Abstract
Background
ABC transporter superfamily is one of the largest and ubiquitous groups of proteins. Because of their role in detoxification, insect ABC transporters have gained more attention in recent years. In this study, we annotated ABC transporters from a newly sequenced sweetpotato whitefly genome. Bemisia tabaci Q biotype is an emerging global invasive species that has caused extensive damages to field crops as well as ornamental plants.
Results
A total of 55 ABC transporters containing all eight described subfamilies (A to H) were identified in the B. tabaci Q genome, including 8 ABCAs, 3 ABCBs, 6 ABCCs, 2 ABCDs, 1 ABCE, 3 ABCFs, 23 ABCGs and 9 ABCHs. In comparison to other species, subfamilies G and H in both phloem- and blood-sucking arthropods are expanded. The temporal expression profiles of these 55 ABC transporters throughout B. tabaci developmental stages and their responses to imidacloprid, a neonicotinoid insecticide, were investigated using RNA-seq analysis. Furthermore, the mRNA expression of 24 ABC transporters (44% of the total) representing all eight subfamilies was confirmed by the quantitative real-time PCR (RT-qPCR). Furthermore, mRNA expression levels estimated by RT-qPCR and RNA-seq analyses were significantly correlated (r = 0.684, p < 0.01).
Conclusions
It is the first genome-wide analysis of the entire repertoire of ABC transporters in B. tabaci. The identification of these ABC transporters, their temporal expression profiles during B. tabaci development, and their response to a neonicotinoid insecticide lay the foundation for functional genomic understanding of their contribution to the invasiveness of B. tabaci.
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-017-3706-6) contains supplementary material, which is available to authorized users.
Keywords: ATP-binding cassette transporter, Bemisia tabaci, Genome, Gene family expansion, Invasiveness
Background
ATP-binding cassette (ABC) transporters constitute one of the largest protein superfamily and exists in all living organisms [1–3]. These transporters share highly conserved nucleotide binding domains (NBDs), which contain Walker A, Walker B and a signature motif, the C loop, linking the two Walker boxes [4, 5]. In addition to NBDs, a eukaryotic ABC transporter typically consists of one or two transmembrane domains (TMDs), which contain 6–11 membrane-spanning α-helices and dictate substrate specificity [1]. A classic domain architecture of a full-transporter has a TMD-NBD-TMD-NBD arrangement from the N- to C-terminus, while half-transporters have only one set of NBD-TMD. Based on the homology of their NBDs, human ABC transporters have been categorized into seven subfamilies, ABCA to ABCG [1]. ABCH, which was first discovered in Drosophila melanogaster, exists in arthropods and zebrafish but not in mammals, plants and fungi [6–12].
Most ABC transporters encodes membrane-bound proteins that carry a wide range of molecules (e.g., amino acids, peptides, vitamins, sugars, lipids, sterols, hormones, endogenous metabolites, inorganics and xenobiotics) across membranes [1]. ABC transporters use energy released by ATP hydrolysis at the NBDs to transport molecular cargoes across the membrane. ABC transporters also function in cell signaling, ribosome assembly and translation [3, 13]. For example, ABCE and ABCF proteins do not function as classic transporters but participate in transcription, translation and ribosome assembly [14, 15]. Furthermore, plant ABC transporters also contribute to osmolality and phytoalexin functions (plant-pathogen interactions) [16].
The advent of Genomics Era has greatly improved our understanding of the diversity and function of invertebrate ABC transporters, including 56 in the fruit fly D. melanogaster [1], 64 in the water flea Daphnia pulex [13], 51 in the silkworm Bombyx mori [8], 73 in the red flour beetle Tribolium castaneum [17] and 103 in the spider mites Tetranychus urticae [10]. Based on the transcriptome data, Epis et al. suggested that ABC transporters were involved in the permethrin resistance in a malaria vector, Anopheles stephensi [18]. Similarly, ABC transporters were implicated in the degradation of plant secondary metabolites in the cotton bollworm, Helicoverpa armigera [19].
The sweetpotato whitefly, Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae), is a phloem-feeding insect pest that cause substantial damages through feeding on over 600 host plants as well as by transmitting over 100 plant viruses [20]. As one of the most invasive pests, B. tabaci is generally considered as a species complex consisting of many biotypes or putative species with different biological and genetic characteristics [21]. The two most invasive and destructive B. tabaci biotypes are the Q (also known as Mediterranean, MED) and B (also known as Middle East-Asia Minor 1, MEAM1) [21]. B. tabaci has demonstrated a remarkable ability to develop resistance to insecticides in the field, including organophosphates, carbamates, pyrethroids, neonicotinoids and juvenile hormone mimics [22–24]. Insecticide was the driving force facilitating the B. tabaci biotype replacement events in China (B replaced indigenous biotypes in early 2000, and the subsequent replacement by Q in 2009) [25]. The overall goal of this research is to understand the involvement of ABC transporters, a family of phase III detoxification enzymes, in the metabolic resistance in B. tabaci. To archive this goal, we 1) provided the first genome-wide analysis of the entire repertoire of ABC transporters in the whitefly B. tabaci Q; 2) investigated the temporal expression profiles of these ABC transporters in different developmental stages; and 3) documented their transcriptomic response to imidacloprid, a neonicotinoid insecticide.
Results and discussion
Identification of ABC transporters in B. tabaci
A total of 55 ABC transporters was identified in B. tabaci Q genome (Table 1). The number of ABC transporters in B. tabaci Q is comparable with other arthropod species; however, subfamilies G and H expanded in phloem- and blood-sucking arthropods (Table 2 and Additional file 1: Figure S1). For example, ABCG and ABCH account for 58% (42 and 16%, respectively) of ABC transporters in the phloem-sucking B. tabaci, and 49% (45 and 4%, respectively) in the blood-sucking Cimex lectularius. The percentage reduces to 21% (18% + 3%) in the red flour beetle, T. castaneum. The expansion of ABCG and ABCH subfamilies in these polyphagous and phloem-sucking arthropods, including the whitefly B. tabaci, the wheat aphid D. noxia and the spider mite T. urticae, suggest the potential contribution of ABC transporters to their adaptability and invasiveness. Phylogenetic analysis categorized the 55 B. tabaci ABC transporters into eight subfamilies (Fig. 1). To further understand the evolutionary placement of B. tabaci ABC transporters, in-depth phylogenetic analyses were conducted for each subfamily. The results of these analyses were described and discussed in the following sections.
Table 1.
ABC transporters identified in the B. tabaci Q genome
| Subfamily | Gene ID | Length (aa) | Location (strand) | Exon | Topology (SMART) | Matched gene (Accession no.) | N-Glca | O-Glcb | CDS statusc |
|---|---|---|---|---|---|---|---|---|---|
| ABCA | Btabq017043.1 | 1655 | Scaffold_3468:1006:32679+ | 20 | TMD-NBD-TMD-NBD | ABCA3 (AIN44094.1) | 10 | 7 | C |
| Btabq003223.3 | 1387 | Scaffold_13:22456:46016- | 18 | TMD-NBD-TMD-NBD | ABCA3 (XP_008199154.1) | 7 | 12 | C | |
| Btabq003232.1 | 1710 | Scaffold_13:231650:263916+ | 22 | TMD-NBD-TMD-NBD | ABCA3 (XP_008580143.1) | 13 | 13 | C | |
| Btabq009375.1 | 1607 | Scaffold_204:588616:628011+ | 24 | TMD-NBD-TMD-NBD | ABCA3 (XP_004627914.1) | 11 | 11 | C | |
| Btabq009376.1 | 1642 | Scaffold_204:635079:664477:- | 24 | TMD-NBD-TMD-NBD | ABCA3 (XP_007499572.1) | 6 | 14 | C | |
| Btabq022409.1 | 1642 | Scaffold_499:371203:397123:+ | 23 | TMD-NBD-TMD-NBD | ABCA3 (XP_007499572.2) | 6 | 16 | C | |
| Btabq022410.1 | 1653 | Scaffold_499:405484:424414:- | 21 | TMD-NBD-TMD-NBD | ABCA3 (XP_007499572.3) | 11 | 15 | C | |
| Btabq008198.1 | 1730 | Scaffold_188:144605:225978+ | 25 | TMD-NBD-TMD-NBD | ABCA5 (XP_015513231.1) | 11 | 25 | C | |
| ABCB | Btabq001304.1 | 837 | Scaffold_11:1763748:1779600:- | 13 | TMD-NBD | ABCB6 (KDR08849.1) | 5 | 0 | C |
| Btabq028971.1 | 684 | Scaffold_89:122752:135916:+ | 13 | TMD-NBD | ABCB7 (XP003707011.1) | 3 | 12 | C | |
| Btabq013065.1 | 903 | Scaffold_266:334207:358911:- | 19 | TMD-NBD | ABCB8 (KDR20884.1) | 2 | 8 | C | |
| ABCC | Btabq000311.1 | 1543 | Scaffold_102:620714:659592:+ | 28 | TMD-NBD-TMD-NBD | ABCC1 (XP_012252456.1) | 8 | 10 | C |
| Btabq019529.2 | 1457 | Scaffold_400:253922:298887:+ | 30 | TMD-NBD-TMD-NBD | ABCC1 (KYM99989.1) | 11 | 5 | C | |
| Btabq026695.3 | 1422 | Scaffold_728:117063:145027:+ | 27 | TMD-NBD-TMD-NBD | ABCC4 (XP_003487856.1) | 8 | 11 | C | |
