Abstract
The fight against infectious diseases is probably one of the greatest public health challenges faced by our society, especially with the emergence of carbapenem-resistant gram-negatives that are in some cases pan-drug resistant. Currently, β-lactamase-mediated resistance does not spare even the newest and most powerful β-lactams (carbapenems), whose activity is challenged by carbapenemases. The worldwide dissemination of carbapenemases in gram-negative organisms threatens to take medicine back into the pre-antibiotic era since the mortality associated with infections caused by these “superbugs” is very high, due to limited treatment options. Clinically-relevant carbapenemases belong either to metallo-β-lactamases (MBLs) of Ambler class B or to serine-β-lactamases (SBLs) of Ambler class A and D enzymes. Class A carbapenemases may be chromosomally-encoded (SME, NmcA, SFC-1, BIC-1, PenA, FPH-1, SHV-38), plasmid-encoded (KPC, GES, FRI-1) or both (IMI). The plasmid-encoded enzymes are often associated with mobile elements responsible for their mobilization. These enzymes, even though weakly related in terms of sequence identities, share structural features and a common mechanism of action. They variably hydrolyse penicillins, cephalosporins, monobactams, carbapenems, and are inhibited by clavulanate and tazobactam. Three-dimensional structures of class A carbapenemases, in the apo form or in complex with substrates/inhibitors, together with site-directed mutagenesis studies, provide essential input for identifying the structural factors and subtle conformational changes that influence the hydrolytic profile and inhibition of these enzymes. Overall, these data represent the building blocks for understanding the structure-function relationships that define the phenotypes of class A carbapenemases and can guide the design of new molecules of therapeutic interest.
Keywords: Biochemical properties, carbapenemase, class A, crystallography, molecular modeling, mutagenesis
1. INTRODUCTION
β-Lactams have been the most successful antimicrobial agents since the beginning of the antibiotic era due largely to their unique combination of efficacy, safety and versatility. β-lactams remain the workhorses for the treatment of several infections (from mild community-acquired infections to severe life-threatening nosocomial infections), either alone or in combination regimens [1]. The expanded-spectrum cephalosporins and the carbapenems, in particular, still represent a major treatment option for serious infections caused by gram-negative bacteria, and are listed among the drugs of choice in guidelines for the empiric treatment of several infections [2].
The expanding problem of resistance to β-lactam antibiotics illustrates the genetic adaptability of bacterial populations over a relatively short period of time. β-Lactamase-mediated resistance to β-lactams may result from selection of bacterial species that naturally produced β-lactamases, from mutation either in structural genes of their regulatory components or from acquisition of specific resistance genes.
Based on the genetic environment and on its genetic support, β-lactamases may be produced in a constitutive or inducible manner [3]. Plasmids and transposons, in particular, play an important role for rapid diffusion of those genes. From a clinical point of view, selection of β-lactamases with extended-spectrum activity has been observed in gram-negatives (Enterobacteriaceae, Pseudomonas spp., Acinetobacter spp.) but not in gram-positives (Staphylococcus, Enterococcus spp.) [4]. β-lactamases play an important role in acquired resistance to β-lactams in Enterobacteriaceae whereas in Pseudomonas aeruginosa and Acinetobacter baumannii other mechanisms are often associated such as impermeability and overexpression of efflux pumps [2, 5-7].
In gram-negative pathogens the major mechanism of acquired β-lactam resistance is the production of β-lactamases; other mechanisms (e.g. impermeability, active efflux, PBP target modification) also contribute in several cases. On the basis of their amino acid sequences and catalytic mechanisms, β-lactamases are divided into 4 molecular classes. Classes A, C and D contain active-site serine β-lactamases (SBLs) whose reaction pathways involve acyl-enzyme adducts while class B contain metallo-β-lactamases (MBLs), which do not form such intermediates [8]. The introduction of new β-lactam classes has always led to the emergence of β-lactamases capable of degrading them, and the evolution of β-lactamases has been a major driving force for β-lactam discovery and development. While a handful of β-lactamases were known in the early 1970’s, the number since then increased exponentially. Currently, β-lactamase-mediated resistance does not spare even the newest and most powerful β-lactams (i.e. the expanded-spectrum cephalosporins and carbapenems), whose activity is challenged by the extended-spectrum β-lactamases (ESBLs, classes A, C and D), the metallo-β-lactamases (MBLs, class B), and the serine-carbapenemases (classes A and D) [2].
Most of the clinically-relevant β-lactamases belong to the Ambler class A serine-active β-lactamases. They operate via a multistep process involving the formation of an intermediate acyl-enzyme and its subsequent hydrolytic destruction (Fig. 1) [9-11]. The steady state kinetic parameters are made of different combinations of microscopic rate constants [11, 12]. Every β-lactamase is thus defined for each substrate by: (i) the turnover number, « kcat », which is the best parameter to describe the hydrolytic activity of β-lactamases, since it depends on the rates of formation and hydrolysis of the acyl–enzyme complex (Fig. 1); (ii) the apparent affinity or half-saturation constant, Km that is frequently referred to as the Michaelis constant because of its equivalence to the half-saturation constant defined for the Michaelis–Menten mechanism (which does not, however, include a covalent intermediate). The Km includes all the rate constants describing the catalytic steps of β-lactam hydrolysis; and (iii) the apparent second-order rate constant, frequently called ‘catalytic efficiency’, kcat ⁄Km, has also been used as to characterize β-lactamases. Class A enzymes (known primarily as penicillinases) tend to hydrolyze penicillins over cephalosporins as substrates although many variants may significantly hydrolyze broad-spectrum cephalosporins and also a few carbapenems [13, 14]. Most class A β-lactamases are inhibited by the clinically-available β-lactamase inhibitors, such as clavulanic acid, tazobactam and sulbactam [15]. Class A β-lactamases are either naturally-produced and chromosome-encoded or acquired and often plasmid-encoded.
Fig. (1).
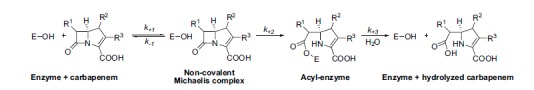
General catalytic pathway of active-site serine β-lactamases. This mechanism is schematically represented by a three-step model. E is the enzyme.
The dissemination of class A carbapenemases among Enterobacteriaceae is a matter of great clinical concern given (i) the major role of these pathogens as causes of nosocomial infections (and, for Escherichia coli, also of community-acquired infections), and (ii) the major role of expanded-spectrum cephalosporins and carbapenems in the treatment of those infections [2]. This review will focus on the clinically-relevant class A carbapenemases and address structure-function relationships, a prerequisite for the design, in a near future, of novel inhibitors of this class of carbapenemases.
2. EPIDEMIOLOGY OF CLASS A CARBAPENEMASES
Class A carbapenemases may be chromosomally-encoded (SME, NmcA, SFC-1, BIC-1, PenA, FPH-1, SHV-38), plasmid-encoded (KPC, GES, FRI-1) or both (IMI). Chromosomally-encoded class A carbapenemases have been identified in rare gram-negative species that appeared sporadically in clinical or environmental samples since their first discovery, more than 20 years ago [13, 16]. Some have been detected in rare Enterobacter cloacae, Serratia marcescens, Serratia fonticola or Pseudomonas fluorescens as single isolates or in small outbreaks [13, 16] while others are linked to the species such as PenA and FPH-1 present in Burkholderia cepacia and Francisella philomiragia, respectively. On the other hand plasmid-encoded enzymes, best exemplified by KPC-2, have spread all around the world and have been isolated in most of the clinically-relevant enterobacterial species, P. aeruginosa and A. baumannii [2, 17]. Usually, class A carbapenemases confer reduced susceptibility to imipenem to bacteria expressing them, but minimal inhibitory concentrations (MICs) can remain in the susceptibility range (e.g., imipenem MICs of < 2 µg/ml). Routine susceptibility testing may therefore not detect these isolates as carbapenemase producers. The mechanism of hydrolysis involves an active-site serine at position 70 of the Ambler class A numbering scheme [18]. Class A carbapenemases have the ability to hydrolyze a broad variety of β-lactams, including carbapenems, cephalosporins, penicillins, and aztreonam, and are more or less inhibited by clavulanate and tazobactam, placing them in the 2f functional subgroup of β-lactamases [19, 20].
2.1. Naturally- and chromosomally-encoded class A carbapenemases
2.1.1. SME and IMI/NmcA
Class A carbapenemases that belong to the SME and IMI/NmcA families are known from tiny numbers of Serratia and Enterobacter isolates [24, 26, 37]. Carbapenem resistance is evident in the IMI/NmcA or SME producing organisms but they usually remain susceptible to expanded-spectrum cephalosporins. In addition, they differ from many plasmid-encoded carbapenemase-producing pathogens in that they generally remain susceptible to all other non-β-lactam antibiotic families (fluoroquinolones, aminoglycosides, …).
SME-1, which stands for “Serratia marcescens enzyme”, was initially detected in two S. marcescens isolates collected in England in 1982 [24, 38]. The SME variants that differ only by one or two amino acid substitutions have so far exclusively been found in S. marcescens. SME-producing isolates have been infrequently and sporadically detected in the UK, across USA, Argentina, Switzerland and Canada [24, 39-45]. SME producing S. marcescens were found as single or small clusters of up to 19 isolates [44]. Pulsed-field gel electrophoresis (PFGE) revealed a great genetic variability among SME-producing S. marcescens isolates collected from different geographical locations, although some degree of clonal spread was evidenced when several isolates were collected from one location [13, 41]. The blaSME gene is not present in all S. marcescens isolates, but only a small subpopulation of the species. Of note, an increasing identification of S. marcescens harbouring blaSME gene has been described in North and South America [44].
IMI/NmcA enzymes (IMIpenemase/Non-metallo-carbapenemaseA), which form two subgroups, IMI and NmcA, respectively [26, 37, 46] have been detected in rare clinical isolates of E. cloacae in the United States, France, Tahiti (French Polynesia), Finland, Ireland, and Argentina [26, 37, 47-51]. NmcA and IMI-1 share 97% amino acid identity and are related to SME-1, with approximately 70% amino acid identity. The chromosomally-encoded NmcA enzyme was initially reported in 1994 in an E. cloacae isolate from France [37]. Since then, it has been identified in rare cases in Finland, Argentina, USA [47, 48, 52]. The chromosomally-located blaIMI-1 gene was initially reported in two E. cloacae isolates from the USA in 1996 [26]. Plasmid-encoded blaIMI-2 gene was subsequently detected in clonally-related E. asburiae isolates recovered from 7 out of 16 rivers in the mid-western regions of the USA [34], and in an E. cloacae clinical isolate from China [53]. IMI carbapenemases are relatively uncommon but their presence in Enterobacter clinical isolates have increasingly been reported in recent years in France, Finland, Singapore, Croatia and Ireland [49, 52, 54-56]. They consist mainly of E. cloacae isolates producing the IMI-1 enzyme or single amino acid variant IMI-3 [57]. However, IMI-producing E. asburiae clinical isolates have also been found in three patients from different cities in France between 2007 and 2011 [58] and in Ireland [49]. In Finland, IMI-positive E. cloacae were found in 2008 and 2010 [52]. Nine IMI-variants have been described, 3 published (IMI-1-3) from the USA, France, Finland, Singapore, Ireland and China, 3 (IMI-4,7,8) sequences are available on GenBank, and three (IMI-5,6,9) are assigned on Lahey Clinic website but not yet released. They differ by 1 to 7 amino acid substitutions from IMI-1, except for IMI-8, recently identified in the UK that has 42 amino acid changes (85% amino acid identity).
2.1.2. The SFC-1 Enzymes
SFC-1 was identified from S. fonticola. The gene coding for this carbapenemase was chromosomally-encoded [59, 60]. The SFC-1 is not ubiquitous in S. fonticola. It was only isolated sofar from a single S. fonticola environmental isolate from Portugal. SFC-1 is related to other class A carbapenemases, in particular KPC-2 from K. pneumoniae (64% identity), NmcA (60%), IMI-1 (60%) and SME-1 (59%) [24, 26, 35, 37]. No putative LysR-type regulator gene was identified upstream of the blaSFC-1 gene, whereas such regulators are present upstream of blaNmcA, blaSME-1, and bla IMI-1 genes [26, 37, 61].
2.1.3. The SHV-38 Enzyme
SHV-38 was isolated from K. pneumoniae isolate displaying reduced susceptibility to imipenem. Its gene was chromosomally-encoded [33]. The SHV-38 enzyme differs by a single substitution, Ala146Val substitution (Ambler numbering [18]), from the broad-spectrum and naturally-occurring β-lactamase SHV-1. Recently, another SHV-38-producing K. pneumoniae clinical isolate was identified from Brazil [62].
2.1.4. BIC-1
BIC-1 carbapenemase was identified in an environmental Pseudomonas fluorescens isolate (PF-1) resistant to carbapenems and recovered from water of the Seine river in Paris (France) [30]. The blaBIC-1 gene was chromosomally-encoded in P. fluorescens PF-1. BIC-1 has 69% and 63% amino acid identities with β-lactamases SFC-1 from S. fonticola and the plasmid-encoded KPC-2, respectively. It was further identified in two other P. fluorescens isolates 3 months later again from water of the Seine river (Paris, France).
2.1.5. FPH-1
FPH-1 carbapenemase was recently characterized from Francisella philomiragia [31]. FPH-1 shares 77% amino acid sequence identity with the FTU-1, a non carbapenem-hydrolyzing β-lactamase from F. tularensis, which is intrinsic in this species [63]. FTU-1 and FPH-like proteins have been found in all F. tularensis and F. philomiragia isolates, respectively sequenced to date, suggesting that these β-lactamases may be naturally-present in the genus Francisella. These enzymes form a new and distinct branch of the class A β-lactamase tree (Fig. 2). Despite their amino acid sequence homologies and their belonging to the genus Francisella, they display different antibiotic resistance profiles. The FTU-1 β-lactamase confers a narrow-spectrum of resistance to β-lactams, limited mainly to penicillins. In contrast, FPH-1 confers a much broader-spectrum of resistance, including expanded-spectrum cephalosporins, aztreonam and carbapenems. Unlike FTU-1, which elevates the MIC of imipenem 2-fold exclusively, FPH-1 elevates the MICs of imipenem, meropenem, ertapenem, and doripenem 8- to 64-fold. The abilities of two closely related enzymes from the genus Francisella, FTU-1 and FPH-1, to confer distinct antibiotic resistance profiles represent another example of the evolutionary plasticity of class A β-lactamases [31, 63].
Fig. (2).
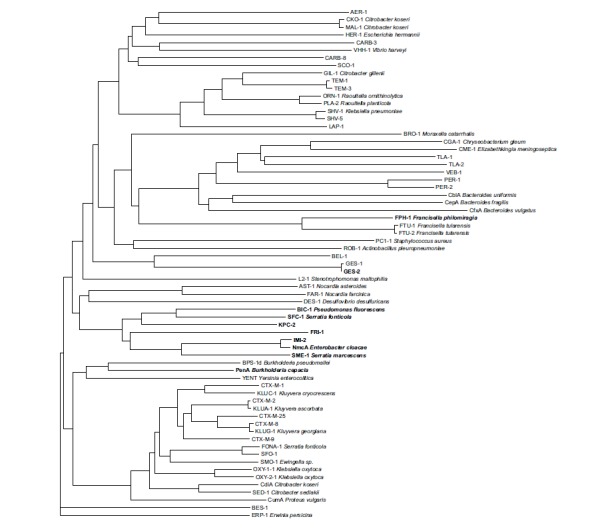
Phylogeny of several chromosomally- and plasmid-encoded class A β-lactamases. For naturally-occurring class A β-lactamases, the bacterial host name is indicated. The carbapenemases are highlighted in bold. The dendrogram was constructed with FigTree [21] using ClustalW [22] aligned protein sequences. The references of all these enzymes can be found in the Beta-Lactamase DataBase at http://www.bldb.eu/BLDB.php?class=A.
