Abstract
Adult-derived human liver stem/progenitor cells (ADHLSCs) are, nowadays, developed as therapeutic medicinal product for the treatment of liver defects. In this study, the impact of hepatogenic differentiation and inflammation priming on the ADHLSCs' immune profile was assessed in vitro and compared to that of mature hepatocytes. The constitutive immunological profile of ADHLSCs was greatly different from that of hepatocytes. Differences in the expression of the stromal markers CD90 and CD105, adhesion molecules CD44 and CD49e, immunoregulatory molecules CD73 and HO-1, and NK ligands CD112 and CD155 were noted. While they globally preserved their immunological profile in comparison to undifferentiated counterparts, differentiated ADHLSCs showed a significant downregulation of CD200 expression as in hepatocytes. This was mainly induced by signals issued from EGF and OSM. On the other hand, the impact of inflammation was quite similar for all studied cell populations with an increased expression level of CD54 and CD106 and induction of that of CD40 and CD274. In conclusion, our immune profiling study suggests CD200 as a key factor in regulating the immunobiology of differentiated ADHLSCs. A better understanding of the molecular and physiological events related to such marker could help in designing the optimal conditions for an efficient therapeutic use of ADHLSCs.
1. Introduction
To date, cell therapy for metabolic liver diseases and hepatic injuries mainly relies on the use of various types of cells including hepatocytes, liver sinusoidal endothelial cells, mesenchymal stem cells (MSCs), endothelial progenitor cells, and macrophages [1]. However, several limitations and problems are associated with these cells that will finally have a critical impact on the efficiency of liver cell therapy [1]. Adult-derived human liver stem/progenitor cells (ADHLSCs) are obtained, in vitro, after primary culture of healthy adult human liver parenchymal cell fraction [2]. These cells exhibit a fibroblastic morphology and a hepatomesenchymal phenotype [2]. Even though considered as MSC-like, much less is known about ADHLSCs in comparison to the classical MSCs. It is reported that ADHLSCs, in their basal state, demonstrate distinct expression and secretion profiles [3, 4]. When exposed to in vitro hepatogenic differentiation, ADHLSCs are capable to differentiate, either in vitro or in vivo, into hepatocyte-like cells [4]. Recently, upon characterizing the immunological profile of ADHLSCs, our group showed that besides their potency in suppressing T cell proliferation, ADHLSCs are nonimmunogenic since they are negative for HLA-DR as well as for costimulatory molecule expression [5]. Altogether, their self-renewal potential, ability to acquire hepatocyte features, and their hypoimmunogenicity highlight ADHLSCs as a potential alternative cell source for liver cell transplantation. However, achieving these goals involves addressing different aspects related to their safety and efficacy. For instance, tracking the changes of ADHLSCs' immunological profile following hepatogenic differentiation and after exposure to inflammation is missing.
Accordingly, the current work was designed to learn more about the ADHLSC immune profile modulation after in vitro hepatogenic differentiation and in an inflamed environment. Flow cytometry analysis demonstrated the dissimilarity between hepatocytes and undifferentiated ADHLSCs as shown by the expression of stromal markers CD90 and CD105, adhesion molecules CD44 and CD49e, immune regulatory molecules CD73 and HO-1, and NK ligands CD112 and CD155. We also confirm that differentiated ADHLSCs do not acquire a complete and similar hepatocyte immune phenotype but rather maintain a profile comparable to that of undifferentiated cells. However, a specific and major downregulation of CD200 expression was highlighted to reach basal levels as those exhibited by hepatocytes. The impact of inflammation was quite similar for all studied cell populations with an increase in the expression level of CD54 and CD106 and induction of that of CD40 and CD274.
Downregulation of CD200 expression occurred early during the in vitro hepatocytic differentiation process and is modulated by the epidermal growth factor (EGF). Oncostatin M is also inhibiting CD200 mRNA expression at the late maturation phase of the hepatogenic differentiation process. Besides identifying CD200 expression level as an important marker to distinguish hepatic differentiated versus undifferentiated ADHLSCs, our observations suggest a potential role for CD200 in regulating the immunobiology of ADHLSCs. The loss of CD200 might have critical repercussions on the immunobiology of the differentiated ADHLSCs and specifically for liver immunology.
2. Materials and Methods
2.1. Isolation and Culture of Hepatocytes and ADHLSC
The present study was accepted by the institution ethical review board for the use of human-derived tissue, under appropriate informed consent of tissue donors. An agreement from the Belgian Ministry of Health was delivered for hepatocytes and hepatic stem cell isolation and banking.
Liver cell suspensions were recovered after a two-step collagenase perfusion technique of livers from healthy cadaveric donors. Following filtration and low-speed centrifugation, the parenchymal fraction, predominantly constituted by hepatocytes, was recovered and seeded as primary cultures. ADHLSCs were then obtained as previously described (Najimi et al. [2]). The cells were cultured using DMEM containing 4.5 g/l glucose (Life Technologies) supplemented with 10% fetal calf serum (FCS) (Life Technologies) and 1% penicillin/streptomycin (Life Technologies), at 37°C in a fully humidified atmosphere (5% CO2). When reaching 80% confluence, cells were lifted with 0.05% Trypsin-EDTA (Life Technologies) and seeded at a density of 5000 cells/cm2. The viability of recovered cells was evaluated using Trypan blue exclusion assay at each passage.
2.2. Hepatogenic Differentiation of ADHLSC
After seeding at 104 cells/cm2 in six-well plates coated with rat tail collagen type I, ADHLSCs were maintained in expansion medium for 48 h. Thereafter, cells were sequentially incubated with hepatogenic differentiation cocktail containing specific growth factors and cytokines as previously described (Najimi et al. [2]). Except for step 1 (one medium change), the differentiation medium was changed each 3 days and the cells were microscopically followed at a regular basis. At the end of the maturation step, cells were harvested for analyses to appreciate the level of their phenotypic and functional differentiation. Control undifferentiated cells were kept during the whole process of differentiation in IMDM medium supplemented with 1% FCS.
2.3. Inflammation Priming
Adherent cells were primed overnight using a cocktail of proinflammatory cytokines: 25 ng/ml IL-1β (Peprotech, Rocky Hill, NJ, USA), 103 U/ml IFN-γ, 50 ng/ml TNF-α, and 3×103 U/ml IFN-α (all from ProSpec Inc., Rehovot, Israel).
2.4. Flow Cytometry
A panel of conjugated monoclonal antibodies (Table 1) was used to assess the phenotype of the cells studied under different culture conditions. After labelling, acquired results were analyzed by using a MACSQuant analyzer (Miltenyi Biotec) [5].
Table 1.
Conjugated monoclonal antibodies used for flow cytometry analysis.
| Primary antibody | Order ID | Species | Dilution | Source |
|---|---|---|---|---|
| Endothelial, stromal, and embryonic markers | ||||
| Anti-CD34-PC5 | 555823 | Mouse | 1/20 | BD |
| Anti-CD90-PE | FAB2067P | Mouse | 1/20 | R&D |
| Anti-CD105-FITC | 326040 | Mouse | 1/20 | AC |
| Anti-SSEA4-PE | FAB1435P | Mouse | 1/20 | R&D |
|
| ||||
| Receptors | ||||
| Anti-CD45-PC7 | 557748 | Mouse | 1/20 | BD |
| Anti-CD95-FITC | 130092415 | Mouse | 1/20 | MB |
| Anti-CD184-PE | 555974 | Mouse | 1/20 | BD |
| Anti-CD200R-PE | 329306 | Mouse | 1/20 | BL |
| Anti-CD210-PE | 556013 | Rat | 1/20 | BD |
| Anti-CD271-PE | 120002227 | Mouse | 1/20 | MB |
| Anti-CD229-PE | 326108 | Mouse | 1/20 | BL |
|
| ||||
| Human leukocyte antigens | ||||
| Anti-HLA-ABC-PE-Cy5 | 15998342 | Mouse | 1/20 | EB |
| Anti-HLA-DR-PerCP | 347402 | Mouse | 1/20 | BD |
| Anti-HLA-G-PE | 1P292C100 | Mouse | 1/20 | ExBIO |
|
| ||||
| Costimulatory molecules | ||||
| Anti-CD27-APC-Cy7 | 302815 | Mouse | 1/20 | BL |
| Anti-CD40-PE | 130094135 | Mouse | 1/20 | MB |
| Anti-CD70-PE | 355104 | Mouse | 1/20 | BL |
| Anti-CD80-FITC | 11080942 | Mouse | 1/20 | EB |
| Anti-CD86-APC | 130094876 | Mouse | 1/20 | MB |
| Anti-CD134-FITC | 350006 | Mouse | 1/20 | BL |
| Anti-CD154-PE-Cy5 | 310808 | Mouse | 1/20 | BL |
| Anti-CD252-PE | 326308 | Mouse | 1/20 | BL |
|
| ||||
| Cell adhesion molecules | ||||
| Anti-CD29-PE-Cy5 | 559882 | Mouse | 1/20 | BD |
| Anti-CD31-PE | 130092653 | Mouse | 1/20 | MB |
| Anti-CD44-FITC | 130095195 | Mouse | 1/20 | MB |
| Anti-CD49e-PE | 555617 | Mouse | 1/20 | BD |
| Anti-CD54-PE | 555511 | Mouse | 1/20 | BD |
| Anti-CD58-FITC | 555920 | Mouse | 1/20 | BD |
| Anti-CD62e-fluorescein | BBA21 | Mouse | 1/20 | R&D |
| Anti-CD102-FITC | 328507 | Mouse | 1/20 | BL |
| Anti-CD106-PE-Cy5 | 551148 | Mouse | 1/20 | BD |
| Anti-CD146-PC5 | A22364 | Mouse | 1/20 | BC |
| Anti-CD166-PE | 559263 | Mouse | 1/20 | BD |
|
| ||||
| Immunoregulatory molecules | ||||
| Anti-CD39-FITC | 328205 | Mouse | 1/20 | BL |
| Anti-CD73-PE | 344003 | Mouse | 1/20 | BD |
| Anti-CD200-APC | 329208 | Mouse | 1/20 | BL |
| Anti-CD274-PE | 557924 | Mouse | 1/20 | BD |
| Anti-HO-1-PE | ADI-OSA-111 | Mouse | 1/20 | ELS |
|
| ||||
| NK ligands | ||||
| Anti-CD112-PE | 337410 | Mouse | 1/20 | BL |
| Anti-CD155-PE | 337508 | Mouse | 1/20 | BL |
| Anti-ULBP-3-PE | FAB1517P | Mouse | 1/20 | R&D |
2.5. RT-qPCR
ADHLSCs' total RNA was extracted using TriPure Isolation Reagent (Roche, Belgium). First-strand cDNA was synthesized using high-capacity cDNA reverse transcription kit according to manufacturer's instructions (Applied Biosystems) and subsequently diluted with nuclease-free water (Invitrogen) to 10 ng/μl cDNA. RT-PCR amplification mixtures (25 μl) containing 25 ng template cDNA, Master Mix buffer (12,5 μl) (Applied Biosystems), and the corresponding TaqMan assay were run in duplicate and performed on StepOnePlus real-time PCR machine (Applied Biosystems). The cycling conditions comprised 10 min polymerase activation at 95°C and 40 cycles at 95°C for 15 s and 60°C for 1 min. Relative quantification was normalized against the house keeping gene PPIA. The Applied Biosystems assays used for the current study are CD200 (Hs01033303_m1) and cyclophilin A (PPIA) (Hs99999904_m1).