| Btabq008004.1 | 1302 | Scaffold_1847:75114:101542:- | 21 | TMD-NBD-TMD-NBD | ABCC4 (XP_015366071.1) | 11 | 4 | C | |
| Btabq004618.1 | 1527 | Scaffold_1440:62283:85796:- | 24 | TMD-NBD-TMD-NBD | ABCC7 (KDR17053.1) | 11 | 3 | C | |
| Btabq003933.1 | 1558 | Scaffold_137:590708:610342:+ | 25 | TMD-NBD-TMD-NBD | ABCC9 (XP_012225332.1) | 9 | 3 | C | |
| ABCD | Btabq026746.1 | 733 | Scaffold_732:28889:99259:+ | 4 | TMD-NBD | ABCD2 (AIN44106.1) | 3 | 5 | C |
| Btabq017051.1 | 670 | Scaffold_347:310155:194947:- | 15 | TMD-NBD | ABCD3 (XP_008186233.1) | 4 | 9 | C | |
| ABCE | BtabqABCE1 | 608 | Scaffold_2889:666:11372:+ | 10 | NBD-NBD | ABCE1 (XP_014275278.1) | 1 | 5 | C |
| ABCF | Btabq009873.1 | 595 | Scaffold_2118:27608:29395:+ | 1 | NBD-NBD | ABCF1 (XP_971562.1) | 5 | 0 | C |
| Btabq014578.1 | 626 | Scaffold_354:220262:229341:+ | 10 | NBD-NBD | ABCF2 (XP_966990.1) | 4 | 2 | C | |
| Btabq016264.1 | 746 | Scaffold_3286:21844:40980:- | 13 | NBD-NBD | ABCF3 (KDR22359.1) | 3 | 5 | C | |
| ABCG | Btabq003568.1 | 679 | Scaffold_133:520218:533101:- | 14 | NBD-TMD | ABCG1 (KDR13078.1) | 6 | 0 | C |
| Btabq006006.1 | 634 | Scaffold_1598:84277:126599:+ | 11 | NBD-TMD | ABCG1 (XP_001943523.1) | 1 | 4 | C | |
| Btabq007377.1 | 594 | Scaffold_176:564047:611847:- | 11 | NBD-TMD | ABCG1 (XP_001947776.2) | 1 | 10 | C | |
| Btabq020594.1 | 703 | Scaffold_434:86892:119931:- | 14 | NBD-TMD | ABCG1 (XP_001942514.1) | 1 | 6 | C | |
| Btabq029952.1 | 654 | Scaffold_973:236:11383:+ | 9 | NBD-TMD | ABCG1 (KDR12258.1) | 1 | 6 | C | |
| Btabq009742.1 | 300 | Scaffold_434:92701:149213:- | 6 | NBD | ABCG1 (XP_008476671.1) | 1 | 3 | P | |
| Btabq029281.1 | 676 | Scaffold_910:266071:279898:- | 12 | NBD-TMD | ABCG4 (XP_003246360.1) | 6 | 0 | C | |
| Btabq007131.3 | 649 | Scaffold_1729:110948:148435:+ | 13 | NBD-TMD | ABCG4 (XP_008472465.1) | 2 | 4 | C | |
| Btabq023008.1 | 590 | Scaffold_523:150957:167280:- | 11 | NBD-TMD | ABCG4 (XP_003246361.1) | 2 | 6 | C | |
| Btabq009608.1 | 590 | Scaffold_208:388361:402826:- | 10 | NBD-TMD | ABCG4 (XP_003246361.1) | 2 | 6 | C | |
| Btabq009611.1 | 590 | Scaffold_208:557522:571601:+ | 11 | NBD-TMD | ABCG4 (XP_003246361.1) | 2 | 6 | C | |
| Btabq023919.1 | 639 | Scaffold_563:13213:51650:- | 10 | NBD-TMD | ABCG4 (XP_008470161.1) | 3 | 9 | C | |
| Btabq013894.1 | 590 | Scaffold_282:256960:283064:+ | 11 | NBD-TMD | ABCG4 (XP_003246361.1) | 2 | 6 | C | |
| Btabq000844.1 | 655 | Scaffold_1057:77963:94241:+ | 10 | NBD-TMD | ABCG4 (XP_015509620.1) | 2 | 0 | C | |
| Btabq026080.1 | 692 | Scaffold_683:129726:137969:+ | 3 | NBD-TMD | ABCG4 (KDR08336.1) | 6 | 11 | C | |
| Btabq020567.1 | 692 | Scaffold_4326:21216:33527:+ | 4 | NBD-TMD | ABCG4 (KDR08336.1) | 6 | 11 | C | |
| Btabq001288.1 | 612 | Scaffold_11:1463363:1478800:- | 10 | NBD-TMD | ABCG5 (KDR20187.1) | 2 | 19 | C | |
| Btabq001290.1 | 952 | Scaffold_11:1493109:1530869:+ | 15 | TMD | ABCG8 (XP_001944045.2) | 4 | 19 | P | |
| Btabq023890.1 | 755 | Scaffold_561:73328:85538:- | 15 | NBD-TMD | ABCG14 (XP_014258500.1) | 3 | 1 | C | |
| Btabq014028.1 | 607 | Scaffold_2842:67736:80804:+ | 12 | NBD-TMD | Scarlet (XP_014276471.1) | 3 | 11 | C | |
| Btabq015484.1 | 645 | Scaffold_3117:65588:78996:+ | 14 | NBD-TMD | Scarlet (XP_014279356.1) | 5 | 9 | C | |
| Btabq002474.1 | 667 | Scaffold_121:146665:158695:- | 11 | NBD-TMD | Scarlet (NP_001306193.1) | 3 | 6 | C | |
| Btabq022510.1 | 700 | Scaffold_50:595327:617912:+ | 13 | NBD-TMD | White (KDR20987.1) | 3 | 8 | C | |
| ABCH | Btabq015123.1 | 685 | Scaffold_304:505925:535800:- | 15 | NBD-TMD | ABCG23 (XP_001945365.2) | 6 | 2 | C |
| Btabq003158.1 | 627 | Scaffold_129:187020:214919:- | 14 | NBD-TMD | ABCG23 (XP_001945365.2) | 4 | 1 | C | |
| Btabq006712.2 | 687 | Scaffold_167:585953:624494:- | 16 | NBD-TMD | ABCG23 (XP_001945365.2) | 6 | 2 | C | |
| Btabq018898.1 | 679 | Scaffold_386:127646:145952:- | 15 | NBD-TMD | ABCG23 (XP_001945365.2) | 4 | 6 | C | |
| Btabq019352.1 | 685 | Scaffold_398:364203:394966:+ | 15 | NBD-TMD | ABCG23 (XP_015364832.1) | 6 | 2 | C | |
| Btabq026264.1 | 647 | Scaffold_7:597935:614293:- | 14 | NBD-TMD | ABCG23 (XP_001942931.1) | 5 | 1 | C | |
| Btabq028063.1 | 749 | Scaffold_82:494722:529828:- | 16 | NBD-TMD | ABCG23 (XP_001945213.1) | 6 | 11 | C | |
| Btabq009745.1 | 695 | Scaffold_21:1506328:1537013:- | 12 | NBD-TMD | ABCG20 (XP_014289229.1) | 5 | 3 | C | |
| BTA027409.1 | 764 | Scaffold_791:944:21302:+ | 16 | NBD-TMD | ABCG20 (XP_001951744.1) | 6 | 8 | C |
aN-glycosylation sites were predicted using the NetNGlyc 1.0 server (http://www.cbs.dtu.dk/services/NetNGlyc/); only N-glycosylation sites with a “potential” score > 0.5 and with a jury agreement were considered
bO-glycosylation sites were predicted using the NetOGlyc 4.0 server (http://www.cbs.dtu.dk/services/NetOGlyc/) [96]; if the G-score was > 0.5, the residue was considered to be O-glycosylated; this table shows the total number of O-glycosylated sites (glycosylated serines and threonines)
cC indicates a complete sequence; P indicates a partial sequence
Table 2.
The distribution of ABC transporter subfamilies in B. tabaci Q and in other arthropod species
| Phloem-sucking | Blood-sucking | Chewing/Sucking | Chewing | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Subfamily | B. tabaci | D. noxia | T. urticae | An. gambiae | P. humanus | C. lectularius | A. mellifera | D. melanogaster | B. mori | H. armicoverpa | T. castaneum |
| ABCA | 8 | 3 | 9 | 9 | 2 | 6 | 3 | 10 | 6 | 7 | 10 |
| ABCB | 3 | 6 | 4 | 5 | 6 | 7 | 7 | 8 | 8 | 11 | 6 |
| ABCC | 6 | 24 | 39 | 13 | 5 | 6 | 9 | 14 | 15 | 11 | 35 |
| ABCD | 2 | 3 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| ABCE | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| ABCF | 3 | 3 | 3 | 3 | 3 | 4 | 3 | 3 | 3 | 3 | 3 |
| ABCG | 23 (42%) | 26 (32%) | 23 (23%) | 16 (31%) | 13 (33%) | 23 (45%) | 15 (35%) | 15 (27%) | 13 (26%) | 16 (30%) | 13 (18%) |
| ABCH | 9 (16%) | 11 (13%) | 22 (21%) | 3 (6%) | 6 (15%) | 2 (3.9%) | 3 (7%) | 3 (5.3%) | 3 (5.8%) | 3 (5.5%) | 3 (4%) |
| Others | 5 | 2 | |||||||||
| Total | 55 (100%) | 82 (100%) | 103 (100%) | 52 (100%) | 40 (100%) | 51 (100%) | 43 (100%) | 56 (100%) | 51 (100%) | 54 (100%) | 73 (100%) |
Species in this survey included Bemisia tabaci, Diuraphis noxia, Tetranychus urticae, Anopheles gambiae, Pediculus humanus humanus, Cimex lectularius, Bombyx mori, Helicoverpa armigera, Apis mellifera, Drosophila melanogaster, and Tribolium castaneum. “Others” refers to uncharacterized genes. The values without parentheses indicate the number of transporters detected; for subfamilies ABCG and ABCH, the values in parentheses indicate the percentage of all ABC transporters represented by the indicated subfamily
Fig. 1.
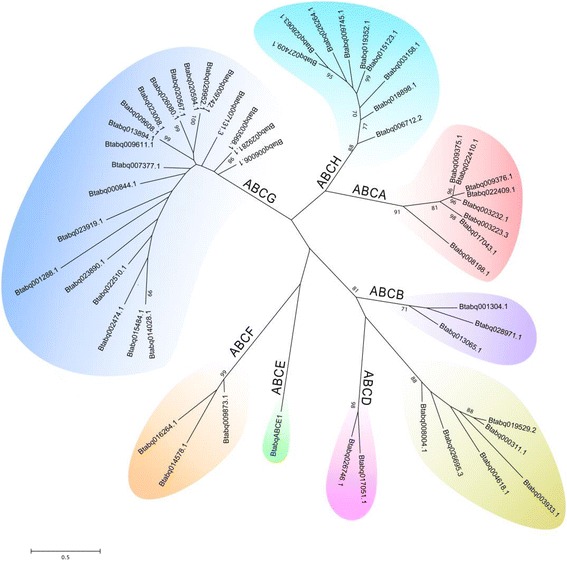
Phylogenetic relationship of 55 ABC transporters in B. tabaci Q. Amino acid sequences of nucleotide-binding domains (NBDs) were aligned using ClustalW and subjected to a maximum likelihood analysis by MEGA5 [97]. Numbers at the branch point of the node represent the value resulting from 1000 replications. All positions with less than 95% site coverage were eliminated
ABCA subfamily
A phylogeny tree of the eight ABCA proteins identified in B. tabaci and other organisms was presented in Additional file 2: Figure S2. All of the B. tabaci ABCA proteins are full-transporters (Table 1), which is consistent with D. pulex and T. urticae [10, 13]. The silkworm ABCA subfamily, in contrast, contains two full-transporters, one half-transporter and three incomplete ABC transporters [8]. Furthermore, no ABCA protein has been identified in yeast [26].
The B. tabaci ABCA subfamily has one of the largest B. tabaci ABC transporters, Btabq008198.1, encoding 1730 amino acids (Table 1). Btabq008198.1 clustered with the ABCA5 group, including Anopheles gambiae AGAp010416-PA, D. noxia DnXP_015375463.1 and H. sapiens ABCA5 cluster (hABCA5, 6, 8, 9 and 10) (Additional file 2: Figure S2). Although these ABCA5-related genes in mammalians (human, dog and mouse) cluster with ABCA5 on the same chromosomes [6, 27], no counterpart of the ABCA5-related genes has been identified in invertebrates, such as B. mori [8] and T. urticae [10]. Besides Btabq008198.1, the other seven B. tabaci ABCA genes (Btabq017043.1, Btabq003223.3, Btabq003232.1, Btabq009375.1, Btabq009376.1, Btabq022409.1, and Btabq022410.1) form a sister-group with a high bootstrap support. This sister-group clustered with D. noxia DnXP_015366480.1 (ABCA3), T. japonicas TjABCA3 and seven T. urticae genes. These genes then clustered with D. melanogaster DmCG31731 with a moderate bootstrap support. DmCG31731 was down-regulated in the salivary glands of a D. melanogaster E93 mutant [28]. As an early ecdysone responsive gene, E93 was a primary regulator of programmed cell death in D. melanogaster development [29]. Injection of dsRNAs synthesized from the two ABCA genes, TcABCA-9A and TcABC A-9B, led to approximately 30% mortality in the red flour beetle, T. castaneum [17].
