Abstract
Despite significant advancements in our understanding of ubiquitin-mediated signaling, the influence of the intracellular environment on formation of transient ubiquitin-partner complexes remains poorly explored. In our work, we introduce macromolecular crowding as a first level of complexity towards the imitation of a cellular environment in the study of such interactions. Using NMR spectroscopy, we find that the stereospecific complex of ubiquitin and ubiquitin-associated domain (UBA) is minimally perturbed by the crowding agent Ficoll. However, in addition to the primary canonical recognition patch on ubiquitin, secondary patches are identified, indicating that in cell-mimicking crowded solution, UBA contacts ubiquitin at multiple sites.
Keywords: biomolecular recognition, macromolecular crowding, NMR spectroscopy, paramagnetic relaxation enhancement, ubiquitin, ubiquitin associated domain
INTRODUCTION
Ubiquitin (Ub) is a prototypical small protein modifier regulating a vast number of fundamental cellular events [1,2]. The addition of a single Ub molecule to a target protein (monoubiquitination) can alter protein activity and localization, while the conjugation of distinct types of Ub chains (polyubiquitination) is implicated in a variety of processes such as proteasomal degradation, DNA repair, and immune signaling [2–5]. Differentially ubiquitinated substrates are recognized by an ample variety of Ub-binding proteins, which propagate the Ub signal eliciting specific cell responses [6,7]. Ub receptors contain modular elements, referred to as Ub-binding domains (UBD), able to specifically associate with the protein modifier. The binding of individual UBD to monoUb is generally weak but high-avidity interactions can be established by polyUb chains [8–11]. Intense work has been carried out during the past two decades to elucidate the structural determinants of Ub-UBD recognition, however further scrutiny is required to obtain a full understanding of the specificity of Ub-mediated signaling.
In spite of the complexity of Ub recognition by effector proteins, the simplicity of monomeric Ub’s architecture is startling. Composed of only 76 amino acids, highly conserved throughout eukaryotes, Ub is among the smallest proteins found in a cell. The polypeptide chain adopts a compact globular β-grasp fold, exposing a surface area of less than 5,000 Å2. A solvent-exposed hydrophobic area, centered around residues Leu8, Ile44, and Val70 (Ile44 patch [4,12]), stands out from the predominantly polar protein surface. The Ub backbone is generally considered rigid, with the exception of the C-terminus and of the β1-β2 loop. However, a certain degree of structural plasticity allows a mechanism of dynamic adaptation to operate during molecular recognition events [13]. Interactions with many ubiquitin-binding proteins involve the Ile44 patch [4], albeit distinct residues form the interaction surface with different protein partners. Furthermore, additional protein recognition sites have been identified, including the C-terminus, the α/β2 groove, the Ile36, the Asp58, and the Phe4 patches [11,14].
Given that a finely tuned surface chemistry dictates the determinants of Ub-UBD recognition, a definitive description of the corresponding modes of interaction requires evaluation of all factors potentially affecting the binding phenomena in the native environment. While our current understanding of Ub-UBD associations relies on interaction studies performed on binary protein mixtures in buffered aqueous solution, it has become clear that the complex cellular environment can exert significant influence on biomolecular properties and interactions [15–17]. A prominent feature of the cellular interior is intense macromolecular crowding caused by elevated concentrations of large biological molecules that occupy a significant fraction (10–40%) of the cell volume [18]. Macromolecular crowders, or cosolutes, alter a protein’s effective concentration, increase solution viscosity, originate excluded-volume effects, and may engage in unspecific or specific chemical interactions with the protein solutes [19–24]. The impact of tight molecular packing on the formation of Ub-UBD complexes has remained unexplored so far.
In our work, we introduced macromolecular crowding as a first level of complexity towards the imitation of a cellular environment in the study of Ub-UBD interactions. Steric repulsions and depletion forces are predicted to perturb conformational equilibria and diffusive dynamics, to shift equilibrium states, and change the probability of intermolecular collisions [19]. However, the extent to which these phenomena reshape the protein-protein interaction landscape, influencing the relative orientations of components in non-covalent assemblies and the specificity of transient associations, remains elusive. Here, we investigated the interaction between Ub and the Ub-associated C-terminal domain (UBA2) of the human homologue of the yeast DNA repair protein RAD23 (HHR23A) in the presence of a non-interacting hydrophilic polymeric crowding agent. UBA2 is a compact three-helix bundle displaying a Ub recognition site, encompassing the non-adjacent helices α1 and α3, that associates weakly (Kd ~0.4 mM) with the Ile44 patch of Ub [25–27]. The Ub-UBA2 interaction can be considered paradigmatic in the study of analogous Ub-UBD pairs.
We used NMR spectroscopy to explore the UBA2 interaction sites on the surface of Ub. NMR allows the exploration of interactions over a broad range of affinities, yielding atomic-resolution insights into dynamic biochemical equilibria [22]. By complementing traditional chemical shift perturbation mapping with solvent paramagnetic relaxation enhancement analysis [28,29], a robust identification of binding surfaces was obtained. In addition to the well-characterized Ile44 patch, secondary patches were found, indicating that in cell-mimicking crowded solution, UBA2 contacts Ub at multiple sites. Identification of such transient specific associations adds further detail to our description of protein-protein interactions in the complex cellular interior.
EXPERIMENTAL SECTION
A description of materials and protein preparation procedures can be found in the Supporting Information.
NMR experiments
All experiments were recorded at 25 °C on a Bruker Avance III spectrometer, operating at 1H Larmor frequency of 600.13 MHz, equipped with a triple resonance TCI cryogenic probe. NMR data were processed with Topspin 3.2 (Bruker) or NMRpipe [30], and analyzed with the software Sparky (T. D. Goddard and D. G. Kneller, University of California, San Francisco).
15N relaxation experiments were performed on samples containing 0.37 mM [15N]-Ub in the absence of crowder, in 200 g/L Ficoll, or in 200 g/L sucrose solution. Longitudinal relaxation rates, 15N-R1, were measured using relaxation delays in the interval 0.01–1.26 s for Ub in uncrowded solution and in the range 0.01–1.44 s for Ub in 200 g/L Ficoll or sucrose. Transverse relaxation rates, 15N-R2, were measured with a CPMG-based pulse program, using relaxation delays in the range 8–208 ms for Ub in simple buffer, 8–176 ms for Ub in 200 g/L sucrose, and 8–104 ms for Ub in 200 g/L Ficoll.
Proton transverse relaxation rates, 1HN-R2, were measured on a 0.5 mM [15N]Ub sample using the pulse program described by Iwahara et al. [31], kindly provided to us by the author. To remove 3JHN-Hα modulation of peak intensities, a selective HN 180° pulse (REBURP of 2 ms) centered at 8.2 ppm was used in the INEPT period. For each titration point (1:0, 1:0.5 and 1:1), measured in the presence or absence of gadodiamide, seven relaxation delays were acquired and the signal intensity decays were fitted to a single exponential function to obtain the corresponding rates. Delays between 11.7 and 52.5 (45.3) ms were used for samples without (with) gadodiamide. Residues affected by signal overlap or with insufficient signal-to-noise ratio were excluded from the analysis.