2.1.6. PenA
PenA is an inhibitor-resistant carbapenemase, most similar to KPC-2 found in Burkholderia multivorans, an opportunistic nosocomial pathogen, primarily infecting patients with cystic fibrosis. Phylogenetic comparisons revealed that PenA is highly related (71% similarity and 62% identity) to PenI (also reported as BPS-1d [64]), a native ESBL of B. pseudomallei, the causative agent of melioidosis [65]. PenA possesses peculiar properties, in the sense that it can hydrolyze inhibitors as well as all classes of β-lactams (i.e. penicillin, cephalosporins, monobactams, and carbapenems). This is worrisome, since β-lactam antibiotics are recommended as a treatment option for infections due to B. cepacia complex and B. pseudomallei and that PenA-β-lactamase inhibitors are not available. These related bacterial species express closely related class A β-lactamases that confer distinct antibiotic resistance profiles.
2.2. Acquired Class A Carbapenemases (Plasmid-Encoded Enzymes)
2.2.1. IMI-2 Enzyme
Unlike IMI-1/NmcA enzymes that are chromosomally-encoded, IMI-2 is so far the only variant that is plasmid-encoded in clonal E. asburiae isolates recovered from several midwestern rivers of the USA [34], and from three isolates identified from patients that had been in contact with an aquatic environment in distantly located French cities, and in clinical E. cloacae isolates from China and France [53, 58] (T. Naas, pers. com.). In addition, a carbapenem-resistant E. coli strain carrying the carbapenemase blaIMI-2 gene located on a similar conjugative plasmid was described in Spain [66], suggesting transfer to other enterobacterial species. With the increased use of carbapenems, it is likely that the isolation of IMI-2-producing Enterobacter spp isolates may increase, thus increasing the risk of transmission of the plasmid carrying blaIMI-2 gene to bacterial species responsible of nosocomial infections.
2.2.2. FRI-1 Enzyme
FRI-1 carbapenemase was identified in a carbapenem-resistant E. cloacae DUB recovered from a rectal swab sample of a patient hospitalized in France [67]. This isolate was resistant to penicillins, 1st and 2nd generation cephalosporins, aztreonam and carbapenems, but remained susceptible to 3rd generation cephalosporins. FRI-1 shares 51-55% amino acid sequence-identity with the other class A carbapenemases but branches off the NmcA/IMI/SME-1 segment. The blaFRI-1 gene was located on a ca. ~ 150 kb untypeable and non self-transferable plasmid carrying no other antibiotic resistance determinant. A gene encoding a putative LysR type regulator was identified upstream of blaFRI-1 gene as observed upstream the blaNmcA/IMI-2 and blaSME carbapenemase genes.
2.2.3. The KPC Enzymes
The KPC (Klebsiella pneumoniae carbapenemase) is the most worrisome functional group 2f carbapenemases because of its location on self-conjugative plasmids, and its frequent association with K. pneumoniae, an organism notorious for its ability to accumulate and transfer resistance determinants. The first member of the KPC family was discovered through the ICARE surveillance project in a K. pneumoniae clinical isolate from North Carolina in 1996 [35]. The geographical distribution of these enzymes in Enterobacteriaceae in general and especially in K. pneumoniae was limited until 2005 to the eastern part of the United States [68], where KPC-producing K. pneumoniae isolates are now frequently identified among nosocomial pathogens and in systemic infections [2, 17]. Within a few years, KPC producers went global and were described in the North and South American countries, in Greece, in Italy, in Israel and in China, where they can be considered as endemic [2]. KPC producers and hospital outbreaks have been described in many European countries and in the Americas. Currently, KPC producers are identified in increasing numbers of countries and very rapidly alarming reports indicate extensive spread in most of the cases linked to direct transfer of hospitalized patients [69]. They have been reported mostly from hospital-acquired K. pneumoniae isolates, but KPC-producing E. coli and other enterobacterial species have also been described. KPC-producing P. aeruginosa and A. baumannii are increasingly reported [2, 17, 70-73]. Until now, KPC-producing A. baumannii are still restricted to Puerto Rico [72]. In K. pneumoniae the worldwide spread of the blaKPC genes is currently linked to a single clone (ST-258) but within a given geographical location, several KPC clones are disseminating, differing by Multi Locus Sequence Typing (MLST) type, by additional β-lactamase content, by size, number and structure of plasmids. Despite of this genetic diversity, the blaKPC genes are generally associated to a single genetic element: Tn4401, a 10-kb Tn3-type transposon [74, 75]. The clonal spread seen in several epidemics underscores the difficulties in outbreak management even with reinforced infection control measures. The most worrisome with these organisms is the multi-drug resistance, leaving very few, if any, treatment options, which results in high mortality rates [17].
SFC-1 carbapenemase from S. fonticola is the nearest phylogenetically-related enzyme of KPC (Fig. 2), with an amino acid sequence identity of 64% (Table 1). Even though the name KPC refers to the species in which the KPC-2 enzymes were originally identified, they have also been found in several other enterobacterial species, in P. aeruginosa and in A. baumannii. Among the 22 KPC-variants assigned to date, the sequences of 19 of them are available and clearly show a low degree of diversity (1 to 5 amino acid differences). For KPC-14, a two amino acid deletion has been identified (Table 5). The increasing number of variants and the increasing reports regarding their occurrence in environmental samples signal the ongoing dispersion of this kind of enzymes [76]. KPC-producing isolates have been recovered from rivers located in Portugal and Brazil, as well as from sewage samples in China, Brazil, and Austria [77-81].
Table 1.
Sequence identity matrix for representative class A carbapenemases.#
| BIC-1 | SFC-1 | KPC-2 | NmcA | IMI-2 | SME-1 | FRI-1 | PenA | GES-2 | FPH-1 | |
|---|---|---|---|---|---|---|---|---|---|---|
| BIC-1 | 100 | 69 | 63 | 56 | 56 | 57 | 52 | 48 | 37 | 28 |
| SFC-1 | 100 | 64 | 60 | 60 | 59 | 54 | 49 | 36 | 32 | |
| KPC-2 | 100 | 55 | 55 | 56 | 53 | 53 | 38 | 30 | ||
| NmcA | 100 | 97 | 70 | 56 | 45 | 35 | 32 | |||
| IMI-2 | 100 | 70 | 56 | 45 | 35 | 33 | ||||
| SME-1 | 100 | 54 | 44 | 39 | 31 | |||||
| FRI-1 | 100 | 43 | 35 | 29 | ||||||
| PenA | 100 | 38 | 29 | |||||||
| GES-2 | 100 | 25 | ||||||||
| FPH-1 | 100 |
# Generated with ClustalW [22]
Table 5.
Sequence alignment for the KPC family.a,b Only the positions with mutations or deletions are shown.
| Positionc | 49 | 92 | 104 | 105 | 120 | 147 | 169 | 202 | 207 | 240 | 242 | 243 | 274 | 286 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| KPC-2 | M | D | P | W | A | G | L | P | F | V | G | T | H | N |
| KPC-3 | Y | |||||||||||||
| KPC-4 | R | G | ||||||||||||
| KPC-5 | R | |||||||||||||
| KPC-6 | G | |||||||||||||
| KPC-7 | I | Y | ||||||||||||
| KPC-8 | G | Y | ||||||||||||
| KPC-9 | A | Y | ||||||||||||
| KPC-10 | R | Y | ||||||||||||
| KPC-11 | L | |||||||||||||
| KPC-12 | M | |||||||||||||
| KPC-13 | G | Y | ||||||||||||
| KPC-14 | –d | –d | ||||||||||||
| KPC-15 | R | L | K | G | Y | |||||||||
| KPC-16 | S | L | ||||||||||||
| KPC-17 | L | |||||||||||||
| KPC-18a | •a | • | • | • | • | • | • | • | • | • | • | • | • | • |
| KPC-19 | Y | T | ||||||||||||
| KPC-20a | •a | • | • | • | • | • | • | • | • | • | • | • | • | • |
| KPC-21 | R | |||||||||||||
| KPC-22 | G | L | ||||||||||||
| KPC-23a | •a | • | • | • | • | • | • | • | • | • | • | • | • | • |
2.2.4. The GES Enzymes
GES-type enzymes (also named IBC) are increasingly reported in gram-negative rods, including in P. aeruginosa, A. baumannii, E. coli, and K. pneumoniae [2, 14]. The GES-1 β-lactamase (for “Guiana extended spectrum”) was first described in 2000 in a K. pneumoniae clinical isolate from French Guiana [83] and subsequently, a point mutant derivative, named IBC-1 (for “integron-borne cephalosporinase”), was isolated from an E. cloacae isolate in Greece [82]. This confusing situation has led to several revisions of the GES family nomenclature in which the IBC names have been converted to the GES nomenclature [14, 84]. BEL-1 ESBL from P. aeruginosa is the closest phylogenetically-related β-lactamase of the GES family members with an amino-acid sequence identity of 50% [85]. GES enzymes are only distantly related to the other class A carbapenemases (Fig. 2), with identities of 38% to KPC-2, 39% to SME-1, and 35% to NmcA (Table 1). GES family members differ from each other by one to four amino acid substitutions. The number of GES-variants known (23, see Table 6) is increasing rapidly, signaling the current worldwide spread of these enzymes. GES-2 was from South Africa, GES-5, GES-6, GES-7 and GES-8 from Greece, and GES-3 and GES-4 from Japan [82, 86-89]. Among the 23 known GES variants (Table 6) eleven with either Gly170Asn or Gly170Ser mutations display carbapenemase activity. The other GES variants are categorised as ESBLs without any carbapenem-hydrolysing activity. One of them, GES-14, has extended its spectrum towards carbapenems, aztreonam and cephamycins and has reduced susceptibility towards inhibitors [90]. These enzymes are worrisome, since extensive use of carbapenems to treat ESBL infections may select for GES-variants capable of hydrolyzing sufficiently carbapenems with a low susceptibility to commercially available inhibitors. On the opposite, it was recently demonstrate that antibiotic pressure with cefotaxime or aztreonam was not able to select resistance to imipenem [91]. The number of point-mutant derivatives of GES-enzymes and their geographic spread signals an ongoing evolution for this family of enzymes. The emergence of GES-type ESBLs sheds a very dark light on the overall picture of antibiotic resistance, as these enzymes may become true carbapenemases by single point mutations.
Table 6.
Sequence alignment for the GES family.a Only the positions with mutations or deletions are shown.
| Positionb | 15 | 16 | 43 | 56 | 62 | 80 | 81 | 104 | 125 | 130 | 167 | 169 | 170 | 237 | 243 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GES-1# | G | I | Q | I | M | V | F | E | A | S | P | M | G | T | G |
| GES-2* | N | ||||||||||||||
| GES-3# | T | K | |||||||||||||
| GES-4* | T | K | S | ||||||||||||
| GES-5* | S | ||||||||||||||
| GES-6* | K | S | |||||||||||||
| GES-7# | K | ||||||||||||||
| GES-8# | L | ||||||||||||||
| GES-9# | S | ||||||||||||||
| GES-10# | T | T | C | ||||||||||||
| Positionb | 15 | 16 | 43 | 56 | 62 | 80 | 81 | 104 | 125 | 130 | 167 | 169 | 170 | 237 | 243 |
| GES-11# | A | ||||||||||||||
| GES-12# | A | A | |||||||||||||
| GES-13# | K | N | |||||||||||||
| GES-14* | S | A | |||||||||||||
| GES-15 | S | S | |||||||||||||
| GES-16 | E | S | |||||||||||||
| GES-17 | K | A | |||||||||||||
| GES-18* | I | S | |||||||||||||
| GES-19# | A | A | |||||||||||||
| GES-20* | A | S | |||||||||||||
| GES-21 | L | S | |||||||||||||
| GES-22# | L | A | |||||||||||||
| GES-23 | L | ||||||||||||||
| GES-24 | T | S | |||||||||||||
| GES-25a | •a | • | • | • | • | • | • | • | • | • | • | • | • | • | • |
| GES-26 | A |
a The sequence of GES-25 is not currently available
b ABL numbering scheme [18].
# Extended-spectrum β-lactamase (ESBL)
* Carbapenemase
Although rare, GES carbapenemases have now been identified worldwide, most frequently associated with single occurrences, in Greece, France, Portugal, South Africa, French Guiana, Brazil, Argentina, Korea, USA, Canada, China, Belgium and Japan [14, 92]. However, several outbreaks of GES-producing gram-negative bacteria have been described: K. pneumoniae in Korea, Portugal, Brazil, S. marcescens in the Netherlands, and P. aeruginosa in South Africa [93-97]. The main source of carbapenem resistance in A. baumannii is the production of carbapenem-hydrolyzing Ambler’s class D β-lactamases (CHDL) [98], however GES-type carbapenemases have been reported for A. baumannii [90, 99-101]. Several GES-type β-lactamases have been identified in A. baumannii: the blaGES-11 gene in France, in Kuwait, in Turkey, in Egypt and Palestinian territories, the blaGES-12 in Egypt, France and Belgium, and the blaGES-14 gene in France, in Turkey and in Kuwait [90, 99, 100, 102, 103] and blaGES-22 in Turkey [104].
GES-5 or GES-13 have also been reported worldwide now in P. aeruginosa from Korea, China, India, Turkey, France, Spain, Mexican and Brazil [14, 105-111]. Thus, similar GES variants have now been identified in different species from distantly located countries. The blaGES-5 carbapenemase gene was found on two plasmids, pRSB113 and pRSB115, and inserted within the variable region of two different class 1 integrons. These plasmids were recovered from an activated sludge of a wastewater treatment plant in Germany [112], had different backbones with different incompatibility groups, and could replicate both in E. coli and in P. aeruginosa. The effluents of wastewater treatment plants run into rivers that flow to oceans, likely contaminating coastal waters with MDR bacteria. The presence of carbapenemase-producing enterobacteria was recently evidenced in water from touristic beaches located in Rio de Janeiro, Brazil. Predominantly environmental Kluyvera sp or Enterobacter sp isolates, producing either KPC-2, GES-5 or GES-16 carbapenemases, were evidenced [113].
3. Genetics of Class A carbapenemases
The genes for SME-like, IMI-1-like, SFC-1 and NmcA are all chromosomally-located, a fact that may have contributed to their limited spread. None is transmissible and there is no evidence of spread, although plasmid-mediated IMI-2 has been found in clonal environmental E. asburiae isolates [34, 53, 58] and in a recent E. coli isolate in Spain [66]. Flanking sequences of the imi-R–blaIMI-1 gene complex have not been investigated in detail, but it may be part of a composite transposon as it is flanked by different mobile elements in E. cloacae and E. asburiae [34, 53]. In E. absuriae the imi-R–blaIMI-2 region is surrounded by an upstream located IS2-like fragment and downstream located transposase gene similar to that of Tn2501 [34]. In E. cloacae, the imi-R–blaIMI-2 genes are flanked on both sides by structures related to the transposase gene of Tn903 and further upstream by an IS2-like element [34, 53]. These mobile elements may have been at the origin of blaIMI gene mobilization, but whether they are still active is unknown. Their possible inactivity might explain the rare occurrence of blaIMI genes in these species. Similarly, the blaFRI-1 gene is plasmid-encoded, and associated with a LysR type regulator suggesting a similar genetic organization as for IMI-2 enzymes. It is likely that FRI-1 is naturally present in an enterobacterial isolate (probably environmental) and that it moved through mobile elements onto the 150-kb plasmid. There is however no indication on mobile elements associated with blaFRI-1 gene. Recent whole-genome sequencing results from three Canadian isolates revealed that the blaSME gene occurred in a novel cryptic prophage genomic island, SmarGI1-1. This genomic island can be excised and circularized, which probably contributes to its dissemination amongst S. marcescens [114]. Between 2010 and 2012, 30 S. marcescens belonging to three widely disseminated pulsotype clusters were predominant in Canada, though, none was associated with known outbreaks suggesting an ongoing spread of this kind of enzymes. All three clusters shared 77% similarity, indicating that there may be at least some clonality in SME-harbouring isolates.