ABCB subfamily
In B. tabaci Q, we identified three ABCB half-transporters but no full-transporters (Table 1). These B. tabaci half-transporters (Btabq001304.1, Btabq013065.1 and Btabq013065.1) belong to three clades, indicating that these genes are homologous to ABCB genes in human, fruit fly and other insects (Additional file 3: Figure S3). Btabq001304.1 clustered with ABCB6 proteins from other species, including D. noxia DnXP_015376167.1, An. gambiae AGAP002278, D. melanogaster DmCG4225, T. japonicas TjABCB6, and H. sapiens hABCB6. Similarly, Btabq028971.1 formed an ABCB7 clade with D. noxia DnXP_015378905.1, D. melanogaster DmCG7955, T. japonicas TjABCB7, and H. sapiens hABCB7. Meanwhile, Btabq013065.1 was clustered with ABCB8 from D. melanogaster (DmCG1824), T. urticae (Tu17g02000), T. japonicas (TjABCB8) and H. sapiens (hABCB8).
In H. sapiens, hABCB6, hABCB7 and hABCB8/MABC1 are mitochondrial transporters with roles in iron metabolism and transportation of Fe/S protein precursors [30, 31]. Drosophila melanogaster DmCG4225 (ABCB6) was tolerant to cadmium [32]. Disruption of C. elegans ABTM- 1, a homologue of ABCB7, induced oxidative stress and premature cell death [33]. Furthermore, a homologous gene of H. sapiens ABCB7 in Aedes aegypti was up-regulated in an insecticide resistant strain [34]. Disruption of mouse ABCB8 decreased mitochondrial iron export and led to cardiomyopathy [35]. Similar to D. pulex, B. tabaci Q do not have homologous genes of human ABCB half-transporters that were associated with antigen processing (hABCB2, hABCB3, and hABCB9) [13].
ABCC subfamily
Insects typically have different number of ABCC transporters (Table 2). In B. tabaci Q, we identified six ABCC full-transporters, including Btabq019529.2, Btabq000311.1, Btabq008004.1, Btabq026695.3, Btabq004618.1 and Btabq003933.1. ABCC subfamily proteins are full-transporters with diverse functions, such as cell-surface receptor, ion transport and translocation of a broad range of substrates including drugs and endogenous compounds [36–39]. Due to their diverse functions, human ABCC subfamily members refer to multidrug resistance associated proteins (MRPs), which function as transporters; the cystic fibrosis transmembrane conductance regulator (CFTR, ABCC7), which forms a chloride channel; and sulfonylurea receptors (SUR1/2, ABCC8/9), which function as regulators of potassium channels [40]. In H. sapiens, “long” MRPs like ABCC1, 2, 3, 6 and 10 (MRP1, 2, 3, 6 and 7, respectively) have a TMD0, the third N-terminal transmembrane-spanning domain, while “short” MRPs like ABCC4, 5, 11 and 12 (MRP4, 5, 8 and 9, respectively) lack TMD0 [40].
In our phylogenetic analysis, two B. tabaci ABCC1 genes, Btabq019529.2 and Btabq000311.1, clustered with D. noxia DnXP_015366208.1, DnXP_015366209.1, D. melanogaster DmCG6214, T. japonicus ABCC1 and a group of H. sapiens “long” MRPs (MRP1, 2, 3 and 6) (Additional file 4: Figure S4). MRP1/ABCC1 transports a structurally diverse range of endogenous substances (e.g. leukotrienes and estrogen conjugates), xenobiotics and their metabolites [41]. These MRPs have been extensively studied as transporters of natural product drugs such as anthracyclines and plant alkaloids [42]. As a homologous gene to H. sapiens MRP1, MRP2, MRP3 and MRP6, DmCG6214 functions as a high capacity ATP-dependent organic anion transporter and acts as a transporter of ecdysteroid and juvenile hormone conjugates [43]. The other two ABCC genes in B. tabaci, Btabq008004.1 and Btabq026695.3, gathered with a large clade of ABCC4 in D. noxia, An. gambiae and a cluster of Drosophila ABCC proteins, including DmCG10505, DmCG14709 and others. MRP4/ABCC4 can remove a wide range of endogenous and exogenous molecules from cells [44]. The ABCC transporter encoded by CG10505 in D. melanogaster was regulated by heavy metals and was involved in biochemical detoxification of zinc and copper [45]. Drosophila DmCG14709 gene, a homologue of H. sapiens MRP4/ABCC4, was tightly regulated by oxygen [46]. The human body louse ABCC4 gene, PhABCC4, was up-regulated after exposed to the pesticide ivermectin. Injection of female lice with PhABCC4 dsRNA significantly increased their sensitivity to ivermectin [47].
Btabq004618.1 clustered with D. noxia DnXP_015373973.1, T. japonicas TjABCC7, D. melanogaster DmCG7806, H. sapien hABCC10/MRP7 and T. urticae Tu03g07840. hABCC10/MRP7 was able to transport amphipathic anions and conferred resistance to some antitumor drugs [48]. In addition, Btabq003933.1 clustered with sulfonylurea receptors (SURs) in D. noxia, D. melanogaster, T. japonicas, T. urticae and H. sapiens. Unlike other ABCC subfamily genes, SURs help form ATP-sensitive potassium (KAT P) channels [38, 49]. Although SUR was considered to be a direct target of chitin synthesis inhibitors [50], it did not express in D. melanogaster epidermis, where chitin disruption was observed [51]. Moreover, data obtained from D. melanogaster suggested that chitin synthesis was independent of SUR [52]. Similarly, RNAi knockdown of SUR homologue in T. castaneum did not result in a phenotype [17]. ABCC subfamily is not only involved in detoxification and multidrug resistance in Manduca sexta but also participate in host plant selection in T. urticae [10, 53].
ABCD subfamily
Bemisia tabaci Q has two ABCD transporters, Btabq026746.1 and Btabq017051.1 (Table 1). The same amount of ABCDs have also been found in other insects [8], while more ABCDs were identified in D. pulex, H. sapiens and C. elegans. Members of ABCD subfamily are half-transporters and involve in the import of fatty acids and acyl-CoAs into this organelle [55]. ABCD transporters participated in peroxisome-related developmental progress in C. elegans [56]. Mutations in hABCD1/ALDP gene resulted in adrenoleukodystrophy, a genetic disorder that occurs primarily in males [57]. Our phylogenetic analysis suggested that three clades of ABCDs in metazoans, with the two B. tabaci ABCDs distributed in two clades (Additional file 5: Figure S5). Btabq026746.1 clustered with D. noxia, DnXP_015374078.1, An. gambiae AGAP002071, D. melanogaster DmCG216, T. japonicas TjABCD2, H. sapiens hABCD2 and hABCD1. Meanwhile, Btabq017051.1 belongs to ABCD3 clade.
ABCE and ABCF subfamilies
ABCE and ABCF subfamilies contain atypical ABC transporters characterized by a pair of linked NBDs with no TMDs [1]. Their structure implied their roles in biological processes other than transportation. ABCE1 is one of the most evolutionarily conserved proteins and expressed in all organisms except eubacteria. Because of its role in translation and ribosome biosynthesis, ABCE1 is essential for all life stages [58]. Homo sapiens ABCE1/RNase L was initially identified as an inhibitor of RNase L [58]. Members of the ABCE subfamily in human and yeast also have a role in translation initiation [59]. However, the ABCE and ABCF transporters have not been well characterized in invertebrates. RNAi knockdown of an ABCE gene, TcABCE-3A, resulted in significant mortality in penultimate T. castaneum larvae [17]. ABCE subfamily has only one member in all metazoans studied to date [9, 60], and B. tabaci follows this rule (BtabqABCE1, Table 1 and Additional file 6: Figure S6).
For ABCF subfamily, three transporters, Btabq009873.1, Btabq014578.1, and Btabq016264.1, were annotated and showed well-supported sister clades (Additional file 7: Figure S7). Btabq009873.1, Btabq014578.1, and Btabq016264.1 were placed within the ABCF1, ABCF2, and ABCF3 clades, respectively. In human and yeast, ABCF proteins participated in gene regulation systems and ribosome assembly [14]. Mutations in the yeast GCN20 gene, which was involved in the initiation and control of translation, reduced Eif2α phosphorylation and translation in the ribosome [61]. Knockdown of TcABCF-2A led to 100% mortality in penultimate T. castaneum larvae [17].
ABCG subfamily
ABCGs have a typical reverse domain architecture, with a NBD localized at the N-terminus and a TMD at the C-terminus (NBD-TMD). The ABCG transporters in metazoans are half-transporters and can be a functional transporter only after dimerization. In plants and fungi, however, ABCGs are full-transporters, also called pleiotropic drug resistance proteins (PDRs) [16, 62]. Because of their duplicated domain structure, yeast PDRs were not included in the phylogenetic analyses in this study.
ABCG proteins represent the largest ABC subfamily (23 members) in B. tabaci (Table 1) and also form the largest ABC subfamily in D. noxia [63], An. gambiae [7], bed bug (Cimex lectularius) [64], human body louse (Pediculus humanus humanus) [65], Drosophila [1] and A. mellifera [8] (Table 2). According to Sturm et al. [13], the expansion of ABCG transporters in D. pulex and D. melanogaster genomes was resulted from extensive lineage specific gene duplications.
Among the 23 ABCG members in B. tabaci Q, 17 are ABCG1 and ABCG4 transporters. These ABCG1 and ABCG4 proteins clustered with those in D. noxia, An. gambiae, T. urticae, and D. melanogaster (DmCG9664, DmCG32091, DmCG4822, DmCG31689, DmCG9663, DmCG17646, DmCG5853 and DmCG3164) (Additional file 8: Figure S8). The ABCG4 gene has the same intron/exon structure as ABCG1, suggesting that they arose by a relatively recent gene duplication event [66]. Similar to ABCG1, ABCG4 can be inducible by oxysterols and retinoids [67]. In H. sapiens, hABCG1 and hABCG4 were involved in cellular cholesterol efflux [68]. In addition, the elevated expression of ABCG4 in An. stephensi suggested its involvement in the degradation of a pyrethroid insecticide [18, 69].
Btabq001288.1 and Btabq001290.1 clustered with DnXP_01579755.1, DnXP_015379710.1, DmCG11069, DmCG31121, Tu01g16280, TjABCG5, hABCG5 and hABCG8. hABCG5 and hABCG8 mediated the intestinal and biliary efflux of cholesterol and sterols [68]. A clear subclass was formed with Btabq023890.1, D. melanogaster DmCG3327, and T. urticae tetur17g02510. DmCG3327 (also named E23), a 20-hydroxyecdysone (20E)-induced ABC transporter, regulated metamorphosis, probably by removing 20E from cells [70]. Similarly, ABCG expression in silkworm midgut was also regulated by 20E [8]. RNAi-based functional study confirmed a similar role in metamorphosis of TcABCG-8A, a homologue of E23 in T. castaneum [17].
Among invertebrates, the functions of ABCGs were first characterized in Drosophila as pigment precursor transporters (brown, scarlet and white genes) [71, 72]. In our phylogenetic analysis, four B. tabaci ABCG proteins, Btabq014028.1, Btabq015484.1, Btabq002474.1 and Btabq022510.1, showed homologous relationships with these pigment precursor transporters in D. noxia, An. gambiae and D. melanogaster. Besides transporting pigment precursors, these transporters also function in courtship behavior [73], transport of biogenic amines [74] and up-take of uric acid [75]. RNAi-mediated suppression and genetic linkage analysis confirmed that down-regulation of Pxwhite gene was tightly linked to Cry1Ac resistance in Plutella xylostella [76].
