ABSTRACT
Prohibitin (PHB) is an evolutionarily conserved protein with multiple functions in both normal and cancer cells. Androgen receptor (AR) was reported to act as a different role in the ER-positive and ER-negative breast cancer. However, little is known about the role of PHB and whether PHB could regulate AR expression in the ER-positive breast cancer. Here, we determined the expression and clinical outcomes of PHB in breast cancer samples using 121 breast cancer tissues and published databases, and investigated the role of PHB in breast cancer cell growth, apoptosis and cell cycle arrest in the ER-positive breast cancer cells. We obtained the expression of PHB is significantly low in breast cancer samples, and low PHB expression positively correlated with poor prognosis of breast cancer. We detected that PHB could inhibit breast cancer cell proliferation, change cell cycle distribution and promote cell apoptosis in the ER-positive breast cancer cells. Moreover, we found PHB could significantly increase AR expression in both mRNA and protein levels in the ER-positive breast cancer cells. Additionally, a significant positive correlation between PHB and AR expression was identified in the 121 breast cancer tissues. PHB and AR expression are associated with prognosis in the ER-positive breast cancer patients. Our results indicate that PHB promotes AR activation in ER-positive breast cancer, making PHB and AR potential molecular targets for ER-positive breast cancer therapy.
KEYWORDS: androgen receptor, AR, breast cancer, PHB, prohibitin
Introduction
Prohibitin (PHB) is an evolutionarily conserved protein with multiple cellular functions, including as a molecular chaperone complex in the inner mitochondrial membrane, activation of the Ras-Raf signaling pathway, and inhibition of various transcription factors (including E2F and steroid receptors), and plays an essential role in the cells through the tumor suppressor, growth-inhibitory and cell-cycle regulation activities.1-4 PHB has been reported to repress E2F via recruitment of the repressive proteins histone deacetylase 1 (HDAC1), nuclear receptor co-repressor (N-CoR) and the chromatin-condensing proteins brahma-related gene-1 (BRG1)/brahma (Brm).5,6 PHB can also inhibit steroid-activated nuclear receptors, such as the androgen receptor (AR) and estrogen receptor (ER).7,8 PHB was reported to act as a potent transcriptional co-repressor of ERα and associate with estrogen-regulated promoters in the absence of hormone, dissociating after hormone treatment in the breast cancer cells.8 PHB was reported to induce the anti-proliferative actions of estrogen antagonists in breast cancer cells and recruit BRG1-containing chromatin-remodeling complex to antagonist-bound AR in prostate cancer cells.9,10 It was reported that PHB could repress AR activity and androgen-stimulated growth of prostate cancer cells and conversely that knockdown of PHB increased AR activity and prostate cancer cell growth in response to androgens, both in vitro and in vivo.11 Dart et al. reported consistently that PHB knockdown increases AR binding to the PSA promoter and therefore increases AR activity in the prostate cancer cells.12 However, Fletcher et al. reported that AR could also downregulate PHB through increasing miR-27a expression, suggesting that AR may inhibit its own repressor PHB in a negative feedback loop.13 Therefore, accumulating evidences support the fact that PHB acts as a potent transcriptional co-repressor of ERα in the breast cancer cells and AR in the prostate cancer cells. However, little is known regarding the role of PHB and whether PHB could regulate AR expression in the ER-positive breast cancer.
AR was reported to act as a different role in the ER-positive and ER-negative breast cancer.14 AR signaling exerts an anti-estrogen, anti-proliferative influence under normal physiologic conditions, and this role may be sustained in ER-positive breast cancer cells.15-17 On the contrary, AR signaling may also promote growth of a subset of ER-negative, AR-positive breast cancers with a molecular apocrine phenotype.18 AR was reported to mediate ligand-dependent activation of Wnt and HER-2 signaling pathways through direct transcriptional induction of WNT7B and HER-3 in the ER-negative breast cancer.18 Specific targeting of AR, Wnt or HER-2 signaling impairs androgen-stimulated tumor cell growth of ER-negative, HER2-positive breast cancers.18 Thus, the present study aims to investigate the role of PHB and whether PHB could regulate AR expression in the ER-positive breast cancer. We obtained the expression of PHB is significantly low in breast cancer samples, and low PHB expression positively correlated with poor prognosis of breast cancer. Moreover, PHB could inhibit breast cancer cell proliferation, change cell cycle distribution and promote cell apoptosis through the activation of AR in the ER-positive breast cancer cells. In addition, we found a significant positive correlation between PHB and AR expression in the breast cancer tissues. PHB and AR expression are also associated with prognosis in the ER-positive breast cancer patients.
Results
The expression of PHB is low in the breast cancer patients
To investigate the expression of PHB in the human breast cancer, we determined the expression status of PHB in 121 breast cancer tissues and corresponding noncancerous tissues (NCTs) by immunohistochemical (IHC) staining. Relationship between the expression of PHB and clinicopathological features of 121 breast cancer patients was determined (Table 1). Of the 121 tumor tissues, 59 cases (48.8%) and 62 cases (51.2%) expressed PHB at high and low levels, respectively (Table 1). However, 86 cases (71.1%) and 35 cases (28.9%) expressed a high and low levels of PHB in the 121 corresponding NCTs, respectively (Table 3). Age, menopausal status, tumor location, tumor size, vascular/perineural invasion, and histological grade were not significantly associated with PHB expression (Table 1). P53, E-Cadherin and Ki-67 expression were also not significantly associated with PHB expression. However, we found high PHB expression was associated with more lymph nodes metastasis, positive ER, PR and negative HER-2 expression, respectively (Table 1). As shown in Fig. 1 and Table 3, the expression of PHB was low in the tumor tissues, while the PHB expression in the corresponding NCTs was high (P < 0.001). Taken together, these data suggest that the expression of PHB is low in the breast cancer patients.
Table 1.