2.6. Immunofluorescence
ADHLSCs from 3 different donors at passage 6 grown on glass labtek were fixed with 4% paraformaldehyde (Sigma) for 15 min at room temperature. Nonspecific immunostaining was prevented by 30 min incubation in PBS containing 3% (w/v) bovine serum albumin (BSA). Cells were incubated for 1 hour with polyclonal goat anti-CD200 primary antibody (AF2724 R&D systems) at room temperature. After washing with PBS, ADHLSCs were incubated for 30 min with donkey anti-goat secondary antibody (A11055, Life Technologies) at room temperature. Nuclei were stained for 5 min with DAPI (Life Technologies). Slides were mounted in fluoromount medium (Sigma). Fluorescence was assessed using Imager A1 fluorescent microscope (Carl Zeiss), and digital images were acquired using AxioVision Software.
2.7. Western Blot Analysis
Total protein lysates were obtained by dissolving ADHLSC pellets in RIPA buffer [50 mM Tris base, pH 8.0, 150 mM NaCl, 1% Triton X-100 0.5% sodium deoxycholate, 0.1% SDS, with protein inhibitors cocktail without EDTA (Roche)]. Protein samples were sonicated shortly and incubated for 30 minutes at 4°C prior to sample clarification by centrifugation (15 min maximum speed). Subsequently, sample supernatants were collected and a total protein quantification was performed (BCA quantification kit, Thermofisher). Twenty μg of total protein extracts were dissolved in loading buffer [Tris-HCl (pH 6.8), glycerol, SDS, DTT, and bromophenol blue], denatured at 95°C for 5 minutes and loaded on a 10% Tris-glycine SDS-PAGE gel for protein separation and transferred overnight at 4°C onto PVDF membranes. Membranes were incubated with 5% BSA blocking solution for 90 min at room temperature. CD200 antibody [0.1 μg/ml] was incubated 90 min at room temperature, and the membranes were thoroughly washed 3× with PBS-T, incubated with fluorescently labelled secondary antibody (Biotium) for 40 min at room temperature, and detected by Li-cor scanner (Odyssey). Quantification analysis was performed by Image Studio Lite Software (Odyssey).
2.8. Construction of CD200 Protein-Protein Interaction (PPI) Network
We have used STRING database v10 (Search Tool for the Retrieval of Interacting Genes, available at http://string-db.org) [6] with CD200 as seed for construction of the protein-protein interaction (PPI) network.
2.9. Statistical Analyses
Results are expressed as mean ± standard error of the mean (SEM). Analyses were done in the GraphPad Prism software program (San Diego, California, USA). Statistical differences were determined by Student's t-test for two groups' comparison or by one-way ANOVA followed by the Tukey post hoc test for multiple comparisons between more than two groups. Differences were considered significant when p values are ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
3. Results
3.1. Flow Cytometry Characterization of ADHLSCs' Immunological Profile after Hepatogenic Differentiation and Inflammation Priming
Flow cytometry analysis was initially carried out to determine the immunologic profile of ADHLSCs under low proliferation conditions and to characterize the impact of hepatogenic differentiation. The quality of hepatogenic differentiation is systematically evaluated at the morphological, expression, and function levels (see Supplementary Figure 1 available online at https://doi.org/10.1155/2017/2679518). The influence of inflammation priming on such profile was also appreciated. Hepatocytes previously isolated from the same donors as ADHLSC were used for comparison as the golden standard reference for all these experimental conditions. The representative FACS histograms (Figure 1) and the related immunopositivity percentages (Table 2) reveal that ADHLSC express a plethora of molecules belonging to several distinct immunological families. Such independent analysis confirmed the mesenchymal profile of ADHLSC and its maintenance in low proliferating and differentiation conditions. Indeed, CD90 was highly and comparably expressed in undifferentiated ADHLSCs (82 ± 3,24%), inflammation-primed undifferentiated ADHLSCs (84 ± 2,52%), differentiated ADHLSC (75,20 ± 7,80%), and inflammation-primed hepatic differentiated ADHLSCs (78,50 ± 8,36%). CD105 expression level exhibited by undifferentiated ADHLSCs (31,83 ± 3,63%) was not significantly changed following inflammation priming (26 ± 2,63%) or after differentiation (29,45 ± 3,46%). Inflammation priming did not significantly impact its expression in differentiated ADHLSCs (28,66 ± 3,47%). Endothelial CD34 and embryonic SSEA4 markers were minimally expressed in the three tested cell types (undifferentiated and differentiated ADHLSCs as well as hepatocytes) without being influenced by inflammation priming.
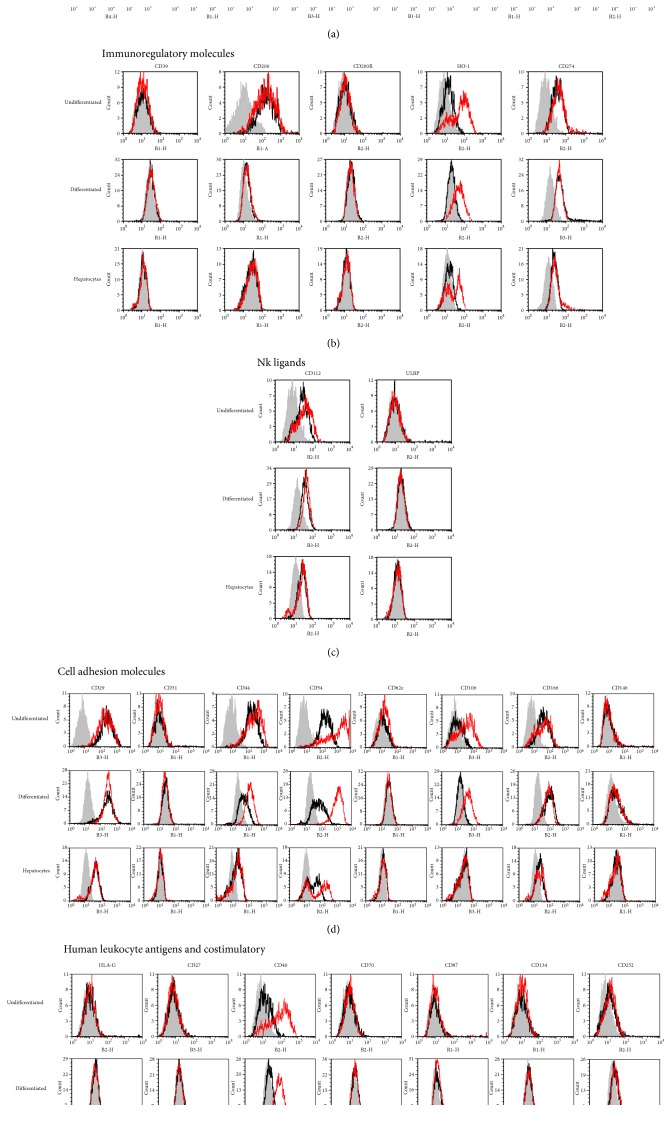
Figure 1.
Characterization of the immunoprofile of ADHLSCs and human hepatocytes. Representative FACS histograms indicate the immunoprofiling of different liver-derived cells as determined and analyzed by flow cytometry. The expression of a panel of immunological markers was evaluated in ADHLSCs before and after hepatogenic differentiation and in comparison to hepatocytes being used as standard reference. These liver-derived cells were investigated under constitutive (black curve) and inflammatory-priming conditions (red curve). The grey curve represents the antibody control. The corresponding monoclonal antibodies used for establishing this immunoprofile are listed in Table 1. The data are also presented and listed in Table 2 as the mean ± SEM percentage of each marker expression. (a) Endothelial, stromal, and embryonic markers (CD34, CD45, CD90, CD95, CD210, and CD271). (b) Immunoregulatory molecules (CD39, CD200, CD200R, CD274, and HO-1). (c) Natural killer ligands (CD112, CD155, and ULBP3). (d) Cell adhesion molecules (CD29, CD31, CD44, CD54, CD62e, CD106, CD166, and CD146). (e) Human leukocyte antigens (HLA-G) and costimulatory molecules (CD27, CD40, CD70, CD86, CD134, and CD252).