Based on a recent review, 80% of the peer-reviewed publications involving arthropod ABC transporters (50) linked ABC transporters with insecticides-resistance [77]. Members of ABCG subfamily were over-expressed in thiamethoxam-resistant B. tabaci, suggesting the involvement of these genes in insecticide resistance [78]. In addition, ABCG subfamily not only participated in multi-pesticide resistance in T. urticae but also reduced the chemical defense from host plants [10]. In B. mori, at least five ABCG genes acted as 20-hydroxyecdysone (20E)-response genes and participated in hormonal regulation [8]. These results suggested that members of the ABCG subfamily play significant roles in the development and the removal of amphiphilic xenobiotics. Although ABC transporters in subfamilies B, C, and G are involved in the metabolic resistance to various xenobiotics, B. tabaci Q, however, contains predominantly G, but few B and C family members (Table 2). This bias reflects the importance of ABCG subfamily in B. tabaci to fend off xenobiotic offences derived from host plants as well as agroecosystems.
ABCH subfamily
As inverse half-transporters, ABCH proteins share the same domain architecture as ABCG transporters. ABCH transporters were first discovered in D. melanogaster and have only annotated in arthropods and zebrafish [6, 8, 10, 11, 13, 17]. ABCH proteins have not been identified in fungi, C. elegans, plants and mammals [1, 16, 54, 62]. ABCH subfamily was documented in zebrafish but not in catfish, fugu or cod [79].
A total of nine ABCH transporters were identified in B. tabaci. Phylogenetic analysis showed that ABCH subfamily members were species-specific. Bemisia tabaci ABCH clustered with genes in D. noxia with a high bootstrap value (Additional file 9: Figure S9). An RNAi screen of D. melanogaster genes revealed that silencing of D. melanogaster CG9990 was lethal [80, 81]. In two insecticide-resistant P. xylostella strains, an ABCH (Px014955), was the most up-regulated ABC transporter [82]. Knockdown of TcABCH-9C, an ABCH in T. castaneum, resulted in 100% larval mortality and a significant reduction of fecundity and hatching rate. In addition, T. castaneum larvae injected with TcABCH-9C dsRNA showed a lack of lipids in their epicuticle, indicating that TcABCH-9C may be required for the formation of a waterproof barrier in the epicuticle [17]. In the cotton bollworm Helicoverpa armigera and Manduca sexta, the expression of ABCH subfamily was highly induced after larvae fed with secondary metabolites [19, 53].
Expression profiles across developmental stages
Based on RNA-seq data, all 55 ABC transporters were expressed with a RPKM (Reads Per Kilobases per Million reads) >1 in at least one of the B. tabaci life stages (Additional file 10: Table S1). Members from ABCE and ABCF subfamilies were ubiquitous and expressed highly throughout B. tabaci developmental stages (Fig. 2). In the polyphagous spider mite, T. urticae, an ABCE (tetur30g01400), showed high expression level throughout the life cycle [10]. Moreover, TcABCE-3A transcripts were abundant in all developmental stages of T. castaneum [17]. The expression profile of ABCE genes in the intertidal copepod, T. japonicas, and sea lamprey, Petromyzon marinus, showed similar trends as well [83, 84]. Btabq014578.1 (ABCF2) had the highest expression level throughout the life cycle. ABCE1, ABCF1 and ABCF2 were highly expressed in all developmental stages of B. tabaci. ABCF3, also ubiquitously expressed, exhibited a lower expression level. These gene expression patterns were consistent with the fundamental functions in ribosome biogenesis and translation regulation [17, 85, 86]. The results suggest that ABCE and ABCF transporters were possibly essential to maintain “housekeeping” function in B. tabaci.
Fig. 2.
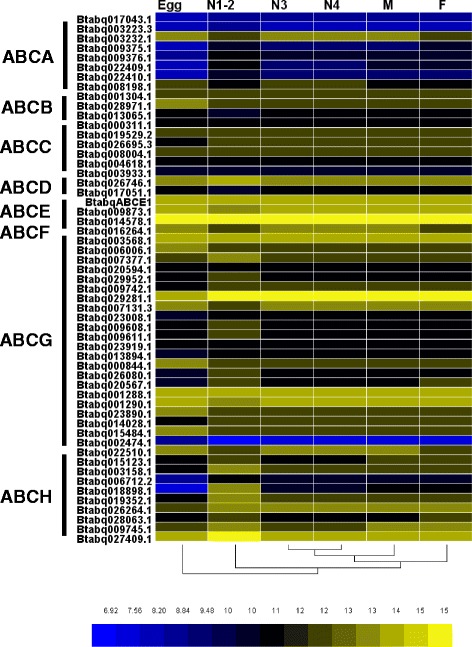
Expression profiles of ABC transporters throughout B. tabaci Q life cycle. The codes on the left are the gene ID numbers of the 55 ABC transporters in B. tabaci Q. The mRNA levels, as represented by log2 (RPKM + 1) values, are shown in the gradient heat map with colors ranging from blue (low expression) to yellow (high expression). For each stage, values are normalized data from three biological replicates. E, egg; N1-2, 1st- and 2nd-instar nymphs; N3, 3rd-instar nymph; N4, 4th-instar nymph; F, adult female; M, adult male
The expression levels for ABCA transporters in B. tabaci were relatively low with the exception of Btabq003232.1 (ABCA3), which was highly expressed in all life stages (Fig. 2). ABCA3 is critical for the proper formation of lamellar bodies and surfactant function and may participate in surfactant phospholipid metabolism [87]. Btabq001304.1 (ABCB6) expressed in all life stages of B. tabaci and highly expressed in eggs and nymphs. Btabq028971.1 (ABCB7) expressed in all developmental stages, especially in the fourth nymph stage and adult. Btabq013065.1 (ABCB8) expressed in all stages but at a lower level than other ABCB genes. Homo sapiens ABCB half-transporters, ABCB6, ABCB7, ABCB8 and ABCB10, were localized at mitochondria and functioned in iron metabolism [88].
Expression levels of all ABCC members were documented in each developmental stage of B. tabaci Q, and the expression patterns were different among these genes (Fig. 2). Only Btabq008004.1 was highly expressed throughout life stages (Fig. 2). The two B. tabaci ABCDs, Btabq026746.1 and Btabq017051.1, showed stable expressional level among different developmental stages (Fig. 2). Btabq026746.1 was highly expressed while Btabq017051.1 showed slight lower expression level. Because the two ABCDs are homologous to those of mammal and insect transporters, similar functions such as roles in fatty acid metabolism can be inferred in B. tabaci Q.
Members of the ABCG subfamily showed different expression patterns in B. tabaci Q. Btabq003568.1, Btabq029281.1 and Btabq007131.3 were ubiquitously with high expression profiles. Expression levels were also high for Btabq006006.1, Btabq007377.1, Btabq029952.1 and Btabq000844.1. Overall, ABCG1 and ABCG4 members in B. tabaci Q had moderate to high expressional levels in all developmental stages. The ABCG4 in An. stephensi showed an increased expressional level in defense against the pyrethroid insecticide [18, 69]. Btabq001288.1 (ABCG5) and Btabq001290.1 (ABCG8) showed high expression in all stages of B. tabaci. Homologues of ABCG5 and ABCG8 in D. melanogaster and T. castaneum function, respectively, in metamorphosis [17] and may have a similar function in B. tabaci. Furthermore, Btabq022510.1 was annotated as a white gene and its expression level in nymph and adult stages was especially high. Recently, a Pxwhite gene was shown to be tightly linked to Cry1Ac resistance in P. xylostella [76].
ABCH transporters differed in expression patterns during B. tabaci development. Btabq027409.1 and Btabq026264.1 were highly expressed in all developmental stages. Moreover, Btabq015123.1, Btabq009745.1, and Btabq019352.1 also showed high expression level, especially in the 4th-instar nymphs and adults. One ABCH subfamily gene, CG9990, was presumed to be involved in mortality [81]. Silencing of T. castaneum TcABCH-9C reduced oviposition and pupal molting [17]. ABCH transporters are highest expressed in the head of H. armigera when the larvae were ingested secondary metabolism [19].
Expansion of ABCG and ABCH subfamilies do exist in B. tabaci Q. Combined with their expressional profiles, we suspect that these ABCG and ABCH genes are involved in development-related metabolism and in the detoxification of xenobiotics including insecticides and secondary metabolites from host plant.
mRNA expression of selected ABC transporters by RT-qPCR
The expression profiles of 24 selected ABC transporters were confirmed by the RT-qPCR (Fig. 3). Within ABCA subfamily, Btabq003232.1 had the highest expressional level, followed by Btabq008198.1 and Btabq017043.1. This trend is consistent with RNA-seq analysis. Similarly, gene expression level of ABCB genes from high to low was Btabq028971.2, Btabq001304.1 and Btabq013065.1. Expression profiles of ABCC, ABCD, ABCE and ABCF subfamilies were also consistent with our RNA-seq data. In ABCG subfamily, the trend persisted. Only two genes in ABCH subfamily, Btabq028063.1 and Btabaq026264.1, exhibited lower expressional level, which is not consistent with the RNA-seq data. Statistical analysis by SPSS demonstrated a significant correlation between transcriptome data and RT-qPCR data (r = 0.684, p < 0.01).
Fig. 3.
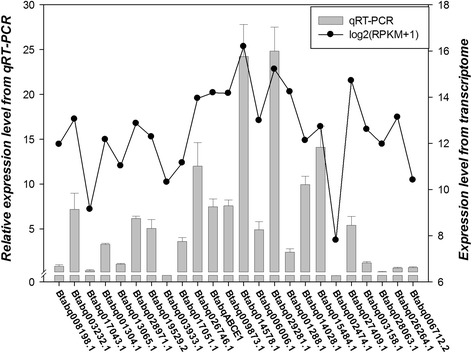
mRNA expressions of selected B. tabaci Q ABC transporters by RT-qPCR. The bars represent expression level of B. tabaci Q ABC transporters relative to the Btabq008198.1. Data are presented as means ± SE. Spots indicated log2 (RPKM + 1) values of ABC transporters in adults. Significant correlation between RT-qPCR and transcriptomic analyses were assessed by SPSS
Transcriptional response to imidacloprid treatments
A total of five genes were significantly up-regulated in imidacloprid-treated B. tabaci (Fig. 4, Additional file 11: Table S4). Btabq008198.1, an ABCA5 gene, can regulate accumulation of cholesterol [89]. The other four belong to ABCG subfamily, including Btabq003568.1 (ABCG1), Btabq020594.1 (ABCG1), Btabq007131.3 (ABCG4), and Btabq029281.1 (ABCG4).
Fig. 4.
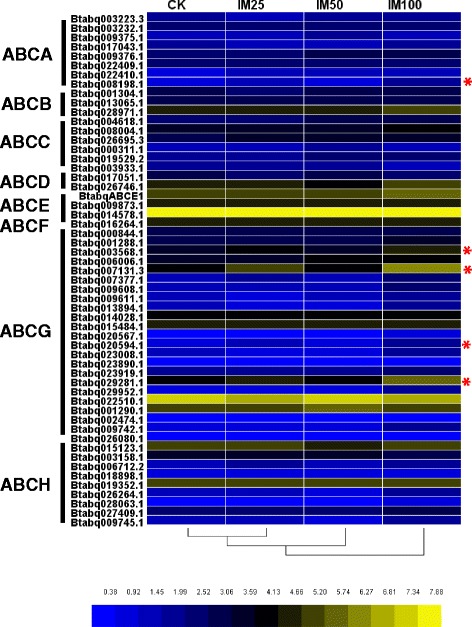
Transcriptional response of ABC transporters in B. tabaci Q adults to imidacloprid. The mRNA expression levels, i.e., log2 (RPKM + 1) values, are represented in a gradient heat map from low (blue) to high (yellow). Values were averaged from the three biological replications. The B. tabaci Q adults were fed with either water (CK) or imidacloprid at 25, 50, and 100 mg/L concentration (IM25, IM50, or IM100, respectively). An asterisk indicates a significant difference
The metabolic detoxification of xenobiotics likely involves a set of detoxification enzymes, which are general classified into three categories. Besides Phase I (e.g., cytochrome P450 monooxygenases) and Phase II (e.g., glutathione S-transferases), ABC transporters represent a major class of Phase III enzymes.
