Abstract
With extremely short generation times and high mutability, many viruses can rapidly evolve and adapt to changing environments. This ability is generally beneficial to viruses as it allows them to evade host immune responses, evolve new behaviours, and exploit ecological niches. However, natural selection typically generates adaptation in response to the immediate selection pressures that a virus experiences in its current host. Consequently, we argue that some viruses, particularly those characterised by long durations of infection and ongoing replication, may be susceptible to short-sighted evolution, whereby a virus’ adaptation to its current host will be detrimental to its onward transmission within the host population. Here we outline the concept of short-sighted viral evolution and provide examples of how it may negatively impact viral transmission among hosts. We also propose that viruses that are vulnerable to short-sighted evolution may exhibit strategies that minimise its effects. We speculate on the various mechanisms by which this may be achieved, including viral life history strategies that result in low rates of within-host evolution, or the establishment of a ‘germline’ lineage of viruses that avoids short-sighted evolution. These concepts provide a new perspective on the way in which some viruses have been able to establish and maintain global pandemics.
Keywords: virus, evolution, transmission
Trends
Adaptive evolutionary change of viruses within hosts can be detrimental to onward transmission (short-sighted evolution).
Loss of transmissibility is likely to be most problematical for rapidly evolving persistent (chronic) viral infections.
Within-host viral populations or subpopulations can exhibit lower rates of evolution that expected.
Selection probably occurs at the point of transmission, often leading to only one or a few viruses establishing new infections.
The preferential transmission of founder-like viruses that have undergone little within-host evolution has been proposed.
Short-Sighted Evolution
On infection of a new host, the genomes of many viruses undergo rapid adaptive evolution, which may result in escape from host immune responses 1, 2, 3, 4, 5, 6, 7, 8 and increases in viral growth rates [9]. Although these genetic changes make viruses superior competitors within their current host, they do not necessarily favour improved transmission between hosts 1, 10, 11. A logical consequence of this process is ‘short-sighted’ evolution (see Glossary), by which adaptation at the within-host level occurs at the expense of the spread of the virus through the host population [12].
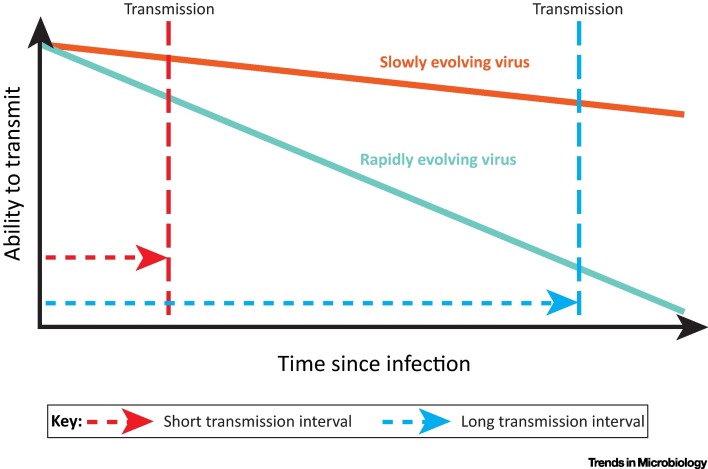
Susceptibility to short-sighted evolution will be influenced by two factors: the rate of viral adaptive evolution and the time between transmission events, which we refer to here as the ‘transmission interval’. For instance, acute viral infections, such as influenza and norovirus, are typically short-lived with little time for within-host adaptation before transmission to a new host. Their strategy is one of ‘smash and grab’: infect a new host, reproduce, and get out before the adaptive immune system removes the infection. Such viruses have short transmission intervals and will exhibit little short-sighted evolution, irrespective of their rate of adaptive evolution. Alternatively, persistent viral infections that use proof-reading polymerases, such as Herpes and Papilloma viruses, are unlikely to suffer short-sighted evolution for a different reason; their low mutation rates constrain the rate of host-specific viral adaptation, regardless of their transmission interval (Figure 1 ). It is common for these viruses to have larger genomes with many genes, which enable the virus to manipulate or hide from host immune responses, for example by persisting in a nonproliferative latent state [13].
Figure 1.
