Abstract
Background
Therapy-related myeloid neoplasms (t-MNs) are often fatal secondary malignancies. Risk factors for t-MNs are not well understood. Recent studies suggested that individuals with clonal hematopoiesis have higher risk of developing hematological malignancies. We hypothesized that cancer patients with clonal hematopoiesis have increased risk of developing t-MNs.
Methods
We conducted a retrospective case-control study to compare the prevalence of clonal hematopoiesis between patients who developed t-MNs (cases) and who did not develop t-MNs (control). For cases, we studied14 patients with various types of cancers who developed t-MNs and whose paired samples of t-MN bone marrow (BM) and peripheral blood (PB) that were previously obtained at the time of primary cancer diagnosis were available. Fifty four patients with lymphoma who received combination chemotherapy and did not develop t-MNs after at least 5 years of follow up were studied as a control. We performed molecular barcode sequencing of 32 genes on the pre-treatment PB samples to detect clonal hematopoiesis. For the t-MN cases, we also performed targeted gene sequencing on t-MN BM samples and investigated clonal evolution from clonal hematopoiesis to t-MNs. To confirm association between clonal hematopoiesis and t-MN development, we also analyzed prevalence of clonal hematopoiesis in a separate cohort of 74 patients with lymphoma. All of these patients were treated under the prospective randomized trial of frontline chemotherapy with cyclophosphamide, doxorubicin, vincristine, and prednisone (CHOP) with or without melatonin and 5 (7%) of them had developed t-MNs.
Findings
In 14 patients with t-MNs, we detected pre-leukemic mutations in 10 of their prior PB samples (71%). In control, clonal hematopoiesis was detected in 17 patients (31%), and the cumulative incidence of t-MNs at 5 years was significantly higher in patients with clonal hematopoiesis (30% [95% CI: 16% – 51%] vs. 7% [95% CI: 2% – 21%], P = 0.016). In the separate cohort, 5 patients (7%) developed t-MNs and 4 (80%) of them had clonal hematopoiesis. The cumulative incidence of t-MNs at 10 years was significantly higher in patients with clonal hematopoiesis (29% [95% CI: 8%–53%] vs. 0% [95% CI: 0%–0%], P = 0.0009). Multivariate Fine and Gray model showed that having clonal hematopoiesis significantly increased the risk of t-MN development (HR = 13.7, P = 0.013).
Interpretation
Pre-leukemic clonal hematopoiesis is frequently detected in patients with t-MNs at the time of their primary cancer diagnosis and before patients were exposed to chemotherapy/radiation therapy. Detection of clonal hematopoiesis significantly increased the risk of t-MN development in patients with lymphoma. These data suggest potential approaches of screening clonal hematopoiesis in cancer patients to identify patients at risk of t-MNs and warrants a validation in prospective trial investigating a role of clonal hematopoiesis as a predictive marker for t-MNs.
Introduction
Therapy-related myeloid neoplasms (t-MNs) are secondary malignancies that develop in patients who have received cytotoxic chemotherapy and/or ionizing radiation therapy.(1, 2) The cumulative incidence of t-MNs is approximately 1–10% of patients at risk, with an incidence that varies significantly among different cancer types and treatment regimens.(3–5) t-MNs usually develop 3–8 years after exposure to chemotherapy and/or radiation therapy, are frequently associated with poor prognostic features, such as complex cytogenetics and TP53 mutations, and respond poorly to conventional chemotherapies.(6) Patients with t-MNs have poor outcomes, with an estimated median overall survival of 8–10 months and a 5-year overall survival rate of 10–20%.(6–9)
Exposure to certain types of chemotherapy is a known treatment-related risk factor for t-MNs. For example, t-MNs occur more frequently in patients who receive alkylating agents and topoisomerase II inhibitors than in patients who receive antimetabolites or taxanes.(4, 10, 11) Use of a granulocyte colony-stimulating factor (G-CSF) in cancer patients is associated with the risk of t-MNs.(12, 13) High-dose chemotherapy followed by autologous stem cell transplant (auto-SCT) has also been shown to increase the risk of t-MNs in lymphoma patients.(14) Furthermore, there is a dose-dependent relationship between the risk of t-MNs and cumulative dose of platinum exposure in patients with ovarian cancer.(15) In contrast, little is known about patient-related risk factors for t-MN susceptibility. Older age has been shown to increase the risk of t-MNs, and although there have been several reports of germline polymorphisms associated with risk, none have been validated.(6, 14, 16–21) In patients with lymphoma who underwent auto-SCT, gene expression signature of 38 genes in pre-SCT samples or accelerated shortening of telomere length in post-SCT myeloid cells were shown to be associated with t-MNs.(22, 23) Despite these efforts, currently, there is no predictive biomarker or risk-stratified approach for early detection or prevention of t-MNs.
Recent studies have reported that pre-leukemic mutations, such as mutations in DNMT3A, TET2, and ASXL1, can be detected in peripheral blood (PB) samples from healthy individuals, a phenomenon referred to as clonal hematopoiesis of indeterminate potential (CHIP).(24–28) Compared to individuals without CHIP, those with CHIP were found to have an increased risk of developing hematological neoplasms. CHIP was also identified in approximately 2% of patients with solid tumors analyzed as part of The Cancer Genome Atlas (TCGA).(29) Furthermore, pre-leukemic TP53 mutations were detectable in peripheral blood (PB) samples that were obtained years before patients developed t-MNs.(30)
These data collectively suggest that t-MNs arise from antecedent clonal hematopoiesis and detection of clonal hematopoiesis at the time of cancer diagnosis could aid in identifying cancer patients at increased risk of developing subsequent t-MNs. We addressed these hypotheses by first studying patients with t-MNs with paired samples of diagnostic BM at the time of t-MN diagnosis and PB that were previously obtained at the time of primary cancer diagnosis. We then compared the prevalence of clonal hematopoiesis between patients who did and did not develop t-MNs and confirmed the association between clonal hematopoiesis and t-MN risk in a separate cohort.