ABC transporters, especially those in the ABCG subfamily, facilitated H. armigera defense against secondary metabolites such as nicotine and tomatine [19]. The high expression of ABCG11 in H. armigera was the results of nicotine ingestion [19]. In addition, ABCG4 in An. stephensi was up-regulated in response to pyrethroid insecticide [18, 69]. The expression levels of three ABC transporters, ABCG20-3, ABCG23-5 and ABCH-B, were higher in field-collected resistant populations of C. lectularius [90]. Then knockdown of ABCG20-3 reduced deltamethrin resistance [90]. ABC transporters, particularly ABCG1 and ABCG4, may have important role in response to xenobiotics. Moreover, ABCG4 gene has the same intron/exon structure as ABCG1, suggesting that they arose by a relatively recent gene duplication event [66]. Overall, feeding on imidacloprid significantly affected the expression of ABC transporters especially those in ABCG subfamily, suggesting the potential involvement of these genes in the B. tabaci metabolic resistance.
Conclusion
In this study, we identified 55 ABC transporters in the sweetpotato whitefly B. tabaci Q genome. ABC transporter subfamilies G and H are expanded in B. tabaci Q genome. The expression profiles of these ABC transporters were validated using both RT-qPCR and RNA-seq analyses. Furthermore, 80% of the significantly up-regulated ABC transporters in imidacloprid-challenged B. tabaci were ABCGs. These combined results imply the potential contribution of ABCGs to the adaptability of this emerging global invasive pest.
Methods
Colony maintenance and sample preparation
Bemisia tabaci Q biotype (Mediterranean, MED) population maintained on cotton plant at 27 ± 1 °C, with a photoperiod of 16 h light: 8 h darkness and 70 ± 10% relative humidity. Every 3–5 generations, the purity of the strain was monitored using a mitochondrial cytochrome oxidase I (mtCOI) marker [91].
The different developmental stage samples (eggs, four nymph stages, females and males) were collected from the Q biotype population described above. Newly emerged (within 5 days) adults of B. tabaci Q on cotton were collected to be treat with imidacloprid following adult leaf-dip bioassay method [92, 93]. In brief, leaf discs (22 mm in diameter) from cotton plants were dipped in imidacloprid at 0 (distilled water control), 25, 50 and 100 mg/L for 10s. When dry, the leaf discs were placed on agar in a flat-bottomed glass tube, 20 B. tabaci adults were added to each tube. After 48 h, adults were collected, snap frozen in liquid nitrogen, and transferred to −80 °C for the long-term storage. Three biological replications for all B. tabaci samples were carried out independently. A total of 100 individuals were collected for each biological replication, and these samples were subjected to RNA-seq analysis.
Identification and annotation of ABC transporters
To identify open reading frames (ORFs) encoding putative ABC transporters, we carried out tblastn searches against the B. tabaci Q genome [94] and transcriptome assembled from SRP064690. We subsequently used the highly conserved NBDs (as defined by InterPro domain IPR003439/PF00005) and full-sequences of all 56 ABC genes in D. melanogaster as queries to search against the updated GLEAN gene in B. tabaci genome with an E-value threshold of 10−5 to identify ABC transporters. Each putative ABC transporter gene was confirmed by subjected it to BLASTX analysis with the non-redundant protein sequence (NR) at GenBank (http://www.ncbi.nlm.nih.gov/).
The exon/intron boundary and start/stop codons of each ABC gene were confirm from the genome and transcriptome. The conserved domains of these identified ABC proteins were predicted using SMART (simple modular architecture research tool, http://smart.embl-heidelberg.de/) [95] and confirmed by using the NCBI conserved domain search service tool. Putative N-glycosylation sites were predicted using NetNGlyc 1.0 server and O-glycosylation sites were predicted using NetOGlyc 4.0 server [96].
Phylogenetic analyses
To analyze the evolutionary placement of ABC transporters in B. tabaci, we collected the readily available ABC transporters from the fruit fly, D. melanogaster, wheat aphid, D. noxia, mosquito, An. gambiae, spider mite, T. urticae, intertidal copepod, T. japonicus, human, H. sapiens, nematode, Caenorhabditis elegans, and yeast, Saccharomyces cerevisiae. NBDs of B. tabaci ABC transporters were used to resolve their phylogenetic relationships at the subfamily level. Within each ABC subfamily, we used the full-length protein sequences to construct their respective phylogeny trees [9, 10]. Additional file 12: Table S5 lists models of the constructed phylogenetic trees. Sequences of entire transporters aligned using ClustalW and then subjected to phylogenetic analysis by MEGA5 with 1000 bootstrap replications [97]. The phylogenetic trees were constructed using the maximum-likelihood method.
Expression profiling of ABC transporters
Expression profiling of ABC transporters was assessed using transcriptome data of different developmental stages and imidacloprid-treated whitefly as described earlier. ABC genes were also verified by quantitative real-time reverse transcription polymerase chain reaction (RT-qPCR). Total RNA was extracted using Trizol reagent according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA, USA). RNA was quantified using a Nanodrop 2000 (Thermo Scientific, Wilmington, DE, USA) meanwhile purity was checked on 1% agarose gels. RNA-seq libraries were constructed as previously described [98] and sequenced on a HiSeq 2500 system according to the manufacturer’s instructions with sequenced at 125 bp (PE125, library size is 280–320 bp). RNA-seq libraries of imidacloprid-treated samples were constructed by the same method and sequenced on a HiSeq 4000 system according to the manufacturer’s instructions with sequenced at 150 bp (PE150, library size is 450–550 bp). The raw reads were filtered with Fastq clean software [99] to trim low quality (Q value < 20) nucleotides on both ends, clipping the adapter and barcode sequences from the 3’ end and discarding the ribosomal RNA (rRNA) sequence. The high-quality cleaned reads were then aligned to the pre-prepared RNA sequence dataset with the Bowtie program [100] allowing one mismatch. After alignments, raw counts for each B. tabaci transcript and each sample were derived and were normalized to Reads Per Kilobase of transcript per Million mapped reads (RPKM). Differentially expressed genes (fold changes > 2 and adjusted P-value < 0.05) were identified by the DESeq package [101–103].
Besides transcriptomic validation, mRNA expressions of ABC transporters in B. tabaci Q biotype were confirmed using RT-qPCR analysis as well. Based on the RPKM value generated from the RNA-seq data, we selected 24 transcripts representing all eight subfamilies of the B. tabaci Q biotype ABC transporters for the RT-qPCR validation study (Additional file 13: Table S2 and Additional file 14: Table S3). RT-qPCR was conducted using an ABI PRISM 7500 Real-time PCR System (Applied Biosystems, Foster, CA, USA), and non-treated B. tabaci adults were subjected to the analysis. All RT-qPCR analyses included three technical replicates for each of three biological replicates. Elongation factor 1 (EF1) and TAF10 RNA polymerase II (TAF) were selected as reference genes [104, 105]. The RT-qPCR was carried out in a 25 μL reaction volume containing 12.5 μL 2 × SuperReal PreMix Plus, 0.5 μL 50 × ROX Reference Dye, 0.75 μL forward primer (10 μM), 0.75 μL reverse primer (10 μM), 1.0 μL cDNA (300 ng/μL), and 9.5 μL RNase-free ddH2O, following the instructions of the SuperReal PreMix Plus (SYBR Green) kit (Tiangen, Beijing, China). The thermal cycling conditions were polymerase activation at 95 °C for 15 min, followed by 40 cycles of denaturation at 95 °C for 10 s, annealing at 60 °C for 30 s and elongation at 72 °C for 32 s. The amplification efficiency was estimated using the equation: E = [10^(−1/slope) − 1] × 100%, in which the slope was derived from plotting the cycle threshold (Ct) value versus six serially diluted template concentrations. The transcript levels of ABC genes were quantified according to the 2−ΔΔCt method [106]. SPSS 19.0 was used to analyze correlations between RT-qPCR data and RNA-seq data.
Additional files
Comparison of gene numbers in each subfamily of ABC transporters between Bemisia tabaci and other organisms. (PDF 23 kb)
Phylogenetic relationship of Bemisia tabaci ABCA subfamily with other organisms. Full-length ABC transporters were aligned using ClustalW and subjected to a maximum likelihood analysis by MEGA5 [97]. Numbers at the branch point of the node represent the values resulting from 1000 replications. Species, abbreviations, and color codes are: Btabq, B. tabaci (red); Dn, D. noxia (blue); AGA, A. gambiae (green); Dm, D. melanogaster (purple); Tu, T. urticae (light blue); Tj, T. japonicus (black); h, H. sapiens (gray); Ce, C. elegans (brick-red); Sc, Saccharomyces cervisiae (orange). (PDF 28 kb)
Phylogenetic relationship of Bemisia tabaci ABCB subfamily with other organisms. See Figure S2 legend for details. (PDF 14 kb)
Phylogenetic relationship of Bemisia tabaci ABCC subfamily with other organisms. See Figure S2 legend for details. (PDF 15 kb)
Phylogenetic relationship of Bemisia tabaci ABCD subfamily with other organisms. See Figure S2 legend for details. (PDF 19 kb)
Phylogenetic relationship of Bemisia tabaci ABCE subfamily with other organisms. See Figure S2 legend for details. (PDF 37 kb)
Phylogenetic relationship of Bemisia tabaci ABCF subfamily with other organisms. See Figure S2 legend for details. (PDF 89 kb)
Phylogenetic relationship of Bemisia tabaci ABCG subfamily with other organisms. See Figure S2 legend for details. (PDF 16 kb)
Phylogenetic relationship of Bemisia tabaci ABCH subfamily with other organisms. See Figure S2 legend for details. (PDF 134 kb)
RPKM values of B. tabaci Q ABC transporters at different developmental stages (DOCX 13 kb)
Fold-change of gene expression following B. tabaci Q exposure to imidacloprid (DOCX 8 kb)
Models used for phylogenetic analysis (DOCX 15 kb)
Selected B. tabaci Q ABC transporters for the RT-qPCR validation study (DOCX 143 kb)
Primers used for the RT-qPCR analysis (DOCX 89 kb)
Acknowledgments
Authors thank anonymous reviewers for their comments on the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (31420103919 and 31672032), and China Agriculture Research System (CARS-26 10). These agencies had no role in study design, data collection/analysis, manuscript preparation, or the decision to publish.
Availability of data and materials
Bemisia tabaci Q ABC transporter dataset will be available from the corresponding authors on reasonable requests. Transcriptome datasets of B. tabaci Q at different development stages and following the exposure to imidacloprid insecticide have been deposited, respectively, in the Sequence Read Archive (SRA) database under the accession number SRP064690 and SRP090405.
Authors’ contributions
LT, XZ and YZ designed this research. LT carried out the experiments, LT, TS, RH analyzed data. XZ and YZ contributed the materials and reagents. LT, WX, QW, SW, XZ and YZ revised the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable
Ethics approval and consent to participate
Not applicable
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- 20E
20-hydroxyecdysone
- ABC
ATP-binding cassette
- CFTR
The cystic fibrosis transmembrane conductance regulator
- MRP
Multidrug resistance associated protein
- NBD
Nucleotide binding domain
- PDR
Pleiotropic drug resistance protein
- RPKM
Reads Per Kilobases of per Million reads
- RT-qPCR
Quantitative real-time reverse transcription polymerase chain reaction
- SUR
Sulfonylurea receptor
- TMD
Transmembrane domain.