The Ability of Viruses to Transmit As Infection Progresses. Adaptation of viruses to the within-host environment is likely to reduce their ability to transmit to new hosts. If the time between infection and onward transmission (the transmission interval) is short, as will be the case for acute viral infections, any losses in the ability to transmit will be minor and predicted to have only a small effect on transmission (red dotted lines). For viruses with longer transmission intervals, within-host adaptation is predicted to result in much greater losses in transmission (blue dotted lines). A fall in the ability to transmit is predicted to be most severe for viruses with a fast rate of within-host evolution and a long transmission interval (teal solid line). If the drop in the ability to transmit is too great, this will prevent the virus from spreading effectively from host to host, and the virus might not be able to persist in the host population over the long term.
In contrast, short-sighted evolution could be problematic for persistent chronic viral infections that use low-fidelity polymerases and which undergo active replication throughout infection, such as human immunodeficiency virus (HIV-1) and hepatitis C virus (HCV). High rates of mutation during replication, large viral population sizes, and long durations of infection combine to create considerable potential for within-host adaptation, enabling these viruses to outpace natural and induced immune responses. However, long transmission intervals mean that this adaptation may come at a cost of reduced transmissibility later in infection (Figure 1).
Chronic viral infections are clearly successful within their natural hosts, so how do those with long transmission intervals avoid the detrimental impacts of short-sighted evolution? Here we suggest that such viruses exhibit life histories that either (i) significantly reduce rates of within-host adaptation, or (ii) lead to the retention of a genetic archive of viruses that are similar to the founder strains that initiated the infection. This archive is analogous to the germline in multicellular animals, which does not carry somatic mutations that accumulate during the lifetime of an individual. We further speculate that mechanisms that limit the effects of short-sighted evolution in chronic viruses could themselves be under viral control and therefore subject to selection (Box 1 ).
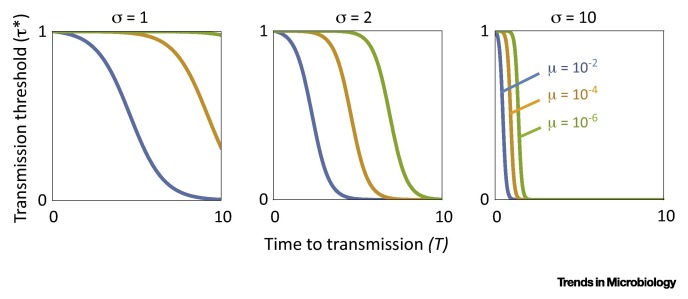
Figure I.
Factors Favouring the Evolution of a Viral Germline. Our mathematical model predicts that allocation of viral particles to a nonreplicating germline is favoured by natural selection if the relative transmission success τ of wild-type virus in the germline exceeds a threshold value τ* describing the threat imposed by mutation. Specifically, increased time to transmission (T), mutation rate (μ), and replicative advantage of the mutant genotype (σ) is associated with increased likelihood of the germline being favoured (lower τ*).
Box 1. Optimal Investment in a Viral Germline.
We construct an idealised mathematical model that can capture investment by a virus into a nonreplicating viral germline compartment, representing, for example, the HIV viral reservoir, or the proviral population of HTLV-1. We consider a viral infection comprising 1 unit of virus particles at the moment that allocation to the viral germline is decided, a proportion 1-μ of which are wild type and a proportion μ having a locally-adapted, mutant genotype – for example, a cytotoxic T-lymphocyte (CTL) escape mutation. A fraction γ of the virus particles are sequestered into the germline compartment, where they do not replicate, and the rest remain in the active compartment in which there is turnover of replicating viral particles, with the mutant viruses replicating at 1+σ times the rate of the wild-type viruses, but no net growth in the overall size of the viral population. We make the assumption of no net growth since, after the first few weeks of infection, viral loads tend to remain approximately stable for the majority of the infection. After a period of time, T, the wild-type virus particles transmit to new hosts, with each virus particle in the germline enjoying a fraction τ of the successful transmission enjoyed by those in the active compartment. The mutant virus particles have an unspecified rate of transmission that is lower than that of the wild type.
The total transmission achieved by the infection’s wild-type virus particles is therefore proportional to w = γ(1–μ)τ + (1–γ)(1–pT), where pT is the proportion of virus particles that are of the mutant type in the active compartment at time T. The first term represents the contribution of wild-type virus from the germline, and the second term is the contribution from the active compartment. The dynamics of the proportion pt of mutant particles in the active compartment, over the course of the infection, are given by dpt/dt = σpt(1–pt) (see [94] equation 3.11b for a derivation). Setting p 0 = μ, this yields pT = (μ exp(σT))/(1+μ(exp(στ)–1)). Wild-type transmission success w is an appropriate measure of Darwinian fitness if the transmission ability of the mutant is sufficiently low that it does not completely displace the wild type from the wider population. This is because any mutant virus particles that do successfully transmit nevertheless enjoy zero long-term reproductive value [95]: owing to the assumed absence of back mutation, all virus particles ultimately derive from wild-type ancestry, and hence no mutant virus particle leaves any descendants in the long term. Accordingly, any transmission achieved by mutant virus particles does not contribute to an infection’s fitness.