The source of blaBIC-1 gene dissemination in urban water is probably also linked to mobile genetic elements, two unrelated insertion sequences located upstream and downstream of the blaBIC-1 gene. The role of these elements in the mobility of the blaBIC-1 gene has not been investigated [30].
The blaGES-1-like and blaKPC genes have been identified onto large sized plasmids of different incompatibility groups varying in size and structure. In most of the cases, plasmids were self-transferable at least to E. coli at high frequency (10-2 -10-5). These plasmids may carry in addition other β-lactamase genes, aminoglycoside resistance genes and qnr resistance determinants. In most cases the blaGES-1-like genes are part of a gene cassette, located in the variable regions of class 1 integrons. However, in rare cases they may be embedded in a class 3 integron or present on integron mobilization units (IMU), that have similar characteristics to those of miniature inverted transposable elements (MITEs) [115]. The blaKPC genes are located on a 10-kb Tn3-type transposon, called Tn4401. Tn4401 possesses two unrelated insertion sequences ISKpn7 and ISKpn6, inserted upstream and downstream of blaKPC, respectively. Several isoforms of Tn4401 (differing by a 100 to 200 bp deletion upstream of blaKPC), have been identified in most KPC-producing bacterial isolates all around the world [116]. In a few KPC-producing enterobacterial isolates other mobile elements have been found immediately upstream of blaKPC gene, but the downstream-located sequence was identical to Tn4401 sequences, suggesting that these novel mobile elements have inserted into the Tn4401 backbone. Transposition assays revealed that Tn4401 is an active transposon capable of mobilizing the carbapenemase blaKPC gene at high frequency (2x10-5 per recipient cell), without apparent preferred insertion site, but always with a 5-bp target site duplication [74]. The very efficient dissemination of blaKPC-producers worldwide is likely associated with spread of strains, plasmids and transposons.
The chromosomal NmcA/IMI/SME β-lactamases can be induced in response to imipenem and cefoxitin, resulting in substantial increases of activity against imipenem. Upstream of blaNmcA, blaIMI-1, blaFRI-1 and blaSME-1 the presence of a divergently transcribed gene related to the LysR family of DNA-binding transcriptional regulatory genes was found [26, 37, 61]. Deletions of the nmcR gene located upstream of blaNmcA eliminated the NmcA inducibility and reduced carbapenem MICs, suggesting that NmcR is a positive regulator of NmcA expression [37]. The ImiR protein shares 97% amino acid sequence identity with NmcR, but an analysis of its regulatory effects has not been published. SmeR is 69% identical to NmcR but has a weaker positive regulatory activity on the blaSME-1 gene expression. NmcA carbapenemase induction was also found to be linked to AmpD as shown for the induction of the chromosomally-encoded cephalosporinase AmpC [117]. The blaIMI-1 gene and its regulator imi-R gene are embedded in the chromosome of the host, whereas the blaIMI-2 genes and their imi-R genes are plasmid-borne. Constitutive expression of blaSME-1 was shown to impart a fitness cost upon E. coli. This SME-1-associated fitness cost may also explain their limited dissemination and the inducible nature of these kind of enzymes [45]. KPC-like, GES-like, SFC-1, BIC-1, PenA and FPH-1 are constitutively expressed.
4. Origin of class A carbapenemases
Imipenem is an N-formimidoyl derivative of thienamycin, a natural carbapenem compound produced by a soil organism called Streptomyces cattleya. Bacteria that produce carbapenem-hydrolyzing enzymes would be well adapted to survive in the soil or more general in the environment. The first class A carbapenemase producing isolates, SME-1-producing S. marcescens in 1982 in London [24, 38] and IMI-1- producing E. cloacae isolate in 1984 in the USA [26], were identified even before imipenem was released for clinical use in 1985 in USA. This strongly suggested that class A carbapenemases of IMI/NmcA, SME, FRI-1, BIC-1, KPC and SFC-1 enzymes, which share a common ancestor (Fig. 2), have existed before the clinical use of imipenem, and are not a consequence of imipenem use driving the evolution of β-lactamases. This situation is likely different with the other class A carbapenemases (Fig. 2), which have quite distinct ancestors (GES enzymes, PenA, FPH-1 and SHV-38) and totally unrelated to each other. For GES-enzymes the use of carbapenems may have played a role in the selection of GES-carbapenemases, as they are point mutant derivatives of GES-1, an ESBL lacking carbapenemase activity. The intrinsic carbapenemase activity of all members of the IMI/NmcA, SME, FRI-1, BIC-1, KPC and SFC-1 cluster suggests that the common ancestor was already capable of carbapenem hydrolysis. The serine β-lactamases are believed to have evolved from the Penicillin Binding Proteins (PBPs: β-lactam target) through changes in the active site, such as inclusion of water molecule and other requisites of the hydrolytic machinery [11]. In addition, the class A carbapenemase genes associated with a LysR regulator gene are those most identical to each other, i.e. the blaIMI, blaNmcA, blaFRI and blaSME genes, further supporting that they originate from a common ancestor (Fig. 2). These genes are, however, far from ubiquitous in the species carrying them.
5. Biochemical properties of class A carbapenemases
Whereas there is no consensus definition of carbapenemases, they have been categorized as enzymes capable of hydrolyzing imipenem with kcat/Vmax > 1 µM-1S-1 [92, 118]. However, this an arbitrary cut-off that does not take into account the fact that not all carbapenems are hydrolysed by all carbapenemases and that the level of resistance may depend on the genetic background of the host organism [92]. Queenan and Bush used the kcat value for distinguishing strong hydrolysis (generally, kcat > 2 s−1), weak hydrolysis (generally, kcat of 0.5 to 2 s−1), and no measurable hydrolysis reported (generally, kcat of < 0.5 s−1) [13]. Using these criteria GES-enzymes, PenA, FPH-1 and SHV-38 would not be carbapenemases (Table 3). By far SHV-38 has the worst kcat (0.01 s-1), and also the lowest catalytic efficiency (kcat /Km: 0.0004 µM-1.s-1). SHV-38 differs from the broad-spectrum penicillinase, SHV-1, by Ala146Val substitution (Ambler numbering [18]) owing to a single transition. Among the known SHV enzymes (more than 191 variants), only SHV-38 possesses a Val at position 146. All the other SHV β-lactamases have an Ala residue and none of them has carbapenemase activity. SHV-38 does not hydrolyse meropenem in measurable amounts, but has a high kcat for ceftazidime (110 s-1) [33].
Table 3.
Substrate and inhibition profiles of the carbapenemases (adapted from reference [13])#,§.
| Hydrolysis Profile | Inh | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Enzyme | Number of Variants | Host | Expression Type | PEN | CEF | CEP-I | CEP-III | AZT | CARBA | CA | Reference |
| Chromosome | |||||||||||
| NmcA | 1 | E. cloacae | inducible LysR | + | + | + | + | + | + | + | [25] |
| IMI-1-like | 8 | E. cloacae | inducible LysR | + | − | + | − | + | + | + | [26] |
| SME-like | 6 | S. marcescens | inducible LysR | + | − | + | ± | + | + | + | [24] |
| SFC-1 | 1 | S. fonticola | constitutive | + | + | + | − | + | + | − | [29] |
| BIC-1 | 1 | P. fluorescens | constitutive | + | ± | + | ± | + | + | − | [30] |
| SHV-38 | 1 | K. pneumoniae | constitutive | + | − | + | + | + | ± | ++ | [33] |
| FPH-1 | 1 | F. philomiragia | constitutive | + | − | + | + | + | + | ± | [31] |
| PenA | 1 | B. multivorans | constitutive | + | − | + | + | + | + | ± | [32] |
| Plasmid | |||||||||||
| IMI-2 | E. asburiae, E. coli | inducible LysR | + | − | + | − | + | + | + | [34] | |
| KPC-like | 22 | Enterobacteriaceae, P. aeruginosa, A. baumannii | constitutive | + | ± | + | + | + | + | ± | [17, 27] |
| GES-like | 23 | Enterobacteriaceae, P. aeruginosa, A. baumannii | constitutive | + | ±a | + | + | −b | ± | ± | [14] |
| FRI-1 | 1 | E. cloacae | inducible LysR | + | − | + | + | + | + | + | [67] |
a + for GES variants displaying a serine in position 170
b Except for GES variants dispalying a serine or alanine in position 243
# Symbols: +: strong hydrolysis (generally, kcat of > 2 s−1); ±: weak hydrolysis (generally, kcat of 0.5 to 2 s−1); '−': no measurable hydrolysis reported (generally, kcat of < 0.5 s−1).
§ Abbreviations: Inh: Inhibition; PEN: Penicillins ; CEF: Cefoxitin ; CEP-I: Early cephalosporins ; CEP-III: Expanded-spectrum cephalosporins ; AZT: Aztreonam ; CARBA: Carbapenems ; CA: Clavulanic acid.
SME, NmcA, and IMI enzymes have a broad spectrum of hydrolysis that includes the penicillins, early cephalosporins, aztreonam, and carbapenems (Tables 2 and 3). Imipenem hydrolysis was easily measurable, with kcat values of >30 s-1 while that of meropenem was at least 10-fold lower. Cefoxitin and expanded-spectrum cephalosporins were inefficiently hydrolyzed, when hydrolysis could be detected at all, and overall cefotaxime was more efficiently hydrolyzed than ceftazidime (Tables 2 and 3). SFC-1 and FRI-1 displayed a similar hydrolytic profile, e.g. significant hydrolysis of penicillins, aztreonam and carbapenems, but not of expanded-spectrum cephalosporins [29, 67]. The IC50 of clavulanic acid and tazobactam were however 10-fold higher than those found for NmcA, IMI and SME, leading to a lower susceptibility towards β-lactamase inhibitors [67]. Unlike IMI/NmcA and SME types, KPC β-lactamases (KPC-1/2 to KPC-23) confer decreased susceptibility or resistance to virtually all β-lactams and are only weakly inhibited by typical class A inhibitors (Tables 2, 3 and 4).
Table 2.
Steady-state kinetic parameters for representative class A carbapenemases#,§.
| Substrate | kcat (s-1) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SME-1 | NmcA | IMI-1 | SFC-1 | BIC-1 | KPC-2 | GES-4 | FPH-1 | PenA | SHV-38 | |
| Benzylpenicillin | 19 | 260 | 36 | 31 | 63 | 130 | 100 | |||
| Cephaloridine | 980 | 2820 | 2000 | 170 | 530 | 490 | 40 | |||
| Cefoxitin | <0.15 | 5.0 | 0.3 | 4.2 | 1 | 0.95 | 85 | 0.26 | – | |
| Cefotaxime | <0.98 | 286 | 3.4 | 8.3 | 20 | 17 | 17 | – | 142 | 1 |
| Ceftazidime | <0.07 | 4.7 | 0.0068 | 2.1 | – | 0.49 | 2.5 | – | 110 | |
| Cefepime | 12 | 3 | ||||||||
| Aztreonam | 108 | 707 | 51 | 162 | 35 | 66 | – | – | 60 | 3 |
| Imipenem | 104 | 1040 | 89 | 54 | 50 | 31 | 0.38 | 0.15 | 0.38 | 0.01 |
| Meropenem | 8.9 | 12 | 10 | 6.5 | 1 | 3.6 | 0.093 | |||
| Ertapenem | 2 | 0.11 | ||||||||
| Km (µM) | ||||||||||
| SME-1 | NmcA | IMI-1 | SFC-1 | BIC-1 | KPC-2 | GES-4 | FPH-1 | PenA | SHV-38 | |
| Benzylpenicillin | 17 | 28 | 64 | 48 | 30 | 2200 | 13 | |||
| Cephaloridine | 770 | 185 | 1070 | 770 | 510 | 160 | 150 | |||
| Cefoxitin | ND | 93 | 45 | 77 | 825 | 140 | 810 | 120 | ||
| Cefotaxime | ND | 956 | 190 | 89 | 2200 | 100 | 700 | – | 295 | 800 |
| Ceftazidime | ND | 90 | 270 | 52 | – | 230 | 1500 | – | 3800 | |
| Cefepime | 540 | 1900 | 1600 | |||||||
| Aztreonam | 260 | 125 | 93 | 484 | 1000 | 420 | – | – | 378 | 5500 |
| Imipenem | 202 | 92 | 170 | 82 | 300 | 90 | 4.7 | 0,42 | 7 | 24 |
| Meropenem | 13 | 4.4 | 26 | 26 | 12 | 13 | 3.1 | |||
| Ertapenem | 10 | 1.4 | ||||||||
| kcat/Km (µM-1.s-1) | ||||||||||
| SME-1 | NmcA | IMI-1 | SFC-1 | BIC-1 | KPC-2 | GES-4 | FPH-1 | PenA | SHV-38 | |
| Benzylpenicillin | 1.1 | 9.3 | 0.56 | 0.65 | 2.1 | 0.81 | 7.7 | |||
| Cephaloridine | 1.3 | 15 | 1.9 | 0.22 | 1 | 0.22 | 0.27 | |||
| Cefoxitin | ND | 0.053 | 0.0067 | 0.054 | 0.0012 | 0.0068 | 0.11 | 0.0002 | ||
| Cefotaxime | ND | 0.30 | 0.018 | 0.093 | 0.0091 | 0.17 | 0.024 | – | 0.48 | 0.0013 |
| Ceftazidime | ND | 0.052 | 0.00003 | 0.04 | – | 0.0021 | 0.0017 | – | 0.029 | |
| Cefepime | 0.022 | 0.6 | 0.0019 | |||||||
| Aztreonam | 0.42 | 5.7 | 0.55 | 0.33 | 0.0035 | 0.16 | – | – | 0.16 | 0.0005 |
| Imipenem | 0.51 | 11 | 0.52 | 0.66 | 0.17 | 0.34 | 0.081 | 0.36 | 0.05 | 0.0004 |
| Meropenem | 0.68 | 2.7 | 0.038 | 0.25 | 0.08 | 0.28 | 0.03 | |||
| Ertapenem | 0.2 | 0.079 | ||||||||
Table 4.
IC50 values of class A carbapenemases.
| Enzyme | Inhibition IC50 (μM) | Reference | ||
|---|---|---|---|---|
| Clavulanic Acid | Tazobactam | Sulbactam | ||
| NmcA | 1.61 | 0.66 | 5.9 | [25] |
| IMI-1 | 0.28 | 0.03 | 1.8 | [26] |
| SME-1 | 0.28 | 0.16 | [24] | |
| SFC-1 | 72.8 | 6.9 | 22.7 | [29] |
| BIC-1 | 30 | 6.5 | [30] | |
| SHV-38 | 0.045 | 0.17 | 5 | [33] |
| FPH-1 | 7.5 | 4 | 220 | [31] |
| PenA | 3.1 | 0.5 | 4.1 | [32] |
| KPC-2 | 10.5 | 0.374 | [17, 35] | |
| GES-4 | 15 | 1.4 | 15 | [14] |
BIC-1 hydrolyzed penicillins, cephalosporins, carbapenems, and monobactams. However, BIC-1 showed no detectable activity against ceftazidime and was weakly inhibited by clavulanic acid (IC50, 30 μM) and tazobactam (IC50, 6.5 μM). These values are in the same range as for those of KPC-2, with 10.5 and 0.37 μM for clavulanic acid and tazobactam, respectively (Table 5).