Contributor Information
Lixia Tian, Email: tianlixia555@126.com.
Tianxue Song, Email: songtianxue921@163.com.
Rongjun He, Email: herongjun@1gene.com.cn.
Yang Zeng, Email: zengyang73@163.com.
Wen Xie, Email: xiewen@caas.cn.
Qingjun Wu, Email: wuqingjun@caas.cn.
Shaoli Wang, Email: wangshaoli@caas.cn.
Xuguo Zhou, Phone: +86-10-8210-9518, Email: xuguozhou@uky.edu.
Youjun Zhang, Phone: +86-10-8210-9518, Email: zhangyoujun@caas.cn.
References
- 1.Dean M, Rzhetsky A, Allikmets R. The human ATP-binding cassette (ABC) transporter superfamily. Genome Res. 2001;11:1156–1166. doi: 10.1101/gr.GR-1649R. [DOI] [PubMed] [Google Scholar]
- 2.Dassa E, Bouige P. The ABC of ABCs: a phylogenetic and functional classification of ABC systems in living organisms. Res Microbiol. 2001;152:211–229. doi: 10.1016/S0923-2508(01)01194-9. [DOI] [PubMed] [Google Scholar]
- 3.Holland IB, Cole SP, Kuchler K, Higgins CF. ABC proteins: from bacteria to man. Academic Press. 2003.
- 4.Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1(8):945. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davidson AL, Dassa E, Orelle C, Chen J. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol Mol Biol Rev. 2008;72:317–364. doi: 10.1128/MMBR.00031-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dean M, Annilo T. Evolution of the ATP-binding cassette (ABC) transporter superfamily in vertebrates. Annu Rev Genom Hum G. 2005;6:123–142. doi: 10.1146/annurev.genom.6.080604.162122. [DOI] [PubMed] [Google Scholar]
- 7.Roth CW, Holm I, Graille M, Dehoux P, Rzhetsky A, Wincker P, Weissenbach J, Brey PT. Identification of the Anopheles gambiae ATP binding cassette transporter superfamily genes. Mol Cells. 2003;15(2):150–158. [PubMed] [Google Scholar]
- 8.Liu S, Zhou S, Tian L, Guo E, Luan Y, Zhang J, Li S. Genome-wide identification and characterization of ATP-binding cassette transporters in the silkworm, Bombyx mori. BMC Genomics. 2011;12:491. doi: 10.1186/1471-2164-12-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dermauw W, Van Leeuwen T. The ABC gene family in arthropods: comparative genomics and role in insecticide transport and resistance. Insect Biochem Mol Biol. 2014;45:89–110. doi: 10.1016/j.ibmb.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 10.Dermauw W, Osborne EJ, Clark RM, Grbić M, Tirry L, Van Leeuwen T. A burst of ABC genes in the genome of the polyphagous spider mite Tetranychus urticae. BMC Genomics. 2013;14:317. doi: 10.1186/1471-2164-14-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Annilo T, Chen ZQ, Shulenin S, Costantino J, Thomas L, Lou H, Stefanov S, Dean M. Evolution of the vertebrate ABC gene family: analysis of gene birth and death. Genomics. 2006;88:1–11. doi: 10.1016/j.ygeno.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 12.Moon S, Jung KH. Genome-wide expression analysis of rice ABC transporter family across spatio-temporal samples and in response to abiotic stresses. J Plant Physiol. 2014;171(14):1276–1288. doi: 10.1016/j.jplph.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 13.Sturm A, Cunningham P, Dean M. The ABC transporter gene family of Daphnia pulex. BMC Genomics. 2009;10:170. doi: 10.1186/1471-2164-10-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tyzack JK, Wang X, Belsham GJ, Proud CG. ABC50 interacts with eukaryotic initiation factor 2 and associates with the ribosome in an ATP-dependent manner. J Biol Chem. 2000;275:34131–34139. doi: 10.1074/jbc.M002868200. [DOI] [PubMed] [Google Scholar]
- 15.Zhao Z, Fang LL, Johnsen R, Baillie DL. ATP-binding cassette protein E is involved in gene transcription and translation in Caenorhabditis elegans. Biochem Biophys Res Commun. 2004;323:104–111. doi: 10.1016/j.bbrc.2004.08.068. [DOI] [PubMed] [Google Scholar]
- 16.Verrier PJ, Bird D, Burla B, Dassa E, Forestier C, Geisler M, Klein M, Kolukisaoglu U, Lee Y, Martinoia E, Murphy A, Rea PA, Samuels L, Schulz B, Spalding EJ, Yazaki K, Theodoulou FL. Plant ABC proteins-a unified nomenclature and updated inventory. Trends Plant Sci. 2008;13:151–159. doi: 10.1016/j.tplants.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 17.Broehan G, Kroeger T, Lorenzen M, Merzendorfer H. Functional analysis of the ATP-binding cassette (ABC) transporter gene family of Tribolium castaneum. BMC Genomics. 2013;14:6. doi: 10.1186/1471-2164-14-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Epis S, Porretta D, Mastrantonio V, Urbanelli S, Sassera D, De Marco L, Mereghetti V, Montagna M, Ricci I, Favia G, Bandi C. ABC transporters are involved in defense against permethrin insecticide in the malaria vector Anopheles stephensi. Parasites Vectors. 2014;7:349. doi: 10.1186/1756-3305-7-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bretschneider A, Heckel DG, Vogel H. Know your ABCs: Characterization and gene expression dynamic s of ABC transporters in the polyphagous herbivore Helicoverpa armigera. Insect Biochem Mol Biol. 2016;72:1–9. doi: 10.1016/j.ibmb.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 20.Brown JK, Coats SA, Bedford ID, Markham PG, Bird J, Frohlich DR. Characterization and distribution of esterase electromorphs in the whitefly, Bemisia tabaci (Genn.) (Homoptera: Aleyrodidae) Biochem Genet. 1995;33(7–8):205–214. doi: 10.1007/BF02401851. [DOI] [PubMed] [Google Scholar]
- 21.De Barro PJ, Liu SS, Boykin LM, Dinsdale AB. Bemisia tabaci: a statement of species status. Annu Rev Entomol. 2011;56:1–19. doi: 10.1146/annurev-ento-112408-085504. [DOI] [PubMed] [Google Scholar]
- 22.Cahill M, Gorman K, Day S, Denholm I. Baseline determination and detection of resistance to imidacloprid in Bemisia tabaci (Homoptera: Aleyrodidae) Bull Entomol Res. 1996;86:343–349. doi: 10.1017/S000748530003491X. [DOI] [Google Scholar]
- 23.Horowitz AR, Kontsedalov S, Ishaaya I. Dynamics of resistance to the neonicotinoids acetamiprid and thiamethoxam in Bemisia tabaci (Homoptera: Aleyrodidae) J Econ Entomol. 2004;97:2051–2056. doi: 10.1093/jee/97.6.2051. [DOI] [PubMed] [Google Scholar]
- 24.Nauen R, Denholm I. Resistance of insect pests to neonicotinoid insecticides: current status and future prospects. Arch Insect Biochem. 2005;58(4):200–215. doi: 10.1002/arch.20043. [DOI] [PubMed] [Google Scholar]
- 25.Pan H, Preisser EL, Chu D, Wang S, Wu Q, Carrière Y, Zhang Y. Insecticides promote viral outbreaks by altering herbivore competition. Ecol Appl. 2015;25(6):1585–1595. doi: 10.1890/14-0752.1. [DOI] [PubMed] [Google Scholar]
- 26.Decottignies A, Goffeau A. Complete inventory of the yeast ABC proteins. Nat Genet. 1997;15:137–145. doi: 10.1038/ng0297-137. [DOI] [PubMed] [Google Scholar]
- 27.Li G, Shi P, Wang Y. Evolutionary dynamics of the ABCA, chromosome 17q24 cluster genes in vertebrates. Genomics. 2007;89(89):385–391. doi: 10.1016/j.ygeno.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 28.Dutta S. Genetic regulation of autophagic cell death in Drosophila melanogaster (doctoral dissertation) 2008. [Google Scholar]
- 29.Lee CY, Wendel DP, Reid P, Lam G, Thummel CS, Baehrecke EH. E93, Directs Steroid-Triggered Programmed Cell Death in Drosophila. Mol Cell. 2000;6(2):433–443. doi: 10.1016/S1097-2765(00)00042-3. [DOI] [PubMed] [Google Scholar]
- 30.Krishnamurthy PC, Du G, Fukuda Y, Sun D, Sampath J, Mercer KE, Wang J, Sosa-Pineda B, Murti KG, Schuetz JD. Identification of a mammalian mitochondrial porphyrin transporter. Nature. 2006;443(7111):586–589. doi: 10.1038/nature05125. [DOI] [PubMed] [Google Scholar]
- 31.Pondarré C, Antiochos BB, Campagna DR, Clarke SL, Greer EL, Deck KM, McDonald A, Han AP, Medlock A, Kutok JL, Anderson SA, Eisenstein RS, Fleming MD. The mitochondrial ATP-binding cassette transporter Abcb7 is essential in mice and participates in cytosolic iron-sulfur cluster biogenesis. Hum Mol Genet. 2006;15:953–964. doi: 10.1093/hmg/ddl012. [DOI] [PubMed] [Google Scholar]
- 32.Sooksa-Nguan T, Yakubov B, Kozlovskyy VI, Barkume CM, Howe KJ, Thannhauser TW, Rutzke MA, Hart JJ, Kochian LV, Rea PA, Vatamaniuk OK. Drosophila ABC transporter, DmHMT-1, confers tolerance to cadmium: DmHMT-1 and its yeast homolog, SpHMT-1, are not essential for vacuolar phytochelatin sequestration. J Biol Chem. 2009;284:354–362. doi: 10.1074/jbc.M806501200. [DOI] [PubMed] [Google Scholar]
- 33.Gonzalez-Cabo P, Bolinches-Amoros A, Cabello J, Ros S, Moreno S, Baylis HA, Palau F, Vazquez-Manrique RP. Disruption of the ATP-binding cassette B7(ABTM-1/ABCB7) induces oxidative stress and premature cell death in Caenorhabditis elegans. J Biol Chem. 2011;286:21304–21314. doi: 10.1074/jbc.M110.211201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bariami V, Jones CM, Poupardin R, Vontas J, Ranson H. Gene amplification, ABC transporters and cytochrome P450s: unraveling the molecular basis of pyrethroid resistance in the dengue vector, Aedes aegypti. PLoS Negl Trop Dis. 2012;6(6):e1692. doi: 10.1371/journal.pntd.0001692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ichikawa Y, Bayeva M, Ghanefar M, Potini V, Sun L, Mutharasan RK, Wu R, Khechaduri A, Jairaj Naik T, Ardehali H. Disruption of ATP-binding cassette B8 in mice leads to cardiomyopathy through a decrease in mitochondrial iron export. Proc Natl Acad Sci U S A. 2012;109:4152–4157. doi: 10.1073/pnas.1119338109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fukuda Y, Schuetz JD. ABC transporters and their role in nucleoside and nucleotide drug resistance. Biochem Pharmacol. 2012;83:1073–1083. doi: 10.1016/j.bcp.2011.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akrouh A, Halcomb SE, Nichols CG, Sala-Rabanal M. Molecular biology of K-ATP channels and implications for health and disease. IUBMB Life. 2009;61:971–978. doi: 10.1002/iub.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jordan IK, Kota KC, Cui G, Thompson CH, McCarty NA. Evolutionary and functional divergence between the cystic fibrosis transmembrane conductance regulator and related ATP-binding cassette transporters. Proc Natl Acad Sci U S A. 2008;105:18865–18870. doi: 10.1073/pnas.0806306105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kruh GD, Belinsky MG. The MRP family of drug efflux pumps. Oncogene. 2003;22:7537–7552. doi: 10.1038/sj.onc.1206953. [DOI] [PubMed] [Google Scholar]