Relationship between the expression of PHB and clinicopathological features of 121 breast cancer patients.
|
Total (n = 121) |
PHB high (n = 59) |
PHB low (n = 62) |
|||||
|---|---|---|---|---|---|---|---|
| Variables | No. | % | No. | % | No. | % | P-value |
| Age | 0.400 | ||||||
| <48 years | 56 | 46.3 | 25 | 42.4 | 31 | 50.0 | |
| ≥ 48 years | 65 | 53.7 | 34 | 57.6 | 31 | 50.0 | |
| Menopausal status | 0.749 | ||||||
| Premenopausal or perimenopausal | 70 | 57.9 | 35 | 59.3 | 35 | 56.5 | |
| Postmenopausal | 51 | 42.1 | 24 | 40.7 | 27 | 43.5 | |
| Tumor location | 0.926 | ||||||
| Left | 61 | 50.4 | 30 | 50.8 | 31 | 50.0 | |
| Right | 60 | 49.6 | 29 | 49.2 | 31 | 50.0 | |
| Tumor size | 0.540 | ||||||
| <2 | 28 | 23.1 | 16 | 27.1 | 12 | 19.4 | |
| 2–5 | 86 | 71.1 | 39 | 66.1 | 47 | 75.8 | |
| ≥ 5 | 7 | 5.8 | 4 | 6.8 | 3 | 4.8 | |
| Vascular invasion | 0.708 | ||||||
| Yes | 35 | 28.9 | 18 | 30.5 | 17 | 27.4 | |
| No | 86 | 71.1 | 41 | 69.5 | 45 | 72.6 | |
| Perineural invasion | 0.870 | ||||||
| Yes | 32 | 26.4 | 16 | 27.1 | 16 | 25.8 | |
| No | 89 | 73.6 | 43 | 72.9 | 46 | 74.2 | |
| Lymph node invasion | 0.178 | ||||||
| Yes | 56 | 46.3 | 31 | 52.5 | 25 | 40.3 | |
| No | 65 | 53.7 | 28 | 47.5 | 37 | 59.7 | |
| Lymph nodes | 0.034* | ||||||
| 0 | 66 | 54.5 | 28 | 47.5 | 38 | 61.3 | |
| 1–3 | 43 | 35.5 | 21 | 35.6 | 22 | 35.5 | |
| ≥ 4 | 12 | 9.9 | 10 | 16.9 | 2 | 3.2 | |
| T | 0.346 | ||||||
| T1 | 50 | 41.3 | 28 | 47.5 | 22 | 35.5 | |
| T2 | 67 | 55.4 | 29 | 49.2 | 38 | 61.3 | |
| T3 | 4 | 3.3 | 2 | 3.4 | 2 | 3.2 | |
| N | 0.059 | ||||||
| N0 | 66 | 54.5 | 28 | 47.5 | 38 | 61.3 | |
| N1 | 43 | 35.5 | 21 | 35.6 | 22 | 35.5 | |
| N2 | 10 | 8.3 | 8 | 13.6 | 2 | 3.2 | |
| N3 | 2 | 1.7 | 2 | 3.4 | 0 | 0 | |
| Histological grade | 0.104 | ||||||
| I | 32 | 26.4 | 16 | 27.1 | 16 | 25.8 | |
| II | 74 | 61.2 | 32 | 54.2 | 42 | 67.7 | |
| III | 15 | 12.4 | 11 | 18.6 | 4 | 6.5 | |
| ER | <0.001* | ||||||
| Positive | 89 | 73.6 | 57 | 96.6 | 32 | 51.6 | |
| Negative | 32 | 26.4 | 2 | 3.4 | 30 | 48.4 | |
| PR | <0.001* | ||||||
| Positive | 78 | 64.5 | 54 | 91.5 | 24 | 38.7 | |
| Negative | 43 | 35.5 | 5 | 8.5 | 38 | 61.3 | |
| HER-2 | 0.009 | ||||||
| Positive | 36 | 29.8 | 11 | 18.6 | 25 | 40.3 | |
| Negative | 85 | 70.2 | 48 | 81.4 | 37 | 59.7 | |
| p53 | 0.063 | ||||||
| Positive | 76 | 62.8 | 42 | 71.2 | 34 | 54.8 | |
| Negative | 45 | 37.2 | 17 | 28.8 | 28 | 45.2 | |
| E-Cadherin | 0.132 | ||||||
| Positive | 78 | 64.5 | 42 | 71.2 | 36 | 58.1 | |
| Negative | 43 | 35.5 | 17 | 28.8 | 26 | 41.9 | |
| Ki-67 | 0.194 | ||||||
| <14% | 31 | 25.6 | 12 | 20.3 | 19 | 30.6 | |
| ≥ 14% | 90 | 74.4 | 47 | 79.7 | 43 | 69.4 | |
Table 3.
PHB and AR expression in the 121 breast cancer tissues and NCTs.
|
Breast cancers (n = 121) |
NCTs (n = 121) |
||||
|---|---|---|---|---|---|
| No. | % | No. | % | P-value | |
| PHB | <0.001 | ||||
| 0 | 30 | 24.8 | 10 | 8.3 | |
| + | 32 | 26.4 | 25 | 20.7 | |
| ++ | 40 | 33.1 | 40 | 33.1 | |
| +++ | 19 | 15.7 | 46 | 38.0 | |
| AR | <0.001 | ||||
| 0 | 36 | 29.8 | 9 | 7.4 | |
| + | 25 | 20.7 | 17 | 14.0 | |
| ++ | 48 | 39.7 | 35 | 28.9 | |
| +++ | 12 | 9.9 | 60 | 49.6 | |
Note. NCTs: corresponding noncancerous tissues.
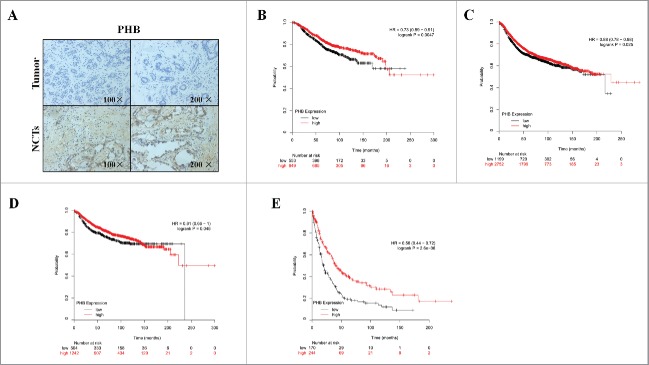
Figure 1.
