; A. wyomingensis, and Afu; A. fumigatus
The RSP types of A. lentulus strains completely match those of the type strain. Because A. lentulus was originally regarded as a sibling species of A. fumigatus, it has proved difficult to discriminate them morphologically [34]. Several mass spectrometric strategies based on mass spectral fingerprinting have been able to discriminate these species [35, 36]; however, these reports do not record the criteria used for discrimination. Our proposed RSP typing, on the other hand, can discriminate these species based on variations in RSPs. The types of 9 RSPs (L40, S30, L38, L42, L32, L26, L27, S23 and S16) differed from that of A. fumigatus, which can be used for discrimination between A. fumigatus and A. lentulus. Interestingly, N. fischeri is very close to the A. lentulus cluster, in which only L42 was different from that of A. lentulus. This species is close to the A. fumigatus cluster in the case of the β-tubulin gene as shown in Fig. 2b. Such differences are likely to be caused by the difference in genes used: the tree compiled using RSP typing (Fig. 3a) is constructed based on a combination of the types of 18 RSPs that reflect 18 different housekeeping genes.
RSP typing has demonstrated that A. udagawae strains form a relatively clear-cut cluster. Although some RSP peaks were not detected clearly, moderate concordance with the reference mass of the type strain was confirmed. Five RSPs (L39, S29, S28, L30, and L34) are further matched completely to those of A. fumigatus, whereas three RSPs (L42, L32, and L26) are totally different from the other species. These RSPs can therefore be used to discriminate A. udagawae from other species. The three strains (IFM 5058, IFM 51744, and IFM 53868) were formerly reported as variant isolates of A. fumigatus (or Aspergillus sp.) but were re-identified, using multiple genes (β-tubulin, hydrophobin, and calmodulin), as A. udagawae [33]. In RSP typing, these strains are included in the A. udagawae cluster, supporting this re-identification.
In contrast to the high conformity of RSPs in A. lentulus and A. udagawae, former A. viridinutans strains (A. felis, A. pseudoviridinutans, and A. wyomingensis) do not form an obvious cluster. Interestingly, the type strain of A. viridinutans (IFM 47045T) is located separately from other former A. viridinutans strains. The diversity of A. viridinutans has already been reported, and this species was divided into some species [37–39]. The strains IFM 55266 and 62075, IFM 59564, 60053 and 62093, and IFM 62083 were re-identified as A. pseudoviridinutans, A. felis and A. wyomingensis, respectively. In each species, the high conformity of RSPs was indicated.
Conclusions
In the context of the ongoing conversion, on the basis of fungal taxonomy, from morphology to molecular phylogeny, molecular biological methods have been adopted for the identification and discrimination of fungal strains. To avoid misidentification of closely-related species, especially within section Fumigati, the reliability of identification results increases on increasing the numbers of evaluation points (i.e., the numbers of genes or proteins). This gives MALDI-TOF MS great potential, since many proteomic peaks that assist with the identification of fungal species are processed. Our proposed RSP typing represents the next generation of mass spectral identification/discrimination of fungal strains, in that it supersedes current mass spectral fingerprinting, which is simple pattern-matching without peak assignment.
The merits of RSP typing are (1) it requires no commercial database and (2) it can be used to perform phylogenetic analysis. As for the first point, RSP typing requires reference to the RSP biomarker lists, via the internet, constructed from the public protein databases. At this time, of course, commercial mass spectral databases are more substantial than available RSP information. However, as whole genome-sequencing of fungal species progresses, information on RSPs is expected to expand exponentially in the near future. Although we initially encountered a confused situation as concerns the protein information registered on the public protein databases, we have successfully corrected the errors in the amino acid sequences and the names of representative Aspergillus RSPs. The sequences and expressed mass of RSPs of other fungal species can now be easily verified and corrected by homology analysis using the sequence list summarized in Supporting Information Additonal file 2: Table SI-2.