The GES enzymes possess an unusual phenotypic variability, similar to that of the TEM and SHV enzymes that can extended their spectrum of activity. Indeed, GES-enzymes may extend their activities against carbapenems, expanded-spectrum β-lactams, cephamycins and/or monobactams by single amino acid changes as compared to GES-1. GES-2 was the first example of an ESBL that has extended its activity against carbapenems and reduced susceptibility to inhibitors, by a single Gly170Asn change inside the omega loop of the catalytic site [86]. A Gly170Ser change was then identified in GES-4, -5, and -6, resulting in slow carbapenem hydrolysis, in weak inhibitor susceptibility (as for GES-2), and additionally, in hydrolysis of cephamycin, and increased ceftazidime hydrolysis, as shown for GES-4 [119]. GES-9, which differs from GES-1 by a Gly243Ser change, does not hydrolyze carbapenems but has extended its activity towards monobactams [85]. Among the 26 assigned GES variants 25 have available sequence (Lahey Clinics, http://www.lahey.org/Studies/other.asp#OTHGES ), two have Gly170Asn (GES-2, GES-13) and 10 Gly170Ser (GES-4,5,6,14,15,16,18,20,21,24) mutations and are likely displaying carbapenemase activity (Table 6). One of them, GES-14 that has Gly170Ser and Gly243Ser amino acid changes, has extended its spectrum towards carbapenems, aztreonam and cephamycins and has reduced susceptibility towards inhibitors [90].
PenA and FPH-1 enzymes have low kcat values for imipenem (comparable to those of GES-carbapenemase-type enzymes). However PenA hydrolyzes cefotaxime with a higher kcat value and both are moderately inhibited by β-lactamase inhibitors.
6. STRUCTURE-FUNCTION relationship
6.1. Structural Features
6.1.1. General Overview
Class A carbapenemases are monomeric enzymes containing 265 to 269 amino acid residues, and with molecular weights from 25 to 32 kDa. The hydrolysis mechanism of class A carbapenemase is similar to other active-site serine β-lactamases, proceeding via a three-step mechanism involving formation of the non-covalent Michaelis complex, acylation and deacylation (Fig. 1).
Class A carbapenemases present the same four conserved motifs that are characteristic to the class A serine β-lactamases (Fig. 3). The first motif is 70SXXK73 (ABL numbering scheme [18]), where X represents a variable residue, Ser70 is the active-site serine and Lys73 is the lysine residue situated in the immediate proximity which was shown to promote Ser70 addition to the β-lactam carbonyl [120]. The second element, 130SDN132, is equivalent to the conserved motifs YA/SN and SXV/I from class C and class D β-lactamases, respectively. The third one is represented by Glu166, which acts as a general base for the deacylation of the acyl-enzyme complex, being part of a more general 166EXXXN170 motif that is generally found in the omega loop. Finally, the 234KTG236 motif is common to most serine-active β-lactamases.
Fig. (3).

Sequence alignment of representative class A carbapenemases. The stars highlight the four conserved motifs that are characteristic of class A β-lactamases: 70SXX73K, 130SD132N, 166E and 234KT236G (ABL numbering scheme [18]). The triangles indicate the positions of cystein residues involved in the 69S238S disulfide bond. The image was generated with ESPript [23].
Cystein residues 69C and 238C form a disulfide bond in the majority of class A carbapenemase (a notable exception being PenA). As this structural feature is not present in other class A β-lactamases it was thought to be directly related to the carbapenem-hydrolyzing activity of these enzymes. It was later shown, with SME-1 as example, that this disulfide bond does not uniquely influence the carbapenemase activity of β-lactamases but is required in general for the enzyme to function as a β-lactamase [121]. Other class A β-lactamases (i.e. GES-1) possess cysteines at positions 69 and 238 without having significant carbapenemase activities, acting as extended-spectrum cephalosporinases instead [121]. However, these enzymes may become true carbapenemases by single amino-acid changes at position 170.
A number of 22 crystal structures are currently available in the Protein Data Bank (PDB) [122] (Table 7). The superposition of all these structures (Fig. 4) shows that they share the same fold, with very well conserved structural features. There are, however, 5 regions (A-E) presenting significant differences in the backbone conformation. These regions, which are situated relatively far from the active site and more likely do not influence directly the activity and the selectivity of class A carbapenemases, will be analyzed in more detail below (see Fig. 4 for more details):
Table 7.
Crystal structures of class A carbapenemases available in the PDB.
| PDB Code | Class A Carbapenemase | Ligand | Mutation | Reference |
|---|---|---|---|---|
| 2OV5 | KPC-2 | – | – | [123] |
| 3C5A | KPC-2 | – | – | [126] |
| 3DW0 | KPC-2 | – | – | [126] |
| 3E2K | KPC-2 | – | G175S | [127] |
| 3E2L | KPC-2 | – | G175S | [127] |
| 3RXW | KPC-2 | penam sulfone PSR-3-226 | – | [128] |
| 3RXX | KPC-2 | 3-nitrophenyl boronic acid | – | [128] |
| 3NI9 | GES-2 | – | – | [129] |
| 3NIA | GES-2 | tazobactam intermediate | – | [129] |
| 4QU3 | GES-2 | ertapenem, pre-isomerized | – | [130] |
| 4GNU | GES-5 | – | – | [131] |
| 4H8R | GES-5 | imipenem, open form | – | [131] |
| 3V3S | GES-18 | – | – | [132] |
| 1BUE | NmcA | – | – | [133] |
| 1BUL | NmcA | penicillanic acid derivative, open form | – | [134] |
| 4EQI | SFC-1 | – | – | [60] |
| 4EUZ | SFC-1 | meropenem, closed form* | S70A | [60] |
| 4EV4 | SFC-1 | meropenem, bound form | E166A | [60] |
| 1DY6 | SME-1 | – | – | [135] |
| 3W4Q | PenA | – | – | [65] |
* Non-covalent, Michaelis complex. All other ligands mentioned in the table are covalently bound.
Fig. (4).
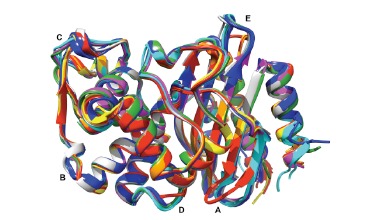
Superposition of all three-dimensional structures of class A carbapenemases available in the PDB (see Table 7 for the corresponding PDB codes): KPC-2 in blue (7), GES-2 in red (3), GES-5 in orange (2), GES-18 in yellow (1), NmcA in green (2), SFC-1 in cyan (3), SME-1 in magenta (1), PenA in gray (1). The image was generated using Chimera [137].
Region A (positions 49 to 57) can be divided in two groups of structures: those corresponding to the GES structures on one hand, and all the other structures on the other. This difference of conformation might be explained by the presence of a Pro51 in all GES structures, whereas the others present a conserved threonine at this position. On this basis we can predict that FRI-1, which also has a proline at the position 51, will adopt a conformation similar to GES in this region;
in the region B (positions 87 to 90), SFC-1, KPC-2 and PenA present one conformation, whereas GES, NmcA and SME-1 adopt a different shape. The presence of a glycine residue at the position 89 in the first group of enzymes and its absence in the second group probably explains this difference in the backbone conformation;
region C is situated between positions 97 and 103. KPC-2, PenA and GES form a group with a characteristic conformation, whereas NmcA, SFC-1 and SME-1 adopt another conformation. The residue responsible for this conformational change is probably the glycine at the position 98, which is present in the first group, but not in the second;
the fourth region, D (positions 195 to 197), present a different conformation for the GES structures compared to all others. This is probably due to the presence of a glycine residue at the position 197 in the GES structures, which is not present in the others;
region E (positions 265 to 276) shows again a different conformation for the GES structures compared to all others. Although it is tempting, on the basis of the sequence
alignment, to suggest that the lysine residue present in position 273 in all structures except GES is responsible for this conformational differentiation, a more in depth analysis of three-dimensional structures shows that this Lys273 is not involved in any stabilizing interaction with neighbouring residues. A more pertinent explanation would be the presence in GES, but not in the other structures, of Ser271 and Glu274 residues that are stabilizing the helix structure in this region, and of Arg275 that is involved in longer distance stabilizing interactions with the backbone of Ala266 and Pro267.
Although a comparison of the active sites of carbapenem-hydrolyzing and carbapenem-inhibited class A β-lactamases does not reveal important structural differences [123, 124], it was reported that a close analysis of the active sites of carbapenem-hydrolyzing (SFC-1, KPC-2, NmcA, SME-1) in comparison with those of carbapenem-inhibited (TEM-1, SHV-1, CTX-M, BlaC) class A β-lactamases can reveal differences in the positions of the conserved amino acids (Ser70, Lys73, Ser130-Asn132, Glu166, Lys234-Gly236) that are characteristic for each group of enzymes (Fig. 5) [60]. However, when this comparison is conducted with a more important number of class A β-lactamases with (KPC-2, GES-2, GES-5, GES-18, NmcA, SFC-1, SME-1, PenA) and without carbapenem-hydrolyzing activity (SHV-2, TEM-1, CTX-M-9, BEL-1, GES-1, PenI, BlaC, BS3), the two groups cannot be discriminated anymore using the position of the above-mentioned amino acids (Fig. 5). Nonetheless, the carbapenemase activity can be predicted qualitatively with good confidence through QM/MM dynamics simulations of acyl-enzyme deacylation, requiring only the 3D structure of the apo-enzyme [125].
Fig. (5).
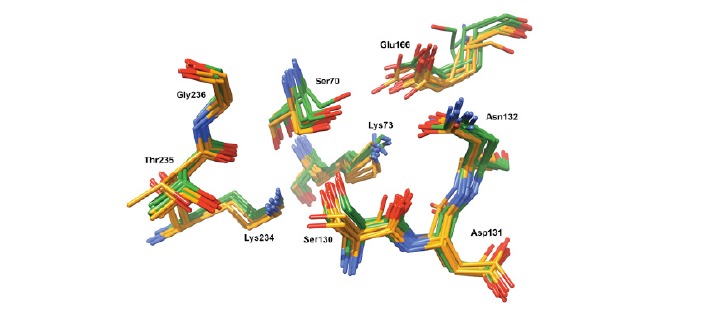
Three-dimensional structures of active site residues of class A β-lactamases with and without carbapenem-hydrolyzing activity, colored in green and orange, respectively (see the main text for details).
6.1.2. KPC-2 Structures
A number of 7 crystal structures of KPC-2 have been described to date (Table 7) [123, 126-128]. The first structure (PDB 2OV5), released in 2007, has a bicine buffer molecule in the active site that interacts via its carboxyl group with conserved active site residues Ser130, Lys234, Thr235, and Thr237. The formed interactions are similar to those made by the β-lactam carboxyl moiety in the Michaelis-Menten complex. This structure evidence shifts in the position of residues Ser70, Asn132 and Asn170, potentially allowing an easier access of bulky substrates [123]. In absence of ligand in the active site, KPC-2 crystallizes as a dimer (PDB 3DW0), with the binding site of a monomer occupied by the C-terminal residues coming from the symmetric KPC-2 monomer. This mode of binding is somewhat similar with the interaction between β-lactamases and the inhibitory protein BLIP. Deletion of the five C-terminal residues of KPC-2 results in a monomeric structure (PDB 3C5A), and in a citrate molecule bound to the active site [126]. The interactions between KPC-2 and BLIP have been evidenced in two structures (PDB 3E2K and 3E2L), thus providing a structural justification for the sub-nanomolar affinity evidenced experimentally for this interaction. The conformational flexibility of the F142 loop of BLIP as observed in these structures seems to contribute to BLIP’s promiscuity, enabling it to respond to mutations at the β-lactamase interface [127]. Both structures used in this study possess a Gly175Ser mutation that was reported as KPC-1 initially [35, 36]. However, Gly175Ser mutation is not located in the BLIP interface, nor does it alter the binding affinity, and therefore the results can be extrapolated for the wild-type KPC-2 [127]. The first structures of KPC-2 with ligands trapped as acyl-enzyme complexes (PDB 3RXW and 3RXX) were recently reported [128]. In the first structure, the 3-nitrophenyl boronic acid ligand is covalently-bound to Ser70, the two boron hydroxyl groups are positioned in the deacylation water pocket and in the oxyanion hole, and the aromatic ring of the ligand provides an edge-to-face interaction with the active site Trp105. The co-crystalization of KPC-2 with PSR-3-226, a penam sulfone, shows a trans-enamine intermediate covalently-bound to Ser70, with the carbonyl oxygen positioned in the oxyanion hole and the carboxyl moiety interacting with the active site residues: Asn132, Asn170 and Glu166 [128].
6.1.3. GES-type Structures
X-ray structures for the GES-2 enzyme in native form (PDB 3NI9) [129] and in complex with the ligands tazobactam (PDB 3NIA) [129] and ertapenem (PDB 4QU3) [130] have been obtained, showing a good similarity of the three-dimensional structures (r.m.s. deviations between 1.1 and 1.4 Å) compared with other class A penicillinases and carbapenemases (e.g. GES-1, NmcA, SME-1, KPC-2, TEM-1, SHV-1) [129]. The structure of tazobactam bound to 70Ser is compatible with either the trans-enamine or imine intermediates [129], whereas the acyl-enzyme complex with ertapenem clearly shows a pre-isomerized form of the ligand, with the pyrroline ring adopting the Δ2 tautomeric form, the C2 atom sp2 hybridized and the S21 sulfur coplanar with the pyrroline ring. The core of ertapenem, comprising the pyrroline, carboxylate and hydroxyethyl moieties, is anchored by a total of eight hydrogen bonds to the surrounding protein molecule, whereas the tail of the molecule, comprising the pyrrolidine and carboxyphenyl rings, makes no direct contacts with the protein [130]. Together with UV spectrometry and mass spectroscopy experiments, these crystallographic studies show that inhibition by tazobactam is not manifested through irreversible inactivation, but via the accumulation of multiple stable intermediates [129].
Two crystal structures were reported for the GES-5 carbapenemase, one in the apo form (PDB 4GNU) and one in complex with the open form of imipenem, which is covalently-bound to Ser70 (PDB 4H8R). The two structures are very similar, but with a significant difference: the presence in the GES-5−imipenem complex of a water molecule that cannot be observed in the apo structure, and occupying a site similar to that assigned to the deacylating water molecule in GES-1 [136]. This water molecule interacts through hydrogen bonds with Ser70 and Glu166, which are also connected by a hydrogen bond. Molecular dynamics simulations showed that this latter interaction is very stable in GES-5 as compared to TEM-1, thus allowing the Glu166 side chain to remain in the right position to interact with the hydrolytic water molecule and thus to promote efficient deacylation step [136].
Similarly, the X-ray structure of GES-18 (PDB 3V3S), obtained in the apo form, was found to be very close to that of GES-11 [132]. The substitution of Gly170Ser leads to the formation of additional hydrogen bonds between the Ser70 and the Glu166 residues with the hydrolytic water molecule, which is in agreement with the hydrolysis mechanism proposed for GES-5 (see above). Interestingly, both Gly170Ser and Gly243Ala mutations (as compared with GES-1) have identical effects on aztreonam and ceftazidime hydrolysis (decreased or increased, respectively). This peculiarity may be explained by the similarity between the structures of these two antibiotics that have the same (C3/C7) side chain, although attached to a different nucleus [132].