Low expression of PHB is associated with poor prognosis of breast cancer. (A) PHB expression in the breast cancer tissues and corresponding noncancerous tissues (NCTs) was detected by immunohistochemical analysis. Kaplan-Meier analyses of association of PHB expression with overall survival (B), relapse-free survival (C), distant metastasis-free survival (D), and post-progression survival (E) of breast cancer patients. P-values were calculated with log-rank (Mantel-Cox) test. Patients were stratified into ‘low’ and ‘high’ PHB expression based on auto select best cutoff.
Low expression of PHB is associated with poor prognosis in patients with breast cancer
To investigate the role of PHB in the breast cancer clinical outcomes, we performed meta-analyses using Kaplan-Meier plotter online breast cancer survival analysis. As shown in Fig. 1, we found low expression of PHB positively correlated with reduced overall survival (Fig. 1B, HR = 0.73, 95%CI = 0.59–0.91, P = 0.0047), relapse-free survival (Fig. 1C, HR = 0.88, 95%CI = 0.78–0.98, P = 0.025), distant metastasis-free survival (Fig. 1D, HR = 0.81, 95%CI = 0.66–1, P = 0.048), and post-progression survival (Fig. 1E, HR = 0.56, 95%CI = 0.44–0.72, P < 0.0001), suggesting a prognostic value of PHB in breast cancers. Taken together, these findings suggest low expression of PHB is associated with poor prognosis in patients with breast cancer.
PHB inhibits breast cancer cell proliferation, induces cell cycle arrest and promotes cell apoptosis
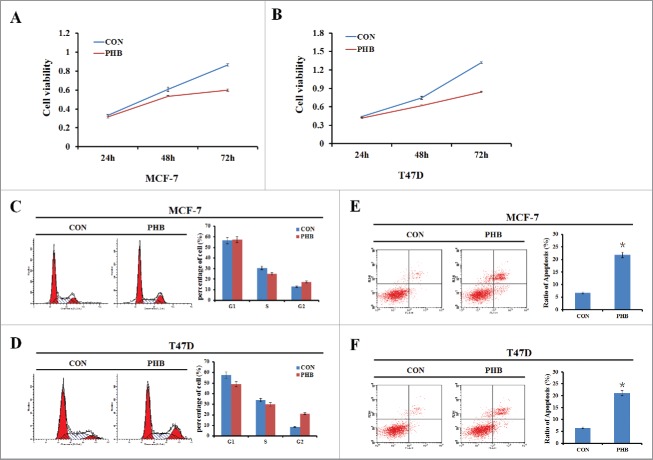
To investigate the role of PHB in the breast cancer cell growth, plasmids encoding PHB were transfected into ER-positive breast cancer cell lines MCF-7 and T47D, and cells were collected at different time points (24, 48, or 72 h) to determine cell survival rates. As shown in Fig. 2A–B, reduction of cell proliferation occurred in a time-dependent manner following overexpression of PHB in both 2 cell lines. Next we sought to determine the effect of PHB on ER-positive breast cancer cell cycle progression by flow cytometry. After transfected with PHB vectors for 48 h, cells in S phase were decreased and G2 phase cells were increased in the both 2 cell lines (Fig. 2C–D). These results showed that PHB could change the cell cycle distribution and induce S/G2 phase cells cycle arrest and therefore inhibit cell proliferation in ER-positive breast cancer cells. Furthermore, cell apoptosis was increased in both MCF-7 and T47D cells following overexpression of PHB (Fig. 2E–F). Collectively, these data indicated that PHB inhibits breast cancer cell proliferation, changes cell cycle distribution and promotes cell apoptosis.
Figure 2.
PHB inhibits breast cancer cell proliferation, induces cell cycle arrest and promotes cell apoptosis. (A, B) Cell viability was measured by MTT assay 24, 48, or 72 h following transfection of plasmids encoding PHB into MCF-7 and T47D cell lines. (C, D) The MCF-7 and T47D cells were transfected PHB vector or control vector for 48 h, and then cell cycle distribution was determined by flow cytometry analysis using PI staining. (E, F) Apoptosis was detected by flow cytometry analysis following transfection of plasmids encoding PHB into MCF-7 and T47D cell lines. Data were represented as the average of 3 independent experiments. *P < 0.05, as compared with control vector transfected cells.
PHB induces AR expression in the ER-positive breast cancer cells
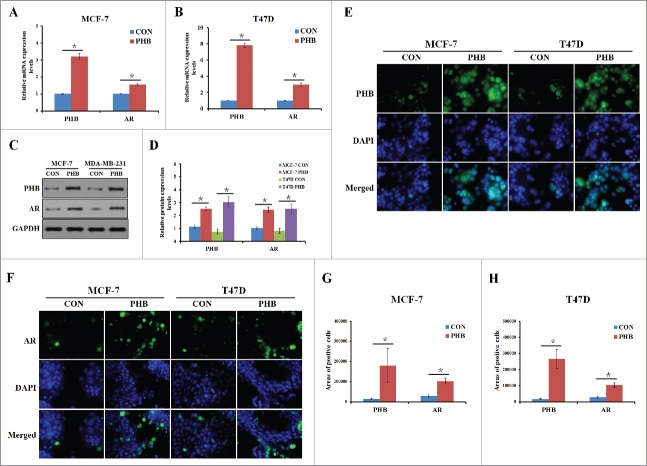
Recent studies have found that the androgen receptor (AR) is expressed in 60–70% of breast tumors.19 However, little is known about whether PHB could regulate AR expression and the underlying mechanism in the ER-positive breast cancer cells. To address it, we transfected MCF-7 and T47D cell lines with PHB plasmids and determined whether PHB contributes to AR expression. As shown in Fig. 3A–B, the expression levels of AR mRNA were significantly increased in the both 2 ER-positive breast cancer after transfection of PHB plasmids. To understand whether PHB also confers AR expression regulation at post-translational level, we detected the protein level of AR in the absence and presence of PHB plasmids. As shown in Fig. 3C–D, PHB plasmids transfection led to an increased AR expression compared with the controls in the MCF-7 and T47D cells. Moreover, immunofluorescence was performed to determine the cellular localization of AR in the cells stimulated with PHB plasmids for 48 h, shown that AR staining was predominantly nuclear and the expression of AR was significantly increased in the nuclear levels in treated ER-positive breast cancer cells (Fig. 3E–H). Taken together, these results suggested that PHB could significantly increase AR expression in mRNA and protein levels in the ER-positive breast cancer cells.