The second benefit of this method is valuable, because the identification results have a phylogenetic rationale: they relate to a combination of more than a dozen housekeeping proteins. This method can eliminate the influence of growth and experimental conditions, if only the RSP peaks are observed. RSPs are one of the most expressed proteins, and RSP fractions are easily collected by cell-grinding and ultracentrifugation. The discriminatory power of the RSP typing appears to be comparable with the conventional β-tubulin gene analysis. This method would therefore be suitable for species identification and discrimination at strain to species level. Because RSP typing can characterize the strains within section Fumigati, this method is potentially a powerful and reliable tool in the field of clinical microbiology.
Methods
Cell culture and preparation of ribosomal protein samples
The strains of the 14 species used in this study are summarized in Table 1. All sample strains were provided by Chiba University’s Medical Mycology Research Center (Chiba, Japan) and were grown in potato dextrose broth (PDB) medium at 25 °C for three days. After incubation, harvesting and the preparation of the ribosomal fractions were similar to the methods described in our previous paper [28]. The cultured mycelia were harvested by centrifugation and ground between zirconia silica beads. After removing the beads and cell debris by centrifugation, the fungus lysates were subjected to ultracentrifugation. The resulting ribosome fraction was solubilized in 20 - 50 μL of 50% acetonitrile containing 1% trifluoroacetic acid (TFA), and then subjected to MALDI–TOF MS measurement. Detailed sample preparation procedures are shown in Supporting Information Additional file 5: Figure SI-15.
MALDI–TOF MS measurements
Sample preparation, apparatus, and MALDI–TOF MS data acquisition methods were similar to those described in our previous papers [18–25]. The ribosomal protein sample solution (ca. 1 μL) was spotted onto the MALDI target. About 1 μL of a sinapinic acid matrix solution at a concentration of 20 mg/mL in 50% acetonitrile with 1% TFA was then overlaid and dried in air. The MALDI–TOF MS measurements were performed using an AXIMA CFR-plus time-of-flight mass spectrometer (Shimadzu/Kratos, Kyoto, Japan) in positive linear mode. At least nine mass spectra for each sample were collected by each of three repeated measurements for each of three sample spots (total 3 spots × 3 measurements). External mass calibration was carried out using three peaks of ACTH (human, 1-24) ([M + H]+, m/z 2932.6) and myoglobin ([M + H]+, m/z 16952.6 and [M + 2H]2+, m/z 8476.8) as references.
Calculation of the theoretical mass of RSPs
The amino acid sequence of each RSP was obtained from UniProtKB [27]. Because the genome sequence of A. viridinutans IFM 54703T, sequenced by Chiba University’s Medical Mycology Research Center, was not registered on the public databases at this time, in-house draft genome sequence data were used. The annotated RSP sequences of A. viridinutans have been deposited at DDBJ under the accession numbers LC213039-LC213063. The amino acid sequences of L39 and S21 of A. niger were not determined from shotgun sequences in the database. Therefore, these sequences were determined and have been deposited at DDBJ under the accession numbers LC255002 and LC215003. The sequence mass of each RSP was predicted using a Compute pI/Mw tool on the ExPASy proteomics server [40]. After taking into account the post-translational modifications as effected in our previous paper [28] (such as N-terminal methionine loss, acetylation, methylation and hydroxylation), the theoretical mass of each expressed RSP was calculated as [M + H]+ ion. Detailed construction procedure of the ribosomal protein biomarker list together with peak assignments are shown in Supporting Information Additional file 5: Figure SI-16.
Phylogenetic analysis of RSPs
The observed masses of each RSP were compared with the reference masses constructed in this study. The matching of the average observed masses to the reference masses was judged from errors within 150 ppm. The results of mass matching were designated as RSP types. The RSP typing profiles for each sample strain were processed using UPGMA to build a dendrogram cluster for analysis employing a categorical coefficient, using BioNumerics software (version 3.5; Applied Maths, Kortrijk, Belgium).
The partial DNA sequence of β-tubulin was obtained from the UniProtKB and the alignment of the sequences was performed using ClustalW [41] software. The dendrogram was constructed using MEGA6 [42] software.