6.1.4. NmcA Structures
Two crystal structures have been described for the NmcA carbapenemase, both in 1998, one of them being in the apo form (PDB 1BUE) [133] and the other in complex with the open form of a penicillanic acid derivative covalently-bound to Ser70 (PDB 1BUL) [134]. In the latter, the ester carbonyl of the ligand that is located within the oxyanion hole, makes hydrogen bonds with the backbone nitrogens of Ser237 and Ser70 residues. The repositioning of helix H5 away from the active site allows the hydroxyl group of the 6R-hydroxypropyl moiety to be at hydrogen-bonding distance to Lys73 Nε, to Asn132 (bifurcated hydrogen bond to Oδ1 and to Nδ2), and to the hydrolytic water. The steric crowding provided by the two geminal methyl groups protects the ester carbonyl from the attack of the hydrolytic water molecule [134]. The overall structures are close to those of other class A enzymes. However, the two domains are in a slightly different orientation, which results in substrate binding and folding variations in the helical domain of the protein that affect the position of helix H5. The hydrolytic water molecule, coordinated by Glu166 and Asn170 residues, is in the same position as in the TEM-1 enzyme. However, Asn132 residue is further away from the active site, thus providing additional space in this region of the protein for the accommodation of poorer substrates of class A β-lactamases (cephamycins and carbapenems).
6.1.5. SFC-1 Structures
Crystal structures of SFC-1 (PDB 4EQI) and of complexes of its Ser70Ala (non-covalent Michaelis complex) and Glu166Ala (acyl-enzyme complex) mutants with meropenem (PDB 4EUZ and 4EV4, respectively) have been obtained [60]. As expected, the acylation-deficient Ser70Ala mutant binds meropenem in the unhydrolyzed form, with a well-defined orientation in which the complete molecule is clearly resolved. Meropenem is in the open form in the acyl-enzyme complex, and bound covalently to Ser70, allowing the 6α-1R-hydroxyethyl group to interact with Asn132, but not with the deacylating water molecule. Comparison of these structures with those of class A β-lactamases inhibited by carbapenems revealed that subtle repositioning of key residues in SFC-1 (Ser70, Ser130, Asn132 and Asn170) enlarges the active site, this allowing rotation of the carbapenem 6α-1R-hydroxyethyl group and thus preventing the deacylating water molecule to interact. These results suggest that SFC-1, and most likely other class A carbapenemases, needs Asn132 to orient the bound carbapenems for efficient deacylation and prevent their interaction with the deacylating water molecule [60].
6.1.6. SME-1 Structure
The overall structure of SME-1 carbapenemase (PDB 1DY6) [135] is superimposable to that of other class A β-lactamases. Active site residues are also conserved, with the exception of positions 104, 105 and 237. In addition, SME-1 has a shorter (1.4 Å) distance between Ser70 and Glu166 side chains, as compared with other class A β-lactamases. This direct hydrogen-bonding interaction between the side chains of Ser70 and Glu166 in SME-1 may strongly enhance the nucleophilicity of the hydroxyl group of the Ser70 residue for the initial attack on the carbonyl C atom of the β-lactam ring of imipenem [135]. Additionally, as the SME-1 active site cannot accommodate the water molecule that is essential for catalytic activity and normally present between Ser70 and Glu166 in the other class A β-lactamases described, SME-1 may undergo significant conformational changes to position properly the water molecule responsible for the hydrolysis of acyl-enzyme intermediate [135].
6.1.7. PenA Structure
The X-ray crystal structure of PenA, a β-lactamase isolated from B. cepacia possesing carbapenemase activity, has been recently obtained (PDB 3W4Q) [65]. This structure reveals important differences that aid in understanding the contrasting phenotype of PenA compared to PenI, a β-lactamase isolated from B. pseudomallei that has 62% sequence identity with PenA and has an extended-spectrum hydrolytic profile. These differences include changes in the positioning of conserved catalytic residues (Lys73, Ser130, Tyr105), as well as altered anchoring and decreased occupancy of the deacylation water. On the other hand, it is noteworthy the lack in PenA of a disulfide bond that is present between Cys69 and Cys238 in KPC-2 as well as other class A carbapenemases, while conserving the carbapenem-hydrolyzing activity. However, PenA is similar to KPC-2 in the five major motifs shared by the class A carbapenemases: (i) 70SXXK73 motif ; (ii) 130SDN132 loop ; (iii) 234KTG236 motif and B3 β-strand ; (iv) 102-110 loop and (v) Ω loop and deacylation water. These similarities between their phenotypes may explain the carbapenemase activity of PenA [65].
6.2. Site-Directed Mutagenesis Investigations
6.2.1. KPC-2
Site-saturation mutagenesis was used to explore the role of residue Trp105 in KPC-2. Four variants (Trp105Phe, Trp105Asn, Trp105Leu and Trp105Val) were studied in more depth. The Trp105Asn mutant showed increased Kms for all tested substrates. In contrast, the Trp105Leu and Trp105Val mutants displayed decreased catalytic efficiencies (kcat/Km) for all substrates. All the three mutants displayed increased Kis and partition ratios for clavulanic acid. Overall, these data demonstrate that the Trp105 residue in KPC-2 is essential for the hydrolysis of substrates [138].
Site-directed mutagenesis at the position R220 of KPC-2 was found to influence selectively the catalytic activity toward substrates, since more than 50% reduction in kcat/Km is observed in mutants of the amino acid. In addition, the R220 seems to play a central role in β-lactamase inhibition/inactivation. Thus, the R220K mutant was relatively “inhibitor susceptible”, being not capable anymore to hydrolyze clavulanic acid as compared to KPC-2. On the other hand, the R220M mutant demonstrated increased Km values for β-lactamase inhibitors, which resulted in inhibitor resistance [139].
Mutagenesis of residue Glu276 into Ala, Asp, Asn and Arg revealed that, in contrast to what is observed for other class A β-lactamases (i.e. TEM-1 and SHV-1), Glu276 has a structural role rather than a kinetic function with substrates or inhibitors, being anchored to a stabilized water molecule, and thus leading to secondary ring opening and protonation of the oxonium ion intermediate to form the imine [139].
The substitution of the Arg164 residue from the Ω loop of KPC-2 led to enhanced ceftazidime hydrolysis in 11 out of 19 variants. The Arg164Ser mutant that was further characterized revealed only a 25% increase in kcat/Km for ceftazidime. Mass spectrometry and pre-steady-state kinetic experiments showed that the rate-limiting step for ceftazidime hydrolysis by KPC-2 was the acylation, while the rate-limiting step for the Arg164Ser variant was the deacylation, thus leading at steady-state to the accumulation of acyl-enzyme forms. Ceftazidime resistance may thus proceed by a novel mechanism that uses “covalent trapping” of the substrate by an enzyme with a lower Km that is impaired in deacylation [140].
In a recent study, the efficacy of ampicillin-avibactam and ceftazidime-avibactam combinations was tested against in vitro generated KPC-2 variants having single amino acid changes at key positions (i.e., Ambler positions 69, 130, 234, 220 and 276 - (see Fig. 6) for a representation of the KPC-2 binding site superposed to the CTX-M-15 complex with avibactam [141]). KPC-2 with substitutions of Ser130Gly, Lys234Arg, and Arg220Met showed increased MICs only for the ampicillin-avibactam combination and steady-state kinetics showed that the Ser130Gly variant was resistant to avibactam inactivation. A possible explanation of this result could be that the protonation of the sulfate nitrogen of avibactam may be slowed in the Ser130Gly variant as the Ser130, which is the likely proton donor, when replaced by Gly cannot fulfill this function anymore, suggesting that this activity could likely be compensated by Lys234 in the Ser130Gly mutant. This mutational analysis revealed the major contribution of Ser130, but also Lys234 and Arg220, to the mechanism of KPC-2 inhibition by avibactam. Of note, emergence in clinic of these KPC variants may not result in treatment failures using ceftazidime-avibactam as the ceftazidime partner remains a potent antibiotic against the E. coli isolates expressing Ser130Gly, Lys234Arg, and Arg220Met [142].
Fig. (6).
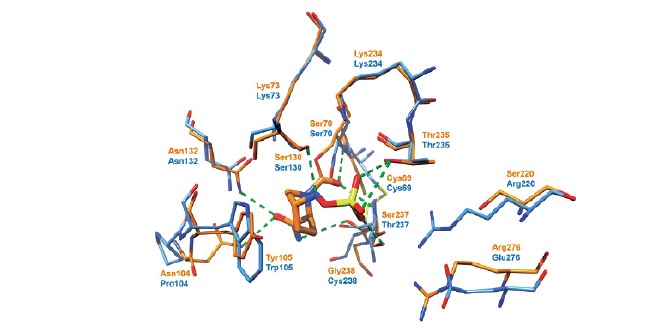
Superposition of X-ray structures of KPC-2 (apo form, blue, PDB 2OV5) and CTX-M-15 (complex with avibactam, orange, PDB 4S2I).
6.2.2. GES
In GES-type β-lactamases, the residues at position 104 and 170 are either Glu/Lys and Gly/Asn/Ser, respectively. These two positions have been suggested to be key amino acids for the interaction with β-lactams. Using site-directed mutagenesis, the following mutants Glu104/Gly170 (GES-1), Glu104/Asn170 (GES-2), Glu104/Ser170 (GES-5), Lys104/ Ser170 (GES-6), Lys104/Gly170 (GES-7) and Lys104/ Asn170 (GES-13) were constructed and their kinetic parameters determined in order to elucidate the precise role of each position. The Glu104Lys-variant enzymes revealed increased oxyimino-cephalosporin and reduced imipenem hydrolysis. The Gly170Asn-containing enzymes displayed reduced rates of penicillin and cephalosporin hydrolysis, whereas the Gly170Ser allowed in addition an increased hydrolysis of cefoxitin, and both Gly170Asn and Gly170Ser had increased carbapenem-hydrolyzing activity with reduced ceftazidime hydrolyzing activity [143]. Similarly, another study showed that Gly170Asn led to stable complexes, such as a cross-linked species and a hydrated aldehyde [144]. In contrast, Gly170Ser led to the formation of a long-lived trans-enamine species. The residue at position 170 was also pointed out as crucial in the clavulanic acid susceptibility of GES enzymes [144]. Pre-steady-state and steady-state kinetics using carbapenems as substrates showed that acylation step was rate-limiting for GES-1 (Gly170), whereas the acylation step was enhanced in GES-2 (Asn170), as compared to GES-1, thus resulting in acylation /deacylation steps being of similar order of magnitude. For GES-5 (Ser170) the acylation rate increased by 5000-fold and the acylation of the enzyme was no longer rate-limiting [145].
6.2.3. NmcA
Site directed mutagenesis was used to investigate the importance of the Cys69 and Cys238 disulfide bridge. These cysteine residues represent a major difference between class A carbapenemases and non-carbapenemases. By replacing them by alanines, it was observed that the removal of the disulfide bridge in the NmcA active site yielded a highly unstable enzyme, which could not be produced and purified to homogeneity [146].
The substitution of Asn132 in NmcA with alanine or glutamine (Asn132Ala and Asn132Gln mutants) led to the severe impairment of the catalytic efficiency (e.g. kcat/Km decreased more than 103 fold for ampicillin). A similar behaviour, strong decrease of the β-lactamase activity, has been observed in the case of the same TEM-1 mutants. These results indicated that Asn132 is an important residue in the catalytic activity of class A β-lactamases, but not typical for the carbapenamase activity [146].
Mutations at positions 141 (Arg141Ala) and 240 (Ala240Val, Ala240Glu) did not affect the NmcA activity profile, whereas substitution of residue 105 (His105Ser) affected the activity profile of NmcA, the mutant being unable to hydrolyse second and third generation cephalosporins, and cephamycins. The catalytic activity against carbapenems was retained, but the specificity was modified. Unlike the wild type enzyme that has a stronger hydrolytic activity against imipenem as compared to meropenem, the mutant enzyme displayed a catalytic efficiency for meropenem (kcat/Km ∼ 106 M-1.s-1) 10-fold higher as compared to imipenem (kcat/Km ∼ 105 M-1.s-1) [146].
6.2.4. SME-1
Ser237Ala mutant of the carbapenemase SME-1 revealed hydrolysis of penicillins and aztreonam in a very similar manner to that of SME-1. However, Ser237Ala mutant displayed reduced catalytic efficiency against cephems (cephalothin and cephaloridine) and cephamycin (cefoxitin), that was almost not detectable with the mutant enzyme. Finally, the Ser237Ala mutant presented a decreased catalytic activity against imipenem, indicating that this serine residue is important for the carbapenemase activity of SME-1 [147].
The role of the disulfide bridge between Cys69 and Cys238 in the hydrolysis of carbapenems and any β-lactams has also been demonstrated for SME-1. The Cys69Ala mutant enzyme in SME-1 resulted in loss of hydrolysis of all the β-lactams tested (amoxicillin, ticarcillin, kanamycin, cefoxitin, imipenem and aztreonam) [135]. In another study, random replacement libraries for these Cys69 and Cys238 positions were created using PCR-based mutagenesis. Only enzymes from these libraries with Cys69 and Cys238 were functional and capable to confer resistance to β-lactams, suggesting that these cysteines and the resulting disulfide bridge are detrimental for the hydrolysis of all β-lactam antibiotics hydrolyzed by SME-1 [121].
Similarly, randomized site-directed mutagenesis of SME-1 residues 104, 105, 132, 167, 237, and 241 was undertaken and functional mutants were selected for their ability to hydrolyze imipenem, ampicillin, and cefotaxime. No single position contributed alone to carbapenem-hydrolysis, although several positions seemed to be important for β-lactam antibiotic hydrolysis in general. These results suggest that SME-1 carbapenemase activity is due to a highly distributed set of interactions that subtly alter the structure of the active-site pocket [124].
6.2.5. SFC-1
The only SFC-1 mutants generated were acylation (Ser70Ala) and deacylation (Glu166Ala) deficient mutants that were used to facilitate the trapping of complexes with β-lactams [60].
6.2.6. PenA
In a study aimed at identifying the factors responsible for the resistance to ceftazidime in clinical isolates, the effects of point mutations, blaPenA gene deletion, and up-regulation were investigated. Deletion of blaPenA gene lowered MICs by at least 4-fold for 6 of the 9 β-lactams tested and by ≥16-fold for ampicillin, amoxicillin, and carbenicillin. Overexpression of PenA resulted for all β-lactams tested in an increase of MICs by 2- to 10-fold. The Cys69Tyr and Pro167Ser mutants of PenA, which were previously observed in resistant clinical isolates, increased MICs for ceftazidime by ≥85- and 5- to 8-fold, respectively. Similarly, the Ser72Phe amino acid substitution resulted in a 4-fold increase in MICs for amoxicillin and clavulanic acid [148].
The sequencing of blaPenA gene in ceftazidime-resistant B. thailandensis isolates identified 516 isolates containing a single point mutation in the coding region of blaPenA gene. A total of 12 amino acid positions were repeatedly found substituted in these mutants (Ambler positions 69, 136, 162, 164, 166, 169, 170, 171, 172, 174, 176, 179), ten of them being located in the Ω loop. All mutations show distinct substitution patterns, distinct MIC levels and different frequencies of occurrence [149].
The ΔGlu168 deletion mutant of class A β-lactamase PenA was able to extend its spectrum of substrates to include ceftazidime. This deletion, that was situated in the Ω loop, allowed increased flexibility at the active site by expanding the space available in the loop [150]. Similarly, three single-amino-acid deletions (ΔThr171, ΔIle173, and ΔPro174) and a two-amino-acid deletion (ΔArg165_Thr167_insP i.e. a three-amino-acid deletion and a proline residue insertion), all of them occurring in the Ω loop, increased the flexibility of the binding cavity and extended the substrate spectrum of the PenA β-lactamase [151].
The substitution of Arg220 to Gly in the B. cepacia PenA β-lactamase resulted in a loss of imipenem resistance and a significant decrease in catalytic activity when measured enzymatically. These results show that the Arg220 residue is critical for the interaction with imipenem [65] and corroborate well with previous studies reporting that the equivalent residue in TEM-1 (Arg224) is able to coordinate a water molecule that is the source of the proton required for carbapenem tautomerization [152].