Figure 3.
PHB induces AR expression in the ER-positive breast cancer cells. PHB and AR mRNA levels were detected by q-PCR following transfection of plasmid encoding PHB into MCF-7 (A) and T47D (B) cells, and normalized to GAPDH expression. (C, D) PHB protein levels were determined by western blotting following transfection of plasmid encoding PHB into MCF-7 and T47D cells, and normalized to GAPDH expression. Localization and expression of PHB (E) and AR (F) were determined by fluorescence microscopy following the same treatment as (A, B). Nuclei were stained with DAPI. The areas of PHB or AR positive cells in the MCF (G) and T47D (H) cells were analyzed.
PHB positively relates to AR expression in the breast cancer patients
To investigate the expression of AR in the human breast cancer, we determined the AR expression status in 121 breast cancer tissues and corresponding NCTs by IHC staining. Relationship between AR expression and clinicopathological features of 121 breast cancer patients was determined (Table 2). Of the 121 tumor tissues, 60 cases (49.6%) and 61 cases (50.4%) expressed AR at high and low levels, respectively (Table 2). However, 95 cases (78.5%) and 26 cases (21.5%) expressed a high and low levels of AR in the 121 corresponding NCTs, respectively (Table 3). Moreover, we found that menopausal status, tumor location, tumor size, vascular/perineural/lymph node invasion, and histological grade were not significantly associated with AR expression (Table 2). PR, HER-2, p53, and Ki-67 expression were also not significantly associated with AR expression. However, we found high AR expression was associated with ages, negative ER and E-Cadherin expression, respectively (Table 2). As shown in Fig. 4 and Table 3, the expression of AR was low in the tumor tissue, while the AR expression in the corresponding NCTs was high (P < 0.001). Moreover, a significant positive correlation between PHB and AR expression was identified in the 121 breast cancer tissues (r = 0.521, P < 0.001) (Table 4). Taken together, these data suggest that PHB positively relates to AR expression in the breast cancer patients.
Table 2.
Relationship between the expression of AR and clinicopathological features of 121 breast cancer patients.
|
Total (n = 121) |
AR high (n = 60) |
AR low (n = 61) |
|||||
|---|---|---|---|---|---|---|---|
| Variables | No. | % | No. | % | No. | % | P-value |
| Age | 0.014* | ||||||
| < 48 years | 56 | 46.3 | 21 | 35.0 | 35 | 57.4 | |
| ≥ 48 years | 65 | 53.7 | 39 | 65.0 | 26 | 42.6 | |
| Menopausal status | 0.172 | ||||||
| Premenopausal or perimenopausal | 70 | 57.9 | 31 | 51.7 | 39 | 63.9 | |
| Postmenopausal | 51 | 42.1 | 29 | 48.3 | 22 | 36.1 | |
| Tumor location | 0.122 | ||||||
| Left | 61 | 50.4 | 26 | 43.3 | 35 | 57.4 | |
| Right | 60 | 49.6 | 34 | 56.7 | 26 | 42.6 | |
| Tumor size | 0.728 | ||||||
| < 2 | 28 | 23.1 | 12 | 20.0 | 16 | 26.2 | |
| 2–5 | 86 | 71.1 | 44 | 73.3 | 42 | 68.9 | |
| ≥ 5 | 7 | 5.8 | 4 | 6.7 | 3 | 4.9 | |
| Vascular invasion | 0.345 | ||||||
| Yes | 35 | 28.9 | 15 | 25.0 | 20 | 32.8 | |
| No | 86 | 71.1 | 45 | 75.0 | 41 | 67.2 | |
| Perineural invasion | 0.441 | ||||||
| Yes | 32 | 26.4 | 14 | 23.3 | 18 | 29.5 | |
| No | 89 | 73.6 | 46 | 76.7 | 43 | 70.5 | |
| Lymph node invasion | 0.779 | ||||||
| Yes | 56 | 46.3 | 27 | 45.0 | 29 | 47.5 | |
| No | 65 | 53.7 | 33 | 55.0 | 32 | 52.5 | |
| Lymph nodes | 0.131 | ||||||
| 0 | 66 | 54.5 | 33 | 55.0 | 33 | 54.1 | |
| 1–3 | 43 | 35.5 | 18 | 30.0 | 25 | 41.0 | |
| ≥ 4 | 12 | 9.9 | 9 | 15.0 | 3 | 4.9 | |
| T | 0.944 | ||||||
| T1 | 50 | 41.3 | 24 | 40.0 | 26 | 42.6 | |
| T2 | 67 | 55.4 | 34 | 56.7 | 33 | 54.1 | |
| T3 | 4 | 3.3 | 2 | 3.3 | 2 | 3.3 | |
| N | 0.202 | ||||||
| N0 | 66 | 54.5 | 33 | 55.0 | 33 | 54.1 | |
| N1 | 43 | 35.5 | 18 | 30.0 | 25 | 41.0 | |
| N2 | 10 | 8.3 | 7 | 11.7 | 3 | 4.9 | |
| N3 | 2 | 1.7 | 2 | 3.3 | 0 | 0 | |
| Histological grade | 0.368 | ||||||
| I | 32 | 26.4 | 15 | 25.0 | 17 | 27.9 | |
| II | 74 | 61.2 | 35 | 58.3 | 39 | 63.9 | |
| III | 15 | 12.4 | 10 | 16.7 | 5 | 8.2 | |
| ER | 0.016* | ||||||
| Positive | 89 | 73.6 | 50 | 83.3 | 39 | 63.9 | |
| Negative | 32 | 26.4 | 10 | 16.7 | 22 | 36.1 | |
| PR | 0.080 | ||||||
| Positive | 78 | 64.5 | 44 | 73.3 | 34 | 55.7 | |
| Negative | 43 | 35.5 | 18 | 30.0 | 27 | 44.3 | |
| HER-2 | 0.648 | ||||||
| Positive | 36 | 29.8 | 19 | 31.7 | 17 | 27.9 | |
| Negative | 85 | 70.2 | 41 | 68.3 | 44 | 72.1 | |
| p53 | 0.212 | ||||||
| Positive | 76 | 62.8 | 41 | 68.3 | 35 | 57.4 | |
| Negative | 45 | 37.2 | 19 | 31.7 | 26 | 42.6 | |
| E-Cadherin | 0.043* | ||||||