Table 2.
Expression levels of different molecules present on undifferentiated and differentiated ADHLSC as well as hepatocytes being cultivated in the presence or absence of inflammation cocktail. The data are presented as the mean ± SEM (standard error of the mean) percentage of each marker expression.
| ADHLSCs | Hepatocytes | |||||
|---|---|---|---|---|---|---|
| Undifferentiated | Differentiated | |||||
| Constitutive | Priming | Constitutive | Priming | Constitutive | Priming | |
| Endothelial, stromal, and embryonic markers | ||||||
| CD34 | 0,96 ± 0,28 | 0,54 ± 0,17 | 0,37 ± 0,15 | 0,9 ± 0,28 | 3,5 ± 0,64 | 1,33 ± 0,92 |
| CD90 | 82 ± 3,24 | 84 ± 2,52 | 75,20 ± 7,80 | 78,50 ± 8,36 | 0,32 ± 0,15 | 0,34 ± 0,22 |
| CD105 | 31,83 ± 3,63 | 26 ± 2,63 | 29,45 ± 3,46 | 28,66 ± 3,47 | 0,40 ± 0,20 | 0,1 ± 0,04 |
| SSEA4 | 1,4 ± 0,38 | 2,52 ± 0,40 | 1,62 ± 0,31 | 1,98 ± 0,50 | 0,57 ± 0,19 | 1,96 ± 0,52 |
|
| ||||||
| Receptors | ||||||
| CD45 | 0,78 ± 0,23 | 1,29 ± 0,46 | 1,38 ± 0,60 | 0,71 ± 0,18 | 0,12 ± 0,08 | 2,08 ± 1,69 |
| CD95 | 2,45 ± 0,58 | 6,80 ± 1,94 | 3,25 ± 1,14 | 7,34 ± 1,92 | 0,20 ± 0,17 | 2 ± 0,86 |
| CD184 | 1,53 ± 0,55 | 2,06 ± 0,68 | 1,13 ± 0,18 | 1,06 ± 0,20 | 0,49 ± 0,23 | 0,73 ± 0,15 |
| CD200R | 1,02 ± 0,27 | 1,23 ± 0,12 | 0,79 ± 0,08 | 1,13 ± 0,33 | 0,32 ± 0,08 | 0,69 ± 0,29 |
| CD210 | 0,73 ± 0,19 | 1,6 ± 0,10 | 1,23 ± 0,13 | 1,30 ± 0,27 | 0,30 ± 0,18 | 0,66 ± 0,24 |
| CD271 | 2,18 ± 0,79 | 2,20 ± 0,34 | 2,33 ± 0,61 | 2,80 ± 0,60 | 0,42 ± 0,18 | 1,36 ± 0,72 |
| CD229 | 0,66 ± 0,17 | 0,53 ± 0,13 | 0,34 ± 0,09 | 0,47 ± 0,14 | 0,28 ± 0,08 | 0,25 ± 0,03 |
|
| ||||||
| Human leukocyte antigens | ||||||
| HLA-ABC | 92,83 ± 2,61 | 93,9 ± 2,88 | 96,16 ± 1,47 | 98,2 ± 0,34 | 89 ± 2,97 | 88 ± 5,02 |
| HLA-DR | 0,78 ± 0,28 | 0,66 ± 0,23 | 0,6 ± 0,33 | 0,4 ± 0,15 | 1,5 ± 0,3 | 1,52 ± 1,49 |
| HLA-G | 2,43 ± 1,13 | 1,62 ± 0,37 | 1,57 ± 0,28 | 1,58 ± 0,6 | 0,29 ± 0,16 | 0,47 ± 0,13 |
|
| ||||||
| Costimulatory molecules | ||||||
| CD27 | 0,27 ± 0,12 | 0,57 ± 0,20 | 1,06 ± 0,34 | 0,3 ± 0,12 | 0,36 ± 0,1 | 0,5 ± 0,07 |
| CD40 | 8,33 ± 0,88 | 70 ± 5,70 | 9,33 ± 0,84 | 68,6 ± 4,84 | 3,5 ± 0,65 | 72,8 ± 5,22 |
| CD70 | 0,84 ± 0,22 | 0,87 ± 0,27 | 0,79 ± 0,30 | 0,57 ± 0,15 | 0,14 ± 0,06 | 0,71 ± 0,20 |
| CD80 | 1,13 ± 0,34 | 1,66 ± 0,77 | 1,06 ± 0,60 | 1,44 ± 0,83 | 1,43 ± 0,07 | 1,53 ± 0,05 |
| CD86 | 0,75 ± 0,18 | 0,67 ± 0,31 | 0,76 ± 0,34 | 1,18 ± 0,44 | 2,34 ± 1,02 | 1,05 ± 0,84 |
| CD134 | 1,56 ± 0,60 | 1,30 ± 0,75 | 1,66 ± 0,59 | 1,58 ± 0,67 | 0,26 ± 0,10 | 0,13 ± 0,05 |
| CD154 | 0,42 ± 0,12 | 1,03 ± 0,33 | 2 ± 0,40 | 1,36 ± 0,33 | 0,45 ± 0,10 | 0,25 ± 0,10 |
| CD252 | 5 ± 1,06 | 4,65 ± 0,20 | 3 ± 0,57 | 4,60 ± 0,71 | 0,43 ± 0,15 | 0,52 ± 0,10 |
|
| ||||||
| Cell adhesion molecules | ||||||
| CD29 | 91 ± 4,80 | 89,60 ± 4,94 | 95 ± 1,38 | 97 ± 0,65 | 84 ± 3 | 89 ± 3,82 |
| CD31 | 1,2 ± 0,45 | 1,7 ± 0,64 | 1,3 ± 0,66 | 1,6 ± 0,93 | 0,26 ± 0,11 | 0,31 ± 0,13 |
| CD44 | 92,83 ± 1,64 | 93,60 ± 2,26 | 59,50 ± 6,23 | 89,60 ± 3,89 | 57,43 ± 2,40 | 53,75 ± 4,53 |
| CD49e | 82 ± 5,43 | 89,75 ± 3,87 | 92,98 ± 3,45 | 97,23 ± 0,51 | 38,75 ± 7,22 | 34,33 ± 5,29 |
| CD54 | 69 ± 5,53 | 97,80 ± 0,67 | 64,20 ± 5,85 | 88,80 ± 4,92 | 63,25 ± 9 | 85,50 ± 5,90 |
| CD58 | 3,12 ± 1,17 | 7,44 ± 2,74 | 2,68 ± 1,47 | 3,06 ± 1,42 | 0,80 ± 0,45 | 3,50 ± 0,65 |
| CD62e | 1,96 ± 0,52 | 2,4 ± 0,98 | 1,56 ± 0,57 | 3,03 ± 1,63 | 0,2 ± 0,09 | 0,9 ± 0,42 |
| CD102 | 1 ± 0,25 | 0,98 ± 0,56 | 0,88 ± 0,30 | 1,40 ± 0,72 | 0,1 ± 0,04 | 0,1 ± 0,01 |
| CD106 | 8,08 ± 1,58 | 48,75 ± 5,54 | 1,09 ± 0,82 | 49,50 ± 5,10 | 0,43 ± 0,02 | 7,75 ± 4,70 |
| CD146 | 4,83 ± 1,51 | 3,64 ± 0,82 | 3,16 ± 0,60 | 4 ± 0,64 | 0,1 ± 0,033 | 0,04 ± 0,038 |
| CD166 | 25,60 ± 1,64 | 24,4 ± 1,31 | 26,83 ± 2,03 | 27,20 ± 3,71 | 30,93 ± 0,05 | 31,67 ± 3,50 |
|
| ||||||
| Immunoregulatory molecules | ||||||
| CD39 | 1,55 ± 0,54 | 1,48 ± 0,64 | 2,68 ± 1,29 | 2,52 ± 1,26 | 0,03 ± 0,02 | 0,09 ± 0,01 |
| CD73 | 88,8 ± 3,99 | 90 ± 2,33 | 71,8 ± 6,72 | 79,75 ± 5,79 | 2,01 ± 0,08 | 1,8 ± 0,52 |
| CD200 | 69,20 ± 2,96 | 60 ± 8,05 | 2,43 ± 1,19 | 1,86 ± 1,05 | 0,14 ± 0,08 | 0,61 ± 0,47 |
| CD274 | 1,50 ± 0,35 | 82,40 ± 5,94 | 0,76 ± 0,13 | 64,80 ± 2,26 | 0,60 ± 0,34 | 66,25 ± 2,98 |
| HO-1 | 65,83 ± 2,21 | 66,25 ± 3,10 | 61,83 ± 4,78 | 68,60 ± 4,22 | 32,55 ± 5,10 | 35,66 ± 2,02 |
|
| ||||||
| NK ligands | ||||||
| CD112 | 50 ± 5,82 | 58,75 ± 4,30 | 46,08 ± 3,35 | 43,60 ± 5,24 | 23,5 ± 1,55 | 25,55 ± 1,93 |
| CD155 | 71,60 ± 4,32 | 70,50 ± 2,63 | 72,80 ± 4,41 | 74 ± 6,55 | 33,75 ± 2,43 | 37,25 ± 3,01 |
| ULBP-3 | 1,07 ± 0,26 | 1,46 ± 0,42 | 0,72 ± 0,16 | 1,20 ± 0,52 | 0,53 ± 0,25 | 1 ± 0,21 |
Comparably to their minimal expression profiles observed in hepatocytes, the cell surface receptors CD45, CD95, CD184, CD200R, CD210, CD229, and CD271 showed a very weak, if any, expression in undifferentiated as well as hepatic-differentiated ADHLSCs. Inflammation priming of the different cell types did not substantially alter the expression level of any of those receptors.