Data analysis
Amide chemical shift perturbation was calculated as: CSP = [(ΔδH)2 + (ΔδN/5)2]0.5, where ΔδH and ΔδN are the chemical shift changes measured in the 1H and 15N frequency dimensions, respectively.
Volume occupancy by Ficoll was calculated from a reported partial specific volume of 650 cm3/g [32], a value in agreement with our experimental verification of volume change upon dissolution of a known mass of solute. The volume fraction occupied by the crowder, ϕC, in a 200 g/L Ficoll solution was determined to be 13%, assuming no volume changes resulted from perturbations of solvent molecules.
Dissociation constant values were obtained by fitting experimental binding isotherms according to a one-site binding model using the Matlab program Kdfit [26]. Titration data were analyzed assuming that the observed CSP is a weighted average between the value corresponding to the free (CSP = 0) and ligand-bound (CSP = CSPbound) states, so that CSP = CSPbound × fbound, where fbound is the relative population of the bound state of the molecule under observation, related to the dissociation constant Kd according to the following expression [26]:
where CP and CL are the total concentrations of Ub and UBA2, respectively. The analytical concentration of the initial Ub solution was 500 μM and that of the titrant stock solution was 10 mM. Dilution-corrected values for CP and CL were used at successive titration steps. Reported values are the average of Kd values determined from seven binding isotherms and the corresponding standard deviation.
Relaxation rate values were obtained from fitting of signal intensity decays with a single exponential function. The rotational correlation time, τR, was estimated with the program ROTDIF [33] from the experimental 15N-R1 and 15N-R2 relaxation rate values, assuming a constant NOE value of 0.74 for all residues displaying secondary structure and using an isotropic model. Relative solution viscosities were estimated using the Stokes-Einstein-Debye equation: τR = 4πηr3/(3kT), where η is the solution viscosity, r the hydrodynamic radius of Ub (assumed constant in all experimental conditions), k the Boltzmann constant and T the temperature. Using the measured τR values, we determined the following ratios: ηFicoll/ηsucrose = 1.19, ηFicoll/ηbuffer = 2.3 for 200 g/L crowder solutions.
Transverse 1HN PRE rates, (1HN-R2p) were obtained as the difference in 1HN-R2 measured on samples containing or not containing 2 mM gadodiamide. In principle, the difference removes contributions to 1HN-R2 relaxation common to both states, including exchange contributions [31]. The deviation of 1HN-R2p from a linear concentration dependence was estimated by:
where
and r1 = 0, r2 = 0.5, r3 = 1, corresponding to the UBA2/Ub molar ratios. The uncertainties of Δ, σΔ, were obtained by propagation of errors on relaxation rates [34].
Protein structures were visualized with PyMOL (The PyMOL Molecular Graphics System, Version 1.1r1, LLC).
RESULTS
The structures of Ub and of the Ub/UBA2 complex are retained in crowded solution
Macromolecular crowding of the interior of cells can be conveniently modelled by use of a variety of crowding agents, including biomacromolecules and synthetic polymers [35,36]. Depending on the chemical nature of cosolutes, macromolecular crowding effects may include both steric repulsions as well as nonspecific chemical interactions. Here, we selected the macromolecular crowder Ficoll (70 kDa), a nearly spherical and densely branched hydrophilic neutral polymer that is reported not to interact with most proteins [17,35] and in particular with Ub [37]. Experiments were conducted after dissolving the protein(s) in 200 g/L Ficoll solution, approximately corresponding to the concentration of macromolecules in the average eukaryotic cytoplasm.
Our preliminary investigation concerned the possible effects of Ficoll on the structural and dynamic properties of Ub. We therefore recorded two-dimensional proton-nitrogen correlation (1H,15N-HSQC) NMR spectra of 15N-enriched Ub, these experiments being exquisitely sensitive to binding events and structural changes. The spectral fingerprint of Ub in Ficoll solution, reporting separate signals for individual amino acid residues of the polypeptide, displayed all peaks in the same positions as those observed in uncrowded solution: amide chemical shift perturbation (CSP) values were close to zero for all residues (Fig. 1A), in agreement with the findings of Cino et al. [38] and Abriata et al. [37]. This observation indicates that Ficoll did not perturb the overall structure of Ub. 15N-spin relaxation rate measurements were then carried out to explore possible changes in Ub main chain dynamics. Data reported in Fig. 1B,C show a significant decrease in longitudinal relaxation rate (15N-R1) values and increase in transverse relaxation rates (15N-R2), due to a slower molecular rotational diffusion resulting from increased viscosity (we calculated a global rotational correlation time, τR, of 4.0 ns and of 9.2 ns for Ub in uncrowded solution and in 200 g/L Ficoll, respectively). It can be noted that 15N-R1 values exhibit lesser variation along the polypeptide sequence in the presence of crowder, and in particular, residues in the β1-β2 loop and in the C-terminus display 15N-R1 values similar to or even larger than those observed in the rigid elements. Although it is possible that the observed trend reflects some reduced local dynamics in crowded solution, an increase of the longitudinal relaxation rate is predicted to take place in flexible regions upon increase in τR if the correlation time for local motion is in the range of hundreds of picoseconds or slower [39].
Figure 1. NMR analysis of 15N-Ub in crowded solution.
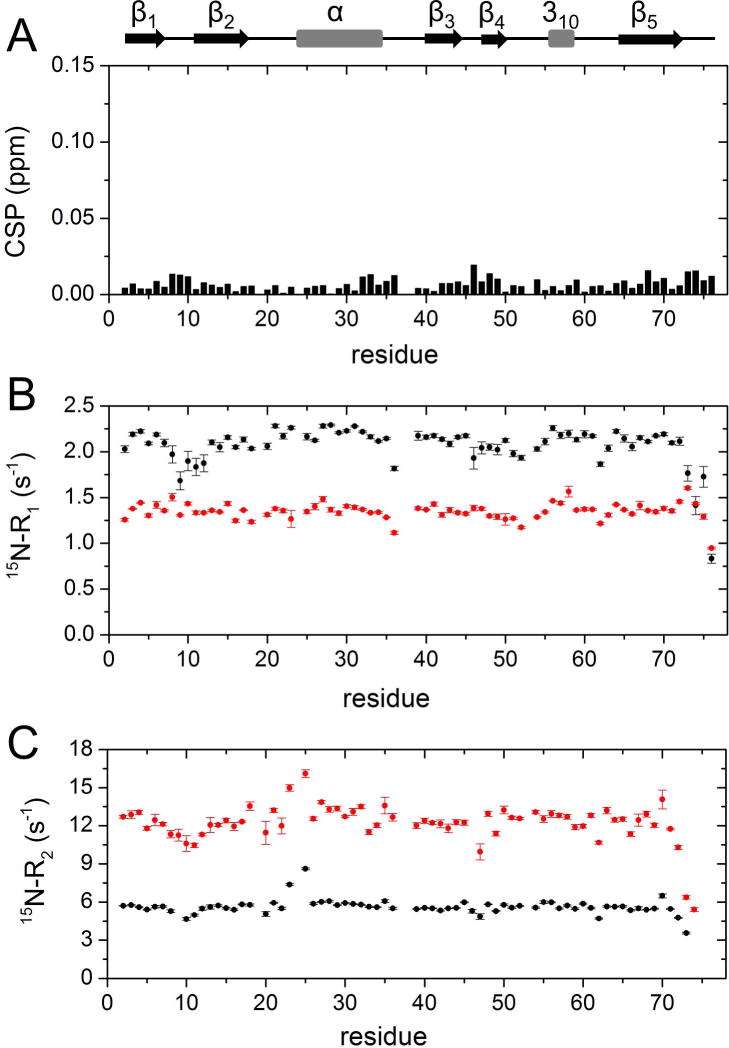
A) Amide chemical shift perturbations (CSP) obtained from 1H,15N-HSQC spectra of [15N]Ub in the presence of 200 g/L of Ficoll with respect to [15N]Ub in buffer. The CSP data are plotted as a function of residue number. B,C) 15N–spin relaxation rates of [15N]Ub with/without Ficoll. Shown are 15N-R1 (B) and 15N-R2 (C) values as a function of residue number, obtained with (red) and without (black) 200 g/L Ficoll at 25 °C.