| Positive | 78 | 64.5 | 44 | 73.3 | 34 | 55.7 | |
| Negative | 43 | 35.5 | 16 | 26.7 | 27 | 44.3 | |
| Ki-67 | 0.877 | ||||||
| < 14% | 31 | 25.6 | 15 | 25.0 | 16 | 26.2 | |
| ≥ 14% | 90 | 74.4 | 45 | 75.0 | 45 | 73.8 | |
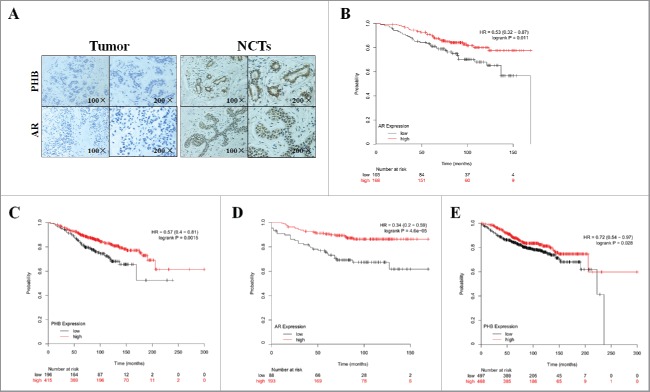
Figure 4.
PHB and AR expression are associated with prognosis in the ER-positive breast cancer patients. (A) PHB and AR expression in the breast cancer tissues and corresponding noncancerous tissues (NCTs) was detected by immunohistochemical analysis. Kaplan-Meier analyses of association of AR expression with overall survival (B) and distant metastasis-free survival (C), PHB expression with overall survival (D) and distant metastasis-free survival (E) in the ER-positive breast cancer patients. P-values were calculated with log-rank (Mantel-Cox) test. Patients were stratified into ‘low’ and ‘high’ AR or PHB expression based on auto select best cutoff.
Table 4.
Correlative analysis of the AR expression with PHB of the 121 breast cancer patients.
| Breast cancer patients (n = 121) |
||
|---|---|---|
| PHB (Positive) | PHB (Negative) | |
| AR (Positive) | 45 | 15 |
| AR (Negative) | 14 | 47 |
| r | 0.521 | |
| P | <0.001 | |
PHB and AR expression are associated with prognosis in the ER-positive breast cancer patients
To investigate the role of PHB and AR in the ER-positive breast cancer clinical outcomes, we then performed meta-analyses using Kaplan-Meier plotter online breast cancer survival analysis. As shown in Fig. 4, we found low expression of AR positively correlated with reduced overall survival (Fig. 4B, HR = 0.53, 95%CI = 0.32–0.87, P = 0.011) and distant metastasis-free survival (Fig. 4D, HR = 0.34, 95%CI = 0.2–0.59, P < 0.0001) in the ER-positive breast cancer patients. In addition, we also determined that low expression of PHB was also positively correlated with reduced overall survival (Fig. 4C, HR = 0.57, 95%CI = 0.4–0.81, P = 0.0015) and distant metastasis-free survival (Fig. 4E, HR = 0.72, 95%CI = 0.54–0.97, P = 0.028) in the ER-positive breast cancer patients, suggesting a prognostic value of PHB and AR in ER-positive breast cancers. Taken together, these findings suggest low expression of PHB and AR is associated with poor prognosis in patients with ER-positive breast cancer.
Discussion
It was reported that PHB could repress AR activity and androgen-stimulated growth of prostate cancer cells and conversely that knockdown of PHB increased AR activity and prostate cancer cell growth in response to androgens, both in vitro and in vivo.11 In the present study, we found PHB could significantly increase AR expression in mRNA and protein levels in the ER-positive breast cancer cells. A significant positive correlation between PHB and AR expression was also identified in 121 breast cancer tissues. However, AR was reported to act as a different role in the ER-positive and ER-negative breast cancer.14 AR signaling exerts an anti-proliferative influence in ER-positive breast cancer cells,15-17 and promotes growth of a subset of ER-negative, AR-positive breast cancers.18 Peters et al. reported that AR inhibits ER-α transactivation activity through binding to an estrogen-responsive element (ERE) in ER-positive breast cancer cells.16 Moreover, He et al. reported that PHB acts as a potent transcriptional co-repressor of ERα and associate with estrogen-regulated promoters in the absence of hormone, dissociating after hormone treatment in the breast cancer cells.8 Collectively, it was postulated that PHB promotes AR activation and AR inhibits ER-α transactivation activity in ER-positive breast cancer.
We used 121 breast cancer tissues and corresponding noncancerous tissues (NCTs) to investigate the expression status of PHB by immunohistochemical (IHC) staining in the human breast cancer. We found that the expression of PHB was low in the tumor tissues, while the PHB expression in the corresponding NCTs was high. The prohibitin (PHB) gene encodes a protein of about 32 kDa molecular mass originally identified as an intracellular, anti-proliferative protein in cancer.20 Our data also showed that PHB could inhibit breast cancer cell proliferation, change cell cycle distribution and promote cell apoptosis in the ER-positive breast cancer cells. As a tumor suppressor, low expression of PHB is associated with poor prognosis in patients with breast cancer. As shown in Fig. 4C and E, we also found that low expression of PHB was positively correlated with reduced overall survival and distant metastasis-free survival in the ER-positive breast cancer patients.