Methods
Study design and participants
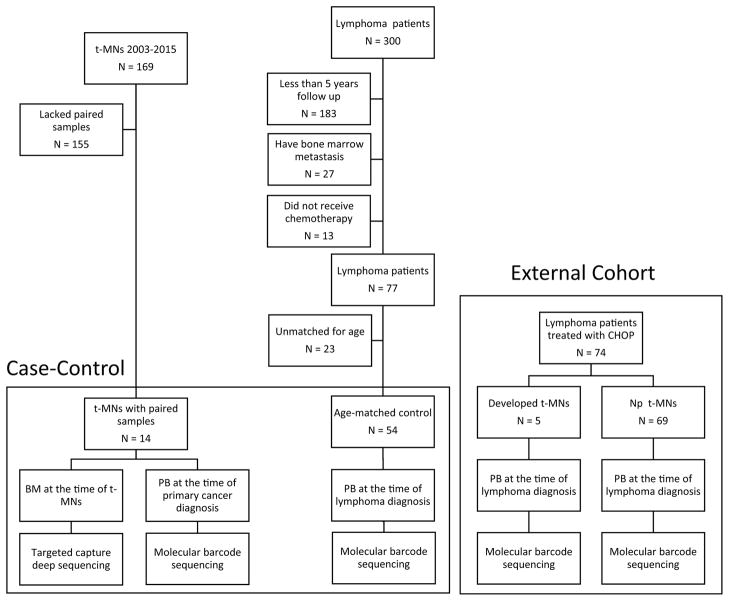
We designed a case-control study to compare prevalence of clonal hematopoiesis between patients who developed t-MNs (cases) and who did not develop t-MNs (control). The flow chart summarizes the study design (Figure 1). For cases, we searched our clinical database for the patients with diagnosis of t-MNs between 2003 and 2015 and identified 169 patients with t-MNs. Of those, 14 patients were found to have BM samples obtained at the time of t-MN diagnosis and PB samples that were previously obtained at the time of primary cancer diagnosis and before they were exposed to chemotherapy and/or radiation therapy. Other 155 t-MN patients did not have prior PB samples available, therefore were not eligible for analysis. The clinical history of these 14 patients is described in the Supplemental Appendix (page 2–3). To create a control group, we searched 300 patients with lymphoma whose pre-treatment PB samples were available for analysis. We first selected the patients who met the following criteria: 1) received combination chemotherapy regimen including alkylating agent, 2) had at least 5 years of follow up and no clinical evidence of t-MN development, and 3) no bone marrow metastasis of lymphoma by bilateral bone marrow biopsy. Seventy seven of 300 patients (26%) met these criteria. After matching the age with cases in 1:3 or grater ratio, 54 patients were matched as a control. We chose lymphoma patients as a control because they are reported to have the highest risk of developing t-MNs(11), they almost always receive alkylating agents with or without topoisomerase II inhibitor containing regimens, and they occasionally undergo auto-SCT, thus have sufficient exposures to treatment-related risk factors. Next, to confirm the association between clonal hematopoiesis and t-MN risk, as a separate cohort, we studied 74 patients with lymphoma who were treated with a frontline randomized trial of cyclophosphamide, doxorubicin, vincristine, and prednisone (CHOP) with or without melatonin (hereafter called external cohort).(31) Pre-treatment PB samples from these patients were available for analysis under an ongoing tissue banking protocol at The University of Texas MD Anderson Cancer Center. Written informed consent for sample collection and analysis was obtained from all patients. All study protocols adhered to the Declaration of Helsinki and were approved by the Institutional Review Board at MD Anderson (PA15-0400).
Figure 1.
Flow chart summarizing the sample selection process.
Procedures
Targeted gene sequencing of t-MN BM samples
We used a SureSelect custom panel of 295 genes (Agilent Technologies, Santa Clara, CA) that are recurrently mutated in hematologic malignancies (Supplemental Appendix, page 5). Full details of sequencing and bioinformatics analyses to detect high-confidence driver mutations are described in the Supplemental Appendix (page 4).
Molecular barcode sequencing of PB samples and detection of clonal hematopoiesis
Because we expected low variant allele frequency (VAF) for mutations in these samples, we used Haloplex High Sensitivity (HS) technology (Agilent Technologies, Santa Clara, CA), an amplicon-based targeted deep sequencing method that incorporates more than 1 million unique molecular barcodes and allows for consensus calls of low frequency alleles. To avoid bias in calling low-VAF variants, we called variants blindly without any bias for driver mutations detected in t-MN BM or clinical outcome of the patients. We applied the same variant calling criteria in all 3 cohorts and bioinformatician who performed variant calling was blinded for clinical outcome. We targeted 32 genes that covered driver mutations detected in t-MN as well as mutations reported as clonal hematopoiesis in previous studies (Supplemental Appendix, page 6).(25, 26, 29) Full details of the sequencing and bioinformatics algorithms are described in Supplemental Appendix (page 4).
Statistical analysis
The chi-square or Fisher exact test was used to assess differences in categorical variables, and the Mann-Whitney U test was used to analyze continuous variables difference after testing normal distribution by Shapiro-Wilk test. Specifically, distribution of VAF did not follow normal distribution (P = 0.001). Cumulative incidence rate of t-MN development over time was compared by Gray test while considering non-t-MN death as a competing event. Fine and Gray proportional hazard regression model was used to evaluate association between t-MN development and other multiple variables.
Since this study was retrospective design and sample size was limited due to the availability of samples, we estimated the power of our study. We expected detection rate of clonal hematopoiesis as 70% for patients who developed t-MNs (cases) and 20% for patients who did not develop t-MNs (control). The first assumption is based on the result of 14 t-MN paired sample analysis. The second assumption is based on the previous study that analyzed TCGA’s data where it reported the prevalence of clonal hematopoiesis in general cancer population to be 2%.(29) Since our molecular barcode sequencing had higher sensitivity than whole exome sequencing used in the TCGA study, we assumed that detection rate of clonal hematopoiesis in control can be up to 20%. Based on this assumption, in case-control study of 68 patients, with 8 years of follow up, hazard function was estimated at 0.082 and 0.012 for patients with clonal hematopoiesis and for patients without, respectively, which yielded 97% power with α error of 0.05. In an external cohort of 74 patients, with 17 years of follow up, hazard function was estimated at 0.015 and 0.001 for patients with clonal hematopoiesis and for patients without, respectively, which yielded 86% power with alpha error of 0.05. All statistical analyses were performed using SPSS (version 22; IBM Corporation, Armonk NY) and R (ver. 3.1.3). The study followed the recommendation by Strengthening The Reporting of Observational Studies in Epidemiology (STROBE) Statement checklist.
Role of the funding sources
The funders of this study had no role in the study design, data collection, data analysis, data interpretation, and manuscript writing. K.T., F.W., and P.A.F. had access to the raw data. The corresponding author had full access to all of the data and the final responsibility to submit for publication.
Results
Clinical characteristics and driver mutations of 14 patients with t-MNs
We first studied 14 patients with t-MNs for whom paired BM and prior PB samples were available. The clinical characteristics of these patients are shown in Table 1. Of the 14 patients, 5 (31%) had therapy-related acute myeloid leukemia (t-AML) and 9 (69%) had therapy-related myelodysplastic syndromes (t-MDS). The median latency period from primary cancer diagnosis to t-MN diagnosis was 3 years (IQR: 2–4 years).
Table 1.