CONCLUSION
The β-lactamases are ancient enzymes that were relatively rare in clinical isolates until β-lactam antibiotics were introduced into medicine and agriculture more that sixty years ago [153]. β-lactamases are the most widespread and important antibiotic resistance determinants in human medicine. The widespread use of all types of β-lactam molecules, including the broad-spectrum β-lactams, cephalosporins and carbapenems, in the last three decades has led to the selection of novel β-lactamases with ever broadened spectrum of hydrolysis. Class A carbapenemases capable of hydrolyzing most if not all β-lactam molecules are currently spreading in many gram-negative isolates, best exemplified with the plasmid located enzymes, such as KPC and in a lesser extend GES-enzymes. The spread of the KPCs in isolates is one of the most frightening issues since it is likely that it may become rapidly uncontrollable due to its appearance in community-acquired pathogens worldwide.
The number of class A β-lactamases displaying carbapenemase activity is still limited and the occurrence of most class A carbapenemases (besides KPC and to a lesser extend GES) is still rare. The most widely dispersed enzymes, KPC enzymes, are encoded by genes located on transferable elements inserted on different broad-host range plasmids facilitating their dissemination, as revealed by the geographical distribution of the KPC enzymes. The KPC carbapenemases initially identified in K. pneumoniae from the Northeastern parts of USA, is now increasingly isolated worlwide. In the light of the global and rapid spread of CTX-M enzymes it is conceivable that a similar scenario may occur for KPC carbapenemases. Carbapenems are drugs resisting hydrolysis by most β-lactamases, including ESBLs and derepressed chromosomal AmpC β-lactamases. Thus with the increased dissemination of ESBLs, increased use of carbapenems is to be expected in order to treat severe infections caused by multidrug-resistant pathogens. This will in turn facilitate the emergence of carbapenemase producers, especially class A β-lactamases that can extend their spectrum of activity towards carbapenems (GES) or with those possessing already a strong carbapenem-hydrolyzing activity (KPC). A wider dissemination of the GES and KPC carbapenemases would therefore pose major therapeutic issues as the treatment options may be seriously limited, possibly with fatal consequences.
The three-dimensional structures of class A carbapenemases share the same fold, with very well conserved structural features. There is no distinctive element that can discriminate the class A carbapenemases from the non-carbapenemases, this difference in phenotypes being the result of multiple small changes in the structure, but also in the dynamics of the active site residues and in the positioning of water molecules. An exception to this rule is maybe the Cys69-Cys238 disulfide bond, which is present in the vast majority of class A carbapenemases and absent in almost all non-carbapenemases. As every exception has its own exceptions, PenA is a carbapenemase even though this disulfide bond is absent (but the carbapenem hydrolysis is weak in PenA compared with KPC-2, and this might be possibly related to the absence of the disulfide bond), and on the other hand this Cys69-Cys238 disulfide bond is conserved throughout the GES family, whereas only a few GES enzymes possess carbapenem-hydrolyzing activity.
The continuously growing number of crystallographic structures available in the PDB (646 β-lactamase structures are available to date, from which 269 class A β-lactamase structures and only 20 class A carbapenemase structures), together with the molecular modelling and site-directed mutagenesis studies which are often associated with the structure determination, will provide essential information for understanding the subtle mechanistic details controlling the activity and selectivity in substrate hydrolysis, as well as the inhibition of class A carbapenemases. As our image of this process will become more and more clear, the design of new classes of efficient antibiotics and β-lactamase inhibitors will be facilitated, bringing some hope in our fight against antibiotic resistance.
ACKNOWLEDGEMENTS
Our laboratories are members of the Laboratory of Excellence in Research on Medication and Innovative Therapeutics (LERMIT) supported by a grant from the French National Research Agency (ANR-10-LABX-33). This work was also funded in part by a grant from Joint Programme Initiative on Antimicrobial Resistance (ANR-14-JAMR-0002).
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
REFERENCES
- 1.Vasoo S., Barreto J.N., Tosh P.K. Emerging issues in Gram-negative bacterial resistance: An update for the practicing clinician. Mayo Clin. Proc. 2015;90:395–403. doi: 10.1016/j.mayocp.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 2.Nordmann P., Naas T., Poirel L. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 2011;17:1791–1798. doi: 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poirel L., Naas T., Nordmann P. Genetic support of extended-spectrum β-lactamases. Clin. Microbiol. Infect. 2008;14(Suppl. 1):75–81. doi: 10.1111/j.1469-0691.2007.01865.x. [DOI] [PubMed] [Google Scholar]
- 4.Page M.G. Extended-spectrum β-lactamases: Structure and kinetic mechanism. Clin. Microbiol. Infect. 2008;14(Suppl. 1):63–74. doi: 10.1111/j.1469-0691.2007.01863.x. [DOI] [PubMed] [Google Scholar]
- 5.Pfeifer Y., Cullik A., Witte W. Resistance to cephalosporins and carbapenems in Gram-negative bacterial pathogens. Int. J. Med. Microbiol. 2010;300:371–379. doi: 10.1016/j.ijmm.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 6.Naas T., Nordmann P. In: Courvalin P., LeClercq R, Rice LB, editors. Washington, DC: In Antibiogram, ASM; 2009. [Google Scholar]
- 7.Nordmann P., Naas T. In: Lactams and Pseudomonas aeruginosa. 3rd ed. Courvalin P., LeClercq R, Rice LB, editors. Washington, DC: Antibiogram, ASM; 2009. [Google Scholar]
- 8.Ambler R.P. The structure of β-lactamases. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1980;289:321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- 9.Buynak J.D. Understanding the longevity of the β-lactam antibiotics and of antibiotic/β-lactamase inhibitor combinations. Biochem. Pharmacol. 2006;71:930–940. doi: 10.1016/j.bcp.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 10.Massova I., Mobashery S. Kinship and diversification of bacterial penicillin-binding proteins and β-lactamases. Antimicrob. Agents Chemother. 1998;42:1–17. doi: 10.1128/aac.42.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Page M.I. The reactivity of β-lactams, the mechanism of catalysis and the inhibition of β-lactamases. Curr. Pharm. Des. 1999;5:895–913. [PubMed] [Google Scholar]
- 12.Matagne A., Lamotte-Brasseur J., Frere J.M. Catalytic properties of class A β-lactamases: Efficiency and diversity. Biochem. J. 1998;330:581–598. doi: 10.1042/bj3300581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Queenan A.M., Bush K. Carbapenemases: The versatile β-lactamases. Clin. Microbiol. Rev. 2007;20:440–458. doi: 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Naas T., Poirel L., Nordmann P. Minor extended-spectrum β-lactamases. Clin. Microbiol. Infect. 2008;14(Suppl. 1):42–52. doi: 10.1111/j.1469-0691.2007.01861.x. [DOI] [PubMed] [Google Scholar]
- 15.Drawz S.M., Bonomo R.A. Three decades of β-lactamase inhibitors. Clin. Microbiol. Rev. 2010;23:160–201. doi: 10.1128/CMR.00037-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walther-Rasmussen J., Høiby N. Class A carbapenemases. J. Antimicrob. Chemother. 2007;60:470–482. doi: 10.1093/jac/dkm226. [DOI] [PubMed] [Google Scholar]
- 17.Nordmann P., Cuzon G., Naas T. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect. Dis. 2009;9:228–236. doi: 10.1016/S1473-3099(09)70054-4. [DOI] [PubMed] [Google Scholar]
- 18.Ambler R.P., Coulson A.F., Frere J.M., et al. A standard numbering scheme for the class A β-lactamases. Biochem. J. 1991;276:269–270. doi: 10.1042/bj2760269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bush K., Jacoby G.A., Medeiros A.A. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 1995;39:1211–1233. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bush K., Jacoby G.A. Updated functional classification of β-lactamases. Antimicrob. Agents Chemother. 2010;54:969–976. doi: 10.1128/AAC.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.FigTree Available from: http://tree.bio.ed.ac.uk/software/figtree/
- 22.Larkin M.A., Blackshields G., Brown N.P., et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 23.Robert X., Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320-4. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Naas T., Vandel L., Sougakoff W., Livermore D.M., Nordmann P. Cloning and sequence analysis of the gene for a carbapenem-hydrolyzing class A β-lactamase, Sme-1, from Serratia marcescens S6. Antimicrob. Agents Chemother. 1994;38:1262–1270. doi: 10.1128/aac.38.6.1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mariotte-Boyer S., Nicolas-Chanoine M.H., Labia R. A kinetic study of NMC-A β-lactamase, an Ambler class A carbapenemase also hydrolyzing cephamycins. FEMS Microbiol. Lett. 1996;143:29–33. doi: 10.1111/j.1574-6968.1996.tb08457.x. [DOI] [PubMed] [Google Scholar]
- 26.Rasmussen B.A., Bush K., Keeney D., et al. Characterization of IMI-1 β-lactamase, a class A carbapenem-hydrolyzing enzyme from Enterobacter cloacae. Antimicrob. Agents Chemother. 1996;40:2080–2086. doi: 10.1128/aac.40.9.2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yigit H., Queenan A.M., Rasheed J.K., et al. Carbapenem-resistant strain of Klebsiella oxytoca harboring carbapenem-hydrolyzing β-lactamase KPC-2. Antimicrob. Agents Chemother. 2003;47:3881–3889. doi: 10.1128/AAC.47.12.3881-3889.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wachino J., Doi Y., Yamane K., et al. Molecular characterization of a cephamycin-hydrolyzing and inhibitor-resistant class A β-lactamase, GES-4, possessing a single G170S substitution in the omega-loop. Antimicrob. Agents Chemother. 2004;48:2905–2910. doi: 10.1128/AAC.48.8.2905-2910.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fonseca F., Sarmento A.C., Henriques I., et al. Biochemical characterization of SFC-1, a class A carbapenem-hydrolyzing β-lactamase. Antimicrob. Agents Chemother. 2007;51:4512–4514. doi: 10.1128/AAC.00491-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Girlich D., Poirel L., Nordmann P. Novel Ambler class A carbapenem-hydrolyzing β-lactamase from a Pseudomonas fluorescens isolate from the Seine River, Paris, France. Antimicrob. Agents Chemother. 2010;54:328–332. doi: 10.1128/AAC.00961-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toth M., Vakulenko V., Antunes N.T., Frase H., Vakulenko S.B. Class A carbapenemase FPH-1 from Francisella philomiragia. Antimicrob. Agents Chemother. 2012;56:2852–2857. doi: 10.1128/AAC.00223-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trépanier S., Prince A., Huletsky A. Characterization of the penA and penR genes of Burkholderia cepacia 249 which encode the chromosomal class A penicillinase and its LysR-type transcriptional regulator. Antimicrob. Agents Chemother. 1997;41:2399–2405. doi: 10.1128/aac.41.11.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poirel L., Heritier C., Podglajen I., Sougakoff W., Gutmann L., Nordmann P. Emergence in Klebsiella pneumoniae of a chromosome-encoded SHV β-lactamase that compromises the efficacy of imipenem. Antimicrob. Agents Chemother. 2003;47:755–758. doi: 10.1128/AAC.47.2.755-758.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aubron C., Poirel L., Ash R.J., Nordmann P. Carbapenemase-producing Enterobacteriaceae, U.S. rivers. Emerg. Infect. Dis. 2005;11:260–264. doi: 10.3201/eid1102.030684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yigit H., Queenan A.M., Anderson G.J., et al. Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2001;45:1151–1161. doi: 10.1128/AAC.45.4.1151-1161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yigit H., Queenan A.M., Anderson G.J., et al. Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2008;52:809. doi: 10.1128/AAC.45.4.1151-1161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Naas T., Nordmann P. Analysis of a carbapenem-hydrolyzing class A β-lactamase from Enterobacter cloacae and of its LysR-type regulatory protein. Proc. Natl. Acad. Sci. USA. 1994;91:7693–7697. doi: 10.1073/pnas.91.16.7693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Y.J., Wu P.J., Livermore D.M. Biochemical characterization of a β-lactamase that hydrolyzes penems and carbapenems from two Serratia marcescens isolates. Antimicrob. Agents Chemother. 1990;34:755–758. doi: 10.1128/aac.34.5.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deshpande L.M., Rhomberg P.R., Sader H.S., Jones R.N. Emergence of serine carbapenemases (KPC and SME) among clinical strains of Enterobacteriaceae isolated in the United States Medical Centers: Report from the MYSTIC Program (1999-2005). Diagn. Microbiol. Infect. Dis. 2006;56:367–372. doi: 10.1016/j.diagmicrobio.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 40.Queenan A.M., Torres-Viera C., Gold H.S., et al. SME-type carbapenem-hydrolyzing class A β-lactamases from geographically diverse Serratia marcescens strains. Antimicrob. Agents Chemother. 2000;44:3035–3039. doi: 10.1128/aac.44.11.3035-3039.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Queenan A.M., Shang W., Schreckenberger P., Lolans K., Bush K., Quinn J. SME-3, a novel member of the Serratia marcescens SME family of carbapenem-hydrolyzing β-lactamases. Antimicrob. Agents Chemother. 2006;50:3485–3487. doi: 10.1128/AAC.00363-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poirel L., Wenger A., Bille J., Bernabeu S., Naas T., Nordmann P. SME-2-producing Serratia marcescens isolate from Switzerland. Antimicrob. Agents Chemother. 2007;51:2282–2283. doi: 10.1128/AAC.00309-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pasteran F., Mendez T., Guerriero L., Rapoport M., Corso A. Sensitive screening tests for suspected class A carbapenemase production in species of Enterobacteriaceae. J. Clin. Microbiol. 2009;47:1631–1639. doi: 10.1128/JCM.00130-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bush K., Pannell M., Lock J.L., et al. Detection systems for carbapenemase gene identification should include the SME serine carbapenemase. Int. J. Antimicrob. Agents. 2013;41:1–4. doi: 10.1016/j.ijantimicag.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 45.Marciano D.C., Karkouti O.Y., Palzkill T. A fitness cost associated with the antibiotic resistance enzyme SME-1 β-lactamase. Genetics. 2007;176:2381–2392. doi: 10.1534/genetics.106.069443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nordmann P., Mariotte S., Naas T., Labia R., Nicolas M.H. Biochemical properties of a carbapenem-hydrolyzing β-lactamase from Enterobacter cloacae and cloning of the gene into Escherichia coli. Antimicrob. Agents Chemother. 1993;37:939–946. doi: 10.1128/aac.37.5.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pottumarthy S., Moland E.S., Juretschko S., Swanzy S.R., Thomson K.S., Fritsche T.R. NmcA carbapenem-hydrolyzing enzyme in Enterobacter cloacae in North America. Emerg. Infect. Dis. 2003;9:999–1002. doi: 10.3201/eid0908.030096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Radice M., Power P., Gutkind G., et al. First class A carbapenemase isolated from Enterobacteriaceae in Argentina. Antimicrob. Agents Chemother. 2004;48:1068–1069. doi: 10.1128/AAC.48.3.1068-1069.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boo T.W., O'Connell N., Power L., et al. First report of IMI-1-producing colistin-resistant Enterobacter clinical isolate in Ireland, March 2013. Euro Surveill. 2013;18(31):20548. doi: 10.2807/1560-7917.es2013.18.31.20548. [DOI] [PubMed] [Google Scholar]