HLA antigens and costimulatory molecules profiling was performed to assess the immunogenicity state of liver-derived cells. As noticed for hepatocytes, both undifferentiated and differentiated ADHLSCs remained nonimmunogenic as the expression of neither HLA-DR and HLA-G nor costimulatory molecules (CD27, CD40, CD70, CD80, CD86, CD134, CD154, and CD252) was significantly induced. On the other hand, and subsequently to inflammation priming, the CD40 expression levels were dramatically increased in both undifferentiated ADHLSCs (from 8,33 ± 0,88 to 70 ± 5,70%) and differentiated ADHLSCs (from 9,33 ± 0,84 to 68,6 ± 4,84) similarly to hepatocytes (from 3,5 ± 0,65 to 72,8 ± 5,22%). HLA-ABC showed high and similar expressions among all the cell groups (undifferentiated ADHLSC (92,83 ± 2,61 versus 93,9 ± 2,88% after inflammation priming), differentiated ADHLSC (96,16 ± 1,47 versus 98,2 ± 0,34 after inflammation priming), and hepatocytes (89 ± 2,97% versus 88 ± 5,02 after inflammation priming).
Profiling of cell adhesion molecules was also evaluated for CD29, CD44, CD49e, CD54, CD58, CD62, CD102, CD106, CD146, and CD166 markers. All exhibited some similarities between undifferentiated ADHLSCs, differentiated ADHLSCs, and hepatocytes. For instance, CD29 and CD166 were highly expressed whilst CD58, CD62, CD102, and CD146 were weakly expressed in all analyzed cell types without striking alterations following both hepatogenic differentiation and inflammation priming. On the other hand, differences were also detected. The CD44 marker was much more expressed in undifferentiated ADHLSC (92,83 ± 1,64%) as compared to differentiated cells (59,50 ± 6,23%) and hepatocytes (57,43 ± 2,40%). Furthermore, CD44 expression was increased following inflammation priming in only differentiated ADHLSC (89,60 ± 3,89%) in contrast to that in undifferentiated cells (93,60 ± 2,26%) and hepatocytes (53,75 ± 4,53%). CD49e expression, part of the fibronectin receptor, was higher in undifferentiated (82 ± 5,43%) as well as in differentiated ADHLSCs (92,98 ± 3,45%) as compared to that in hepatocytes (38,75 ± 7,22%). However, inflammation priming did not exert any striking effect on this integrin expression level in all analyzed cell types. CD54 or intercellular adhesion molecule-1 (ICAM-1) expression was comparable in undifferentiated ADHLSCs (69 ± 5,53%), differentiated ADHLSCs (64,20 ± 5,85%), and hepatocytes (63,25 ± 9%). Inflammation priming of each of those cell types showed a similar elevated CD54 expression in all analyzed cell types (97,80 ± 0,67%, 88,80 ± 4,92%, and 85,50 ± 5,90%, resp.). Although minimally expressed in hepatocytes and differentiated ADHLSC (0,43 ± 0,02% and 1,09 ± 0,82%, respectively) and weak in undifferentiated cells (8,08 ± 1,58%), CD106 expression was substantially induced following inflammation priming of either undifferentiated (48,75 ± 5,54%) or differentiated ADHLSCs (49,50 ± 5,10%) but to a lesser extent in hepatocytes (7,75 ± 4,70%).
As NK cells play a critical role in liver pathology, we determined the expression of the major activating ligands of NK cytolytic activity. The expression pattern of the NK ligands CD112, CD155, and ULBP-3 showed no significant alterations following hepatogenic differentiation or inflammation priming. For instance, the high and comparable CD112 and CD155 expression levels exhibited in undifferentiated ADHLSCs (50 ± 5,82% for CD112; 71,60 ± 4,32% for CD155) and differentiated ADHLSC (46,08 ± 3,35 for CD112; 72,80 ± 4,41 for CD155) were not modulated after inflammation priming of undifferentiated ADHLSCs (58,75 ± 4,30% for CD112; 70,50 ± 2,63% for CD155) as well as differentiated ADHLSCs (43,60 ± 5,24% for CD112; 74 ± 6,55% for CD155). This maintained expression was also noticed in hepatocytes (23,5 ± 1,55% for CD112; 33,75 ± 2,43% for CD155) versus inflammation-primed hepatocytes (25,55 ± 1,93% for CD112; 37,25 ± 3,01% for CD155). On the other hand, ULBP-3 was minimally expressed in all these three cell types without being influenced by inflammation.
Immune regulation and its underlying mechanisms are an important component of liver immunity. The immune-regulatory molecules CD39 and CD274 were constitutively negative in all cell types whereas only CD274 was strongly upregulated after inflammation priming of undifferentiated ADHLSCs (82,40 ± 5,94%), differentiated ADHLSC (64,80 ± 2,26%), and hepatocytes (66,25 ± 2,98%). CD73 and HO-1 molecules exhibited a high constitutive expression level in undifferentiated ADHLSCs (88,8 ± 3,99% for CD73; 65,83 ± 2,21% for HO-1) which remains unchangeable both after hepatogenic differentiation (71,8 ± 6,72% for CD73; 61,83 ± 4,78% for HO-1) and after inflammation priming (90 ± 2,33% for CD73; 66,25 ± 3,10% for HO-1). On the other hand, although HO-1 was substantially and similarly expressed in hepatocytes (32,55 ± 5,10%) and inflammation-primed hepatocytes (35,66 ± 2,02%), CD73 showed only minimal expression under these conditions.
Intriguingly, CD200 being highly expressed in undifferentiated ADHLSCs (69,20 ± 2,96%) was dramatically downregulated following hepatogenic differentiation (2,43 ± 1,19%). Similarly, its expression was very low in mature hepatocytes (0,14 ± 0,08%). In all these experimental cell groups, inflammation priming has any significant impact on CD200 protein expression level.
3.2. Modulation of CD200 Expression following Hepatogenic Differentiation
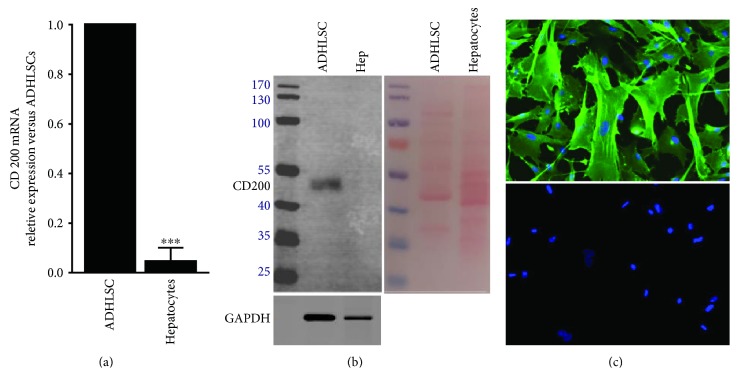
Firstly, quantitative real-time PCR (RT-qPCR) was carried out to estimate the CD200 mRNA level in naïve ADHLSC (expansion culture conditions) and hepatocytes. In agreement with flow cytometry results, RT-qPCR analysis revealed that CD200 transcript expression was extremely low in hepatocytes (Figure 2(a)). This observation is also confirmed by using western blotting (Figure 2(b)). CD200 protein expression in ADHLSC was also confirmed using immunofluorescence and is homogenously distributed at the membrane level (Figure 2(c)).
Figure 2.
CD200 expression in naïve ADHLSC and mature nonplated hepatocytes. (a) Total RNA extracted from both naïve ADHLSCs and nonplated hepatocytes was retrotranscribed, the cDNA-synthesized and PPIA-normalized CD200 mRNA levels were evaluated by RT-qPCR. A very significant low level of this marker is noticed in mature hepatocytes (n = 4) (∗∗∗p < 0.001 versus naïve ADHLSC, unpaired t-test). (b) Cell extracts were prepared and immunoblotted with anti-CD200 and anti-GAPDH antibodies as described in Materials and Methods. Low CD200 protein expression in mature hepatocytes (n = 3) is correlated to mRNA expression as compared to ADHLSC. Ponceau S staining shows the quality and the quantity of loading. (c) Immunofluorescence on fixed naïve ADHLSCs was also performed to appreciate the CD200 protein expression. CD200-positive immunoreactivity was significantly confirmed. The lower picture shows that no immunofluorescence signal was observed when cells were incubated with only secondary antibody. Pictures from 3 different donors were taken at a magnification of 200x.