Clinical characteristics of the 14 patients with therapy-related myeloid neoplasms (t-MNs). UID indicates de-identified unique patient identification number.
| UID | Age at primary cancer | Gender | Primary Cancer | Chemotherapy | Radiation therapy | Latency to t-MN (years) | Age at t-MN | t-MN diagnosis | Cytogenetics in t-MN bone marrow |
|---|---|---|---|---|---|---|---|---|---|
| UID12766 | 55 | Male | Colon adenocarcinoma | 5-FU, Oxaliplatin | - | 4 | 59 | t-AML | Normal |
| UID984 | 63 | Male | Esophageal adenocarcinoma | 5-FU, Docetaxel | 50 Gy | 7 | 70 | t-MDS | 46~47,XY,+X,del(7)( q11.2),r(7),add(9)(q12) |
| UID10164 | 50 | Female | Malignant peripheral nerve sheath tumor | Doxorubicin, Ifosphamide | - | 1 | 51 | t-MDS | 46,XX,+1,der(1;7)(q10;p10) |
| UID6982 | 70 | Male | Small Cell Lung Cancer | Cisplatin, Etoposide | 70 Gy | 3 | 73 | t-AML | 46,XY,del(5)(q15q33) |
| UID488 | 47 | Female | NSCLC | Carboplatin, Paclitaxel, Gemcitabine, Vinorelbine | 50 Gy | 8 | 55 | t-MDS | Normal |
| UID36491 | 62 | Male | Small cell lung and NSCLC | Cisplatin, Etoposide, Carboplatin, Paclitaxel | 70 Gy | 4 | 66 | t-MDS | 42,X,-Y,del(1)(q21),del(7)(q 22q34),del(14)(q12q21),−16,−18,−21,−22,− 22, |
| UID4473 | 44 | Male | Rectal adenocarcinoma | Capecitabine | 50 Gy | 6 | 50 | t-MDS | Normal |
| UID17285 | 69 | Male | Hodgkin lymphoma | Doxorubicin, Bleomycin, Vinblastine, Dacarbazine | - | 2 | 71 | t-MDS | 45,XY,add(2)(p12),−5,−7,t(11;17)(q13;p11.2),+mar |
| UID19684 | 63 | Male | Follicular lymphoma | Rituximab, Cyclophosphamide, Doxorubicin, Vincristine, Prednisone | - | 3 | 66 | t-MDS | 46,XY,−7,+22 |
| UID7394 | 74 | Male | Penile Squamous cell cancer | Paclitaxel, Ifosphamide, Cisplatin, Capecitabine | 65 Gy | 3 | 77 | t-MDS | 46,XY,del(5)(q13q33), 46,XY,der(3;5)(q10;p10),+8 |
| UID49278 | 63 | Male | Lung adenocarcinoma | Carboplatin, pemetrexed | 66 Gy | 1 | 64 | t-AML | 45,X,-Y |
| UID12484 | 64 | Male | Mantle Cell Lymphoma | Bortezomib, Rituximab, Cyclophosphamide, Vincristine, Doxorubicin, Cytarabine, Methotrexate, Ibrutinib | - | 2 | 66 | t-AML | 45,XY,der(7;17)(p10; q10) |
| UID19304 | 25 | Female | Gliobastoma multiforme | Temozolamide | 60 Gy | 3 | 28 | t-MDS | 46,XX,inv(3)(q21q26.2) |
| UID31000** | 40 | Male | Rhabdomyos arcoma | Ifosphamide, Adriamycin | 50 Gy | 3 | 43 | t-AML | 44,XY,del(5)(q13),add(7)(q11.2),−11,−12,−17,−17,+r,+mar |
UID: Unique patient ID,
NSCLC: Non small cell lung cancer, t-MN: therapy-related myeloid neoplasms.
The case was later confirmed to have Li-Fraumeni Syndrome
Chemotherapy for primary cancers included alkylating agent-containing regimens in 10 of 14 patients (71%) and topoisomerase II inhibitor-containing regimens in 6 of 14 patients (43%). A high proportion of the patients had high-risk cytogenetic abnormalities such as del 5q/−5 in 4 of 14 patients (29%), del 7q/−7 in 6 of 14 patients (43%), or complex karyotype in 5 of 14 patients (36%).
Targeted gene sequencing of 295 genes of the t-MN BM samples (median 383x coverage, IQR: 224–584x coverage) revealed 29 driver mutations in 16 genes in 13 patients (Figure 2). We did not detect any driver point mutations in patient UID984. Consistent with the prior data, the most frequently detected driver mutation in t-MN BM was TP53 mutation in 5 of 14 patients (36%).(30) The median VAF of driver mutations detected in t-MN BM samples was 26.2% (IQR: 18–41%).
Figure 2.
Landscape of high-confidence driver mutations detected in diagnostic BM samples from patients with t-MNs. Only 13 cases are shown because UID984 did not have any detectable driver mutations. Asterix indicates double mutations in one gene.
Pre-leukemic driver mutations are detectable at the time of cancer diagnosis and before therapy
We next studied whether t-MN driver mutations could be detected in the PB samples that were previously obtained at the time of primary cancer diagnosis. PB samples obtained at the time of primary cancer diagnosis were sequenced using molecular barcode deep sequencing of 32 genes (median coverage 1,446x, IQR: 315–3,138x coverage). Among 29 driver mutations detected in 13 t-MN BM samples, 21 mutations (72%) were detectable as pre-leukemic clonal hematopoiesis in 10 patients’ prior PB samples (77%, Table 2). The median VAF of driver mutations detected in prior PB samples was 8.5% (IQR: 3.9–19.9%). Patient UID31000 had two TP53 mutations with stable VAF around 50% in both samples, and these were also detected in skin fibroblasts, which confirmed a germline origin (this case was confirmed as Li-Fraumeni Syndrome and is described elsewhere(32)). Therefore, these 2 TP53 mutations were removed from further analysis. Of note, this patient also had a somatic TET2 mutation as clonal hematopoiesis, and it later became a driver in the t-MN BM.
Table 2.
Summary of the changes of variant allele frequency (VAF) of driver mutations from the time of primary cancer diagnosis to the time of of t-MN diagnosis. Only 13 cases are shown because UID984 did not have any driver mutations detected. Eleven of 13 patients (85%) had evidence of detectable driver mutations in PB samples obtained at the time of primary cancer diagnosis. Variants that were not detected in prior PB samples are indicated as ND (not detected). For each variant detected in the prior PB samples, depth of sequencing at the given allele and binomial P value of the variant are listed.