Functional studies have suggested that PHB could contribute to the control of the G1 to S transition in the breast cancer cells, which was later found to be associated with p53 gene.21,22 However, there is also study reported that PHB could contribute to the control of the S to G2 transition in the prostate cancer cells and cells containing PHB siRNA had an increase in cell population entering the S/G2 M phase.23 Specifically 2% of these cells were detected in this phase when compared with control cells, which increased to 10% after the stimulation of dihydrotestosterone (DHT).23 In the present study, we found that cells in S phase were decreased and G2 phase cells were increased in the both 2 cell lines after transfected with PHB vectors for 48 h. Moreover, we detected that p53 expression was slightly associated with PHB expression in the 121 breast cancer patients' tissues (P = 0.063). PHB was also reported to physically interact with Rb family proteins in inhibiting E2F-mediated transcription to inhibit cell proliferation in breast cancer.6,22 As shown in Fig. 2A, reduction of cell proliferation occurred in a time-dependent manner following overexpression of PHB in the both 2 ER-positive breast cancer cell lines.
Collectively, our data showed that the expression of PHB is significantly low in breast cancer samples, and low PHB expression positively correlated with poor prognosis of breast cancer. Overexpression of PHB could inhibit breast cancer cell proliferation, change cell cycle distribution and promote cell apoptosis in the ER-positive breast cancer cells. Moreover, we found PHB could significantly increase AR expression in the ER-positive breast cancer cells. A significant positive correlation between PHB and AR expression was identified in the breast cancer patient tissues. Our results indicated that PHB could promote AR activation in ER-positive breast cancer, making PHB and AR potential molecular targets for ER-positive breast cancer therapy. To the best of our knowledge, this is the first report revealing that the association of PHB and AR expression in the breast cancer. However, additional larger studies and more detailed investigations of the regulatory mechanisms underlying the influence of PHB on the expression of AR in the ER-positive breast cancer are needed to validate our findings.
Materials and methods
Clinical samples and survival analysis
Breast cancer tissues section was collected from Wuhu Second People's Hospital (Anhui, China) between March 2010 and March 2013. None of the patients received chemotherapy or radiotherapy before surgery. Experiments were approved by the Ethics Committee of Jinling Hospital and were conducted in compliance with the Helsinki Declaration. Histological parameters were determined in accordance with the criteria of the World Health Organization. Pathologic staging was performed in accordance with the current International Union against Cancer tumor-lymph node metastasis classification. Kaplan-Meier survival analyses for disease outcomes were performed using the online database (www.kmplot.com) and the percentiles of the patients between the upper and lower quartiles were auto-selected based on the computed best performing thresholds as cutoffs.24
Immunohistochemistry
One hundred and 21 breast tumor tissue samples were deparaffinized in xylene, followed by heat-mediated antigen retrieval using citrate buffer (BioGenex Laboratories, San Ramon, CA, USA). Antibody staining was visualized with DAB (Sigma, D-5637) and hematoxylin counterstain. The H-score method was used in this trial; we multiplied the percentage score by the staining intensity score. The percentage of positively stained cells was scored as ‘−’ (0%), ‘+’ (1–25%), ‘++’ (26–50%), or ’+++’ (51–100%). Intensity was scored as ‘−’ (negative), ‘+’ (weak), ‘++’ (moderate), or ’+++’ (strong). Immunohistochemical scoring was performed without prior knowledge of the clinical response. Immunostained sections were scanned using a microscope (Aiovert 200; Carl Zeiss).
Cell lines and cell culture
Breast cancer cell lines MCF-7 and T47D were purchased from the American Type Culture Collection (Manassas, VA, USA), and cultured in RPMI 1640 or DMEM medium (GIBCO, Gaithersburg, MD, USA) supplemented with 10% FBS and 1% penicillin/streptomycin at 37°C in a humidified atmosphere containing 5% CO2.
Plasmids and transient transfection
PHB plasmids were generously provided by D. Johnson.25 Cells (1 × 106 cells/well) were plated in 6-well plates 24 h before transfection. Plasmids were then transfected into cells using TurboFect Transfection Reagent (Thermo Scientific) according to the manufacturer's protocol. Following incubation at 37°C for 24 h, cells were collected and lysed to verify the expression of related proteins by western blot analysis.
Cell survival (MTT) assay
A total of 1 × 104 cells per well were seeded into 96-well plates and incubated with various concentrations of resveratrol or transfected with required plasmids for 48 h. Following addition of 20 µL of 0.5 mg/mL MTT solution (Sigma) to each well, the medium was replaced with 200 µL DMSO after 4 h and vortexed for 10 min. Absorbance was measured at 490 nm with a microplate reader (BIO-RAD, USA) to determine the relative numbers of viable cells. Assays were performed independently 3 times.
Cell cycle and apoptosis analysis
Cells were treated with plasmids for 48 h, then harvested by trypsinization (no EDTA) and washed with phosphate-buffered saline (PBS). Analysis of the cell cycle and apoptosis was performed as described previously.26-29 Each sample was tested in triplicate and untreated cells were used as controls.
RNA isolation and quantitative RT-PCR
Total RNA was extracted from cultured cells using Trizol (Invitrogen, CA, USA) according to the manufacturer's protocol. Reverse transcription was conducted using a PrimeScript 1st Strand cDNA synthesis kit (TaKaRa, Dalian, China) according to the manufacturer's instructions. Primer sequences (forward and reverse, respectively) were as follows: PHB, 5′- CCATGATGTGCACTTTGGGC -3′ and 5′- CACTCCACGGAATCGGTCAA -3′; AR, 5′-GACGACCAGATGGCTGTCATT-3′ and 5′-GGGCGAAGTAGAGCATCCT-3′; GAPDH, 5′-CATCTTCTTTTGCGTCGCCA-3′ and 5′- TTAAAAGCAGCCCTGGTGACC -3′. PCR analysis was performed in a 20-μL volume with amplification conditions: 95°C for 2 min [94°C for 10 s, 60°C for 10 s, and 72°C for 40 s], 40 cycles. All reactions were performed in triplicate. Threshold cycles (CT) were determined using fixed threshold settings. All PCR assays included no template controls and were run in triplicate.