- 40.Deeley RG, Westlake C, Cole SPC. Transmembrane transport of endo- and xenobiotics by mammalian ATP-binding cassette multidrug resistance proteins. Physiol Rev. 2006;86:849–899. doi: 10.1152/physrev.00035.2005. [DOI] [PubMed] [Google Scholar]
- 41.He SM, Li R, Kanwar JR, Zhou SF. Structural and functional properties of human multidrug resistance protein 1 (MRP1/ABCC1) Curr Med Chem. 2011;18(3):439–481. doi: 10.2174/092986711794839197. [DOI] [PubMed] [Google Scholar]
- 42.Borst P, Evers R, Kool M, Wijnholds J. A family of drug transporters: the multidrug resistance-associated proteins. J Natll Cancer I. 2000;92(16):1295–1302. doi: 10.1093/jnci/92.16.1295. [DOI] [PubMed] [Google Scholar]
- 43.Tarnay JN, Szeri F, Ilias A, Annilo T, Sung C, Le Saux O, Robinow S. The dMRP/CG6214 gene of Drosophila is evolutionarily and functionally related to the human multidrug resistance-associated protein family. Insect Mo Biol. 2004;13(5):539–548. doi: 10.1111/j.0962-1075.2004.00512.x. [DOI] [PubMed] [Google Scholar]
- 44.Russel FGM, Koenderink JB, Masereeuw R. Multidrug resistance protein 4 (MRP4/ABCC4): a versatile efflux transporter for drugs and signalling molecules. Trends Pharmacol Sci. 2008;29(4):200–207. doi: 10.1016/j.tips.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 45.Yepiskoposyan H, Egli D, Fergestad T, Selvaraj A, Treiber C, Multhaup G, Georgiev O, Schaffner W. Transcriptome response to heavy metal stress in Drosophila reveals a new zinc transporter that confers resistance to zinc. Nucleic Acids Res. 2006;34:4866–4877. doi: 10.1093/nar/gkl606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang H, Haddad GG. Drosophila dMRP4 regulates responsiveness to O2 deprivation and development under hypoxia. Physiol Genomics. 2007;29(29):260–266. doi: 10.1152/physiolgenomics.00166.2006. [DOI] [PubMed] [Google Scholar]
- 47.Yoon KS, Strycharz JP, Baek JH, Sun W, Kim JH, Kang JS, Pittendrigh BR, Lee SH, Clark JM. Brief exposures of human body lice to sublethal amounts of ivermectin over-transcribes detoxification genes involved in tolerance. Insect Mol Biol. 2011;20:687–699. doi: 10.1111/j.1365-2583.2011.01097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hopper-Borge E, Xu X, Shen T, Shi Z, Chen ZS, Kruh GD. Human multidrug resistance protein 7 (ABCC10) is a resistance factor for nucleoside analogues and epothilone B. Cancer Res. 2009;69(1):178–184. doi: 10.1158/0008-5472.CAN-08-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bryan J, Muñoz A, Zhang X, Düfer M, Drews G, Krippeit-Drews P, Aguilar-Bryan L. ABCC8 and ABCC9: ABC transporters that regulate K+ channels. Pflügers Archiv - Eur J Physiol. 2007;453(5):703–718. doi: 10.1007/s00424-006-0116-z. [DOI] [PubMed] [Google Scholar]
- 50.Abo-Elghar GE, Fujiyoshi P, Matsumura F. Significance of the sulfonylurea receptor (SUR) as the target of diflubenzuron in chitin synthesis inhibition in Drosophila melanogaster and Blattella germanica. Insect Biochem Mol Biol. 2004;34:743–752. doi: 10.1016/j.ibmb.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 51.Gangishetti U, Breitenbach S, Zander M, Saheb SK, Mueller U, Schwarz H, Moussian B. Effects of benzoylphenylurea on chitin synthesis and orientation in the cuticle of the Drosophila larva. Eur J Cell Biol. 2009;88:167–180. doi: 10.1016/j.ejcb.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 52.Meyer F, Flötenmeyer M, Moussian B. The sulfonylurea receptor SUR is dispensable for chitin synthesis in Drosophila melanogaster embryos. Pest Manag Sci. 2013;69(10):1136–1140. doi: 10.1002/ps.3476. [DOI] [PubMed] [Google Scholar]
- 53.Koenig C, Bretschneider A, Heckel DG, Grosse-Wilde E, Hansson BS, Vogel H. The plastic response of Manduca sexta to host and non-host plants. Insect Biochem Mol Biol. 2015;63:72–85. doi: 10.1016/j.ibmb.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 54.Sheps JA, Ralph S, Zhao Z, Baille DL, Ling V. The ABC transporter gene family of Caenorhabditis elegans has implications for the evolutionary dynamics of multidrug resistance in eukaryotes. Genome Biol. 2004;5(3):1. doi: 10.1186/gb-2004-5-3-r15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Theodoulou FL, Holdsworth M, Baker A. Peroxisomal ABC transporters. FEBS Lett. 2006;580:1139–1155. doi: 10.1016/j.febslet.2005.12.095. [DOI] [PubMed] [Google Scholar]
- 56.Petriv OI, Pilgrim DB, Rachubinski RA, Titorenko VI. RNA interference of peroxisome-related genes in C. elegans: a new model for human peroxisomal disorders. Physiol Genomics. 2002;10:79–91. doi: 10.1152/physiolgenomics.00044.2002. [DOI] [PubMed] [Google Scholar]
- 57.Berger J, Gartner J. X-linked adrenoleukodystrophy: clinical, biochemical and pathogenetic aspects. Biochim Biophys Acta. 2006;1763(12):1721–1732. doi: 10.1016/j.bbamcr.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 58.Barthelme D, Dinkelaker S, Albers SV, Londei P, Ermler U, Tampé R. Ribosome recycling depends on a mechanistic link between the FeS cluster domain and a conformational switch of the twin-ATPase ABCE1. Proc Natl Acad Sci U S A. 2011;108(8):3228–3233. doi: 10.1073/pnas.1015953108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou A, Hassel BA, Silverman RH. Expression cloning of 2-5A-dependent RNAase: a uniquely regulated mediator of interferon action. Cell. 1993;72(5):753–765. doi: 10.1016/0092-8674(93)90403-D. [DOI] [PubMed] [Google Scholar]
- 60.Chen ZQ, Dong J, Ishimura A, Daar I, Hinnebusch AG, Dean M. The essential vertebrate ABCE1 protein interacts with eukaryotic initiation factors. J Biol Chem. 2006;281(11):7452–7457. doi: 10.1074/jbc.M510603200. [DOI] [PubMed] [Google Scholar]
- 61.Marton MJ, Vazquez de Aldana CR, Qu H, Chakraburtty K, Hinnebusch AG. Evidence that GCN1 and GCN20, translational regulators of GCN4, function on elongating ribosomes in activation of eIF2 α kinase GCN2. Mol Cell Biol. 1997;17(8):4474–4489. doi: 10.1128/MCB.17.8.4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kovalchuk A, Driessen AJ. Phylogenetic analysis of fungal ABC transporters. BMC Genomics. 2010;11(1):177. doi: 10.1186/1471-2164-11-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nicholson SJ, Nickerson ML, Dean M, Song Y, Hoyt PR, Rhee H, Kim C, Puterka GJ. The genome of Diuraphis noxia, a global aphid pest of small grains. BMC Genomics. 2015;16(1):1. doi: 10.1186/s12864-015-1525-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Benoit JB, Adelman ZN, Reinhardt K, et al. Unique features of a global human ectoparasite identified through sequencing of the bed bug genome. Nat Commun. 2016;7. [DOI] [PMC free article] [PubMed]
- 65.Lee SH, Kang JS, Min JS, Yoon KS, Strycharz JP, Johnson R, Mittapalli O, Margam VM, Sun W, Li HM, Xie J, Wu J, Kirkness, Berenbaum MR, Pittendrigh BR, Clark JM. Decreased detoxification genes and genome size make the human body louse an efficient model to study xenobiotic metabolism. Insect Mol Biol. 2010;19(5):599–615. doi: 10.1111/j.1365-2583.2010.01024.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Annilo T, Tammur J, Hutchinson A, Rzhetsky A, Dean M, Allikmets R. Human and mouse orthologs of a new ATP-binding cassette gene, ABCG4. Cytogenet Cell Genet. 2002;94(2–3):196–201. doi: 10.1159/000048816. [DOI] [PubMed] [Google Scholar]
- 67.Engel T, Lorkowski S, Lueken A, Rust S, Schluter B, Berger G, Cullen P, Assmann G. The human ABCG4 gene is regulated by oxysterols and retinoids in monocyte-derived macrophages. Biochem Biophys Res Commun. 2001;288(2):483–488. doi: 10.1006/bbrc.2001.5756. [DOI] [PubMed] [Google Scholar]
- 68.Kusuhara H, Sugiyama Y. ATP-binding cassette, subfamily G (ABCG family) Pflugers Arch. 2007;453(5):735–744. doi: 10.1007/s00424-006-0134-x. [DOI] [PubMed] [Google Scholar]
- 69.Epis S, Porretta D, Mastrantonio V, Urbanelli S, Sassera D, Marco L, Mereghetti V, Montagna M, Ricci I, Favia G, Bandi C. Temporal dynamics of the ABC transporter response to insecticide treatment: insights from the malaria vector Anopheles stephensi. Sci Rep. 2014;4:7435. doi: 10.1038/srep07435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hock T, Cottrill T, Keegan J, Garza D. The E23 early gene of Drosophila encodes an ecdysone-inducible ATP-binding cassette transporter capable of repressing ecdysone-mediated gene activation. Proc Natl Acad Sci U S A. 2000;97(17):9519–9524. doi: 10.1073/pnas.160271797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sullivan DT, Grillo SL, Kitos RJ. Subcellular localization of the first three enzymes of the ommochrome synthetic pathway in Drosophila melanogaster. J Exp Zool. 1974;188(2):225–233. doi: 10.1002/jez.1401880210. [DOI] [PubMed] [Google Scholar]
- 72.Mackenzie SM, Brooker MR, Gill TR, Cox GB, Howells AJ, Ewart GD. Mutations in the white gene of Drosophila melanogaster affecting ABC transporters that determine eye colouration. Biochim Biophys Acta. 1999;1419:173–185. doi: 10.1016/S0005-2736(99)00064-4. [DOI] [PubMed] [Google Scholar]
- 73.Zhang SD, Odenwald WF. Misexpression of the white (omega) gene triggers male-made courtship in Drosophila. Proc Natl Acad Sci U S A. 1995;92:5525–5529. doi: 10.1073/pnas.92.12.5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Borycz J, Borycz JA, Kubow A, Lloyd V, Meinertzhagen IA. Drosophila ABC transporter mutants white, brown and scarlet have altered contents and distribution of biogenic amines in the brain. J Exp Biol. 2008;211:3454–3466. doi: 10.1242/jeb.021162. [DOI] [PubMed] [Google Scholar]
- 75.Tatematsu K, Yamamoto K, Uchino K, Narukawa J, Iizuka T, Banno Y, Katsuma S, Shimada T, Tamura T, Sezutsu H, Daimon T. Positional cloning of silkworm white egg2 (w-2) locus shows functional conservation and diversification of ABC transporters for pigmentation in insects. Genes Cells. 2011;16:331–342. doi: 10.1111/j.1365-2443.2011.01490.x. [DOI] [PubMed] [Google Scholar]