| Patient ID | Gene | AA Change | VAF Primary Cacner (%) | VAF t-MNs (%) | Depth Primary Cancer (x) | Binomial P value |
|---|---|---|---|---|---|---|
| UID12766 | WT1 | p.S381X | ND | 14.75 | 2197 | NA |
| UID10164 | RUNX1 | p.L98fs | 3.69 | 23.27 | 1463 | 2.20E-16 |
| UID6982 | IDH2 | p.R140Q | 15.83 | 45.51 | 2502 | 2.20E-16 |
| SRSF2 | p.P95delinsRP | 13.76 | 25.32 | 872 | 2.20E-16 | |
| UID488 | DNMT3A | p.R882P | 19.93 | 33.14 | 1612 | 2.20E-16 |
| UID36491 | IDH2 | p.R172K | ND | 15.46 | 2300 | NA |
| TP53 | p.H193R | 22.31 | 73.09 | 2627 | 2.20E-16 | |
| UID4473 | TET2 | p.L1212X | 5.29 | 45.34 | 1286 | 2.20E-16 |
| UID17285 | TET2 | p.Y1255X | 8.45 | 18.06 | 568 | 2.20E-16 |
| TP53 | p.Y205C | 8.57 | 22.31 | 2008 | 2.20E-16 | |
| U2AF1 | p.Q157P | 3.92 | 11.73 | 2578 | 2.20E-16 | |
| UID19684 | DNMT3A | p.R882C | 18.85 | 47.06 | 1920 | 2.20E-16 |
| NRAS | p.G13V | 7.65 | 8.82 | 1424 | 2.20E-16 | |
| PTPN11 | p.G60V | 4.31 | 14.81 | 881 | 2.20E-16 | |
| UID7393 | KRAS | p.G12A | ND | 10.28 | 389 | NA |
| NRAS | p.G13R | ND | 17.1 | 1715 | NA | |
| TP53 | p.Y107X | 0.92 | 97.16 | 3058 | 2.20E-16 | |
| UID49278 | FLT3 | p.D593delinsEAPGEVD | 0.98 | 22.45 | 1636 | 2.20E-16 |
| GNB1 | p.K57E | 28.53 | 37.79 | 1623 | 2.20E-16 | |
| KDM6A | p.R658X | 1.19 | 72.12 | 672 | 6.36E-12 | |
| RAD21 | p.E553X | ND | 41.2 | 198 | NA | |
| RUNX1 | p.G165fs | ND | 20.66 | 939 | NA | |
| RUNX1 | p.R204X | 0.68 | 30.67 | 1629 | 1.23E-12 | |
| SRSF2 | p.P85H | 36.92 | 33.58 | 1154 | 2.20E-16 | |
| UID12484 | TP53 | p.L194H | ND | 41.97 | 1723 | NA |
| UID19304 | GATA2 | p.Y322_M325de linsW | ND | 33.7 | 3172 | NA |
| UID31000 | TET2 | p.H1380Y | 8.74 | 21.21 | 2093 | 2.20E-16 |
| TP53 | p.R156H | 54.14 | 58.64 | 1620 | 2.20E-16 | |
| TP53 | p.R267Q | 42.11 | 52.46 | 1850 | 2.20E-16 |
We also expanded our analysis to other mutations including non-drivers and mutations that were lost from prior PB samples (Supplemental Appendix, page 14). We detected 12 mutations in prior PB samples that were not detected in t-MN BM samples. There were also 8 other mutations in prior PB samples that were detected in t-MN BM but with small VAF and not designated as drivers. The VAF was significantly higher in the mutations that became drivers than in the mutations that did not become drivers (8.5% [IQR: 3.9–19.9%] vs. 1.2% [IQR: 0.6–1.2%], P < 0.0001; Supplemental Appendix, page 15). These results implicate the clonal selection process under the selective pressure of chemotherapy with or without radiation therapy.
All 14 patients had normal blood counts and no clinical evidence of leukemia at the time of primary cancer diagnosis. Of note, 3 patients (UID17285, UID19684, and UID12484) had bilateral bone marrow biopsies as part of their lymphoma staging work-up, and none of them showed morphological evidence of leukemia at the time of their lymphoma diagnosis, despite the presence of clonal hematopoiesis with relatively high VAF in UID17285 and UID19684.
Prevalence of clonal hematopoiesis in the control cohort
A high prevalence of pre-leukemic driver mutations in patients who developed t-MNs (10 of 14, 71%) suggests that detection of clonal hematopoiesis at the time of primary cancer diagnosis may be useful in identifying patients at increased risk of t-MNs. To further explore this, we examined the prevalence of clonal hematopoiesis in pre-treatment PB samples from age-matched control cohort of patients with lymphoma who did not develop t-MNs after therapy. The clinical characteristics of the 54 patients in the control cohort are described in Supplemental Appendix (page 7) and are compared with those of 14 t-MN cases. The median age was 58 years (IQR: 49–63 years) and was similar to that of the 14 t-MN cases (median 62 years [IQR: 46–65 years], P = 0.62). Because control cohort is comprised of patients with lymphoma, there were some obvious differences in clinical characteristics including the difference in primary cancer diagnosis, proportion of patients who received alkylating agents or topoisomerase II inhibitors, and radiation therapy. Follow up duration was also shorter in control cohort (cases vs. control, median 8.9 years [95% CI: 6.3–11.5 years, IQR: 7.2–9.1 years] vs. 6.1 years [95% CI: 6.0–6.3 years, IQR: 5.8–6.2 years], P = 0.001). Although there were significant difference in each of the clinical characteristics, overall, this control cohort had sufficient treatment-related risk factors for t-MNs, because all the patients received multiple cycles of alkylating agents, 14 of 54 (26%) patients received radiation therapy, and 6 of 53 (11%) patients underwent for auto-SCT.
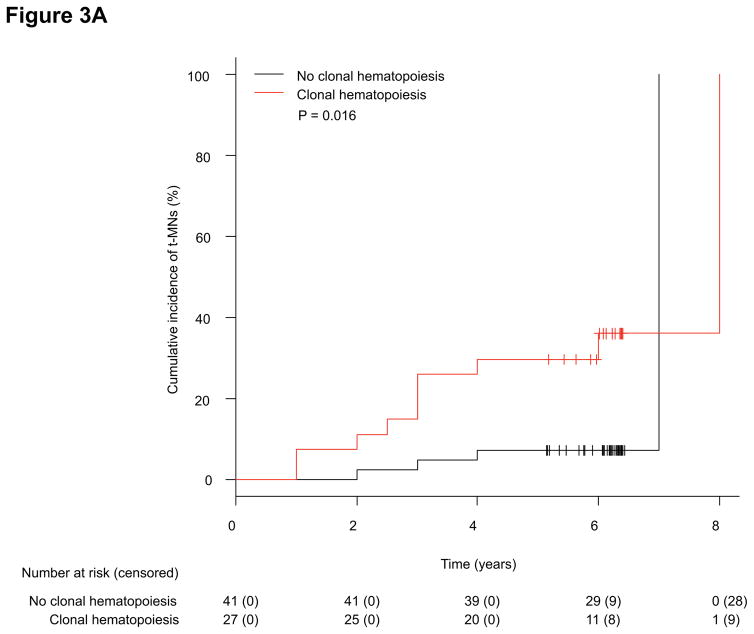
Using the same molecular barcode sequencing, we detected 22 mutations in 17 of 54 (31%) patients’ pre-treatment PB samples (Supplemental Appendix, page 8). Overall, patients who developed t-MNs had significantly higher incidence of clonal hematopoiesis at the time of cancer diagnosis (71% vs. 31%, P = 0.008). The cumulative incidence of t-MNs at 5 years was significantly higher in patients with clonal hematopoiesis than in patients without (30% [95% CI: 16–51%] vs. 7% [95% CI: 2–21%], P = 0.016; Figure 3A). The median VAF of the mutations detected as clonal hematopoiesis was significantly higher in the t-MN cases than in the control (t-MN cases vs. control, median 2.4% [IQR: 1%–8.5%] vs. 0.8% [IQR: 0.5%–1.3%], P = 0.001; Figure 3B).