Western blotting
Total protein was extracted using RIPA buffer supplemented with protease and phosphatase inhibitors, with protein concentrations determined using a BCA kit (Thermo Scientific). Approximately 20 µg protein was loaded per lane and separated on a sodium dodecylsulfate-polyacrylamide gel and blotted onto nitrocellulose. Blots were blocked with 5% dry milk in tris-buffered saline/0.1% tween-20 and incubated overnight with a diluted solution of primary antibody at 4°C, followed by incubation with a horseradish peroxidase-conjugated secondary antibody (1:5000) for 2 h. Antibodies used for western blot were: rabbit anti-PHB antibody (1:100, Ls-B7282), and rabbit anti-AR antibody (1:100, ab74272). Bands were normalized to GAPDH expression, which was used as an internal loading control. Results from at least 2 separate experiments were analyzed.
Immunofluorescence analysis
Immunofluorescence staining was used to verify protein expression and examine the subcellular localization of PHB and AR. Cells were plated onto glass coverslips in 6-well plates and transfected with PHB plasmids for 48 h. Cells were then washed with PBS and fixed in 4% paraformaldehyde for 20 min, permeabilized with 0.1% TritonX-100 for 10 min, and incubated for 1 h at 37°C with the following antibodies: rabbit anti-PHB antibody (1:100, Ls-B7282), and rabbit anti-AR antibody (1:100, ab74272). Cells were then washed with PBS and incubated for 30 min at 37°C with mouse anti-rabbit IgG conjugated with FITC (Invitrogen; 1:200). Subsequently, nuclei were counterstained with 4′, 6-diamidino-2-phenylindole (DAPI; Sigma) for 10 min. Finally, cells were mounted with mounting solution (DAKO, Glostrup, Denmark) and examined under a LSM510 confocal microscope (Carl Zeiss, Gottingen, Germany).
Statistical analysis
SPSS Statistics 19.0 (SPSS Inc.) was used for statistical analysis. Data were analyzed using one-way ANOVA or a Student's t-test. Data are presented as means ± SD of 3 independent experiments. The log-rank test was used to assess statistical significance of Kaplan-Meier plots. The chi-square test was used for IHC data. The correlation between PHB and AR expression level was estimated using Spearman's correlation analysis, *P < 0.05, or **P < 0.001.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This research was supported by National Natural Science Foundation of China (No.81470357), a Foundation for Clinical Medicine Science and Technology Special Project of the Jiangsu Province, China (No. BL2014071).
References
- [1].Nijtmans LG, Artal SM, Grivell LA, Coates PJ. The mitochondrial PHB complex: Roles in mitochondrial respiratory complex assembly, ageing and degenerative disease. Cell Mol Life Sci 2002; 59:143-55; PMID:11852914; http://dx.doi.org/ 10.1007/s00018-002-8411-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Rajalingam K, Wunder C, Brinkmann V, Churin Y, Hekman M, Sievers C, Rapp UR, Rudel T. Prohibitin is required for Ras-induced Raf-MEK-ERK activation and epithelial cell migration. Nat Cell Biol 2005; 7:837-43; PMID:16041367; http://dx.doi.org/ 10.1038/ncb1283 [DOI] [PubMed] [Google Scholar]
- [3].Coates PJ, Jamieson DJ, Smart K, Prescott AR, Hall PA. The prohibitin family of mitochondrial proteins regulate replicative lifespan. Curr Biol 1997; 7:607-10; PMID:9259555; http://dx.doi.org/ 10.1016/S0960-9822(06)00261-2 [DOI] [PubMed] [Google Scholar]
- [4].McClung JK, Jupe ER, Liu XT, Dell'Orco RT. Prohibitin: Potential role in senescence, development, and tumor suppression. Exp Gerontol 1995; 30:99-124; PMID:8591812; http://dx.doi.org/ 10.1016/0531-5565(94)00069-7 [DOI] [PubMed] [Google Scholar]
- [5].Choi D, Lee SJ, Hong S, Kim IH, Kang S. Prohibitin interacts with RNF2 and regulates E2F1 function via dual pathways. Oncogene 2008; 27:1716-25; PMID:17873902; http://dx.doi.org/ 10.1038/sj.onc.1210806 [DOI] [PubMed] [Google Scholar]
- [6].Wang S, Fusaro G, Padmanabhan J, Chellappan SP. Prohibitin co-localizes with Rb in the nucleus and recruits N-CoR and HDAC1 for transcriptional repression. Oncogene 2002; 21:8388-96; PMID:12466959; http://dx.doi.org/ 10.1038/sj.onc.1205944 [DOI] [PubMed] [Google Scholar]
- [7].Gamble SC, Odontiadis M, Waxman J, Westbrook JA, Dunn MJ, Wait R, Lam EW, Bevan CL. Androgens target prohibitin to regulate proliferation of prostate cancer cells. Oncogene 2004; 23:2996-3004; PMID:14968116; http://dx.doi.org/ 10.1038/sj.onc.1207444 [DOI] [PubMed] [Google Scholar]
- [8].He B, Feng Q, Mukherjee A, Lonard DM, DeMayo FJ, Katzenellenbogen BS, Lydon JP, O'Malley BW. A repressive role for prohibitin in estrogen signaling. Mol Endocrinol 2008; 22:344-60; PMID:17932104; http://dx.doi.org/ 10.1210/me.2007-0400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wang S, Zhang B, Faller DV. BRG1/BRM and prohibitin are required for growth suppression by estrogen antagonists. EMBO J 2004; 23:2293-303; PMID:15141164; http://dx.doi.org/ 10.1038/sj.emboj.7600231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Dai Y, Ngo D, Jacob J, Forman LW, Faller DV. Prohibitin and the SWI/SNF ATPase subunit BRG1 are required for effective androgen antagonist-mediated transcriptional repression of androgen receptor-regulated genes. Carcinogenesis 2008; 29:1725-33; PMID:18487222; http://dx.doi.org/ 10.1093/carcin/bgn117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Dart DA, Spencer-Dene B, Gamble SC, Waxman J, Bevan CL. Manipulating prohibitin