In order to obtain mechanistic information on UBA2 binding to Ub under macromolecular crowding conditions, titration experiments were conducted by acquiring a series of 1H,15N-HSQC spectra in the presence and absence of 200 g/L Ficoll. Unlabelled UBA2 was added stepwise to [15N]Ub, and individual peaks were monitored throughout the titration (Fig. 2). The directions of peak movements along the titration in crowding conditions were unchanged compared to those in dilute solution (Fig. 2A,B), indicating that Ficoll did not perturb the mode of binding of Ub to UBA2. Despite the increase in solution viscosity when the polymer was present, we noted that binding still occurred in the fast exchange regime on the chemical shift time scale, indicating that the dissociation rate was not slowed down considerably in crowded solution. The CSP patterns along the protein sequence in the presence of Ficoll closely resembled those measured in the absence of crowder (Fig. 2C), suggesting that the average structure of the complex was not affected by the presence of crowder molecules. The backbone regions most affected by UBA2-binding in both solution conditions contain residues surrounding Leu8, Ile44 and Val70, the key residues forming the well-characterized Ile44 hydrophobic patch [12,14,26].
Figure 2. Ub-UBA2 titration monitored by 1H,15N-HSQC.
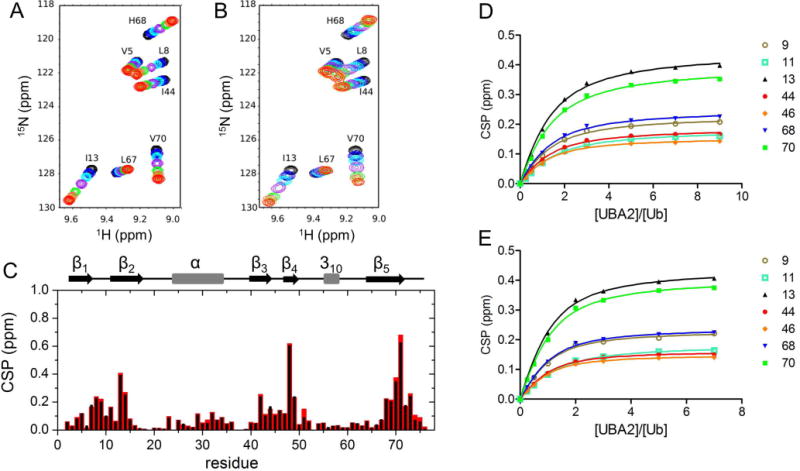
A, B) Overlay of portions of 15N-Ub spectra collected in buffer without (A) and with (B) 200 g/L Ficoll, upon successive additions of UBA2. The displayed spectra correspond to UBA2/Ub molar ratios of 0 (black), 0.25 (blue), 0.5 (cyan), 1 (purple), 2 (green), 3 (tomato), 5 (orange), 7 (red). C) Plot of the chemical shift perturbation (CSP) data from 1H,15N-HSQC spectra of [15N]Ub in the presence of 7-fold molar excess UBA2 with respect to free [15N]Ub, in the absence (black) and presence (red) of 200 g/L Ficoll. D,E) Ub/UBA2 binding isotherms for selected Ub residues based on CSP data collected in buffer without (D) and with (E) 200 g/L Ficoll. Residue numbers are indicated in the legend.
Titration experiments in 200 g/L Ficoll solution pointed out that binding site saturation was reached at a lower nominal concentration of UBA2 than in uncrowded solution. Indeed, by fitting the binding isotherms of selected residues (Fig. 2D,E) we determined Kd = 412 ± 52 μM for the Ub/UBA2 interaction in buffer, while in the presence of Ficoll the corresponding value was 226 ± 48 μM. Accounting for the actual protein concentrations (corrected for the volume occupied by Ficoll particles, see Materials and Methods for details) would result in at most ~10–15% increase of the Kd. Therefore, our data indicate that the apparent affinity of the complex increased in the presence of crowder. After repeating the titration experiment in 200 g/L sucrose (the monomeric counterpart of Ficoll) (Fig. S1), the Kd was found to be 381 ± 52 μM, indicating that the macromolecular nature of Ficoll was responsible for the increased affinity.
Solvent paramagnetic relaxation enhancement analysis reveals secondary UBA2 footprints on the surface of Ub
After assessment of formation of the stereospecific Ub/UBA2 complex in crowded solution, we set to perform a more comprehensive evaluation of the protein-protein contacts, possibly including less represented binding sites. To this aim we measured solvent paramagnetic relaxation enhancements (PRE), a sensitive NMR approach based on the use of soluble paramagnetic relaxation agents to reveal changes in macromolecular surface accessibility occurring during dynamic events such as protein complex formation. Due to their inherently large sensitivity (caused by the large magnetic moment of unpaired electrons), paramagnetic probes have been used successfully to characterize the structure of protein-protein complexes [40,41], formation of encounter complexes [42], and transient interactions [43]. In particular, Johansson et al. [34], were able to identify residues of human growth hormone that are involved in either unspecific or specific protein self-interactions based on the PRE profile of amide protons induced by gadodiamide (a gadolinium-based paramagnetic relaxation agent, also referred to as Gd-DTPA-BMA). Gadodiamide-induced PRE effects on Ub were previously interpreted according to a relaxation model where the relaxation agent forms an unspecific, yet rotationally correlated, complex with the protein [44].