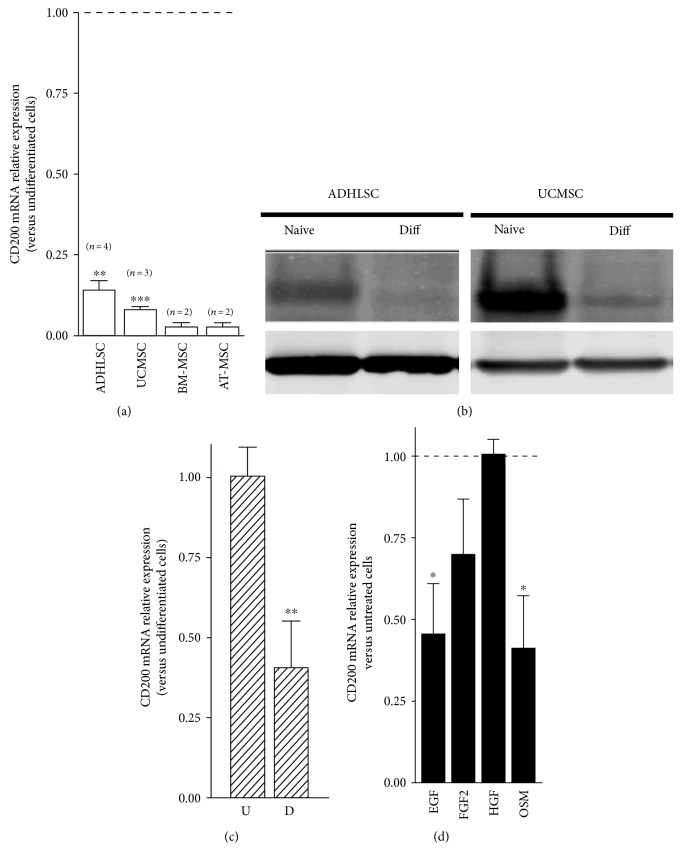
Upon hepatogenic differentiation, we demonstrated that the CD200 mRNA expression level was clearly abolished in differentiated ADHLSCs as compared to undifferentiated cells (Figure 3(a)). Such CD200 downregulation is also observed in MSC from other tissues like the umbilical cord, bone marrow, and adipose tissue and submitted to the same hepatogenic differentiation protocol. Western blot analysis was performed to further assess, at the protein level, the alteration of CD200 expression noticed following hepatogenic differentiation. Consistently with flow cytometry results, CD200 protein expression level exhibited by the undifferentiated ADHLSCs is dramatically diminished after in vitro hepatogenic differentiation. The same results were obtained in both ADHLSCs and UCMSCs (Figure 3(b)).
Figure 3.
Downregulation of CD200 expression after in vitro hepatogenic differentiation of ADHLSC. ADHLSCs were submitted to an in vitro hepatogenic differentiation protocol and processed for CD200 expression analysis. (a) Total RNA extracted from both undifferentiated and differentiated MSCs (the umbilical cord MSC (UCMSC), bone marrow MSC (BM-MSC), and adipose tissue MSC (AT-MSC)) was retrotranscribed, the cDNA synthesized, and RT-qPCR applied for CD200 gene marker. Hepatogenic differentiation was correlated with a strong downregulation of this marker. (b) Total proteins extracted from differentiated and undifferentiated ADHLSCs and UCMSC were analyzed using western blotting. Hepatogenic differentiation was associated with a significant decrease of CD200 protein expression. (c) Downregulation of CD200 mRNA occurs early at the first step of the hepatogenic differentiation protocol as demonstrated using RT-qPCR. (d) Downregulation of CD200 expression in differentiated ADHLSCs occurs under EGF and oncostatin M treatment. ADHLSCs were treated with one or the other growth factor/cytokine used in the hepatogenic differentiation protocol (epidermal growth factor (EGF), basic fibroblast growth factor (bFGF), hepatocyte growth factor (HGF), or oncostatin M (OSM)). Cells were treated for 3 days using the same basal medium as for differentiation experiments and under serum-free conditions. Total RNA for each group was extracted and retrotranscribed for CD200 mRNA analysis using RT-qPCR. Data shown represent the mean ± SEM of three different experiments as indicated in the graph (∗p < 0.05; ∗∗p < 0.01 versus corresponding untreated cells, unpaired t-test).
During hepatogenic differentiation, the downregulation of CD200 looks to be dependent on culture steps. Given that the hepatogenic differentiation protocol is a multistep process, we investigated the kinetic of CD200 downregulation. Our data demonstrated that CD200-decreased mRNA expression happens starting from the first step (Figure 3(c)). Thereafter, we treated ADHLSC separately with each of the growth factors used in the cocktail of differentiation to gain insights regarding the potential signaling events behind the dramatic shutdown of CD200 expression following hepatogenic differentiation. As shown in Figure 3(d), data presented as a relative CD200 mRNA level in treated versus untreated ADHLSC (serum-free conditions) clearly shows that HFG and bFGF exerted no effect and a slightly nonsignificant effect, respectively. Nevertheless, both EGF and OSM treatments resulted in a significant decrease of about half of the CD200 mRNA level. Such diminution is in line with the decreased CD200 mRNA levels observed after the first and the last steps of the hepatogenic differentiation protocol.
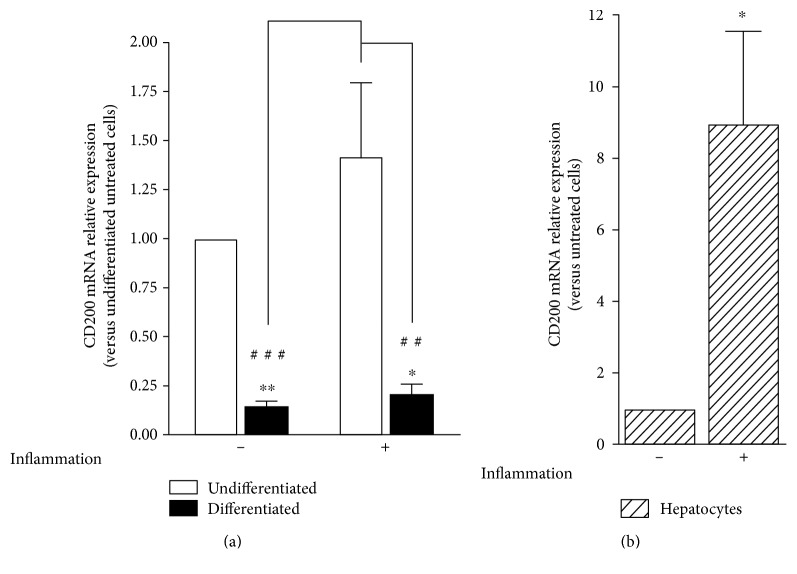
Finally, we checked if CD200 mRNA expression was also modulated after inflammation priming. As shown in Figure 4, this parameter was investigated in undifferentiated and differentiated ADHLSCs as well as in hepatocytes. Whilst no effect was observed in ADHLSCs (Figure 4(a)), inflammation priming of hepatocytes strongly induced the CD200 mRNA level (Figure 4(b)).
Figure 4.
CD200 mRNA expression after inflammation priming. Undifferentiated ADHLSCs, (a) differentiated ADHLSCs as well as (b) hepatocytes were grown under basic or inflammatory-priming conditions. Total RNA was isolated and PPIA-normalized CD200 mRNA levels were quantified by RT-qPCR. Data shown represent the mean ± SEM of three different experiments (∗p < 0.05; ∗∗p < 0.01 versus corresponding untreated cells, unpaired t-test; ##p < 0.01; ###p < 0.001 versus corresponding treated undifferentiated cells, one-way ANOVA followed by the Tukey post hoc test).
The functional network of CD200 and its interacting proteins was analyzed to better highlight which pathways might be altered during hepatogenic differentiation. Due to its great immunological importance, CD200 protein network was analyzed by using STRING version10 database. This analysis was selected for its many advantages among which extensive collection of precomputed interaction data derived from various sources, such as, high-throughput experimental data, literature data, and computational predictions. This tool was used to query, retrieve, and analyze the CD200 protein interaction network with the interactions restricted to those available for Homo sapiens. Using CD200 as a query, and choosing the prediction methods to include neighborhood, gene fusion, co-occurrence, coexpression, experiments, databases, and text mining, a network of eleven interacting proteins was constructed (Figure 5). This interaction network is visualized in the form of a graph with the protein molecules forming the nodes of the graph and the interactions forming the edges. Among those eleven predicted interacting proteins, four were already confirmed to interact with CD200 among which three (CD200R1, HCRTR2, and CD200R1L) were experimentally determined, and one (CD200R1) was determined from a curated database whilst the remaining seven were predicted based on text mining and were verified using PubMed literature database. Indeed, CD200R1 and CD200R1L (CD200R2) are receptors for CD200 whilst HCRTR2 is a hypocretin (orexin) receptor 2 implicated in neuropeptide signaling pathway [7]. Orexins and orexin receptors, a family of hypothalamic neuropeptides and G protein-coupled receptors are involved in the regulation of feeding behavior [7] in addition to their ability to trigger phospholipase C signaling via activating Ca2+ influx [8].
Figure 5.
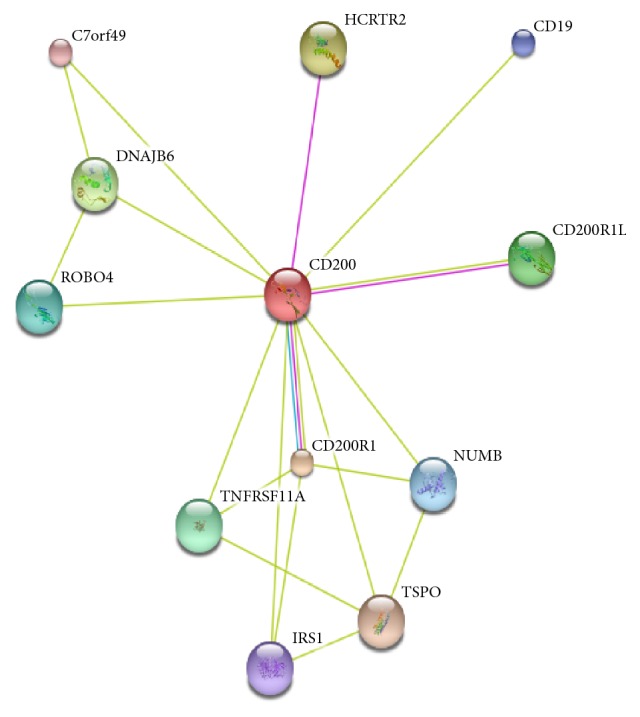
STRING database-generated protein interaction network generated using CD200 protein name as query. The blue and pink colours correspond to known interactions from curated and experimental work, respectively, whilst the light-green colour corresponds to interaction prediction based on text mining.