levels provides evidence for an in vivo role in androgen regulation of prostate tumours. Endocr Relat Cancer 2009; 16:1157-69; PMID:19635783; http://dx.doi.org/ 10.1677/ERC-09-0028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Dart DA, Brooke GN, Sita-Lumsden A, Waxman J, Bevan CL. Reducing prohibitin increases histone acetylation, and promotes androgen independence in prostate tumours by increasing androgen receptor activation by adrenal androgens. Oncogene 2012; 31:4588-98; PMID:22179832; http://dx.doi.org/ 10.1038/onc.2011.591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Fletcher CE, Dart DA, Sita-Lumsden A, Cheng H, Rennie PS, Bevan CL. Androgen-regulated processing of the oncomir miR-27a, which targets Prohibitin in prostate cancer. Hum Mol Genet 2012; 21:3112-27; PMID:22505583; http://dx.doi.org/ 10.1093/hmg/dds139 [DOI] [PubMed] [Google Scholar]
- [14].Hickey TE, Robinson JL, Carroll JS, Tilley WD. Minireview: The androgen receptor in breast tissues: Growth inhibitor, tumor suppressor, oncogene? Mol Endocrinol 2012; 26:1252-67; PMID:22745190; http://dx.doi.org/ 10.1210/me.2012-1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Margolis RN, Evans RM, O'Malley BW, Consortium NA. The nuclear receptor signaling atlas: Development of a functional atlas of nuclear receptors. Mol Endocrinol 2005; 19:2433-6; PMID:16051673; http://dx.doi.org/ 10.1210/me.2004-0461 [DOI] [PubMed] [Google Scholar]
- [16].Peters AA, Buchanan G, Ricciardelli C, Bianco-Miotto T, Centenera MM, Harris JM, Jindal S, Segara D, Jia L, Moore NL, et al.. Androgen receptor inhibits estrogen receptor-alpha activity and is prognostic in breast cancer. Cancer Res 2009; 69:6131-40; PMID:19638585; http://dx.doi.org/ 10.1158/0008-5472.CAN-09-0452 [DOI] [PubMed] [Google Scholar]
- [17].Szelei J, Jimenez J, Soto AM, Luizzi MF, Sonnenschein C. Androgen-induced inhibition of proliferation in human breast cancer MCF7 cells transfected with androgen receptor. Endocrinology 1997; 138:1406-12; PMID:9075695; http://dx.doi.org/ 10.1210/endo.138.4.5047 [DOI] [PubMed] [Google Scholar]
- [18].Ni M, Chen Y, Lim E, Wimberly H, Bailey ST, Imai Y, Rimm DL, Liu XS, Brown M. Targeting androgen receptor in estrogen receptor-negative breast cancer. Cancer Cell 2011; 20:119-31; PMID:21741601; http://dx.doi.org/ 10.1016/j.ccr.2011.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Niemeier LA, Dabbs DJ, Beriwal S, Striebel JM, Bhargava R. Androgen receptor in breast cancer: Expression in estrogen receptor-positive tumors and in estrogen receptor-negative tumors with apocrine differentiation. Mod Pathol 2010; 23:205-12; PMID:19898421; http://dx.doi.org/ 10.1038/modpathol.2009.159 [DOI] [PubMed] [Google Scholar]
- [20].Terashima M, Kim KM, Adachi T, Nielsen PJ, Reth M, Kohler G, Lamers MC. The IgM antigen receptor of B lymphocytes is associated with prohibitin and a prohibitin-related protein. EMBO J 1994; 13:3782-92; PMID:8070406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Roskams AJ, Friedman V, Wood CM, Walker L, Owens GA, Stewart DA, Altus MS, Danner DB, Liu XT, McClung JK. Cell cycle activity and expression of prohibitin mRNA. J Cell Physiol 1993; 157:289-95; PMID:8227162; http://dx.doi.org/ 10.1002/jcp.1041570211 [DOI] [PubMed] [Google Scholar]
- [22].Fusaro G, Dasgupta P, Rastogi S, Joshi B, Chellappan S. Prohibitin induces the transcriptional activity of p53 and is exported from the nucleus upon apoptotic signaling. J Biol Chem 2003; 278:47853-61; PMID:14500729; http://dx.doi.org/ 10.1074/jbc.M305171200 [DOI] [PubMed] [Google Scholar]
- [23].Feldman BJ, Feldman D. The development of androgen-independent prostate cancer. Nat Rev Cancer 2001; 1:34-45; PMID:11900250; http://dx.doi.org/ 10.1038/35094009 [DOI] [PubMed] [Google Scholar]
- [24].Gyorffy B, Surowiak P, Budczies J, Lanczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One 2013; 8:e82241; PMID:24367507; http://dx.doi.org/ 10.1371/journal.pone.0082241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Chander H, Halpern M, Resnick-Silverman L, Manfredi JJ, Germain D. Skp2B attenuates p53 function by inhibiting prohibitin. EMBO Rep 2010; 11:220-5; PMID:20134482; http://dx.doi.org/ 10.1038/embor.2010.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Lai J, Nie W, Zhang W, Wang Y, Xie R, Wang Y, Gu J, Xu J, Song W, Yang F. Transcriptional regulation of the p73 gene by Nrf-2 and promoter CpG methylation in human breast cancer. Oncotarget 2014; 5:6909-22; PMID:25071010; http://dx.doi.org/ 10.18632/oncotarget.2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zhang W, Luo J, Chen F, Yang F, Song W, Zhu A, Guan X. BRCA1 regulates PIG3-mediated apoptosis in a p53-dependent manner. Oncotarget 2015; 6:7608-18; PMID:25797244; http://dx.doi.org/ 10.18632/oncotarget.3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zhang W, Luo J, Yang F, Wang Y, Yin Y, Strom A, JÅ Gustafsson, Guan X. BRCA1 inhibits AR-mediated proliferation of breast cancer cells through the activation of SIRT1. Sci Rep 2016; 6:22034; PMID:26902145; http://dx.doi.org/ 10.1038/srep22034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Zhang W, Cao L, Sun Z, Xu J, Tang L, Chen W, Luo J, Yang F, Wang Y, Guan X. Skp2 is over-expressed in breast cancer and promotes breast cancer cell proliferation. Cell Cycle 2016; 15:1344-51; PMID:27111245; http://dx.doi.org/ 10.1080/15384101.2016.1160986 [DOI] [PMC free article] [PubMed] [Google Scholar]