In our work, we measured the amide proton transverse relaxation rates, 1HN-R2, on Ub in isolation, and in the presence of UBA2 at 1:0.5 and 1:1 molar ratios. To determine solvent PRE for 1HN of Ub, we repeated the same set of measurements in the presence of gadodiamide. A relaxation agent concentration of 2 mM was chosen to allow measurement of sufficiently large 1HN-R2 without excessively compromising the quality of signals for samples containing 200 g/L of Ficoll (where resonances were already broadened compared to dilute solution conditions). Transverse 1HN PRE rates, 1HN-R2p, were calculated as the difference in 1HN-R2 between samples with and without gadodiamide.
The values of 1HN-R2p determined for Ub display a significant variability along the protein sequence (Fig. 3, main panel), in qualitative agreement with the solvent accessibility along the Ub backbone (Fig. 3, top panel). In particular, residues 8–14, located in or adjacent to the β1-β2 loop, residues 45–49 forming the β3-β4 loop, and the C-terminal tail showed the largest 1HN-R2p values, while residues 20–30 displayed the smallest PRE effects. We then examined the PRE trends upon subsequent additions of UBA2. In Ficoll solution, at Ub:UBA2 1:0.5 molar ratio, 1HN-R2p values increased for all of the residues (Fig. 4A,B, red plots). As explained by Johansson et al. for their system [34], the observed increase in PRE with increasing total protein concentration (that of UBA2 in the present case) can be attributed to a reduced rate of diffusion of the relaxation agent along the protein surface, which in turn is caused by increased molecular crowding associated with more frequent non-specific transient collisions. However, at a higher Ub:UBA2 molar ratio (1:1), the 1HN-R2p values were found to consistently increase for a subset of residues (Fig. 4A, red plots), while another set of residues displayed no increase or an increase smaller than that observed between 1:0 and 1:0.5 Ub:UBA2 molar ratios (Fig. 4B, red plots). The non-linear trend of 1HN-R2p values (which we refer to as a ‘roof’ pattern) can be explained by the progressive formation of long-lived specific protein-protein associations that reduce access of gadodiamide to residues at or near the binding interface. To quantify this effect, we calculated the deviation of 1HN-R2p values from a linear concentration dependence (Δ), as described previously [34]. We excluded from this analysis all of the residues that in free Ub displayed a 1HN-R2p value smaller than 8 s−1 (Fig. S3), indicative of a buried position. Δ values were considered significant if the parameter exceeded twice its uncertainty (i.e. Δ/σΔ > 2, [34]).
Figure 3. Solvent PRE on Ub.
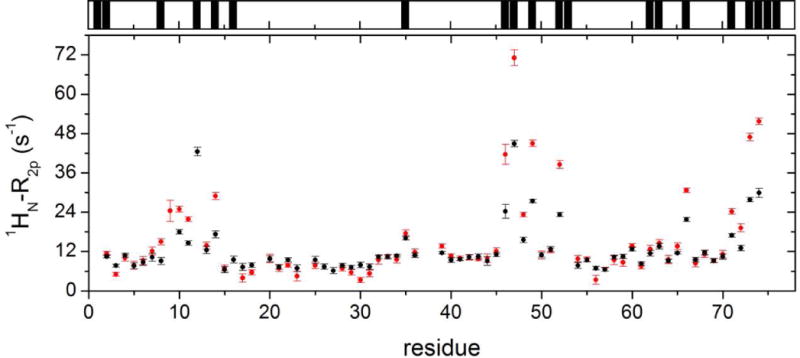
Main panel: 1HN-R2p rates of Ub in phosphate buffer solution without (black) and with 200 g/L Ficoll (red). Top panel: black bars indicate Ub residues whose backbone nitrogen atoms display significant (> 0.3 Å2) solvent accessible surface area (calculations performed using the software GetArea [61] with the protein structure, PDB: 1ubq [62]).
Figure 4. Solvent PRE on Ub upon UBA2 addition.
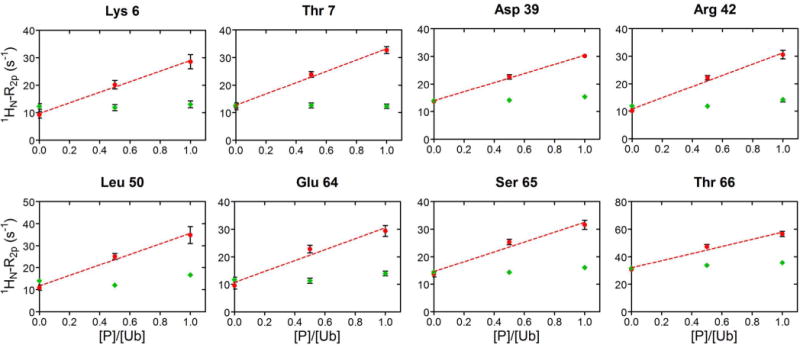
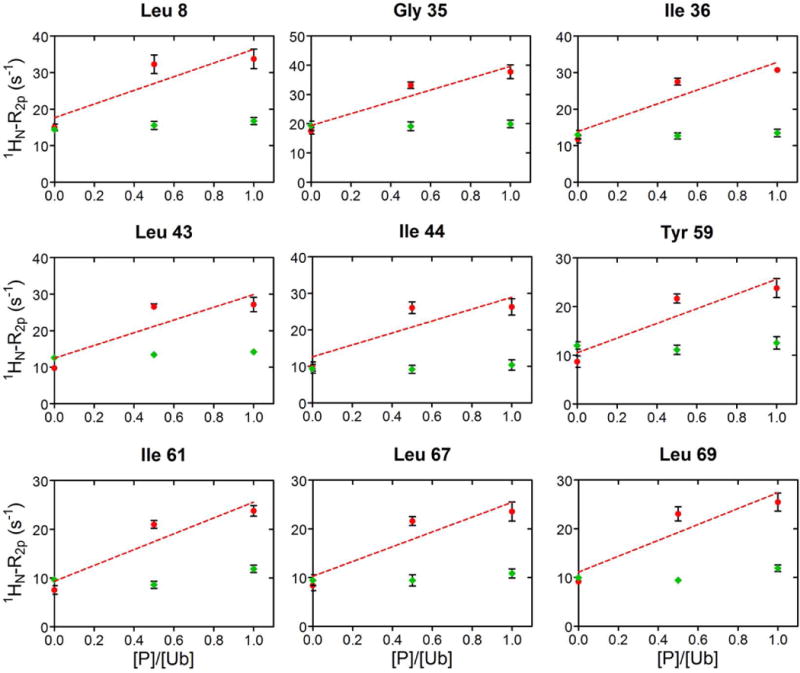
A) In red, 1HN-R2p rates of a representative group of Ub residues, that increase linearly by addition of UBA2 to Ub. The dashed line corresponds to the least-squares linear fit over three experimental points. As control, 1HN-R2p rates measured upon addition of GB1 to Ub, are reported in green. Rates are plotted versus the protein/Ub molar ratio. B) 1HN-R2p rates of representative Ub residues, whose value at 1:0.5 was above the linear trace connecting points 1:0 and 1:1, are shown in red. The dashed line corresponds to the least-squares linear fit over three experimental points. 1HN-R2p rates measured upon addition of GB1 to Ub are shown in green. Rates are plotted versus the protein/Ub molar ratio.