Additional files
The accession number (TrEMBL) of ribosomal protein biomarkers of genome-sequenced strains used in this study. (DOCX 22 kb)
Corrected amino acid sequences and relating information of RSPs. (DOCX 80 kb)
Mass spectra of genome sequenced sample strains used in this study. Figure SI-1. Mass spectra of RSPs of N. fischeri NRRL 181T. Figure SI-2. Mass spectra of RSPs of A. lentulus IFM 54703T. Figure SI-3. Mass spectra of RSPs of A. viridinutans IFM 47045T. Figure SI-4. Mass spectra of RSPs of A. udagawae IFM 46973T. Figure SI-5. Mass spectra of RSPs of A. clavatus NRRL 1NT. Figure SI-6. Mass spectra of RSPs of A. niger CBS 513.88. Figure SI-7. Mass spectra of RSPs of A. kawachii IFO 4308. Figure SI-8. Mass spectra of RSPs of A. flavus NRRL 3357. Figure SI-9. Mass spectra of RSPs of A. oryzae RIB 40. Figure SI-10. Mass spectra of RSPs of A. nidulans FGSC A4. (PPTX 472 kb)
Post-translational modifications. Figure SI-11. Peak shift (+42 Da) of S24 with acetylation. (a) A. fumigatus A1163, (b) A. viridinutans IFM 47045T, (c) A. clavatus NRRL 1NT, (d) A. niger CBS 513.88, (e) A. flavus NRRL 3357, and (f) A. nidulans FGSC A4. Figure SI-12. Methylation of L42. (1) Peak shift of +14 Da from sequence mass in L42 of the Aspergillus species; (a) A. fumigatus A1163, (b) N. fischeri NRRL 181T, (c) A. lentulus IFM 54703T, (d) A. viridinutans IFM 47045T, (e) A. udagawae IFM 46973T, (f) A. clavatus NRRL 1NT, (g) A. niger CBS 513.88, (h) A. flavus NRRL 3357, and (i) A. nidulans FGSC A4. (2) Amino acid sequences around Lys-55. Figure SI-13. Dihydroxylation of S23. (1) Peak shift of +32 Da from sequence mass in L23 of Aspergillus species; (a) A. fumigatus A1163, (b) N. fischeri NRRL 181T, (c) A. niger CBS 513.88, (d) A. flavus NRRL 3357, and (e) A. nidulans FGSC A4. (2) Amino acid sequences around Pro-64. Figure SI-14. Peak shift (+28 Da) of S27 with dimethylation. (a) A. fumigatus A1163, (b) A. niger CBS 513.88, (c) A. flavus NRRL 3357, and (d) A. nidulans FGSC A4. (DOCX 187 kb)
Detailed experimental protocols. Detailed sample preparation procedures. Figure SI-16. Detailed construction procedure of the ribosomal protein biomarker list together with peak assignments. (PPTX 61 kb)
Acknowledgements
This work was supported in part by a research grant from the Institute for Fermentation, Osaka (IFO), JSPS Kakenhi Grant Number 25430198, and the National Bioresource Project (Pathogenic Microbes) in Japan (http://www.nbrp.jp/).
Availability of data and materials
The annotated RSP sequences of A. viridinutans have been deposited at DDBJ under the accession numbers LC213039-LC213063. The amino acid sequences of L39 and S21 of A. niger have been deposited at DDBJ under the accession numbers LC255002 and LC215003.
Authors’ contribution
SN, HS, RT, and TY conceived the study. RT, and TY cultured fungal strains and prepared the RSP samples. SN and HS operated MALDI-TOFMS and did the mass spectral data analysis. RT, YK, HT, and TY did gene sequencing and all authors participated gene data analysis. SN and HS drafted the manuscript. RT, HT, and TY helped to draft the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- CDS
Coding DNA sequences
- MALDI-TOF MS
Matrix-assisted laser desorption ionization time-of-flight mass spectrometry
- MLST
Multi-locus sequence typing
- RSP
Ribosomal subunit protein
- UPGMA
Unweighted pair group method with arithmetic mean
Contributor Information
Sayaka Nakamura, Email: sayaka-nakamura@aist.go.jp.