- 76.Guo Z, Shi K, Xun Z, Xia J, Wu Q, Wang S, Xie W, Zhang Y. Down-regulation of a novel ABC transporter gene (Pxwhite) is associated with Cry1Ac resistance in the diamondback moth, Plutella xylostella (L.) Insect Biochem Mol Biol. 2015;59:30–40. doi: 10.1016/j.ibmb.2015.01.009. [DOI] [PubMed] [Google Scholar]
- 77.Porretta D, Epis S, Mastrantonio V, Ferrari M, Bellini R, Favia G, Urbanelli S. How heterogeneous is the involvement of ABC transporters against insecticides? Acta Trop. 2016;157:131–135. doi: 10.1016/j.actatropica.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 78.Yang NN, Xie W, Jones CM, Bass C, Jiao XG, Yang X, Liu BM, Li RM, Zhang YJ. Transcriptome profiling of the whitefly Bemisia tabaci, reveals stage-specific gene expression signatures for thiamethoxam resistance. Insect Mol Biol. 2013;22(5):485–496. doi: 10.1111/imb.12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu S, Li Q, Liu Z. Genome-wide identification, characterization and phylogenetic analysis of 50 catfish ATP-binding cassette (ABC) transporter genes. PLoS One. 2013;8:e63895. doi: 10.1371/journal.pone.0063895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mummery-Widmer JL, Yamazaki M, Stoeger T, Novatchkova M, Bhalerao S, Chen D, Dietzl G, Dickson BJ, Knoblich JA. Genome-wide analysis of Notch signalling in Drosophila by transgenic RNAi. Nature. 2009;458:987–992. doi: 10.1038/nature07936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang S, Feany MB, Saraswati S, Littleton JT, Perrimon N. Inactivation of Drosophila Huntingtin affects long-term adult functioning and the pathogenesis of a Huntington’s disease model. Dis Model Mech. 2009;2:247–266. doi: 10.1242/dmm.000653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.You M, Yue Z, He W, Yang X, Yang G, Xie M, Zhan D, Baxter SW, Vasseur L, Gurr GM, et al. A heterozygous moth genome provides insights intoherbivory and detoxification. Nat Genet. 2013;45:220–225. doi: 10.1038/ng.2524. [DOI] [PubMed] [Google Scholar]
- 83.Jeong CB, Kim BM, Lee JS, Rhee JS. Genome-wide identification of whole ATP-binding cassette (ABC) transporters in the intertidal copepod Tigriopus japonicus. BMC Genomics. 2014;15(1):1–15. doi: 10.1186/1471-2164-15-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ren J, Chung-Davidson YW, Yeh CY, Scott C, Brown T, Li WM. Genome-wide analysis of the ATP-binding cassette (ABC) transporter gene family in sea lamprey and Japanese lamprey. BMC Genomics. 2015;16(1):1–14. doi: 10.1186/1471-2164-16-S7-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pisarev AV, Skabkin MA, Pisareva VP, Skabkina OV, Rakotondrafara AM, Hentze MW. The Role of ABCE1 in Eukaryotic Posttermination Ribosomal Recycling. Mol Cell. 2010;37(2):196–210. doi: 10.1016/j.molcel.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Paytubi S, Wang X, Lam YW, Izquierdo L, Hunter MJ, Jan E, Hundal HS, Proud CG. ABC50 promotes translation initiation in mammalian cells. J Biol Chem. 2009;284:24061–24073. doi: 10.1074/jbc.M109.031625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shulenin S, Nogee LM, Annilo T, Wert SE, Whitsett JA, Dean M. ABCA3 gene mutations in newborns with fatal surfactant deficiency. New Engl J Med. 2004;350(13):1296–1303. doi: 10.1056/NEJMoa032178. [DOI] [PubMed] [Google Scholar]
- 88.Zutz A, Gompf S, Schägger H, Tampé R. Mitochondrial ABC proteins in health and disease. BBA-Bioenergetics. 2009;1787(6):681–690. doi: 10.1016/j.bbabio.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 89.DeStefano GM, Kurban M, Anyane-Yeboa K, Dall’Armi C, Di Paolo G, Feenstra H, Fantauzzo KA, et al. Mutations in the cholesterol transporter gene ABCA5 are associated with excessive hair overgrowth. PLoS Genet. 2014;10(5):e1004333. doi: 10.1371/journal.pgen.1004333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhu F, Gujar H, Gordon JR, Haynes KF, Potter MF, Palli SR. Bed bugs evolved unique adaptive strategy to resist pyrethroid insecticides. Sci Rep. 2013;3:1456. doi: 10.1038/srep01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang LP, Zhang YJ, Zhang WJ, Wu QJ, Xu BY, Chu D. Analysis of genetic diversity among different geographical populations and determination of biotypes of Bemisia tabaci in China. J Appl Entomol. 2005;129:121–128. doi: 10.1111/j.1439-0418.2005.00950.x. [DOI] [Google Scholar]
- 92.Feng YT, Wu QJ, Xu BY, Wang SL, Chang XL, Xie W, Zhang YJ. Fitness costs and morphological change of laboratory-selected thiamethoxam resistance in the B-type Bemisia tabaci (Hemiptera: Aleyrodidae) J Appl Entomol. 2009;133:466–472. doi: 10.1111/j.1439-0418.2009.01383.x. [DOI] [Google Scholar]
- 93.Feng YT, Wu QJ, Wang SL, Chang XL, Xie W, Xu BY, Zhang YJ. Cross-resistance study and biochemical mechanisms of thiamethoxam resistance in B-biotype Bemisia tabaci (Hemiptera: Aleyrodidae) Pest Manag Sci. 2010;66:313–318. doi: 10.1002/ps.1877. [DOI] [PubMed] [Google Scholar]
- 94.Xie W, Chen C, Yang Z, Guo L, Yang X, Wang D, Chen M, Huang J, Wen Y, Zeng Y, Liu Y, Xia J, Tian L, Cui H, et al.: Genome sequencing of the sweetpotato whitefly Bemsia tabaci MED/Q. GigaScience. 2017;(in press). [DOI] [PMC free article] [PubMed]
- 95.Letunic L, Doerks T, Bork P. SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 2015;43(D1):D257–D260. doi: 10.1093/nar/gku949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Steentoft C, Vakhrushev SY, Joshi HJ, Kong Y, Vester-Christensen MB, Schjoldager KT, Lavrsen K, Dabelsteen S, Pedersen NB, Marcos-Silva L, Gupta R, Bennett EP, Mandel U, Brunak S, Wandall HH, Levery SB, Clausen H. Precision mapping of the human O-GalNAc glycoproteome through Simple Cell technology. EMBO J. 2013;32(10):1478–1488. doi: 10.1038/emboj.2013.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Boil Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Xie W, Guo L, Jiao X, Yang N, Yang X, Wu Q, Wang S, Zhou X, Zhang Y. Transcriptomic dissection of sexual differences in Bemisia tabaci, an invasive agricultural pest worldwide. Sci Rep. 2014;4:4088. doi: 10.1038/srep04088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang M, Sun H, Fei Z, Zhan F, Gong X, Gao S. Fastq clean: An optimized pipeline to clean the Illumina sequencing data with quality control. Bioinformatics and Biomedicine (BIBM), 2014 IEEE International Conference on. IEEE. 2014;44–48.
- 100.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):1. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Xu Y, Gao S, Yang Y, Huang M, Cheng L, Wei Q, Fei Z, Gao J, Hong B. Transcriptome sequencing and whole genome expression profiling of chrysanthemum under dehydration stress. BMC Genomics. 2013;14(1):662. doi: 10.1186/1471-2164-14-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gao S, Ou J, Xiao K. R language and Bioconductor in bioinformatics applications (Chinese Edition) Tianjin: Tianjin Science and Technology Translation and Publishing Co.; 2014. [Google Scholar]
- 103.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Li RM, Xie W, Wang S, Wu QJ, Yang NN, Yang X, Pan HP, Zhou XM, Bai LY, Xu BY, Zhou XG, Zhang YJ. Reference gene selection for qRT-PCR analysis in the sweetpotato whitefly, Bemisia tabaci, (Hemiptera: Aleyrodidae) Plos One. 2013;8(1):e53006. doi: 10.1371/journal.pone.0053006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Su YL, He WB, Wang J, Li JM, Liu SS, Wang XW. Selection of endogenous reference genes for gene expression analysis in the mediterranean species of the Bemisia tabaci (Hemiptera: Aleyrodidae) complex. J Econ Entomol. 2013;106(3):1446–1455. doi: 10.1603/EC12459. [DOI] [PubMed] [Google Scholar]
- 106.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of gene numbers in each subfamily of ABC transporters between Bemisia tabaci and other organisms. (PDF 23 kb)
Phylogenetic relationship of Bemisia tabaci ABCA subfamily with other organisms. Full-length ABC transporters were aligned using ClustalW and subjected to a maximum likelihood analysis by MEGA5 [97]. Numbers at the branch point of the node represent the values resulting from 1000 replications. Species, abbreviations, and color codes are: Btabq, B. tabaci (red); Dn, D. noxia (blue); AGA, A. gambiae (green); Dm, D. melanogaster (purple); Tu, T. urticae (light blue); Tj, T. japonicus (black); h, H. sapiens (gray); Ce, C. elegans (brick-red); Sc, Saccharomyces cervisiae (orange). (PDF 28 kb)
Phylogenetic relationship of Bemisia tabaci ABCB subfamily with other organisms. See Figure S2 legend for details. (PDF 14 kb)
Phylogenetic relationship of Bemisia tabaci ABCC subfamily with other organisms. See Figure S2 legend for details. (PDF 15 kb)
Phylogenetic relationship of Bemisia tabaci ABCD subfamily with other organisms. See Figure S2 legend for details. (PDF 19 kb)
Phylogenetic relationship of Bemisia tabaci ABCE subfamily with other organisms. See Figure S2 legend for details. (PDF 37 kb)
Phylogenetic relationship of Bemisia tabaci ABCF subfamily with other organisms. See Figure S2 legend for details. (PDF 89 kb)
Phylogenetic relationship of Bemisia tabaci ABCG subfamily with other organisms. See Figure S2 legend for details. (PDF 16 kb)
Phylogenetic relationship of Bemisia tabaci ABCH subfamily with other organisms. See Figure S2 legend for details. (PDF 134 kb)
RPKM values of B. tabaci Q ABC transporters at different developmental stages (DOCX 13 kb)
Fold-change of gene expression following B. tabaci Q exposure to imidacloprid (DOCX 8 kb)
Models used for phylogenetic analysis (DOCX 15 kb)
Selected B. tabaci Q ABC transporters for the RT-qPCR validation study (DOCX 143 kb)
Primers used for the RT-qPCR analysis (DOCX 89 kb)
Data Availability Statement
Bemisia tabaci Q ABC transporter dataset will be available from the corresponding authors on reasonable requests. Transcriptome datasets of B. tabaci Q at different development stages and following the exposure to imidacloprid insecticide have been deposited, respectively, in the Sequence Read Archive (SRA) database under the accession number SRP064690 and SRP090405.