As a control, we repeated the experiment using protein GB1 (not a partner of Ub) in place of UBA2 (Fig. 4A,B, green plots). In this case, we observed modest variations of Ub’s 1HN-R2p values, attributable to less frequent protein-protein collisions at the surface of Ub, when in the presence of a non-interacting protein.
Residues displaying a significant Δ value were: 2, 4, 8, 10, 11, 14, 20, 32–36, 40, 41, 43–46, 51, 54, 55, 58–60, 63, 67–69. These residues were mapped onto the Ub structure with distinct colors representing different surface patches (Fig. 5A,B). Notably, residues displaying the largest Δ values (> 5 s−1) were residues 8, 14, 35, 36, 43, 44, 46, 59, 63, 67, 69, which include those forming the Ile44 patch. Therefore, this analysis revealed additional specific contact surfaces containing residues Phe4, Ile36, and Asp58, which were identified in alternative binding interfaces used by Ub to recognize different binding partners [11,45,46]. Some of the identified contact residues in the secondary patches (e.g. 32–36) also show a small CSP (Fig. 2C, 5C). Others, occupying positions 58–60 in a region between strands β4 and β5, do not exhibit significant CSP (Fig. 2C, 5C), probably due to the small population of the corresponding complex.
Figure 5. Mapping of interaction surfaces on Ub.
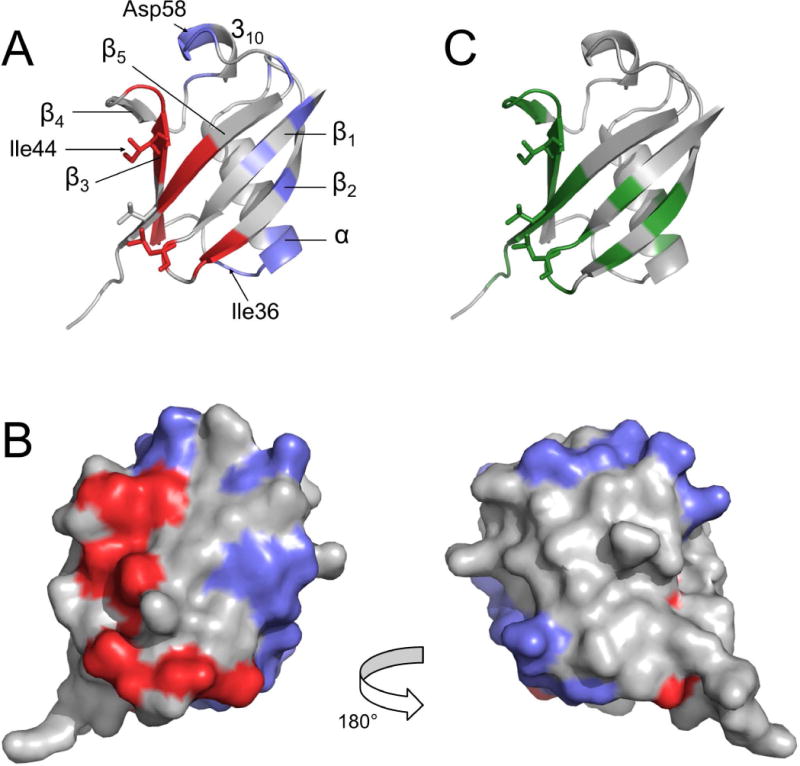
Surface residues (1HN-R2p(0) > 8 s−1) displaying Δ/σΔ > 2 are mapped onto the Ub structure (PDB: 1ubq [62]), represented with a ribbon (A) or surface (B) models; two groups are identified: the red area surrounding the Ile44 patch and the blue surfaces comprising the additional contact sites. C) CSP mapping: Ub residues displaying CSP > 0.1 ppm upon addition of a seven-fold molar excess of UBA2 are colored in green. All data refer to experiments performed in 200 g/L Ficoll solution. Residues Leu8, Ile44, and Val70, belonging to the canonical Ile44 patch, are represented in sticks in panels A and C.
It must be noted that the approach described above proved unsuccessful in identifying specific binding patches in the uncrowded solution. Indeed, 1HN-R2p values obtained upon addition of UBA2 in the same concentration range as explored with Ficoll, did not display the typical ‘roof’ pattern even in the case of residues belonging to the Ile44 patch (Fig. S4). Given the small size of the chosen test proteins, it is possible that Ub/UBA2 collisions and reorientations in dilute conditions are too fast to be detectable by our PRE method.
DISCUSSION
Ubiquitin function is determined by covalent modification of protein substrates as well as by non-covalent recognition of partner protein surfaces. Owing to its small size, a large part of Ub’s amino acid residues are exposed to the exterior and are potentially available for biomolecular recognition. Among these, a prominent role is played by residues forming the Ile44 patch, which centralizes most of the interactions with Ub binding domains [4,11]. Remarkably, this region is able to attract UBDs displaying highly divergent structural traits, such as single helices, multiple helices, and even β sheets [14]. The interactive capacity and binding versatility of the Ile44 patch is ensured by a finely tuned combination of rigidity and plasticity [13,47]. In contrast to the high level of detail obtained in the description of protein binding at the Ile44 region, the role of the remainder of Ub’s surface in protein-protein association events remains less well characterized.
Monomeric Ub and individual Ub moieties in polyUb chains offer limited contact size for binding, and most Ub-UBD interactions are characterized by low affinity in vitro. Weak transient protein-protein interactions can be influenced by the heterogeneous and crowded intracellular environment to a larger extent than high-affinity associations [48], however investigations of Ub-UBD binding in cell-mimicking media are lacking. In our work, we aimed at assessing the effect of macromolecular crowding on the interaction between Ub and a representative UBD, UBA2, that was previously shown to target the canonical Ile44 patch on Ub [26,41]. Because of the multiple possible perturbations originating from macromolecular crowders, we focused our study on a hydrophilic sucrose polymer, Ficoll 70, eliciting viscosity and excluded-volume effects [35], but not establishing chemical interactions with the test proteins.
Ub experienced minimal perturbations in the presence of up to 200 g/L Ficoll (a concentration close to the total macromolecular concentration in the average eukaryotic cytoplasm), except for viscosity-dependent hydrodynamic properties. Similarly, the overlapping features of site-resolved NMR spectral fingerprints collected with saturating amounts of UBA2 in dilute and in crowded solutions indicated that the structure of the Ub/UBA2 complex was retained. Thus, the Ile44 region mediates specific Ub/UBA2 recognition in buffer as well as in crowded solution. The most prominent crowding-induced perturbation concerns the apparent binding affinity, which was found increased with respect to uncrowded solution. By comparing the results obtained in polymeric (Ficoll) and non-polymeric (sucrose) crowder solutions, we found that the macromolecular nature of the cosolute was responsible for the increased complex stability. Protein-protein associations are predicted to be favored under macromolecular crowding conditions according to excluded volume and depletion force theories [49–53]. Depletion forces stabilize equilibrium-state and transition-state complexes, enhancing binding affinity and association rates [48,51]. Macromolecular crowding may also decrease dissociation rates by increasing rebinding probability from the encounter complex [48]. On the other hand, diffusion and collision between associating molecules are slower under macromolecular crowding (the viscosity of a Ficoll solution is higher than that of a sucrose solution at equal mass concentration, see Materials and Methods, and Fig. 1, S2) resulting in decreased association rate if the reaction is diffusion-limited, or unperturbed on-rate if it is under transition-state control [48]. Although the precise influence of viscosity on formation of the Ub/UBA2 complex remains unknown, it appears that depletion attraction is a major contributor to the observed increase of binding affinity in Ficoll solution.