Hiroaki Sato, Email: sato-hiroaki@aist.go.jp.
Reiko Tanaka, Email: moru@faculty.chiba-u.jp.
Yoko Kusuya, Email: y-kusuya@chiba-u.jp.
Hiroki Takahashi, Email: hiroki.takahashi@chiba-u.jp.
Takashi Yaguchi, Email: yaguchi@chiba-u.jp.
References
- 1.Gibbons JG, Rokas A. The function and evolution of the Aspergillus genome. Trends Microbiol. 2013;21(1):14–22. doi: 10.1016/j.tim.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Latge JP. Aspergillus fumigatus and aspergillosis. Clin Microbiol Rev. 1999;12(2):310–354. doi: 10.1128/cmr.12.2.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buzina W. Aspergillus - Classification and Antifungal Susceptibilities. Curr Pharm Des. 2013;19(20):3615–3628. doi: 10.2174/1381612811319200005. [DOI] [PubMed] [Google Scholar]
- 4.McClenny N. Laboratory detection and identification of Aspergillus species by microscopic observation and culture: the traditional approach. Med Mycol. 2005;43:S125–S128. doi: 10.1080/13693780500052222. [DOI] [PubMed] [Google Scholar]
- 5.Hinrikson HP, Hurst SF, Lott TJ, Warnock DW, Morrison CJ. Assessment of ribosomal large-subunit D1-D2, internal transcribed spacer 1, and internal transcribed spacer 2 regions as targets for molecular identification of medically important Aspergillus species. J Clin Microbiol. 2005;43(5):2092–2103. doi: 10.1128/JCM.43.5.2092-2103.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glass NL, Donaldson GC. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol. 1995;61(4):1323–1330. doi: 10.1128/aem.61.4.1323-1330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hong SB, Go SJ, Shin HD, Frisvad JC, Samson RA. Polyphasic taxonomy of Aspergillus fumigatus and related species. Mycologia. 2005;97(6):1316–1329. doi: 10.1080/15572536.2006.11832738. [DOI] [PubMed] [Google Scholar]
- 8.Serrano R, Gusmao L, Amorim A, Araujo R. Rapid identification of Aspergillus fumigatus within the section Fumigati. BMC Microbiol. 2011;11:7. doi: 10.1186/1471-2180-11-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peterson SW. Phylogenetic analysis of Aspergillus species using DNA sequences from four loci. Mycologia. 2008;100(2):205–226. doi: 10.1080/15572536.2008.11832477. [DOI] [PubMed] [Google Scholar]
- 10.Bain JM, Tavanti A, Davidson AD, Jacobsen MD, Shaw D, Gow NAR, Odds FC. Multilocus sequence typing of the pathogenic fungus Aspergillus fumigatus. J Clin Microbiol. 2007;45(5):1469–1477. doi: 10.1128/JCM.00064-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chalupova J, Raus M, Sedlarova M, Sebela M. Identification of fungal microorganisms by MALDI-TOF mass spectrometry. Biotechnol Adv. 2014;32(1):230–241. doi: 10.1016/j.biotechadv.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 12.De Carolis E, Posteraro B, Lass-Florl C, Vella A, Florio AR, Torelli R, Girmenia C, Colozza C, Tortorano AM, Sanguinetti M, et al. Species identification of Aspergillus, Fusarium and Mucorales with direct surface analysis by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Microbiol Infect. 2012;18(5):475–484. doi: 10.1111/j.1469-0691.2011.03599.x. [DOI] [PubMed] [Google Scholar]