Natural selection favours an increase in allocation to the germline γ if dw/dγ > 0, which is equivalent to τ > 1/(1+μ(exp(σT)–1)). Denoting the right-hand side of the condition τ* yields a threshold level of germline transmission above which allocation to the germline is favoured, and below which it is not. Specifically, since the condition for increase is independent of the value of γ, natural selection favours full investment into the germline when the condition is satisfied (γ* = 1 when τ > τ*) and favours zero investment into the germline when the condition is not satisfied (γ* = 0 when τ < τ*). The threshold level of germline transmission τ* is a monotonically decreasing function of mutation rate μ, mutant replicative advantage σ, and time to transmission T (Figure I), such that increasing the values of these three parameters (and increasing the level of germline transmission, τ) makes it more likely that the germline will be favoured by natural selection.
For example, if only a short period of replication occurs prior to transmission (small T, left-hand side of each panel in Figure I), then allocation to the germline is only favoured if transmission from the germline is high (large τ*), whereas a longer period of replication prior to transmission (large T, right-hand side of each panel in Figure I) means that allocation to the germline can be favoured even if transmission of wild-type virus from the germline is greatly impaired (small τ*).
Alt-text: Box 1
To begin, we outline the evidence that within-host adaption can reduce viral transmissibility. We then discuss mechanisms by which viruses may avoid short-sighted evolution, before examining the evidence that, in rapidly evolving viruses, germline lineages are preferentially transmitted. Because they have been much more widely studied, most of the examples we use come from viruses that infect humans, but the general principles apply to all viruses.
Within-Host Adaptation Can Limit Transmission
Short-sighted evolution occurs when within-host adaptation reduces viral spread among hosts, either by reducing the per-contact transmissibility of the virus, or by reducing the contact rate of infected hosts due to increased pathogenicity, including host death. Here, we summarise the evidence for the accumulation of viral mutations during chronic infections that not only confer fitness advantages within hosts, but also limit the ability to transmit among hosts.
Adaptation to a Changing Within-Host Environment
The host environment presents a shifting landscape of selection pressures, created by a dynamic immune response and changes in the availability of target cells. Viral adaptation to these changes may result in short-sighted evolution. Unsurprisingly, most evidence for the loss of transmissibility following within-host adaptation comes from HIV-1. In the bid to develop a vaccine, there has been intense interest in characterising the viruses that are successfully transmitted and which initiate new infections (so-called transmitted/founder, or TF, viruses). Crucially, many of the characteristics of TF viruses appear to be selected against during the course of infection. Perhaps the most frequently cited of these is the switch from using the CCR5 to the CXCR4 coreceptor during late-stage infection in some patients, enabling the virus to infect naïve CD4+ T cells; in late infection there is a fall in the number of activated CD4+ CCR5+ T cells that can support highly productive viral infection, making it advantageous for the virus to infect naïve CD4+ CXCR4+ T cells, even though infection of these cells is less productive (reviewed in [14]). However, CXCR4 viruses are rarely transmitted, probably because their between-host transmissibility is severely diminished, although a competing explanation for the lack of CXCR4 TF viruses is simply because they are uncommon in the donor population 15, 16.
Another well established characteristic of HIV-1 TF viruses is the reduced number of N-linked glycosylation sites encoded by the env gene compared to viruses circulating during later chronic infection (reviewed in [17]). It is hypothesised that heavy glycosylation evolves during the course of infection because it increases viral resistance to neutralising antibodies [18], but is detrimental at the point of transmission because viruses are more easily trapped or inhibited by agents in the transmission fluid, and/or are more likely to be targeted by the innate immune system. Other characteristics of HIV-1 TF viruses include high densities of the Env protein compared to viruses circulating during later infection. This might increase infection of cells in the genital tract and enhance binding to dendritic cells, thereby enabling efficient transport of the virus from the genital tract to the gut (reviewed in [17], although a recent study of transmission pairs gives more equivocal results [19]).