In addition to the identification of the primary recognition interface, we explored the possibility to detect UBA2 binding to alternative surface patches. Indeed, transient complexes play an important role in macromolecular associations, and repositioning of binding partners after preliminary collisions efficiently leads to formation of longer-lived assemblies [54]. Furthermore, it is expected that alternative encounters are particularly evident in low-affinity protein-protein complexes [55], such as Ub-UBD, due to the high concentration of UBD necessary to obtain saturation of the primary binding site. Paramagnetic relaxation enhancement approaches have established capability to detect ultra-weak interactions [42,43,56]. Here, we adopted the solvent PRE method in place of covalent conjugation by paramagnetic tags in order to avoid potential artifacts due to chemical modification of a small-sized protein. From the systematic analysis of 1H PREs of Ub at increasing concentrations of UBA2 in Ficoll solution, a differential behaviour of surface residues was observed. Specific interactions, revealed by a non-linear increase in PRE attributed to the exclusion of the paramagnetic molecular probe from the protein-protein interface, were detected for residues of the primary binding site at the Ile44 patch and for residues previously identified in non-canonical binding surfaces on Ub [11,14,45,46]. For example, a ‘polar’ surface centered on Asp58, involving Ub residues 51, 54, 55, and 57–60, was recognized as a novel interaction interface in Ub-Rabex5 complex in addition to the canonical Ile44 hydrophobic area [57,58]. Also, in Ubch5b-Ub-NEDD4L complex, Ub contacts the E3 partner via the Ile36 patch (Ile36-Leu71-Leu73) and other residues including Gly35, Gln40 and Leu69 [59]. Binding of linear ubiquitin chains by NEMO involves distinct patches on the Ub surface: while the distal Ub binds via its C-terminal tail and the Ile44 area, the proximal Ub employs residues Gln2, Phe4, Lys6, Gly10, Thr12, Ile13, Thr14, Glu16, Glu64, and Thr66 [60].
Thus, our data suggest that, in equilibrium conditions, low-populated specific UBA2/Ub associations coexist with the main stereospecific complex. It remains to be established whether these interactions are unintended (noise) or productive (part of the signal, i.e. functional encounter complexes), and whether they are specific of the investigated UBA2 or paradigmatic for all UBDs. By investigation of further (larger) protein pairs it should become possible to determine if macromolecular crowding affects the relative free energies of primary and secondary interactions, a notion that could be exploited to select among alternative bound conformations.
CONCLUSIONS
Weak protein-protein interactions, such as those between Ub and UBD, are potentially prone to be influenced by macromolecular crowding, a distinctive feature of the intracellular environment. In our work we found that high concentrations of a crowding agent did not influence the preferential binding of UBA2 to the canonical Ile44 patch of Ub. On the other hand, from a more comprehensive exploration of Ub/UBA2 contacts based on a solvent PRE approach, secondary contact surfaces were detected. The regions were classified as specific based on the non-linear PRE trend observed at varying UBA2 concentration, although the absence of strong concomitant CSP hinted at ultra-weak affinity of the corresponding interactions and/or a significant heterogeneity of the ensemble of complex conformations. Thus, Ub/UBA2 complexes populate high energy local minima of the free energy landscape which may be in equilibrium with the low energy minimum corresponding to the stereospecific complex. Alternatively, the identified patches may mediate formation of futile complexes that are in competition with the main conformation. It is probably not accidental that other UBD target these regions, which can thus be considered pivotal for biomolecular recognition. In the broader context, identification of weak secondary interaction surfaces in cell-mimicking crowded solutions by use of PRE methods could improve our understanding of dynamic protein-protein interaction networks and, ultimately, of the molecular-level structural organization of the intracellular milieu.
Supplementary Material
Acknowledgments
This work was supported by the Italian Ministry for Education and Research via departmental funding. We thank “Centro Piattaforme Tecnologiche” of the University of Verona for access to NMR instrumentation. SZ and FM received a fellowship grant (Assegno di Ricerca) from the Department of Biotechnology. DF was supported by NIH grant GM065334.
Abbreviations
- CSP
chemical shift perturbation
- PRE
paramagnetic relaxation enhancement
- Ub
ubiquitin
- UBA
ubiquitin associated
- UBD
ubiquitin binding domain
Footnotes
AUTHOR CONTRIBUTION
MA conceived and supervised the study; AB prepared protein samples; AB and FM performed experiments and analyzed data; MD and SZ analyzed data; DF critically revised the manuscript; MA and FM wrote the manuscript.