- 13.Iriart X, Lavergne RA, Fillaux J, Valentin A, Magnaval JF, Berry A, Cassaing S. Routine Identification of Medical Fungi by the New Vitek MS Matrix-Assisted Laser Desorption Ionization-Time of Flight System with a New Time-Effective Strategy. J Clin Microbiol. 2012;50(6):2107–2110. doi: 10.1128/JCM.06713-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Del Chierico F, Masotti A, Onori M, Fiscarelli E, Mancinelli L, Ricciotti G, Alghisi F, Dimiziani L, Manetti C, Urbani A, et al. MALDI-TOF MS proteomic phenotyping of filamentous and other fungi from clinical origin. J Proteome. 2012;75(11):3314–3330. doi: 10.1016/j.jprot.2012.03.048. [DOI] [PubMed] [Google Scholar]
- 15.Ranque S, Normand AC, Cassagne C, Murat JB, Bourgeois N, Dalle F, Gari-Toussaint M, Fourquet P, Hendrickx M, Piarroux R. MALDI-TOF mass spectrometry identification of filamentous fungi in the clinical laboratory. Mycoses. 2014;57(3):135–140. doi: 10.1111/myc.12115. [DOI] [PubMed] [Google Scholar]
- 16.Hettick JM, Green BJ, Buskirk AD, Kashon ML, Slaven JE, Janotka E, Blachere FM, Schmechel D, Beezhold DH. Discrimination of Aspergillus isolates at the species and strain level by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry fingerprinting. Anal Biochem. 2008;380(2):276–281. doi: 10.1016/j.ab.2008.05.051. [DOI] [PubMed] [Google Scholar]
- 17.Welker M. Proteomics for routine identification of microorganisms. Proteomics. 2011;11(15):3143–3153. doi: 10.1002/pmic.201100049. [DOI] [PubMed] [Google Scholar]
- 18.Sun L, Teramoto K, Sato H, Torimura M, Tao H, Shintani T. Characterization of ribosomal proteins as biomarkers for matrix-assisted laser desorption/ionization mass spectral identification of Lactobacillus plantarum. Rapid Commun Mass Spectrom. 2006;20(24):3789–3798. doi: 10.1002/rcm.2801. [DOI] [PubMed] [Google Scholar]
- 19.Teramoto K, Sato H, Sun L, Torimura M, Tao H, Yoshikawa H, Hotta Y, Hosoda A, Tamura H. Phylogenetic classification of Pseudomonas putida by MALDI-MS using ribosomal proteins as biomarkers. Anal Chem. 2007;79(22):8712–8719. doi: 10.1021/ac701905r. [DOI] [PubMed] [Google Scholar]
- 20.Teramoto K, Sato H, Sun L, Torimura M, Tao H. A simple intact protein analysis by MALDI-MS for characterization of ribosomal proteins of two genome-sequenced lactic acid bacteria and verification of their amino acid sequences. J Proteome Res. 2007;6(10):3899–3907. doi: 10.1021/pr070218l. [DOI] [PubMed] [Google Scholar]
- 21.Hotta Y, Teramoto K, Sato H, Yoshikawa H, Hosoda A, Tamura H. Classification of genus Pseudomonas by MALDI-TOF MS based on ribosomal protein coding in S10-spc-alpha operon at strain level. J Proteome Res. 2010;9(12):6722–6728. doi: 10.1021/pr100868d. [DOI] [PubMed] [Google Scholar]
- 22.Hotta Y, Sato J, Sato H, Hosoda A, Tamura H. Classification of the genus Bacillus based on MALDI-TOF MS analysis of ribosomal proteins coded in S10 and spc operons. J Agric Food Chem. 2011;59(10):5222–5230. doi: 10.1021/jf2004095. [DOI] [PubMed] [Google Scholar]
- 23.Sato H, Teramoto K, Ishii Y, Watanabe K, Benno Y. Ribosomal protein profiling by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry for phylogenety-based subspecies resolution of Bifidobacterium longum. Syst Appl Microbiol. 2011;34(1):76–80. doi: 10.1016/j.syapm.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 24.Hotta Y, Sato H, Hosoda A, Tamura H. MALDI-TOF MS analysis of ribosomal proteins coded in S10 and spc operons rapidly classified the Sphingomonadaceae as alkylphenol polyethoxylate-degrading bacteria from the environment. FEMS Microbiol Lett. 2012;330:23–29. doi: 10.1111/j.1574-6968.2012.02525.x. [DOI] [PubMed] [Google Scholar]