4. Discussion
By maintaining vital metabolic homeostasis, good functioning of the liver is essential for survival. Due to its high ability to regenerate and to its continuous extrahepatic cellular supply, cell therapy is currently appreciating the potential of exogenous cell suspensions in replenishing the liver and correcting its functional and/or structural defects. Extensive preclinical work has been conducted to study the mechanistic pathways that govern genesis of the major liver cells, hepatocytes. Nowadays, different stem cells are being investigated to hopefully increase the diversity of cell pools and availability of cell-based therapy products for patients who can no longer regenerate their altered hepatic tissue [9]. In addition to adult bone marrow-derived and embryonic stem cells that have been well described for their potential to transdifferentiate into mature hepatocyte-like cells [9], ADHLSCs have recently emerged as a novel progenitor reservoir of hepatocytes [2, 4, 10]. ADHLSC in vitro and in vivo hepatocytic differentiation potential [2, 4] and hypoimmunogenic profile [5] support their usefulness for liver regeneration [2–5, 10].
Acceptance of engrafted ADHLSCs could be a result of the surrounding inflammatory condition sensing as well as the modulation of the host immune responses like to what has been reported for other MSCs [11, 12]. Based on the interesting expression and secretion profiles of naïve ADHLSC [3], the current study aimed at appreciating the impact of hepatogenic differentiation and inflammation on those features. In agreement with our recent observations [5], we confirmed that ADHLSCs are negative for CD34 and SSEA-4 but positive for CD90 and CD105 with no influence of inflammation and differentiation on that expression profile. Of note, CD90, which is absent in mature hepatocytes, remains expressed in differentiated ADHLSCs. This constitutive CD90 expression could be advantageous since decreased expression of CD90 has been reported to impair MSC immunosuppressive capacity [13].
Consistent with the previously reported ADHLSCs' hypoimmunogenicity [2, 5], we observed that, regardless of inflammation priming, undifferentiated and differentiated ADHLSCs as well as hepatocytes are negative for HLA-DR, HLA-G, and the costimulatory molecules CD27, CD70, CD80, CD86, CD134, CD154, and CD252 but positive for HLA-ABC molecules. The absence of CD134 and CD252 in differentiated ADHLSCs is of special interest because of the ability of these molecules to impair the suppressive action of regulatory T cells and thus to trigger host immune responses [14]. The increased CD40 expression, following inflammation priming, may thus not be sufficient to activate T cells since such activation involves other costimulatory molecules that appeared to be negative in our case [15].
Furthermore, our data showed that undifferentiated and differentiated ADHLSCs as well as hepatocytes are all negative for the surface receptors CD45 (a marker used to identify and isolate human hepatic progenitor cells) [16], CD95 (a marker that triggers apoptosis in liver cells) [17], CD184 (involved in repairing liver injury upon triggering MSCs to migrate, transdifferentiate, and fuse with hepatocytes) [18], CD200R (involved in mediating anti-inflammatory hepatic responses) [19], CD210 (playing important anti-inflammatory and immunosuppressive roles in liver) [20], CD229 (involved in the control of hepatocyte proliferation) [21], and CD271 (a marker of human hepatic stellate cells) [22]. This expression profile is not influenced after inflammation priming.
Regarding cell adhesion molecules (CAMs), undifferentiated ADHLSCs, differentiated ADHLSCs, and hepatocytes are negative for CD31 (with anti-inflammatory properties in the liver) [23], CD58 (proposed to augment cell mediated immunity against hepatitis B virus and thus leading to hepatocyte destruction) [24], CD62e, CD102 (involved in recruitment of lymphocytes to the liver) [25], and CD146 (although poorly characterized but described to be involved in angiogenesis) [26]. However, all cells are positive to CD29 (essential for hepatocyte survival) [27], CD49e, and CD166 (playing an antiapoptotic role for liver cells) [28] regardless of presence or absence of inflammatory signals. Our observations for CD102, CD146, and CD166 expression levels in undifferentiated ADHLSCs are in line with our previous observations [5]. CD44 (involved in regulating liver inflammation) [29] and CD54 (involved in the recruitment of hepatic-natural killer cells in the liver) [30] exhibit high levels in undifferentiated and differentiated ADHLSCs as well as hepatocytes. Inflammatory signals reinforce CD54 expression in these different cells and increase CD44 expression only in differentiated ADHLSCs. Remarkably, the minimal CD106 expression (involved in enabling neutrophil migration within the liver) [31] exhibited by both undifferentiated and differentiated ADHLSCs was strongly induced by inflammatory signals to levels higher than those exhibited by inflammation-primed hepatocytes. These observations indicate that the different examined CAMs are heterogeneously expressed in the different tested cell types and exhibit different sensitivities to inflammation.
As a key component of the liver's innate immunity, NK cells are primarily involved in host defence but also have regulatory effects during their interactions with other types of liver cells [32]. Regardless of inflammation priming, both differentiated ADHLSCs and hepatocytes were positive for CD112 and CD155 but negative for ULBP-3, in a manner comparable to that exhibited by undifferentiated ADHLSCs. NK cells' lytic activity could be triggered upon interaction of CD112 or CD155 with the same NK-activating receptor DNAM-1 [33] or ULBP-3 with NKG2D receptor [34]. Although the absence of ULBP-3 seems advantageous, the presence of CD112 and CD155 might be risky in terms of allowing NK-mediated lysis of those liver-derived cells.
Given that ADHLSCs could exhibit immunomodulatory properties, it was then important to evaluate the expression and modulation of several relevant immune regulatory molecules. Undifferentiated and differentiated ADHLSCs were positive for CD73 and HO-1 but negative for CD39, regardless of inflammation priming. Interestingly, although initially negative for CD274, inflammation strikingly increased its expression on all the studied cells. CD274 expression is known to be upregulated in response to proinflammatory cytokines where it exerts an inhibitory role on the costimulatory pathway of T cells [35]. CD274 upregulation could then compensate the effect of upregulated CD40 expression observed after inflammation priming, thus inhibiting T cell activation.
A major output of this study is the downregulation of CD200 expression that takes place during hepatogenic differentiation. This impaired CD200 expression in differentiated ADHLSCs is consistent with the absence of CD200 expression in hepatocytes. CD200 is a transmembrane surface glycoprotein which, upon engagement with its receptor (CD200R), delivers inhibitory signals leading to immunosuppression, inhibition of inflammation, and tolerance to allografts [33]. Given the important inflammation-inhibitory potential of CD200, the observed loss of its expression in differentiated ADHLSCs could limit their anti-inflammatory capacity, thus imposing undesired consequences during the application of ADHLSC-based liver therapy. CD200 is widely expressed in many cell types, among which are extrahepatic MSCs. This expression is uneven and can be modulated depending on the tissue origin and growth conditions. For instance, CD200 expression is not equal in the bone marrow, umbilical cord blood, Wharton's jelly, and adipose tissue-derived MSCs [36, 37]. Of great importance, hepatic progenitor cells (HPCs) isolated from the adult human liver are highly similar to cultured primary hepatocytes in their transcriptional profiles where both cell types are negative for CD200 expression [16]. Expression of several MSC markers including CD73, CD146, and CD200 is downregulated after adipogenic and osteogenic differentiation [38] whilst only CD200 expression is completely abolished following chondrogenic differentiation [38]. Identifying specific markers to evaluate the multilineage differentiation potential of stem cells is thus highly important for the improvement of stem-cell-based therapies. CD105 expression was demonstrated to be significantly downregulated following adipogenic, chondrogenic, and osteogenic differentiation of umbilical cord blood MSCs [39]. In addition, to be used as a specific marker for a determined cell population or for a defined cell differentiation status, the observations made in this study might suggest CD200 as a marker for the immunomodulatory potential of ADHLSCs.
Protein-protein interaction (PPI) network revealed that several proteins may interact with CD200. These proteins may be expressed by different liver-derived cells and may also have implications in various cell biology features such as for roundabout, axon guidance receptor, homolog 4 (Robo4) in cell migration and angiogenesis during endothelial inflammatory responses [40, 41], and insulin receptor substrate 1 (IRS-1) that may mediate the control of various cellular processes related to insulin and insulin-like growth factor-1 (IGF-1) receptors [42]. Following the engagement of CD200 with CD200 receptor 1, the tumor necrosis factor receptor superfamily member 11a (TNFRSF11A) can interact with the TNF receptor-associated factors (TRAFs) [43] which modulate the immune and inflammatory responses particularly during the interaction between T cells and dendritic cells [43]. The potential interaction of CD200 with translocator protein (TSPO) could have important consequences for ADHLSC therapeutic applications because of its involvement in the regulation of cellular metabolic energy. TSPO, a mitochondrial outer membrane protein that belongs to a family of tryptophan-rich sensory, modulates critical biological functions, such as cellular bioenergetics and metabolism, immunomodulation, and apoptosis [44]. The juxtaposition of TSPO at the cytosolic/mitochondrial interface and the existence of endogenous ligands that are regulated by metabolism suggest that TSPO functions to adapt mitochondrial to cellular metabolism [45]. Indeed, TSPO has been shown to affect mitochondrial energy homeostasis through modulation of fatty acid oxidation (FAO). Such function positively correlates with high levels of TSPO expression as observed in cell types active in lipid storage/metabolism [46]. From a pharmacological perspective, the specific upregulation of TSPO in inflammatory and injury conditions makes TSPO an interesting, druggable target of mitochondrial metabolism [47].