References
- 1.Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta. 2004;1695:55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 2.Komander D, Rape M. The Ubiquitin Code. Annu Rev Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 3.Hicke L. Protein regulation by monoubiquitin. Nat Rev Mol Cell Biol. 2001;2:195–201. doi: 10.1038/35056583. [DOI] [PubMed] [Google Scholar]
- 4.Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol. 2004;8:610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 5.Alfano C, Faggiano S, Pastore A. The Ball and Chain of Polyubiquitin Structures. Trends Biochem Sci. 2016;41:371–385. doi: 10.1016/j.tibs.2016.01.006. [DOI] [PubMed] [Google Scholar]
- 6.Hicke L, Schubert HL, Hill CP. Ubiquitin-binding domains. Nat Rev Mol Cell Biol. 2005;6:610–621. doi: 10.1038/nrm1701. [DOI] [PubMed] [Google Scholar]
- 7.Dikic I, Wakatsuki S, Walters KJ. Ubiquitin-binding domains — from structures to functions. Nat Rev Mol Cell Biol. 2009;10:659–671. doi: 10.1038/nrm2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Varadan R, Assfalg M, Raasi S, Pickart C, Fushman D. Structural determinants for selective recognition of a Lys48-linked polyubiquitin chain by a UBA domain. Mol Cell. 2005;18:687–698. doi: 10.1016/j.molcel.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 9.Sims JJ, Haririnia A, Dickinson BC, Fushman D, Cohen RE. Avid interactions underlie the Lys63-linked polyubiquitin binding specificities observed for UBA domains. Nat Struct Mol Biol. 2009;16:883–889. doi: 10.1038/nsmb.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fushman D, Wilkinson K. Structure and recognition of polyubiquitin chains of different lengths and linkage. F1000 Biol Rep. 2011;3 doi: 10.3410/B3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Husnjak K, Dikic I. Ubiquitin-Binding Proteins: Decoders of Ubiquitin-Mediated Cellular Functions. Annu Rev Biochem. 2012;81:291–322. doi: 10.1146/annurev-biochem-051810-094654. [DOI] [PubMed] [Google Scholar]
- 12.Beal R, Deveraux Q, Xia G, Rechsteiner M, Pickart C. Surface hydrophobic residues of multiubiquitin chains essential for proteolytic targeting. Proc Natl Acad Sci U S A. 1996;93:861–866. doi: 10.1073/pnas.93.2.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lange OF, Lakomek N-A, Fares C, Schroder GF, Walter KFA, Becker S, Meiler J, Grubmuller H, Griesinger C, de Groot BL. Recognition Dynamics Up to Microseconds Revealed from an RDC-Derived Ubiquitin Ensemble in Solution. Science. 2008;320:1471–1475. doi: 10.1126/science.1157092. [DOI] [PubMed] [Google Scholar]
- 14.Winget JM, Mayor T. The Diversity of Ubiquitin Recognition: Hot Spots and Varied Specificity. Mol Cell. 2010;38:627–635. doi: 10.1016/j.molcel.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 15.Ellis RJ, Minton AP. Cell biology: join the crowd. Nature. 2003;425:27–28. doi: 10.1038/425027a. [DOI] [PubMed] [Google Scholar]
- 16.Gierasch LM, Gershenson A. Post-reductionist protein science, or putting Humpty Dumpty back together again. Nat Chem Biol. 2009;5:774–777. doi: 10.1038/nchembio.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou H-X. Influence of crowded cellular environments on protein folding, binding, and oligomerization: Biological consequences and potentials of atomistic modeling. FEBS Lett. 2013;587:1053–1061. doi: 10.1016/j.febslet.2013.01.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Theillet F-X, Binolfi A, Frembgen-Kesner T, Hingorani K, Sarkar M, Kyne C, Li C, Crowley PB, Gierasch L, Pielak GJ, Elcock AH, Gershenson A, Selenko P. Physicochemical Properties of Cells and Their Effects on Intrinsically Disordered Proteins (IDPs) Chem Rev. 2014;114:6661–6714. doi: 10.1021/cr400695p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Minton AP. Quantitative assessment of the relative contributions of steric repulsion and chemical interactions to macromolecular crowding. Biopolymers. 2013;99:239–244. doi: 10.1002/bip.22163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miklos AC, Li C, Sharaf NG, Pielak GJ. Volume exclusion and soft interaction effects on protein stability under crowded conditions. Biochemistry. 2010;49:6984–6991. doi: 10.1021/bi100727y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuznetsova I, Turoverov K, Uversky V. What Macromolecular Crowding Can Do to a Protein. Int J Mol Sci. 2014;15:23090–23140. doi: 10.3390/ijms151223090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crowley PB, Brett K, Muldoon J. NMR Spectroscopy Reveals Cytochromec–Poly(ethylene glycol) Interactions. ChemBioChem. 2008;9:685–688. doi: 10.1002/cbic.200700603. [DOI] [PubMed] [Google Scholar]
- 23.Ceccon A, Busato M, Pérez Santero S, D’Onofrio M, Musiani F, Giorgetti A, Assfalg M. Transient Interactions of a Cytosolic Protein with Macromolecular and Vesicular Cosolutes: Unspecific and Specific Effects. ChemBioChem. 2015;16:2633–2645. doi: 10.1002/cbic.201500451. [DOI] [PubMed] [Google Scholar]
- 24.Pérez Santero S, Favretto F, Zanzoni S, Chignola R, Assfalg M, D’Onofrio M. Effects of macromolecular crowding on a small lipid binding protein probed at the single-amino acid level. Arch Biochem Biophys. 2016;606:99–110. doi: 10.1016/j.abb.2016.07.017. [DOI] [PubMed] [Google Scholar]
- 25.Ryu K-S, Lee K-J, Bae S-H, Kim B-K, Kim K-A, Choi B-S. Binding surface mapping of intra- and interdomain interactions among hHR23B, ubiquitin, and polyubiquitin binding site 2 of S5a. J Biol Chem. 2003;278:36621–36627. doi: 10.1074/jbc.M304628200. [DOI] [PubMed] [Google Scholar]
- 26.Varadan R, Assfalg M, Haririnia A, Raasi S, Pickart C, Fushman D. Solution conformation of Lys63-linked di-ubiquitin chain provides clues to functional diversity of polyubiquitin signaling. J Biol Chem. 2004;279:7055–7063. doi: 10.1074/jbc.M309184200. [DOI] [PubMed] [Google Scholar]
- 27.Mueller TD, Kamionka M, Feigon J. Specificity of the interaction between ubiquitin-associated domains and ubiquitin. J Biol Chem. 2004;279:11926–11936. doi: 10.1074/jbc.M312865200. [DOI] [PubMed] [Google Scholar]
- 28.Clore GM, Iwahara J. Theory, Practice, and Applications of Paramagnetic Relaxation Enhancement for the Characterization of Transient Low-Population States of Biological Macromolecules and Their Complexes. Chem Rev. 2009;109:4108–4139. doi: 10.1021/cr900033p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Göbl C, Madl T, Simon B, Sattler M. NMR approaches for structural analysis of multidomain proteins and complexes in solution. Prog Nucl Magn Reson Spectrosc. 2014;80:26–63. doi: 10.1016/j.pnmrs.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 30.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 31.Iwahara J, Tang C, Marius Clore G. Practical aspects of 1H transverse paramagnetic relaxation enhancement measurements on macromolecules. J Magn Reson. 2007;184:185–195. doi: 10.1016/j.jmr.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Groen J, Foschepoth D, te Brinke E, Boersma AJ, Imamura H, Rivas G, Heus HA, Huck WTS. Associative Interactions in Crowded Solutions of Biopolymers Counteract Depletion Effects. J Am Chem Soc. 2015;137:13041–13048. doi: 10.1021/jacs.5b07898. [DOI] [PubMed] [Google Scholar]