Figure 3.
(A) Cumulative incidence of t-MN development between patients with or without clonal hematopoiesis in a case-control study. (B) Box plot comparing the VAF of mutations detected as clonal hematopoiesis between patients who developed t-MNs and those who did not (control) (median 2.4% [IQR: 1%–8.5%] vs. 0.8% [IQR: 0.5%–1.3%], P = 0.001).
Clonal hematopoiesis increases the risk of t-MNs
To confirm the association between clonal hematopoiesis and t-MN development, we sequenced the mononuclear cells of pre-treatment PB (PBMC) samples from 74 patients with lymphoma who received frontline CHOP-based chemotherapy as part of a clinical trial (external cohort). The clinical characteristics of these patients are summarized in Supplemental Appendix (page 9). The median age of this group was 56 years (IQR: 44–64 years). All patients received CHOP with or without melatonin as a frontline therapy, and 35 of 74 (47%) and 16 of 74 (22%) patients received radiation therapy and high-dose chemotherapy followed by autologous SCT, respectively. The median follow-up duration for this cohort was 14.8 years (95% CI: 14.5–15.1 years, IQR: 12.2–15.2 years), and 5 of 74 patients (7%) developed t-MNs (clinical characteristics are summarized in Supplemental Appendix, page 10) with a median latency period of 5.4 years (IQR: 3.3–8.3 years).
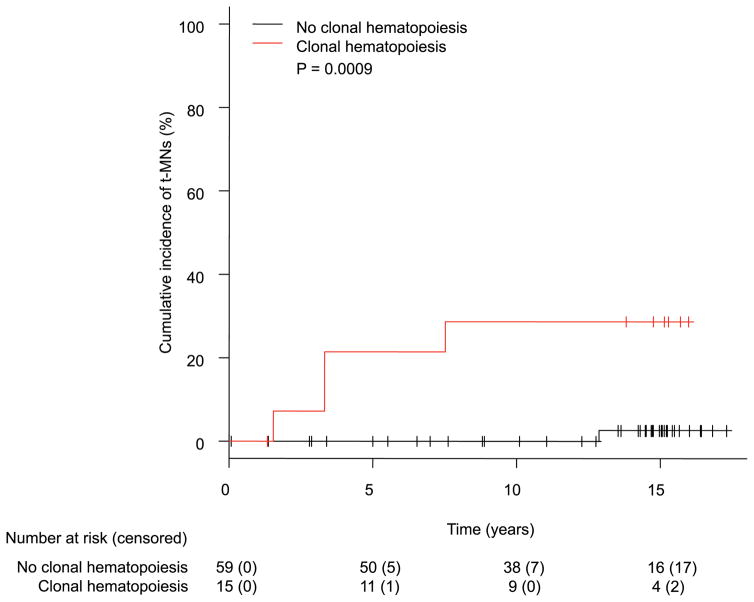
Molecular barcode sequencing of pre-treatment PBMC samples detected a total of 17 mutations as clonal hematopoiesis in 15 of 74 patients (23%; Supplemental Appendix, page 11). Clonal hematopoiesis was detected in 4 of 5 patients (80%) who developed t-MNs, whereas it was detected in only 11 of 69 patients (16%) who did not develop t-MNs (P = 0.005). Of note, one patient (UID800699) who developed t-MDS but did not have clonal hematopoiesis had an unusually long latency period (12.8 years), raising the possibility of a de novo MDS. Nonetheless, the positive predictive value (PPV) and negative predictive value (NPV) of clonal hematopoiesis in t-MN development were 26.7% (95% CI: 7.8–55.1%) and 98.3% (95% CI: 90.9–99.9%), respectively. The cumulative incidence of t-MN development at 10 years was significantly higher in patients with clonal hematopoiesis than in patients without (29% [95% CI: 8–53%] vs. 0% [95% CI: 0–0%], P = 0.0009; Figure 4). Consistent with prior studies, patients who underwent auto-SCT had higher rates of t-MNs at 10 years than patients who did not (19% [95% CI: 4–41%] vs. 2% [95% CI: 1–9%], P = 0.003; Supplemental Appendix, page 16). Age at the time of lymphoma diagnosis (60 years or older vs. younger) and radiation treatment (yes vs. no) did not affect the rate of t-MNs at 10 years (Supplemental Appendix, page 17–18). In a Fine and Gray model for t-MN development considering clonal hematopoiesis and auto-SCT, both variables significantly increased the risk of t-MNs (clonal hematopoiesis: HR 13.7 [95% CI: 1.7–108.7], P = 0.013; auto-SCT: HR 10.8 [95% CI: 1.1–107.9], P = 0.043; Table 3). There was no correlation between the two variables (P = 0.29, Supplemental Appendix, page 12). In the external cohort, there was no difference in VAF of clonal hematopoiesis between patients who developed t-MNs and those who did not (median VAF 0.4% [IQR: 0.3–1.1%] vs. 0.9% [IQR: 0.2–1.9%], P = 0.56).
Figure 4.
Cumulative incidence of t-MN development between patients with or without clonal hematopoiesis in an external cohort.
Table 3.
Fine-Gray proportional hazard regression model for t-MN development.
| Full model | HR (95% CI) | P – value | Reduced model | HR (95% CI) | P-value |
|---|---|---|---|---|---|
| Clonal hematopoiesis (vs. no) | 14.0 (1.4 – 136.4) | 0.023 | Clonal hematopoiesis (vs. no) | 13.7 (1.7– 108.7) | 0.013 |
| Auto-SCT (vs. no) | 9.2 (0.6–150.5) | 0.12 | Auto-SCT (vs. no) | 10.8 (1.1– 107.9) | 0.043 |
| XRT (vs. no) | 0.6 (0.08–4.0) | 0.56 | |||
| Age ≥ 60 years (vs. < 60 years) | 0.7 (0.07–6.3) | 0.73 |
HR: Hazard ratio, CI: Confidence interval, SCT: Stem cell transplant, XRT: Radiation therapy
Overall, in the entire cohort including patients from the case-control and external cohort (N = 142), patients with clonal hematopoiesis were older than patients without (median 60 years [IQR: 50–66 years] vs. 56 years [IQR: 45–62 years], P = 0.028; Supplemental Appendix, page 19). Furthermore, RUNX1, TP53, SRSF2 and TET2 were more frequently mutated as clonal hematopoiesis in patients who developed t-MNs compared to the patients who did not develop t-MNs (Supplemental Appendix, page 20).