The potential interactions of CD200 with Numb, a cell fate determinant with a critical role in the regulation of pluripotency and stem cell division [48], are of importance. Indeed, Numb phosphorylation by pluripotency-associated transcription factor NANOG and subsequent p53 degradation drive self-renewability and proliferation of tumor-initiating cells, which results in higher liver oncogenesis [49]. Second, Numb inhibits the Notch pathway, a crucial regulator of stem cell behavior [50]. Several mechanisms of actions are proposed [50] including the ubiquitination of Notch1 receptor and the subsequent degradation of the Notch intracellular domain following receptor activation [51] and the inhibition of Notch endosomal trafficking and recycling [52]. In the context of ADHLSC cell therapy, CD200 and Numb interactions could play a crucial role to maintain the proper homeostasis of both undifferentiated and differentiated cells.
In conclusion, we suggest that the modulation of the CD200 pathway might be involved in the control of the differentiated ADHLSC immunosuppressive function. Whilst requiring further investigations, the current data are helpful for tackling the interest of deeply exploring the immune biology of liver cells to transplant for an efficient management of liver diseases by cell therapy-based approaches. Thanks to their ability to functionally differentiate into hepatocyte-like cells and to maintain the major immunological profile of mature hepatocytes, differentiated ADHLSCs might support their development for liver cell therapy.
Supplementary Material
ADHLSCs were sequentially incubated with specific growth factors/cytokines and processed for the evaluation of the hepatogenic differentiation quality. A) Differentiated ADHLSC display significant morphological changes with polygonal epithelial-like shape. Pictures were taken at magnification of 200x. Presented data are representative of at least three different experiments. B) RT-PCR analysis of hepatocyte specific gene expression profile of differentiated (Diff) compared to undifferentiated ADHLSCs (Und) confirms a positive correlation with the morphological changes. Data shown are agarose gel electrophoresis of amplification products corresponding to hepatic markers: MRP2, multidrug resisting protein-2; TDO, tryptophan 2,3-dioxygenase; CYP3A4, cytochrome P450, family 3, subfamily A, polypeptide 4; GAPDH, glyceraldehyde-3-phosphate dehydrogenase is used as house-keeping control. Presented data are representative of at least three different experiments. C) Forty μg of total protein extracted from differentiated ADHLSCs and isolated hepatocytes were analyzed using western blotting. Hepatogenic differentiation was supported, by demonstrating the expression of CYP3A4 and hepatocyte nuclear factor-4 alpha (HNF4a) proteins in differentiated ADHLSC (Diff) as compared to hepatocytes (Hep). D) After the hepatogenic differentiation process, undifferentiated (U) and differentiated (D) ADHLSCs were recovered for CYP3A4 activity analysis using P450-GloTM assay a Victor3 luminometer (PerkinElmer). Data shown are the mean ± SEM of three independent experiments (T-test ∗∗∗ p< 0.001 vs undifferentiated ADHLSC)
Acknowledgments
This project was supported by BRUSTEM, an impulse program of the Institute for the encouragement of Scientific Research and Innovation of Brussels (ISRIB). Hoda El-Kehdy is a postdoc grant holder from IREC (Institut de Recherche Expérimentale and Clinique). Mustapha Najimi is a Principal Investigator of IREC. Mehdi Najar is a Belgian Fonds National de la Recherche Scientifique (FNRS) postdoctoral researcher.
Abbreviations
- ADHLSC:
Adult-derived human liver stem/progenitor cells
- bFGF:
Basic fibroblast growth factor
- EGF:
Epidermal growth factor
- HGF:
Hepatocyte growth factor
- NK:
Natural killer
- OSM:
Oncostatin M.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Authors' Contributions
Mustapha Najimi and Mehdi Najar contributed equally to this work.
References
- 1.Forbes S. J., Gupta S., Dhawan A. Cell therapy for liver disease: from liver transplantation to cell factory. Journal of Hepatology. 2015;62(1):S157–S169. doi: 10.1016/j.jhep.2015.02.040. [DOI] [PubMed] [Google Scholar]
- 2.Najimi M., Khuu D. N., Lysy P. A., et al. Adult-derived human liver mesenchymal-like cells as a potential progenitor reservoir of hepatocytes? Cell Transplantation. 2007;16(7):717–728. doi: 10.3727/000000007783465154. [DOI] [PubMed] [Google Scholar]
- 3.Berardis S., Lombard C., Evraerts J., et al. Gene expression profiling and secretome analysis differentiate adult-derived human liver stem/progenitor cells and human hepatic stellate cells. PLoS One. 2014;9(1, article e86137) doi: 10.1371/journal.pone.0086137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khuu D. N., Scheers I., Ehnert S., et al. In vitro differentiated adult human liver progenitor cells display mature hepatic metabolic functions: a potential tool for in vitro pharmacotoxicological testing. Cell Transplantation. 2011;20(2):287–302. doi: 10.3727/096368910X516655. [DOI] [PubMed] [Google Scholar]
- 5.Raicevic G., Najar M., Najimi M., et al. Influence of inflammation on the immunological profile of adult-derived human liver mesenchymal stromal cells and stellate cells. Cytotherapy. 2015;17(2):174–185. doi: 10.1016/j.jcyt.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 6.Szklarczyk D., Franceschini A., Kuhn M., et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Research. 2011;39(supplement 1):D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sakurai T. In Hypocretins: Integrators of Physiological Functions, de Lecea, L. and Sutcliffe, J. G., Eds. New York: Springer; 2005. Orexin and orexin receptors; pp. 13–23. [DOI] [Google Scholar]
- 8.Lund P. E., Shariatmadari R., Uustare A., et al. The orexin OX1 receptor activates a novel Ca2+ influx pathway necessary for coupling to phospholipase C. The Journal of Biological Chemistry. 2000;275(40):30806–30812. doi: 10.1074/jbc.M002603200. [DOI] [PubMed] [Google Scholar]
- 9.Lee S. Y., Kim H. J., Choi D. Cell sources, liver support systems and liver tissue engineering: alternatives to liver transplantation. International Journal of Stem Cells. 2015;8(1):36–47. doi: 10.15283/ijsc.2015.8.1.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khuu D. N., Nyabi O., Maerckx C., Sokal E., Najimi M. Adult human liver mesenchymal stem/progenitor cells participate in mouse liver regeneration after hepatectomy. Cell Transplantation. 2013;22(8):1369–1380. doi: 10.3727/096368912X659853. [DOI] [PubMed] [Google Scholar]
- 11.Pluchino S., Cossetti C. How stem cells speak with host immune cells in inflammatory brain diseases. Glia. 2013;61(9):1379–1401. doi: 10.1002/glia.22500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y., Chen X., Cao W., Shi Y. Plasticity of mesenchymal stem cells in immunomodulation: pathological and therapeutic implications. Nature Immunology. 2014;15(11):1009–1016. doi: 10.1038/ni.3002. [DOI] [PubMed] [Google Scholar]
- 13.Campioni D., Rizzo R., Stignani M., et al. A decreased positivity for CD90 on human mesenchymal stromal cells (MSCs) is associated with a loss of immunosuppressive activity by MSCs. Cytometry. Part B, Clinical Cytometry. 2009;76(3):225–230. doi: 10.1002/cyto.b.20461. [DOI] [PubMed] [Google Scholar]
- 14.Vu M. D., Xiao X., Gao W., et al. OX40 costimulation turns off Foxp3+ Tregs. Blood. 2007;110(7):2501–2510. doi: 10.1182/blood-2007-01-070748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nasef A., Mathieu N., Chapel A., et al. Immunosuppressive effects of mesenchymal stem cells: involvement of HLA-G. Transplantation. 2007;84(2):231–237. doi: 10.1097/01.tp.0000267918.07906.08. [DOI] [PubMed] [Google Scholar]
- 16.Li J., Xin J., Zhang L., et al. Human hepatic progenitor cells express hematopoietic cell markers CD45 and CD109. International Journal of Medical Sciences. 2014;11(1):65–79. doi: 10.7150/ijms.7426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malhi H., Gores G. J., Lemasters J. J. Apoptosis and necrosis in the liver: a tale of two deaths? Hepatology. 2006;43(S1):31–44. doi: 10.1002/hep.21062. [DOI] [PubMed] [Google Scholar]
- 18.Hao N., Li C., Lü M., et al. SDF-1/CXCR4 axis promotes MSCs to repair liver injury partially through trans-differentiation and fusion with hepatocytes. Stem Cells International. 2015;2015:p. 10. doi: 10.1155/2015/960387.960387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan E. P., Wells R. G. Today’s hepatic stellate cells: not your father’s Sternzellen. Hepatology. 2007;45(5):1326–1327. doi: 10.1002/hep.21725. [DOI] [PubMed] [Google Scholar]
- 20.Shen X., Hong F., Nguyen V. A., Gao B. IL-10 attenuates IFN-α-activated STAT1 in the liver: involvement of SOCS2 and SOCS3. FEBS Letters. 2000;480(2–3):132–136. doi: 10.1016/S0014-5793(00)01905-0. [DOI] [PubMed] [Google Scholar]
- 21.Bouhlal H., Nyga R., Cartier F., et al. Molecular and functional characterisation of hepatic slam CD229 and ITS implication in the development of hepatocellular carcinoma. Gut. 2011;60(Supplement 1):A226–A227. doi: 10.1136/gut.2011.239301.477. [DOI] [Google Scholar]