- 25.Sato H, Torimura M, Kitahara M, Ohkuma M, Hotta Y, Tamura H. Characterization of the Lactobacillus casei group based on the profiling of ribosomal proteins coded in S10-spc-alpha operons as observed by MALDI-TOF MS. Syst Appl Microbiol. 2012;35(7):447–454. doi: 10.1016/j.syapm.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 26.Ryzhov V, Fenselau C. Characterization of the protein subset desorbed by MALDI from whole bacterial cells. Anal Chem. 2001;73(4):746–750. doi: 10.1021/ac0008791. [DOI] [PubMed] [Google Scholar]
- 27.UniProt Knowledgebase (UniProtKB) [http://www.uniprot.org/]
- 28.Nakamura S, Sato H, Tanaka R, Yaguchi T. Verification of Ribosomal Proteins of Aspergillus fumigatus for use as Biomarkers in MALDI-TOF MS identification. Mass Spectrometry (Tokyo) 2016;5:A0049. doi: 10.5702/massspectrometry.A0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.National Center for Biotechnology Information (NCBI) The protein database [https://www.ncbi.nlm.nih.gov/protein/]
- 30.Mager WH, Planta RJ, Ballesta JPG, Lee JC, Mizuta K, Suzuki K, Warner JR, Woolford J. A new nomenclature for the cytoplasmic ribosomal proteins Saccharomyces cerevisiae. Nucleic Acids Res. 1997;25(24):4872–4875. doi: 10.1093/nar/25.24.4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lamoth F. Aspergillus fumigatus-Related Species in Clinical Practice. Front Microbiol. 2016;7:8. doi: 10.3389/fmicb.2016.00683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Payne GA, Nierman WC, Wortman JR, Pritchard BL, Brown D, Dean RA, Bhatnagar D, Cleveland TE, Machida M, Yu J. Whole genome comparison of Aspergillus flavus and A-oryzae. Med Mycol. 2006;44:S9–S11. doi: 10.1080/13693780600835716. [DOI] [PubMed] [Google Scholar]
- 33.Yaguchi T, Horie Y, Tanaka R, Matsuzawa T, Ito J, Nishimura K. Molecular Phylogenetics of Multiple Genes on Aspergillus Section Fumigati Isolated from Clinical Specimens in Japan. Medical Mycology Journal. 2007;48(1):37–46. doi: 10.3314/jjmm.48.37. [DOI] [PubMed] [Google Scholar]
- 34.Balajee SA, Gribskov JL, Hanley E, Nickle D, Marr KA. Aspergillus lentulus sp nov., a new sibling species of A-fumigatus. Eukaryot Cell. 2005;4(3):625–632. doi: 10.1128/EC.4.3.625-632.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Verwer PEB, van Leeuwen WB, Girard V, Monnin V, van Belkum A, Staab JF, Verbrugh HA, Bakker-Woudenberg I, van de Sande WWJ. Discrimination of Aspergillus lentulus from Aspergillus fumigatus by Raman spectroscopy and MALDI-TOF MS. Eur J Clin Microbiol Infect Dis. 2014;33(2):245–251. doi: 10.1007/s10096-013-1951-4. [DOI] [PubMed] [Google Scholar]
- 36.Pinel C, Arlotto M, Issartel JP, Berger F, Pelloux H, Grillot R, Symoens F. Comparative proteomic profiles of Aspergillus fumigatus and Aspergillus lentulus strains by surface-enhanced laser desorption ionization time-of-flight mass spectrometry (SELDI-TOF-MS) BMC Microbiol. 2011;11:10. doi: 10.1186/1471-2180-11-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Novakova A, Hubka V, Dudova Z, Matsuzawa T, Kubatova A, Yaguchi T, Kolarik M. New species in Aspergillus section Fumigati from reclamation sites in Wyoming (USA) and revision of A. viridinutans complex. Fungal Divers. 2014;64(1):253–274. doi: 10.1007/s13225-013-0262-5. [DOI] [Google Scholar]