- 50.Blanco V.M., Rojas L.J., De La Cadena E., et al. First report of a nonmetallocarbapenemase class A carbapenemase in an Enterobacter cloacae isolate from Colombia. Antimicrob. Agents Chemother. 2013;57:3457. doi: 10.1128/AAC.02425-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Naas T., Cuzon G., Levy M., Nordmann P. IMI-1 producing Enterobacter cloacae clinical isolate from Tahiti, French Polynesia.; 53rd Interscience Conference on Antimicrobial Agents and Chemotherapy; Denver, CO, USA. 2013. pp. C1–055. [DOI] [PubMed] [Google Scholar]
- 52.Österblad M., Kirveskari J., Hakanen A.J., Tissari P., Vaara M., Jalava J. Carbapenemase-producing Enterobacteriaceae in Finland: The first years (2008-11). J. Antimicrob. Chemother. 2012;67:2860–2864. doi: 10.1093/jac/dks299. [DOI] [PubMed] [Google Scholar]
- 53.Yu Y.S., Du X.X., Zhou Z.H., Chen Y.G., Li L.J. First isolation of blaIMI-2 in an Enterobacter cloacae clinical isolate from China. Antimicrob. Agents Chemother. 2006;50:1610–1611. doi: 10.1128/AAC.50.4.1610-1611.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Naas T., Cattoen C., Bernusset S., Cuzon G., Nordmann P. First identification of blaIMI-1 in an Enterobacter cloacae clinical isolate from France. Antimicrob. Agents Chemother. 2012;56:1664–1665. doi: 10.1128/AAC.06328-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Teo J.W., La M.V., Krishnan P., Ang B., Jureen R., Lin R.T. Enterobacter cloacae producing an uncommon class A carbapenemase, IMI-1, from Singapore. J. Med. Microbiol. 2013;62:1086–1088. doi: 10.1099/jmm.0.053363-0. [DOI] [PubMed] [Google Scholar]
- 56.Bejuk D., Novkoski M., Juranko V., et al. A report of rarely observed resistance pattern to carbapenems in a clinical isolate of Enterobacter cloacae. Lijec. Vjesn. 2013;135:316–321. [PubMed] [Google Scholar]
- 57.Chu Y.W., Tung V.W., Cheung T.K., et al. Carbapenemases in Enterobacteria, Hong Kong, China, 2009. Emerg. Infect. Dis. 2011;17:130–132. doi: 10.3201/eid1701.101443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bernusset S., Naas T., Tandé D., et al. Characterisation of carbapenemase IMI-2 in Enterobacter spp. clinical isolates from France.; 22nd European Congress of Clinical Microbiology and Infectious Diseases; 31 March - 3 April 2012; London, United Kingdom. p. 1238. [Google Scholar]
- 59.Henriques I., Moura A., Alves A., Saavedra M.J., Correia A. Molecular characterization of a carbapenem-hydrolyzing class A β-lactamase, SFC-1, from Serratia fonticola UTAD54. Antimicrob. Agents Chemother. 2004;48:2321–2324. doi: 10.1128/AAC.48.6.2321-2324.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fonseca F., Chudyk E.I., van der Kamp M.W., Correia A., Mulholland A.J., Spencer J. The basis for carbapenem hydrolysis by class A β-lactamases: A combined investigation using crystallography and simulations. J. Am. Chem. Soc. 2012;134:18275–18285. doi: 10.1021/ja304460j. [DOI] [PubMed] [Google Scholar]
- 61.Naas T., Livermore D.M., Nordmann P. Characterization of an LysR family protein, SmeR from Serratia marcescens S6, its effect on expression of the carbapenem-hydrolyzing β-lactamase Sme-1, and comparison of this regulator with other β-lactamase regulators. Antimicrob. Agents Chemother. 1995;39:629–637. doi: 10.1128/AAC.39.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tollentino F.M., Polotto M., Nogueira M.L., et al. High prevalence of blaCTX-M extended spectrum β-lactamase genes in Klebsiella pneumoniae isolates from a tertiary care hospital: First report of blaSHV-12, blaSHV-31, blaSHV-38, and blaCTX-M-15 in Brazil. Microb. Drug Resist. 2011;17:7–16. doi: 10.1089/mdr.2010.0055. [DOI] [PubMed] [Google Scholar]
- 63.Antunes N.T., Frase H., Toth M., Vakulenko S.B. The class A β-lactamase FTU-1 is native to Francisella tularensis. Antimicrob. Agents Chemother. 2012;56:666–671. doi: 10.1128/AAC.05305-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cheung T.K., Ho P.L., Woo P.C., Yuen K.Y., Chau P.Y. Cloning and expression of class A β-lactamase gene blaA(BPS) in Burkholderia pseudomallei. Antimicrob. Agents Chemother. 2002;46:1132–1135. doi: 10.1128/AAC.46.4.1132-1135.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Papp-Wallace K.M., Taracila M.A., Gatta J.A., Ohuchi N., Bonomo R.A., Nukaga M. Insights into β-lactamases from Burkholderia species, two phylogenetically related yet distinct resistance determinants. J. Biol. Chem. 2013;288:19090–19102. doi: 10.1074/jbc.M113.458315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rojo-Bezares B., Martin C., Lopez M., Torres C., Saenz Y. First detection of blaIMI-2 gene in a clinical Escherichia coli strain. Antimicrob. Agents Chemother. 2012;56:1146–1147. doi: 10.1128/AAC.05478-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dortet L., Poirel L., Abbas S., Oueslati S., Nordmann P. Genetic and Biochemical Characterization of FRI-1, a Carbapenem-hydrolyzing class A β-lactamase from Enterobacter cloacae. Antimicrob. Agents Chemother. 2015;59:7420–7425. doi: 10.1128/AAC.01636-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bratu S., Landman D., Haag R., et al. Rapid spread of carbapenem-resistant Klebsiella pneumoniae in New York City: A new threat to our antibiotic armamentarium. Arch. Intern. Med. 2005;165:1430–1435. doi: 10.1001/archinte.165.12.1430. [DOI] [PubMed] [Google Scholar]
- 69.Canton R., Akova M., Carmeli Y., et al. Rapid evolution and spread of carbapenemases among Enterobacteriaceae in Europe. Clin. Microbiol. Infect. 2012;18:413–431. doi: 10.1111/j.1469-0691.2012.03821.x. [DOI] [PubMed] [Google Scholar]
- 70.Naas T., Bonnin R.A., Cuzon G., Villegas M.V., Nordmann P. Complete sequence of two KPC-harbouring plasmids from Pseudomonas aeruginosa. J. Antimicrob. Chemother. 2013;68:1757–1762. doi: 10.1093/jac/dkt094. [DOI] [PubMed] [Google Scholar]
- 71.Cuzon G., Naas T., Villegas M.V., Correa A., Quinn J.P., Nordmann P. Wide dissemination of Pseudomonas aeruginosa producing β-lactamase blaKPC-2 gene in Colombia. Antimicrob. Agents Chemother. 2011;55:5350–5353. doi: 10.1128/AAC.00297-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Robledo I.E., Aquino E.E., Vazquez G.J. Detection of the KPC gene in Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter baumannii during a PCR-based nosocomial surveillance study in Puerto Rico. Antimicrob. Agents Chemother. 2011;55:2968–2970. doi: 10.1128/AAC.01633-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Davies T.A., Marie Queenan A., Morrow B.J., et al. Longitudinal survey of carbapenem resistance and resistance mechanisms in Enterobacteriaceae and non-fermenters from the USA in 2007-09. J. Antimicrob. Chemother. 2011;66:2298–2307. doi: 10.1093/jac/dkr290. [DOI] [PubMed] [Google Scholar]
- 74.Cuzon G., Naas T., Nordmann P. Functional characterization of Tn4401, a Tn3-based transposon involved in blaKPC gene mobilization. Antimicrob. Agents Chemother. 2011;55:5370–5373. doi: 10.1128/AAC.05202-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Naas T., Cuzon G., Villegas M.V., Lartigue M.F., Quinn J.P., Nordmann P. Genetic structures at the origin of acquisition of the β-lactamase blaKPC gene. Antimicrob. Agents Chemother. 2008;52:1257–1263. doi: 10.1128/AAC.01451-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Woodford N., Wareham D.W., Guerra B., Teale C. Carbapenemase-producing Enterobacteriaceae and non-Enterobacteriaceae from animals and the environment: An emerging public health risk of our own making? J. Antimicrob. Chemother. 2014;69:287–291. doi: 10.1093/jac/dkt392. [DOI] [PubMed] [Google Scholar]
- 77.Poirel L., Barbosa-Vasconcelos A., Simoes R.R., Da Costa P.M., Liu W., Nordmann P. Environmental KPC-producing Escherichia coli isolates in Portugal. Antimicrob. Agents Chemother. 2012;56:1662–1663. doi: 10.1128/AAC.05850-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Oliveira S., Moura R.A., Silva K.C., et al. Isolation of KPC-2-producing Klebsiella pneumoniae strains belonging to the high-risk multiresistant clonal complex 11 (ST437 and ST340) in urban rivers. J. Antimicrob. Chemother. 2014;69:849–852. doi: 10.1093/jac/dkt431. [DOI] [PubMed] [Google Scholar]
- 79.Zhang X., Lu X., Zong Z. Enterobacteriaceae producing the KPC-2 carbapenemase from hospital sewage. Diagn. Microbiol. Infect. Dis. 2012;73:204–206. doi: 10.1016/j.diagmicrobio.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 80.Picao R.C., Cardoso J.P., Campana E.H., et al. The route of antimicrobial resistance from the hospital effluent to the environment: Focus on the occurrence of KPC-producing Aeromonas spp. and Enterobacteriaceae in sewage. Diagn. Microbiol. Infect. Dis. 2013;76:80–85. doi: 10.1016/j.diagmicrobio.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 81.Galler H., Feierl G., Petternel C., et al. KPC-2 and OXA-48 carbapenemase-harbouring Enterobacteriaceae detected in an Austrian wastewater treatment plant. Clin. Microbiol. Infect. 2014;20:O132–O134. doi: 10.1111/1469-0691.12336. [DOI] [PubMed] [Google Scholar]
- 82.Giakkoupi P., Tzouvelekis L.S., Tsakris A., Loukova V., Sofianou D., Tzelepi E. IBC-1, a novel integron-associated class A β-lactamase with extended-spectrum properties produced by an Enterobacter cloacae clinical strain. Antimicrob. Agents Chemother. 2000;44:2247–2253. doi: 10.1128/aac.44.9.2247-2253.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Poirel L., Le Thomas I., Naas T., Karim A., Nordmann P. Biochemical sequence analyses of GES-1, a novel class A extended-spectrum β-lactamase, and the class 1 integron In52 from Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2000;44:622–632. doi: 10.1128/aac.44.3.622-632.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Poirel L., Naas T., Nordmann P. Pyrosequencing as a rapid tool for identification of GES-type extended-spectrum β-lactamases. J. Clin. Microbiol. 2006;44:3008–3011. doi: 10.1128/JCM.02576-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Poirel L., Brinas L., Verlinde A., Ide L., Nordmann P. BEL-1, a novel clavulanic acid-inhibited extended-spectrum β-lactamase, and the class 1 integron In120 in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2005;49:3743–3748. doi: 10.1128/AAC.49.9.3743-3748.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Poirel L., Weldhagen G.F., Naas T., De Champs C., Dove M.G., Nordmann P. GES-2, a class A β-lactamase from Pseudomonas aeruginosa with increased hydrolysis of imipenem. Antimicrob. Agents Chemother. 2001;45:2598–2603. doi: 10.1128/AAC.45.9.2598-2603.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wachino J., Doi Y., Yamane K., et al. Nosocomial spread of ceftazidime-resistant Klebsiella pneumoniae strains producing a novel class a β-lactamase, GES-3, in a neonatal intensive care unit in Japan. Antimicrob. Agents Chemother. 2004;48:1960–1967. doi: 10.1128/AAC.48.6.1960-1967.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vourli S., Giakkoupi P., Miriagou V., Tzelepi E., Vatopoulos A.C., Tzouvelekis L.S. Novel GES/IBC extended-spectrum β-lactamase variants with carbapenemase activity in clinical enterobacteria. FEMS Microbiol. Lett. 2004;234:209–213. doi: 10.1016/j.femsle.2004.03.028. [DOI] [PubMed] [Google Scholar]
- 89.Mavroidi A., Tzelepi E., Tsakris A., Miriagou V., Sofianou D., Tzouvelekis L.S. An integron-associated β-lactamase (IBC-2) from Pseudomonas aeruginosa is a variant of the extended-spectrum β-lactamase IBC-1. J. Antimicrob. Chemother. 2001;48:627–630. doi: 10.1093/jac/48.5.627. [DOI] [PubMed] [Google Scholar]
- 90.Bogaerts P., Naas T., El Garch F., et al. GES extended-spectrum β-lactamases in Acinetobacter baumannii isolates in Belgium. Antimicrob. Agents Chemother. 2010;54:4872–4878. doi: 10.1128/AAC.00871-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bontron S., Poirel L., Nordmann P. In vitro prediction of the evolution of GES-1 β-lactamase hydrolytic activity. Antimicrob. Agents Chemother. 2015;59:1664–1670. doi: 10.1128/AAC.04450-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Walsh T.R. Emerging carbapenemases: A global perspective. Int. J. Antimicrob. Agents. 2010;36(Suppl. 3):S8–S14. doi: 10.1016/S0924-8579(10)70004-2. [DOI] [PubMed] [Google Scholar]
- 93.Jeong S.H., Bae I.K., Kim D., et al. First outbreak of Klebsiella pneumoniae clinical isolates producing GES-5 and SHV-12 extended-spectrum β-lactamases in Korea. Antimicrob. Agents Chemother. 2005;49:4809–4810. doi: 10.1128/AAC.49.11.4809-4810.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Poirel L., Weldhagen G.F., De Champs C., Nordmann P. A nosocomial outbreak of Pseudomonas aeruginosa isolates expressing the extended-spectrum β-lactamase GES-2 in South Africa. J. Antimicrob. Chemother. 2002;49:561–565. doi: 10.1093/jac/49.3.561. [DOI] [PubMed] [Google Scholar]
- 95.de Vries J.J., Baas W.H., van der Ploeg K., Heesink A., Degener J.E., Arends J.P. Outbreak of Serratia marcescens colonization and infection traced to a healthcare worker with long-term carriage on the hands. Infect. Control Hosp. Epidemiol. 2006;27:1153–1158. doi: 10.1086/508818. [DOI] [PubMed] [Google Scholar]
- 96.Duarte A., Boavida F., Grosso F., et al. Outbreak of GES-1 β-lactamase-producing multidrug-resistant Klebsiella pneumoniae in a university hospital in Lisbon, Portugal. Antimicrob. Agents Chemother. 2003;47:1481–1482. doi: 10.1128/AAC.47.4.1481-1482.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Picao R.C., Poirel L., Gales A.C., Nordmann P. Diversity of β-lactamases produced by ceftazidime-resistant Pseudomonas aeruginosa isolates causing bloodstream infections in Brazil. Antimicrob. Agents Chemother. 2009;53:3908–3913. doi: 10.1128/AAC.00453-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bonnin R.A., Nordmann P., Poirel L. Screening and deciphering antibiotic resistance in Acinetobacter baumannii: A state of the art. Expert Rev. Anti Infect. Ther. 2013;11:571–583. doi: 10.1586/eri.13.38. [DOI] [PubMed] [Google Scholar]