- 22.Patil P. B., Joshi M., Kuna V. K., et al. CD271 identifies functional human hepatic stellate cells, which localize in peri-sinusoidal and portal areas in liver after partial hepatectomy. Cytotherapy. 2014;16(7):990–999. doi: 10.1016/j.jcyt.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 23.Malik I. A., Stange I., Martius G., et al. Role of PECAM-1 in radiation-induced liver inflammation. Journal of Cellular and Molecular Medicine. 2015;19(10):2441–2452. doi: 10.1111/jcmm.12630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang P., Qi B., Chen P., et al. CD58 expression of liver tissue in patients with chronic hepatitis B virus infection. Chinese Medical Journal. 2008;121(6):557–560. [PubMed] [Google Scholar]
- 25.Lalor P. F., Shields P., Grant A., Adams D. H. Recruitment of lymphocytes to the human liver. Immunology and Cell Biology. 2002;80(1):52–64. doi: 10.1046/j.1440-1711.2002.01062.x. [DOI] [PubMed] [Google Scholar]
- 26.Sers C., Riethmüller G., Johnson J. P. MUC18, a melanoma-progression associated molecule, and its potential role in tumor vascularization and hematogenous spread. Cancer Research. 1994;54(21):5689–5694. [PubMed] [Google Scholar]
- 27.Pinkse G. G. M., Voorhoeve M. P., Noteborn M., Terpstra O. T., Bruijn J. A., De Heer E. Hepatocyte survival depends on beta1-integrin-mediated attachment of hepatocytes to hepatic extracellular matrix. Liver International. 2004;24(3):218–226. doi: 10.1111/j.1478-3231.2004.0914.x. [DOI] [PubMed] [Google Scholar]
- 28.Yu W., Wang J., Ma L., et al. CD166 plays a pro-carcinogenic role in liver cancer cells via inhibition of FOXO proteins through AKT. Oncology Reports. 2014;32(2):677–683. doi: 10.3892/or.2014.3226. [DOI] [PubMed] [Google Scholar]
- 29.Kimura K., Nagaki M., Kakimi K., et al. Critical role of CD44 in hepatotoxin-mediated liver injury. Journal of Hepatology. 2008;48(6):952–961. doi: 10.1016/j.jhep.2008.01.033. [DOI] [PubMed] [Google Scholar]
- 30.Luo D., Vanderkerken K., Bouwens L., et al. The role of adhesion molecules in the recruitment of hepatic natural killer cells (pit cells) in rat liver. Hepatology. 1996;24(6):1475–1480. doi: 10.1002/hep.510240629. [DOI] [PubMed] [Google Scholar]
- 31.Essani N. A., Bajt M. L., Farhood A., Vonderfecht S. L., Jaeschke H. Transcriptional activation of vascular cell adhesion molecule-1 gene in vivo and its role in the pathophysiology of neutrophil-induced liver injury in murine endotoxin shock. Journal of Immunology. 1997;158(12):5941–5948. [PubMed] [Google Scholar]
- 32.Tian Z., Chen Y., Gao B. Natural killer cells in liver disease. Hepatology. 2013;57(4):1654–1662. doi: 10.1002/hep.26115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gorczynski R. M. CD200:CD200R-mediated regulation of immunity. ISRN Immunology. 2012;2012:1–18. doi: 10.5402/2012/682168.682168 [DOI] [Google Scholar]
- 34.Mistry A. R., O’Callaghan C. A. Regulation of ligands for the activating receptor NKG2D. Immunology. 2007;121(4):439–447. doi: 10.1111/j.1365-2567.2007.02652.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wiendl H., Mitsdoerffer M., Schneider D., et al. Human muscle cells express a B7-related molecule, B7-H1, with strong negative immune regulatory potential: a novel mechanism of counterbalancing the immune attack in idiopathic inflammatory myopathies. The FASEB Journal. 2003;17(13):1892–1894. doi: 10.1096/fj.03-0039fje. [DOI] [PubMed] [Google Scholar]
- 36.Pietilä M., Lehtonen S., Tuovinen E., et al. CD200 positive human mesenchymal stem cells suppress TNF-alpha secretion from CD200 receptor positive macrophage-like cells. PLoS One. 2012;7(2, article e31671) doi: 10.1371/journal.pone.0031671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Najar M., Raicevic G., Jebbawi F., et al. Characterization and functionality of the CD200-CD200R system during mesenchymal stromal cell interactions with T-lymphocytes. Immunology Letters. 2012;146(1–2):50–56. doi: 10.1016/j.imlet.2012.04.017. [DOI] [PubMed] [Google Scholar]
- 38.Delorme B., Ringe J., Gallay N., et al. Specific plasma membrane protein phenotype of culture-amplified and native human bone marrow mesenchymal stem cells. Hematop Stem Cells. 2008;111(5):2631–2635. doi: 10.1182/blood-2007-07-099622.The. [DOI] [PubMed] [Google Scholar]
- 39.Jin H. J., Park S. K., Oh W., Yang Y. S., Kim S. W., Choi S. J. Down-regulation of CD105 is associated with multi-lineage differentiation in human umbilical cord blood-derived mesenchymal stem cells. Biochemical and Biophysical Research Communications. 2009;381(4):676–681. doi: 10.1016/j.bbrc.2009.02.118. [DOI] [PubMed] [Google Scholar]
- 40.Suchting S., Heal P., Tahtis K., Stewart L. M., Bicknell R. Soluble Robo4 receptor inhibits in vivo angiogenesis and endothelial cell migration. The FASEB Journal. 2005;19(1):121–123. doi: 10.1096/fj.04-1991fje. [DOI] [PubMed] [Google Scholar]
- 41.Zhao H., Anand A. R., Ganju R. K. Slit2-Robo4 pathway modulates lipopolysaccharide-induced endothelial inflammation and its expression is dysregulated during endotoxemia. Journal of Immunology. 2014;192(1):385–393. doi: 10.4049/jimmunol.1302021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mardilovich K., Pankratz S. L., Shaw L. M. Expression and function of the insulin receptor substrate proteins in cancer. Cell Communication and Signaling: CCS. 2009;7(1):p. 14. doi: 10.1186/1478-811X-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Inoue J. I., Ishida T., Tsukamoto N., et al. Tumor necrosis factor receptor-associated factor (TRAF) family: adapter proteins that mediate cytokine signaling. Experimental Cell Research. 2000;254(1):14–24. doi: 10.1006/excr.1999.4733. [DOI] [PubMed] [Google Scholar]
- 44.Liu G. J., Middleton R. J., Kam W. W., et al. Functional gains in energy and cell metabolism after TSPO gene insertion. Cell Cycle. 2017;19(5):436–447. doi: 10.1080/15384101.2017.1281477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gut P. Targeting mitochondrial energy metabolism with TSPO ligands. Biochemical Society Transactions. 2015;43(4):537–542. doi: 10.1042/BST20150019. [DOI] [PubMed] [Google Scholar]
- 46.Tu L. N., Zhao A. H., Hussein M., Stocco D. M., Selvaraj V. Translocator protein (TSPO) affects mitochondrial fatty acid oxidation in steroidogenic cells. Endocrinology. 2016;157(3):1110–11121. doi: 10.1210/en.2015-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Faraldo M. M., Glukhova M. A. Regulating the regulator: Numb acts upstream of p53 to control mammary stem and progenitor cell. The Journal of Cell Biology. 2015;211(4):737–739. doi: 10.1083/jcb.201510104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Siddique H. R., Feldman D. E., Chen C. L., Punj V., Tokumitsu H., Machida K. NUMB phosphorylation destabilizes p53 and promotes self-renewal of tumor-initiating cells by a NANOG-dependent mechanism in liver cancer. Hepatology. 2015;62(5):1466–1479. doi: 10.1002/hep.27987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu J., Sato C., Cerletti M., Wagers A. Notch signaling in the regulation of stem cell self-renewal and differentiation. Current Topics in Developmental Biology. 2010;92:367–409. doi: 10.1016/S0070-2153(10)92012-7. [DOI] [PubMed] [Google Scholar]
- 50.Kandachar V., Roegiers F. Endocytosis and control of Notch signaling. Current Opinion in Cell Biology. 2012;24(4):534–540. doi: 10.1016/j.ceb.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McGill M. A., McGlade C. J. Mammalian numb proteins promote Notch1 receptor ubiquitination and degradation of the Notch1 intracellular domain. The Journal of Biological Chemistry. 2003;278(25):23196–23203. doi: 10.1074/jbc.M302827200. [DOI] [PubMed] [Google Scholar]
- 52.Couturier L., Mazouni K., Schweisguth F. Inhibition of Notch recycling by Numb: relevance and mechanism(s) Cell Cycle. 2013;12(11):1647–1648. doi: 10.4161/cc.24983. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ADHLSCs were sequentially incubated with specific growth factors/cytokines and processed for the evaluation of the hepatogenic differentiation quality. A) Differentiated ADHLSC display significant morphological changes with polygonal epithelial-like shape. Pictures were taken at magnification of 200x. Presented data are representative of at least three different experiments. B) RT-PCR analysis of hepatocyte specific gene expression profile of differentiated (Diff) compared to undifferentiated ADHLSCs (Und) confirms a positive correlation with the morphological changes. Data shown are agarose gel electrophoresis of amplification products corresponding to hepatic markers: MRP2, multidrug resisting protein-2; TDO, tryptophan 2,3-dioxygenase; CYP3A4, cytochrome P450, family 3, subfamily A, polypeptide 4; GAPDH, glyceraldehyde-3-phosphate dehydrogenase is used as house-keeping control. Presented data are representative of at least three different experiments. C) Forty μg of total protein extracted from differentiated ADHLSCs and isolated hepatocytes were analyzed using western blotting. Hepatogenic differentiation was supported, by demonstrating the expression of CYP3A4 and hepatocyte nuclear factor-4 alpha (HNF4a) proteins in differentiated ADHLSC (Diff) as compared to hepatocytes (Hep). D) After the hepatogenic differentiation process, undifferentiated (U) and differentiated (D) ADHLSCs were recovered for CYP3A4 activity analysis using P450-GloTM assay a Victor3 luminometer (PerkinElmer). Data shown are the mean ± SEM of three independent experiments (T-test ∗∗∗ p< 0.001 vs undifferentiated ADHLSC)