Discussion
In this study, we have demonstrated that in 10 of 14 patients (71%) with t-MNs, pre-leukemic driver mutations were detectable as clonal hematopoiesis at the time of primary cancer diagnosis. Although clonal hematopoiesis was detected in the control patients who did not develop t-MNs, the prevalence was significantly lower (26%). This finding was also confirmed in an external cohort, in which the presence of clonal hematopoiesis significantly increased the risk of t-MNs in a multivariate model, which suggests the potential usefulness of clonal hematopoiesis as a clinical biomarker for risk prediction, surveillance, and early detection of t-MNs.
Early detection of myeloid neoplasms has been a challenging task due to the lack of a clearly identifiable pre-malignant state. However, the recent discovery of CHIP in healthy individuals suggested that myeloid neoplasms have a pre-malignant condition characterized by clonal hematopoiesis and its detection may help identify individuals at risk of developing myeloid neoplasms.(25–27, 29) We explored this hypothesis in the context of t-MNs because exposures to chemotherapy may further increase the risk of developing t-MNs in patients with antecedent clonal hematopoiesis, which could allow us to identify a population at significant risk of myeloid neoplasms.
In a case-control study, we found that the VAF of the detected mutations was significantly higher in patients who went on to develop t-MNs compared to patients in the control, which is consistent with the findings in healthy individuals.(25) Similarly, the mutations detected as clonal hematopoiesis that became drivers in t-MN BM had higher VAF than the mutations that did not become drivers. These findings were not supported in an external cohort, which may be a result of the sampling strategy (PBMC) enriching for non-myeloid cells (the presumptive lineage of origin for clonal hematopoiesis) as opposed to buffy coats in the first two cohorts. Although, the recent study that compared VAF of clonal hematopoiesis in different cellular compartments (myeloid and lymphoid) suggested that clonal hematopoiesis originates from a long term hematopoietic stem and progenitor cells and VAF was similar between myeloid and lymphoid cells.(33)
Nonetheless, these results suggest that each instance of clonal hematopoiesis possesses a different degree of risk for the development of t-MNs, and the risk is influenced by both the mutation and its VAF. In fact, genes such as RUNX1 and TP53 were more frequently mutated as clonal hematopoiesis in patients who developed t-MNs than in patients who did not. Clonal hematopoiesis characterized by the above mutations and high VAF may carry higher risk of developing into t-MNs. Further study will be required to better understand the risk stratification of clonal hematopoiesis. In our study, the PPV of clonal hematopoiesis was 26.7% (95% CI: 7.8–55.1%), whereas the NPV was 98.3% (95% CI: 90.9–99.9%). The low PPV of clonal hematopoiesis in this study currently limits its clinical utility as a predictive marker. This is likely because we included all detected mutations as clonal hematopoiesis. Future studies that define the best predictive VAF cut-off and high-risk mutations may improve the predictive value of clonal hematopoiesis.
The prevalence of clonal hematopoiesis in our series was much higher than what was reported in TCGA patients (approximately 2%).(29) This is likely attributed to the higher sensitivity of the molecular barcode deep sequencing method used in our study compared to the whole exome sequencing used in the TCGA data set. It highlights not only how frequent clonal hematopoiesis is when sensitive sequencing methods are used(30, 33) but also the importance of defining clinically relevant clonal hematopoiesis (i.e., high-risk clonal hematopoiesis).
Not surprisingly, patients with clonal hematopoiesis in our series were significantly older than those without. Prior studies have indicated that older age is a risk factor for t-MNs.(6, 14, 16) However, in our study, age did not affect risk of t-MNs but clonal hematopoiesis did. It is currently unclear whether age itself or the increased risk of clonal hematopoiesis in the elderly plays a larger role in the development of t-MNs.
Previously, Wong et al. analyzed 7 patients with t-MNs with TP53 mutations and found that 4 of them had had identical TP53 mutations years before t-MN development.(30) In particular, 2 patients had evidence of TP53 mutations prior to therapy. The results presented here confirm and expand upon these data. A substantial fraction of t-MN patients had evidence of driver mutations as clonal hematopoiesis before they were treated with chemotherapy. These mutations included TP53 as well as other driver genes. Our results raise fundamental questions about the role of chemotherapy and radiation therapy in t-MN development. The prevailing hypothesis has been that genotoxic insult from chemotherapy and radiation therapy induces mutations in hematopoietic cells, and accumulation of these mutations leads to t-MNs.(34) However, these data from our group and others suggest that ancestral driver mutations pre-exist as clonal hematopoiesis before exposure to therapy. In the mice model of chimeric bone marrow transplantation, Bondar et al. and Marusyk et al. previously showed that Tp53+/− hematopoietic cells had selective advantage over wild type cells in response to irradiation, which appeared to be mediated by decreased radiation induced cellular senescence in the Tp53+/− cells.(35, 36) Similarly, Wong et al. showed that Tp53+/− hematopoietic cells had competitive expansion over wild type cells after exposure to N-ethyl-N-nitrosourea.(30) In the absence of irradiation or chemotherapy, the competitive advantage of mutant cells over wild type cells was minimal. It is likely that clonal hematopoiesis with pre-leukemic driver mutations has clonal advantage over wild type cells but in non-stress physiological state, such advantage is marginal. However, upon exposure to chemotherapy and/or radiation therapy, mutated clone demonstrates grater clonal advantage over wild type cells which leads to clonal expansion of mutant cells. In addition to the clonal expansion, secondary acquisition of other driver mutations or chromosomal aberrations likely contributes to t-MN transformation. In the current study, we could not identify longitudinal samples between the time from primary cancer and t-MN development. Future work in a prospective trial monitoring clonal trajectory over time may provide further insight into the role of chemo-radiation therapy exposure in instigating clonal expansion and secondary alterations that further drive clonal hematopoiesis to full transformation and ultimately to t-MNs.
Our study has several limitations. First, both control and an external cohorts are comprised of patients with lymphoma and the prevalence of clonal hematopoiesis in these cohorts may not accurately reflect that of patients with other cancers. Based on the analysis of TCGA’s data, Xie et al. reported that prevalence of clonal hematopoiesis was not significantly different among patients with different cancer types.(29) Although this study did not include patients with lymphoma, we assume that the prevalence of clonal hematopoiesis in our patients with lymphoma is not significantly different from that of patients with other cancer types. Second, case-control study lacked formal matching procedure and control cohort had relatively short follow up duration. As stated above, difference in primary cancer types would likely not affect the prevalence of clonal hematopoiesis as long as the age is matched. Furthermore, overall treatment-related risk factor should be equivalent or even higher in the control cohort because lymphoma patients almost uniformly receive alkylating agents and some will even undergo auto-SCT, Additionally, longer follow up of the control cohort would likely not yield different conclusion, as the difference in prevalence of clonal hematopoiesis is substantial (70% vs. 26%) and additional follow up would likely add only 1–2 t-MN development, if it occurs. From these reasons, we believe that the current control cohort can function as a reasonable control. Plus, we believe that the confirmation of the findings in an external cohort which has much longer follow up duration and homogeneous clinical courses would complement these limitations. Third, t-MN sequencing was conducted on whole bone marrow aspirate. Due to the inevitable contamination of normal cells, VAF of driver mutations may not accurately reflect actual clonality of the mutation. This may explain why some of the driver mutations detected in t-MN samples had less than 50% VAF. Fourth, we could not analyze association between G-CSF use and t-MNs risk in this study. Use of G-CSF in cancer patients has been shown as an important risk factor for t-MNs.(12,13) We attempted to retrospectively collect data on G-CSF use in the second cohort. However, because the trial was conducted 17 years ago and did not prospectively collect the data on G-CSF use and many patients received chemotherapy at outside institution, we could not capture all the data of G-CSF use in this study. It is of interest to analyze interaction between clonal hematopoiesis and G-CSF use and its effect on t-MN risk.