- 99.Bonnin R.A., Nordmann P., Potron A., Lecuyer H., Zahar J.R., Poirel L. Carbapenem-hydrolyzing GES-type extended-spectrum β-lactamase in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2011;55:349–354. doi: 10.1128/AAC.00773-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Moubareck C., Bremont S., Conroy M.C., Courvalin P., Lambert T. GES-11, a novel integron-associated GES variant in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2009;53:3579–3581. doi: 10.1128/AAC.00072-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zowawi H.M., Balkhy H.H., Walsh T.R., Paterson D.L. β-Lactamase production in key gram-negative pathogen isolates from the Arabian Peninsula. Clin. Microbiol. Rev. 2013;26:361–380. doi: 10.1128/CMR.00096-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bonnin R.A., Rotimi V.O., Al Hubail M., et al. Wide dissemination of GES-type carbapenemases in Acinetobacter baumannii isolates in Kuwait. Antimicrob. Agents Chemother. 2013;57:183–188. doi: 10.1128/AAC.01384-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cicek A.C., Saral A., Iraz M., et al. OXA- and GES-type β-lactamases predominate in extensively drug-resistant Acinetobacter baumannii isolates from a Turkish University Hospital. Clin. Microbiol. Infect. 2014;20:410–415. doi: 10.1111/1469-0691.12338. [DOI] [PubMed] [Google Scholar]
- 104.Castanheira M., Costello S.E., Woosley L.N., Deshpande L.M., Davies T.A., Jones R.N. Evaluation of clonality and carbapenem resistance mechanisms among Acinetobacter baumannii-Acinetobacter calcoaceticus complex and Enterobacteriaceae isolates collected in European and Mediterranean countries and detection of two novel β-lactamases, GES-22 and VIM-35. Antimicrob. Agents Chemother. 2014;58:7358–7366. doi: 10.1128/AAC.03930-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Maurya A.P., Choudhury D., Talukdar A.D., Dhar Chanda A., Chakravarty A., Bhattacharjee A. A report on the presence of GES-5 extended spectrum β-lactamase producing Pseudomonas aeruginosa associated with urinary tract infection from north-east India. Indian J. Med. Res. 2014;140:565–567. [PMC free article] [PubMed] [Google Scholar]
- 106.Garza-Ramos U., Barrios H., Reyna-Flores F., et al. Widespread of ESBL- and carbapenemase GES-type genes on carbapenem-resistant Pseudomonas aeruginosa clinical isolates: A multicenter study in Mexican hospitals. Diagn. Microbiol. Infect. Dis. 2015;81:135–137. doi: 10.1016/j.diagmicrobio.2014.09.029. [DOI] [PubMed] [Google Scholar]
- 107.Sepehri S., Poliquin G., Alfattoh N., et al. Osteomyelitis due to multiple carbapenemase-producing Gram-negative bacteria: The first case report of a GES-13-producing Pseudomonas aeruginosa isolate in Canada. Can. J. Infect. Dis. Med. Microbiol. 2014;25:229–231. doi: 10.1155/2014/657047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Botelho J., Grosso F., Sousa C., Peixe L. Characterization of a new genetic environment associated with GES-6 carbapenemase from a Pseudomonas aeruginosa isolate belonging to the high-risk clone ST235. J. Antimicrob. Chemother. 2015;70:615–617. doi: 10.1093/jac/dku391. [DOI] [PubMed] [Google Scholar]
- 109.Iraz M., Duzgun A.O., Cicek A.C., et al. Characterization of novel VIM carbapenemase, VIM-38, and first detection of GES-5 carbapenem-hydrolyzing β-lactamases in Pseudomonas aeruginosa in Turkey. Diagn. Microbiol. Infect. Dis. 2014;78:292–294. doi: 10.1016/j.diagmicrobio.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 110.Viedma E., Juan C., Acosta J., et al. Nosocomial spread of colistin-only-sensitive sequence type 235 Pseudomonas aeruginosa isolates producing the extended-spectrum β-lactamases GES-1 and GES-5 in Spain. Antimicrob. Agents Chemother. 2009;53:4930–4933. doi: 10.1128/AAC.00900-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Walsh T.R. Clinically significant carbapenemases: An update. Curr. Opin. Infect. Dis. 2008;21:367–371. doi: 10.1097/QCO.0b013e328303670b. [DOI] [PubMed] [Google Scholar]
- 112.Girlich D., Poirel L., Szczepanowski R., Schluter A., Nordmann P. Carbapenem-hydrolyzing GES-5-encoding gene on different plasmid types recovered from a bacterial community in a sewage treatment plant. Appl. Environ. Microbiol. 2012;78:1292–1295. doi: 10.1128/AEM.06841-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Montezzi L.F., Campana E.H., Correa L.L., et al. Occurrence of carbapenemase-producing bacteria in coastal recreational waters. Int. J. Antimicrob. Agents. 2015;45:174–177. doi: 10.1016/j.ijantimicag.2014.10.016. [DOI] [PubMed] [Google Scholar]
- 114.Mataseje L.F., Boyd D.A., Delport J., et al. Serratia marcescens harbouring SME-type class A carbapenemases in Canada and the presence of blaSME on a novel genomic island, SmarGI1-1. J. Antimicrob. Chemother. 2014;69:1825–1829. doi: 10.1093/jac/dku040. [DOI] [PubMed] [Google Scholar]
- 115.Poirel L., Carrer A., Pitout J.D., Nordmann P. Integron mobilization unit as a source of mobility of antibiotic resistance genes. Antimicrob. Agents Chemother. 2009;53:2492–2498. doi: 10.1128/AAC.00033-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Naas T., Cuzon G., Truong H.V., Nordmann P. Role of ISKpn7 and deletions in blaKPC gene expression. Antimicrob. Agents Chemother. 2012;56:4753–4759. doi: 10.1128/AAC.00334-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Naas T., Massuard S., Garnier F., Nordmann P. AmpD is required for regulation of expression of NmcA, a carbapenem-hydrolyzing β-lactamase of Enterobacter cloacae. Antimicrob. Agents Chemother. 2001;45:2908–2915. doi: 10.1128/AAC.45.10.2908-2915.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Giske C.G., Sundsfjord A.S., Kahlmeter G., et al. Redefining extended-spectrum β-lactamases: balancing science and clinical need. J. Antimicrob. Chemother. 2009;63:1–4. doi: 10.1093/jac/dkn444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang C., Cai P., Chang D., Mi Z. A Pseudomonas aeruginosa isolate producing the GES-5 extended-spectrum β-lactamase. J. Antimicrob. Chemother. 2006;57:1261–1262. doi: 10.1093/jac/dkl116. [DOI] [PubMed] [Google Scholar]
- 120.Meroueh S.O., Fisher J.F., Schlegel H.B., Mobashery S. Ab initio QM/MM study of class A β-lactamase acylation: Dual participation of Glu166 and Lys73 in a concerted base promotion of Ser70. J. Am. Chem. Soc. 2005;127:15397–15407. doi: 10.1021/ja051592u. [DOI] [PubMed] [Google Scholar]
- 121.Majiduddin F.K., Palzkill T. Amino acid sequence requirements at residues 69 and 238 for the SME-1 β-lactamase to confer resistance to β-lactam antibiotics. Antimicrob. Agents Chemother. 2003;47:1062–1067. doi: 10.1128/AAC.47.3.1062-1067.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Berman H.M., Westbrook J., Feng Z., et al. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ke W., Bethel C.R., Thomson J.M., Bonomo R.A., van den Akker F. Crystal structure of KPC-2: Insights into carbapenemase activity in class A β-lactamases. Biochemistry. 2007;46:5732–5740. doi: 10.1021/bi700300u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Majiduddin F.K., Palzkill T. Amino acid residues that contribute to substrate specificity of class A β-lactamase SME-1. Antimicrob. Agents Chemother. 2005;49:3421–3427. doi: 10.1128/AAC.49.8.3421-3427.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chudyk E.I., Limb M.A., Jones C., Spencer J., van der Kamp M.W., Mulholland A.J. QM/MM simulations as an assay for carbapenemase activity in class A β-lactamases. Chem. Commun. (Camb.) 2014;50:14736–14739. doi: 10.1039/c4cc06495j. [DOI] [PubMed] [Google Scholar]
- 126.Petrella S., Ziental-Gelus N., Mayer C., Renard M., Jarlier V., Sougakoff W. Genetic and structural insights into the dissemination potential of the extremely broad-spectrum class A β-lactamase KPC-2 identified in an Escherichia coli strain and an Enterobacter cloacae strain isolated from the same patient in France. Antimicrob. Agents Chemother. 2008;52:3725–3736. doi: 10.1128/AAC.00163-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hanes M.S., Jude K.M., Berger J.M., Bonomo R.A., Handel T.M. Structural and biochemical characterization of the interaction between KPC-2 β-lactamase and β-lactamase inhibitor protein. Biochemistry. 2009;48:9185–9193. doi: 10.1021/bi9007963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ke W., Bethel C.R., Papp-Wallace K.M., et al. Crystal structures of KPC-2 β-lactamase in complex with 3-nitrophenyl boronic acid and the penam sulfone PSR-3-226. Antimicrob. Agents Chemother. 2012;56:2713–2718. doi: 10.1128/AAC.06099-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Frase H., Smith C.A., Toth M., Champion M.M., Mobashery S., Vakulenko S.B. Identification of products of inhibition of GES-2 β-lactamase by tazobactam by X-ray crystallography and spectrometry. J. Biol. Chem. 2011;286:14396–14409. doi: 10.1074/jbc.M110.208744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Stewart N.K., Smith C.A., Frase H., Black D.J., Vakulenko S.B. Kinetic and structural requirements for carbapenemase activity in GES-type β-lactamases. Biochemistry. 2015;54:588–597. doi: 10.1021/bi501052t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Smith C.A., Frase H., Toth M., et al. Structural basis for progression toward the carbapenemase activity in the GES family of β-lactamases. J. Am. Chem. Soc. 2012;134:19512–19515. doi: 10.1021/ja308197j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bebrone C., Bogaerts P., Delbruck H., et al. GES-18, a new carbapenem-hydrolyzing GES-type β-lactamase from Pseudomonas aeruginosa that contains Ile80 and Ser170 residues. Antimicrob. Agents Chemother. 2013;57:396–401. doi: 10.1128/AAC.01784-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Swarén P., Maveyraud L., Raquet X., et al. X-ray analysis of the NMC-A β-lactamase at 1.64-A resolution, a class A carbapenemase with broad substrate specificity. J. Biol. Chem. 1998;273:26714–26721. doi: 10.1074/jbc.273.41.26714. [DOI] [PubMed] [Google Scholar]
- 134.Mourey L., Miyashita K., Swarén P., Bulychev A., Samama J-P., Mobashery S. Inhibition of the NMC-A β-lactamase by a penicillanic acid derivative and the structural bases for the increase in substrate profile of this antibiotic resistance enzyme. J. Am. Chem. Soc. 1998;120:9382–9383. [Google Scholar]
- 135.Sougakoff W., L'Hermite G., Pernot L., et al. Structure of the imipenem-hydrolyzing class A β-lactamase SME-1 from Serratia marcescens. Acta Crystallogr. D Biol. Crystallogr. 2002;58:267–274. doi: 10.1107/s0907444901019606. [DOI] [PubMed] [Google Scholar]
- 136.Smith C.A., Caccamo M., Kantardjieff K.A., Vakulenko S. Structure of GES-1 at atomic resolution: Insights into the evolution of carbapenamase activity in the class A extended-spectrum β-lactamases. Acta Crystallogr. D Biol. Crystallogr. 2007;63:982–992. doi: 10.1107/S0907444907036955. [DOI] [PubMed] [Google Scholar]
- 137.Pettersen E.F., Goddard T.D., Huang C.C., et al. UCSF Chimera - a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 138.Papp-Wallace K.M., Taracila M., Wallace C.J., et al. Elucidating the role of Trp105 in the KPC-2 β-lactamase. Protein Sci. 2010;19:1714–1727. doi: 10.1002/pro.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Papp-Wallace K.M., Taracila M.A., Smith K.M., Xu Y., Bonomo R.A. Understanding the molecular determinants of substrate and inhibitor specificities in the carbapenemase KPC-2: Exploring the roles of Arg220 and Glu276. Antimicrob. Agents Chemother. 2012;56:4428–4438. doi: 10.1128/AAC.05769-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Levitt P.S., Papp-Wallace K.M., Taracila M.A., et al. Exploring the role of a conserved class A residue in the omega loop of KPC-2 β-lactamase: A mechanism for ceftazidime hydrolysis. J. Biol. Chem. 2012;287:31783–31793. doi: 10.1074/jbc.M112.348540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lahiri S.D., Mangani S., Durand-Reville T., et al. Structural insight into potent broad-spectrum inhibition with reversible recyclization mechanism: Avibactam in complex with CTX-M-15 and Pseudomonas aeruginosa AmpC β-lactamases. Antimicrob. Agents Chemother. 2013;57:2496–2505. doi: 10.1128/AAC.02247-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Papp-Wallace K.M., Winkler M.L., Taracila M.A., Bonomo R.A. Variants of the KPC-2 β-lactamase which are resistant to inhibition by avibactam. Antimicrob. Agents Chemother. 2015;59:3710–3717. doi: 10.1128/AAC.04406-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kotsakis S.D., Miriagou V., Tzelepi E., Tzouvelekis L.S. Comparative biochemical and computational study of the role of naturally occurring mutations at Ambler positions 104 and 170 in GES β-lactamases. Antimicrob. Agents Chemother. 2010;54:4864–4871. doi: 10.1128/AAC.00771-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Frase H., Toth M., Champion M.M., Antunes N.T., Vakulenko S.B. Importance of position 170 in the inhibition of GES-type β-lactamases by clavulanic acid. Antimicrob. Agents Chemother. 2011;55:1556–1562. doi: 10.1128/AAC.01292-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Frase H., Shi Q., Testero S.A., Mobashery S., Vakulenko S.B. Mechanistic basis for the emergence of catalytic competence against carbapenem antibiotics by the GES family of β-lactamases. J. Biol. Chem. 2009;284:29509–29513. doi: 10.1074/jbc.M109.011262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Mahy F. Étude par mutagenèse dirigée de la carbapénèmase de classe A NMCA.PhD thesis, University of Liège, 2002. [Google Scholar]
- 147.Sougakoff W., Naas T., Nordmann P., Collatz E., Jarlier V. Role of Ser-237 in the substrate specificity of the carbapenem-hydrolyzing class A β-lactamase Sme-1. Biochim. Biophys. Acta. 1999;1433:153–158. doi: 10.1016/s0167-4838(99)00138-7. [DOI] [PubMed] [Google Scholar]
- 148.Rholl D.A., Papp-Wallace K.M., Tomaras A.P., Vasil M.L., Bonomo R.A., Schweizer H.P. Molecular investigations of PenA-mediated β-lactam resistance in Burkholderia pseudomallei. Front. Microbiol. 2011;2:139. doi: 10.3389/fmicb.2011.00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Yi H., Cho K.H., Cho Y.S., Kim K., Nierman W.C., Kim H.S. Twelve positions in a β-lactamase that can expand its substrate spectrum with a single amino acid substitution. PLoS One. 2012;7:e37585. doi: 10.1371/journal.pone.0037585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Yi H., Kim K., Cho K.H., Jung O., Kim H.S. Substrate spectrum extension of PenA in Burkholderia thailandensis with a single amino acid deletion, Glu168del. Antimicrob. Agents Chemother. 2012;56:4005–4008. doi: 10.1128/AAC.00598-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hwang J., Cho K.H., Song H., Yi H., Kim H.S. Deletion mutations conferring substrate spectrum extension in the class A β-lactamase. Antimicrob. Agents Chemother. 2014;58:6265–6269. doi: 10.1128/AAC.02648-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zafaralla G., Manavathu E.K., Lerner S.A., Mobashery S. Elucidation of the role of arginine-244 in the turnover processes of class A β-lactamases. Biochemistry. 1992;31:3847–3852. doi: 10.1021/bi00130a016. [DOI] [PubMed] [Google Scholar]
- 153.Abraham E.P., Chain E. An enzyme from bacteria able to destroy penicillin. Nature. 1940;146:837. [PubMed] [Google Scholar]