- 33.Walker O, Varadan R, Fushman D. Efficient and accurate determination of the overall rotational diffusion tensor of a molecule from (15)N relaxation data using computer program ROTDIF. J Magn Reson. 2004;168:336–345. doi: 10.1016/j.jmr.2004.03.019. [DOI] [PubMed] [Google Scholar]
- 34.Johansson H, Jensen MR, Gesmar H, Meier S, Vinther JM, Keeler C, Hodsdon ME, Led JJ. Specific and Nonspecific Interactions in Ultraweak Protein–Protein Associations Revealed by Solvent Paramagnetic Relaxation Enhancements. J Am Chem Soc. 2014;136:10277–10286. doi: 10.1021/ja503546j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou H-X, Rivas G, Minton AP. Macromolecular Crowding and Confinement: Biochemical, Biophysical, and Potential Physiological Consequences. Annu Rev Biophys. 2008;37:375–397. doi: 10.1146/annurev.biophys.37.032807.125817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benton LA, Smith AE, Young GB, Pielak GJ. Unexpected Effects of Macromolecular Crowding on Protein Stability. Biochemistry. 2012;51:9773–9775. doi: 10.1021/bi300909q. [DOI] [PubMed] [Google Scholar]
- 37.Abriata LA, Spiga E, Peraro MD. Molecular Effects of Concentrated Solutes on Protein Hydration, Dynamics, and Electrostatics. Biophys J. 2016;111:743–755. doi: 10.1016/j.bpj.2016.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cino EA, Karttunen M, Choy W-Y. Effects of Molecular Crowding on the Dynamics of Intrinsically Disordered Proteins. PLoS ONE. 2012;7:e49876. doi: 10.1371/journal.pone.0049876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fushman D, Cahill S, Cowburn D. The main-chain dynamics of the dynamin pleckstrin homology (PH) domain in solution: analysis of 15N relaxation with monomer/dimer equilibration. J Mol Biol. 1997;266:173–194. doi: 10.1006/jmbi.1996.0771. [DOI] [PubMed] [Google Scholar]
- 40.Camacho-Zarco AR, Munari F, Wegstroth M, Liu W-M, Ubbink M, Becker S, Zweckstetter M. Multiple paramagnetic effects through a tagged reporter protein. Angew Chem Int Ed Engl. 2015;54:336–339. doi: 10.1002/anie.201408615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Varadan R, Assfalg M, Fushman D. Using NMR spectroscopy to monitor ubiquitin chain conformation and interactions with ubiquitin-binding domains. Methods Enzymol. 2005;399:177–192. doi: 10.1016/S0076-6879(05)99012-5. [DOI] [PubMed] [Google Scholar]
- 42.Volkov AN, Ubbink M, van Nuland NAJ. Mapping the encounter state of a transient protein complex by PRE NMR spectroscopy. J Biomol NMR. 2010;48:225–236. doi: 10.1007/s10858-010-9452-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clore GM, Tang C, Iwahara J. Elucidating transient macromolecular interactions using paramagnetic relaxation enhancement. Curr Opin Struct Biol. 2007;17:603–616. doi: 10.1016/j.sbi.2007.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pintacuda G, Otting G. Identification of protein surfaces by NMR measurements with a pramagnetic Gd(III) chelate. J Am Chem Soc. 2002;124:372–373. doi: 10.1021/ja016985h. [DOI] [PubMed] [Google Scholar]
- 45.Searle MS, Garner TP, Strachan J, Long J, Adlington J, Cavey JR, Shaw B, Layfield R. Structural insights into specificity and diversity in mechanisms of ubiquitin recognition by ubiquitin-binding domains. Biochem Soc Trans. 2012;40:404–408. doi: 10.1042/BST20110729. [DOI] [PubMed] [Google Scholar]
- 46.Kulathu Y, Komander D. Atypical ubiquitylation — the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat Rev Mol Cell Biol. 2012;13:508–523. doi: 10.1038/nrm3394. [DOI] [PubMed] [Google Scholar]
- 47.Fenwick RB, Esteban-Martín S, Richter B, Lee D, Walter KFA, Milovanovic D, Becker S, Lakomek NA, Griesinger C, Salvatella X. Weak Long-Range Correlated Motions in a Surface Patch of Ubiquitin Involved in Molecular Recognition. J Am Chem Soc. 2011;133:10336–10339. doi: 10.1021/ja200461n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Phillip Y, Sherman E, Haran G, Schreiber G. Common crowding agents have only a small effect on protein-protein interactions. Biophys J. 2009;97:875–885. doi: 10.1016/j.bpj.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Asakura S, Oosawa F. Interaction between particles suspended in solutions of macromolecules. J Polym Sci. 1958;33:183–192. [Google Scholar]
- 50.Minton AP. Molecular crowding: analysis of effects of high concentrations of inert cosolutes on biochemical equilibria and rates in terms of volume exclusion. Methods Enzymol. 1998;295:127–149. doi: 10.1016/s0076-6879(98)95038-8. [DOI] [PubMed] [Google Scholar]
- 51.Minton AP. The effect of volume occupancy upon the thermodynamic activity of proteins: some biochemical consequences. Mol Cell Biochem. 1983;55:119–140. doi: 10.1007/BF00673707. [DOI] [PubMed] [Google Scholar]
- 52.Phillip Y, Schreiber G. Formation of protein complexes in crowded environments - From in vitro to in vivo. FEBS Lett. 2013;587:1046–1052. doi: 10.1016/j.febslet.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marenduzzo D, Finan K, Cook PR. The depletion attraction: an underappreciated force driving cellular organization. J Cell Biol. 2006;175:681–686. doi: 10.1083/jcb.200609066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tang C, Iwahara J, Clore GM. Visualization of transient encounter complexes in protein-protein association. Nature. 2006;444:383–386. doi: 10.1038/nature05201. [DOI] [PubMed] [Google Scholar]
- 55.Blundell TL, Fernández-Recio J. Cell biology: brief encounters bolster contacts. Nature. 2006;444:279–280. doi: 10.1038/nature05306. [DOI] [PubMed] [Google Scholar]
- 56.Liu Z, Zhang W-P, Xing Q, Ren X, Liu M, Tang C. Noncovalent Dimerization of Ubiquitin. Angew Chem Int Ed. 2012;51:469–472. doi: 10.1002/anie.201106190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Penengo L, Mapelli M, Murachelli AG, Confalonieri S, Magri L, Musacchio A, Di Fiore PP, Polo S, Schneider TR. Crystal structure of the ubiquitin binding domains of rabex-5 reveals two modes of interaction with ubiquitin. Cell. 2006;124:1183–1195. doi: 10.1016/j.cell.2006.02.020. [DOI] [PubMed] [Google Scholar]
- 58.Lee S, Tsai YC, Mattera R, Smith WJ, Kostelansky MS, Weissman AM, Bonifacino JS, Hurley JH. Structural basis for ubiquitin recognition and autoubiquitination by Rabex-5. Nat Struct Mol Biol. 2006;13:264–271. doi: 10.1038/nsmb1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kamadurai HB, Souphron J, Scott DC, Duda DM, Miller DJ, Stringer D, Piper RC, Schulman BA. Insights into ubiquitin transfer cascades from a structure of a UbcH5B approximately ubiquitin-HECT(NEDD4L) complex. Mol Cell. 2009;36:1095–1102. doi: 10.1016/j.molcel.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rahighi S, Ikeda F, Kawasaki M, Akutsu M, Suzuki N, Kato R, Kensche T, Uejima T, Bloor S, Komander D, Randow F, Wakatsuki S, Dikic I. Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell. 2009;136:1098–1109. doi: 10.1016/j.cell.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 61.Fraczkiewicz R, Braun W. Exact and efficient analytical calculation of the accessible surface areas and their gradients for macromolecules. J Comput Chem. 1998;19:319–333. [Google Scholar]
- 62.Vijay-Kumar S, Bugg CE, Cook WJ. Structure of ubiquitin refined at 1.8 A resolution. J Mol Biol. 1987;194:531–544. doi: 10.1016/0022-2836(87)90679-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