In summary, pre-leukemic clonal hematopoiesis was frequently detected in patients with t-MNs at the time of primary cancer diagnosis and before exposure to therapy. Detection of clonal hematopoiesis significantly increased the risk of t-MN development. These data suggest potential approaches of screening for clonal hematopoiesis in cancer patients to identify patients at risk of t-MN development and warrants validation in a prospective trial investigating a role of clonal hematopoiesis as a risk factor for t-MNs.
Supplementary Material
Research in context.
Evidence before this study
We searched PubMed for reviews and research articles published in English before June, 2016, about therapy-related myeloid neoplasms (t-MNs) and their risk factors. We used the following keywords: “therapy-related myeloid neoplasms”, “t-MNs”, “t-MDS”, “t-AML”, and “risk factors”. There are several known treatment-related risk factors for t-MNs, including exposures to alkylating agents, topoisomerase 2 inhibitors, and high dose chemotherapy with autologous stem cell transplant. In contrast, little is known about host susceptibility. Older age was shown to increase the risk of t-MNs. Several germline polymorphisms have also been associated with the risk, but none of them were validated. As such, there is no predictive biomarker for t-MNs.
Added value of this study
Pre-leukemic clonal hematopoiesis was frequently detected in cancer patients who subsequently developed t-MNs, and it was detected at the time of their primary cancer diagnosis and before any therapy was given. Patients with pre-leukemic clonal hematopoiesis had significantly higher risk of developing t-MNs than patients without clonal hematopoiesis.
Implications of all the available evidence
Cancer patients with pre-leukemic clonal hematopoiesis have increased risk of developing t-MNs. Clonal hematopoiesis may function as a potential biomarker for risk prediction and early detection of t-MNs and may be considered as a future therapeutic target to prevent t-MN development.
Acknowledgments
Funding: Cancer Prevention Research Institute of Texas, Welch Foundation, UT System STARS Award, Edward P. Evans Foundation, Fundacion Ramon Areces, Red and Charline McCombs Institute for the Early Detection and Treatment of Cancer, Institutional Research Grant at the MD Anderson Cancer Center, NCI Leukemia SPORE, Khalifa Scholar Award, NIH through MD Anderson Cancer Center Support Grant, and MD Anderson’s MDS/AML Moon Shot Program.
This study was supported by the Cancer Prevention Research Institute of Texas (R120501: PAF and RP100202 GGM), the Welch Foundation (G-0040, PAF), the UT System STARS Award (PS100149: PAF), the Edward P. Evans Foundation (GGM), the Fundacion Ramon Areces (GGM), the Red and Charline McCombs Institute for the Early Detection and Treatment of Cancer Award (KT), an Institutional Research Grant at the MD Anderson Cancer Center (KT), NCI Leukemia SPORE Career Development Grant (KT), Khalifa Scholar Award (KT), National Institute of Health (NIH) through the MD Anderson Cancer Center Support Grant P30 CA016672, and by generous philanthropic contributions to MD Anderson’s MDS/AML Moon Shot Program (KT, HK, GGM, and PAF). We would like to thank Zachary Bohannan and Joseph Munch for their professional input on the manuscript.
Footnotes
Conflict of interest disclosures
Authors have no conflicts of interest to declare.
Author contributions
KT, GGM, and AF designed the study, analyzed data, and wrote the manuscript. FW and JZ performed bioinformatics analysis and wrote the manuscript. LZ and XH assisted statistical analysis. KT, CDD, FR, HK, FS and GGM treated patients and collected patients’ sample. DD, KK, ET and CG performed sequencing. CBR and KPT performed pathological diagnosis. KP, SP, YW, FS and XW provided samples. SC analyzed data and wrote manuscript. HK, GGM and AF provided leadership and managed the study team. All authors read and approved the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Leone G, Pagano L, Ben-Yehuda D, Voso MT. Therapy-related leukemia and myelodysplasia: susceptibility and incidence. Haematologica. 2007;92(10):1389–98. doi: 10.3324/haematol.11034. [DOI] [PubMed] [Google Scholar]
- 2.Travis LB. The epidemiology of second primary cancers. Cancer Epidemiol Biomarkers Prev. 2006;15(11):2020–6. doi: 10.1158/1055-9965.EPI-06-0414. [DOI] [PubMed] [Google Scholar]
- 3.Smith MR, Neuberg D, Flinn IW, et al. Incidence of therapy-related myeloid neoplasia after initial therapy for chronic lymphocytic leukemia with fludarabine-cyclophosphamide versus fludarabine: long-term follow-up of US Intergroup Study E2997. Blood. 2011;118(13):3525–7. doi: 10.1182/blood-2011-03-342485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leone G, Fianchi L, Pagano L, Voso MT. Incidence and susceptibility to therapy-related myeloid neoplasms. Chem Biol Interact. 2010;184(1–2):39–45. doi: 10.1016/j.cbi.2009.12.013. [DOI] [PubMed] [Google Scholar]
- 5.Armitage JO, Carbone PP, Connors JM, Levine A, Bennett JM, Kroll S. Treatment-related myelodysplasia and acute leukemia in non-Hodgkin’s lymphoma patients. J Clin Oncol. 2003;21(5):897–906. doi: 10.1200/JCO.2003.07.113. [DOI] [PubMed] [Google Scholar]
- 6.Smith SM, Le Beau MM, Huo D, et al. Clinical-cytogenetic associations in 306 patients with therapy-related myelodysplasia and myeloid leukemia: the University of Chicago series. Blood. 2003;102(1):43–52. doi: 10.1182/blood-2002-11-3343. [DOI] [PubMed] [Google Scholar]
- 7.Kayser S, Dohner K, Krauter J, et al. The impact of therapy-related acute myeloid leukemia (AML) on outcome in 2853 adult patients with newly diagnosed AML. Blood. 2011;117(7):2137–45. doi: 10.1182/blood-2010-08-301713. [DOI] [PubMed] [Google Scholar]
- 8.Abdelhameed A, Pond GR, Mitsakakis N, et al. Outcome of patients who develop acute leukemia or myelodysplasia as a second malignancy after solid tumors treated surgically or with strategies that include chemotherapy and/or radiation. Cancer. 2008;112(7):1513–21. doi: 10.1002/cncr.23325. [DOI] [PubMed] [Google Scholar]
- 9.Quintas-Cardama A, Daver N, Kim H, et al. A prognostic model of therapy-related myelodysplastic syndrome for predicting survival and transformation to acute myeloid leukemia. Clinical lymphoma, myeloma & leukemia. 2014;14(5):401–10. doi: 10.1016/j.clml.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leone G, Fianchi L, Voso MT. Therapy-related myeloid neoplasms. Current opinion in oncology. 2011;23(6):672–80. doi: 10.1097/CCO.0b013e32834bcc2a. [DOI] [PubMed] [Google Scholar]