- 38.Barrs VR, van Doorn TM, Houbraken J, Kidd SE, Martin P, Pinheiro MD, Richardson M, Varga J, Samson RA. Aspergillus felis sp nov., an Emerging Agent of Invasive Aspergillosis in Humans, Cats, and Dogs. Plos One. 2013;8(6):11. doi: 10.1371/journal.pone.0064871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sugui JA, Peterson SW, Figat A, Hansen B, Samson RA, Mellado E, Cuenca-Estrella M, Kwon-Chung KJ. Genetic Relatedness versus Biological Compatibility between Aspergillus fumigatus and Related Species. J Clin Microbiol. 2014;52(10):3707–3721. doi: 10.1128/JCM.01704-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Compute pI/Mw tool [http://www.expasy.org/tools/pi_tool.html]
- 41.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 42.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The accession number (TrEMBL) of ribosomal protein biomarkers of genome-sequenced strains used in this study. (DOCX 22 kb)
Corrected amino acid sequences and relating information of RSPs. (DOCX 80 kb)
Mass spectra of genome sequenced sample strains used in this study. Figure SI-1. Mass spectra of RSPs of N. fischeri NRRL 181T. Figure SI-2. Mass spectra of RSPs of A. lentulus IFM 54703T. Figure SI-3. Mass spectra of RSPs of A. viridinutans IFM 47045T. Figure SI-4. Mass spectra of RSPs of A. udagawae IFM 46973T. Figure SI-5. Mass spectra of RSPs of A. clavatus NRRL 1NT. Figure SI-6. Mass spectra of RSPs of A. niger CBS 513.88. Figure SI-7. Mass spectra of RSPs of A. kawachii IFO 4308. Figure SI-8. Mass spectra of RSPs of A. flavus NRRL 3357. Figure SI-9. Mass spectra of RSPs of A. oryzae RIB 40. Figure SI-10. Mass spectra of RSPs of A. nidulans FGSC A4. (PPTX 472 kb)
Post-translational modifications. Figure SI-11. Peak shift (+42 Da) of S24 with acetylation. (a) A. fumigatus A1163, (b) A. viridinutans IFM 47045T, (c) A. clavatus NRRL 1NT, (d) A. niger CBS 513.88, (e) A. flavus NRRL 3357, and (f) A. nidulans FGSC A4. Figure SI-12. Methylation of L42. (1) Peak shift of +14 Da from sequence mass in L42 of the Aspergillus species; (a) A. fumigatus A1163, (b) N. fischeri NRRL 181T, (c) A. lentulus IFM 54703T, (d) A. viridinutans IFM 47045T, (e) A. udagawae IFM 46973T, (f) A. clavatus NRRL 1NT, (g) A. niger CBS 513.88, (h) A. flavus NRRL 3357, and (i) A. nidulans FGSC A4. (2) Amino acid sequences around Lys-55. Figure SI-13. Dihydroxylation of S23. (1) Peak shift of +32 Da from sequence mass in L23 of Aspergillus species; (a) A. fumigatus A1163, (b) N. fischeri NRRL 181T, (c) A. niger CBS 513.88, (d) A. flavus NRRL 3357, and (e) A. nidulans FGSC A4. (2) Amino acid sequences around Pro-64. Figure SI-14. Peak shift (+28 Da) of S27 with dimethylation. (a) A. fumigatus A1163, (b) A. niger CBS 513.88, (c) A. flavus NRRL 3357, and (d) A. nidulans FGSC A4. (DOCX 187 kb)
Detailed experimental protocols. Detailed sample preparation procedures. Figure SI-16. Detailed construction procedure of the ribosomal protein biomarker list together with peak assignments. (PPTX 61 kb)
Data Availability Statement
The annotated RSP sequences of A. viridinutans have been deposited at DDBJ under the accession numbers LC213039-LC213063. The amino acid sequences of L39 and S21 of A. niger have been deposited at DDBJ under the accession numbers LC255002 and LC215003.