- 11.Morton LM, Dores GM, Tucker MA, et al. Evolving risk of therapy-related acute myeloid leukemia following cancer chemotherapy among adults in the United States, 1975–2008. Blood. 2013;121(15):2996–3004. doi: 10.1182/blood-2012-08-448068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Relling MV, Boyett JM, Blanco JG, et al. Granulocyte colony-stimulating factor and the risk of secondary myeloid malignancy after etoposide treatment. Blood. 2003;101(10):3862–7. doi: 10.1182/blood-2002-08-2405. [DOI] [PubMed] [Google Scholar]
- 13.Lyman GH, Dale DC, Wolff DA, et al. Acute myeloid leukemia or myelodysplastic syndrome in randomized controlled clinical trials of cancer chemotherapy with granulocyte colony-stimulating factor: a systematic review. J Clin Oncol. 2010;28(17):2914–24. doi: 10.1200/JCO.2009.25.8723. [DOI] [PubMed] [Google Scholar]
- 14.Krishnan A, Bhatia S, Slovak ML, et al. Predictors of therapy-related leukemia and myelodysplasia following autologous transplantation for lymphoma: an assessment of risk factors. Blood. 2000;95(5):1588–93. [PubMed] [Google Scholar]
- 15.Travis LB, Holowaty EJ, Bergfeldt K, et al. Risk of leukemia after platinum-based chemotherapy for ovarian cancer. The New England journal of medicine. 1999;340(5):351–7. doi: 10.1056/NEJM199902043400504. [DOI] [PubMed] [Google Scholar]
- 16.Bhatia S, Ramsay NK, Steinbuch M, et al. Malignant neoplasms following bone marrow transplantation. Blood. 1996;87(9):3633–9. [PubMed] [Google Scholar]
- 17.Ding Y, Sun CL, Li L, et al. Genetic susceptibility to therapy-related leukemia after Hodgkin lymphoma or non-Hodgkin lymphoma: role of drug metabolism, apoptosis and DNA repair. Blood Cancer J. 2012;2(3):e58. doi: 10.1038/bcj.2012.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Felix CA, Walker AH, Lange BJ, et al. Association of CYP3A4 genotype with treatment-related leukemia. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(22):13176–81. doi: 10.1073/pnas.95.22.13176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larson RA, Wang Y, Banerjee M, et al. Prevalence of the inactivating 609C-->T polymorphism in the NAD(P)H:quinone oxidoreductase (NQO1) gene in patients with primary and therapy-related myeloid leukemia. Blood. 1999;94(2):803–7. [PubMed] [Google Scholar]
- 20.Sasai Y, Horiike S, Misawa S, et al. Genotype of glutathione S-transferase and other genetic configurations in myelodysplasia. Leukemia research. 1999;23(11):975–81. doi: 10.1016/s0145-2126(99)00119-8. [DOI] [PubMed] [Google Scholar]
- 21.Seedhouse C, Faulkner R, Ashraf N, Das-Gupta E, Russell N. Polymorphisms in genes involved in homologous recombination repair interact to increase the risk of developing acute myeloid leukemia. Clinical cancer research: an official journal of the American Association for Cancer Research. 2004;10(8):2675–80. doi: 10.1158/1078-0432.ccr-03-0372. [DOI] [PubMed] [Google Scholar]
- 22.Li L, Li M, Sun C, et al. Altered hematopoietic cell gene expression precedes development of therapy-related myelodysplasia/acute myeloid leukemia and identifies patients at risk. Cancer cell. 2011;20(5):591–605. doi: 10.1016/j.ccr.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chakraborty S, Sun CL, Francisco L, et al. Accelerated telomere shortening precedes development of therapy-related myelodysplasia or acute myelogenous leukemia after autologous transplantation for lymphoma. J Clin Oncol. 2009;27(5):791–8. doi: 10.1200/JCO.2008.17.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Busque L, Patel JP, Figueroa ME, et al. Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nature genetics. 2012;44(11):1179–81. doi: 10.1038/ng.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jaiswal S, Fontanillas P, Flannick J, et al. Age-related clonal hematopoiesis associated with adverse outcomes. The New England journal of medicine. 2014;371(26):2488–98. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Genovese G, Kahler AK, Handsaker RE, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. The New England journal of medicine. 2014;371(26):2477–87. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steensma DP, Bejar R, Jaiswal S, et al. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 2015;126(1):9–16. doi: 10.1182/blood-2015-03-631747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McKerrell T, Park N, Moreno T, et al. Leukemia-associated somatic mutations drive distinct patterns of age-related clonal hemopoiesis. Cell Rep. 2015;10(8):1239–45. doi: 10.1016/j.celrep.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie M, Lu C, Wang J, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nature medicine. 2014;20(12):1472–8. doi: 10.1038/nm.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wong TN, Ramsingh G, Young AL, et al. Role of TP53 mutations in the origin and evolution of therapy-related acute myeloid leukaemia. Nature. 2015;518(7540):552–5. doi: 10.1038/nature13968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sarma A, Rodriguez MA, Cabanillas F, et al. A randomized trial of CHOP chemotherapy with or without melatonin in patients with favorable prognosis large B-cell lymphoma. Journal of Clinical Oncology. 2004;22(14):745s-s. [Google Scholar]
- 32.DiNardo CD, Bannon SA, Routbort M, et al. Evaluation of patients and families with concern for predispositions to hematologic malignancies within the Hereditary Hematologic Malignancy Clinic (HHMC) Clinical Lymphoma Myeloma and Leukemia. 2016;16(7):417–28. doi: 10.1016/j.clml.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Young AL, Challen GA, Birmann BM, Druley TE. Clonal haematopoiesis harbouring AML-associated mutations is ubiquitous in healthy adults. Nat Commun. 2016;7:12484. doi: 10.1038/ncomms12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bhatia S. Therapy-related myelodysplasia and acute myeloid leukemia. Semin Oncol. 2013;40(6):666–75. doi: 10.1053/j.seminoncol.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bondar T, Medzhitov R. p53-mediated hematopoietic stem and progenitor cell competition. Cell Stem Cell. 2010;6(4):309–22. doi: 10.1016/j.stem.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marusyk A, Porter CC, Zaberezhnyy V, DeGregori J. Irradiation selects for p53-deficient hematopoietic progenitors. PLoS Biol. 2010;8(3):e1000324. doi: 10.1371/journal.pbio.1000324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.