A recent detailed study of eight transmission pairs suggests that TF viruses are resistant to type-1 interferons, and that this feature correlates with high particle infectivity and ability to replicate [19]. In contrast to TF viruses, isolates from chronically infected donors were generally interferon sensitive, suggesting that HIV-1 within-host adaptation results in increased susceptibility to restriction by innate immune responses. In support of this, a recent report indicates that TF viruses are resistant to interferon-induced transmembrane proteins (IFITMs), which are believed to restrict cell entry of various viruses, including HIV-1 [1]. However, within the first 6 months of infection neutralising antibody responses select for specific escape mutations in HIV-1 Env that result in susceptibility to IFITMs [1]. Taken together, these reports suggest that within-host adaptation to adaptive immune responses increase the sensitivity of HIV-1 to interferon-stimulated genes, which in turn is detrimental to onward transmission.
Transmissibility-reducing adaptations that have evolved in response to adaptive immune responses occur in other chronic viruses. During chronic hepatitis B virus (HBV) infection, viral variants that do not produce HBeAg antigen often emerge, which is likely driven by the appearance of anti-HBeAg antibody responses and/or enhanced cytotoxic T lymphocyte (CTL) killing of HBeAg-positive cells 20, 21. Although HBeAg is dispensable for ongoing infection, it is important for the establishment of immmunotolerance in neo/antenatal infections [22]. Consequently, data from small animal models and human transmission studies suggest that HbeAg-negative virus is much less likely to transmit and establish chronic infections 22, 23, 24.
Adaptation to Different Host Genotypes
Transmissibility-reducing mutations can also occur in response to the genetic composition of individual hosts. The necessity to escape or avoid the host adaptive immune response can result in the accumulation of CTL and/or antibody escape mutations that are tailored to the specific host genotype, such as human leukocyte antigen (HLA) type. Whilst these mutations will be advantageous in the current individual, evidence suggests that antigenic escape in HIV-1, HCV, and HBV can have substantial fitness costs when measured in the absence of specific immune responses 25, 26, 27. This can put viruses harbouring escape mutations at a disadvantage in hosts with different genotypes, potentially hindering transmission. This is supported by a number of findings. First, HIV-1 variants matching the population consensus are more likely to be transmitted, even if these variants are in a minority within the donor at the time of transmission [28]. Second, the rapid reversion of some CTL escape mutations if they infect HLA mismatched hosts [29]. Third, the frequency of CTL escape mutations that carry a large cost is proportional to the frequency of the corresponding HLA alleles in the host population, but CTL escape mutations with little or no cost tend to accumulate at the population level [30].
A study looking at maternal transmission of HCV is also supportive of a transmission bias: transient immunodeficiency during pregnancy relaxes the selection pressure on HCV CTL escape variants, enabling the emergence of viruses that do not harbour these escape mutations. It is these viruses that preferentially transmit from mother to child, rather than the variants carrying CTL escape mutations specific to the mother [11].
Adaptations That Increase Within-Host Competitive Ability
At the within-host level, mutations that enhance viral competitive ability will have a selective advantage. If these mutations also increase transmission they will rapidly spread throughout the viral population at the epidemiological level, as was observed during the 2014/15 West Africa Ebola outbreak [31]. However, within-host adaptive mutations can increase the pathogenicity (virulence) of viruses, and if this results in fewer potential transmission events then within-host viral adaptation will reduce overall rates of transmission (even if transmissibility per contact is increased). Perhaps the clearest example of this is found in bovine viral diarrhoea virus (BVDV), a pestivirus of cows. Although BVDV can cause acute infection when transmitted horizontally, persistent infection can be established only via vertical in utero transmission; persistently infected animals are thought to be essential for the maintenance of BVDV within a herd. Chronically infected cows typically exhibit mild symptoms, but within-host evolution can lead to the emergence of a cytopathic BVDV biotype that escapes viral control of the rate of within-host virus replication 32, 33. This highly virulent form replicates quickly, leading to high viral loads, and is invariably fatal after a couple of weeks. Notably, cytopathic BVDV is incapable of establishing persistent infection, and therefore its emergence almost certainly limits the ability of BVDV to spread among hosts and it is generally regarded as an evolutionary dead-end [34]. A similar spontaneous emergence is thought to give rise to feline infectious peritonitis virus (FIPV) in cats persistently infected with feline enteric coronavirus. In this case viral mutations enable FIPV to efficiently replicate in monocytic cells, resulting in systemic infection and very high mortality 35, 36, 37.
In a less extreme example, a high HIV-1 replicative capacity (broadly defined as the ability of the virus to replicate in the absence of an immune response) is associated with high viral loads in untreated infection 9, 38, 39. Although higher viral loads are correlated with higher rates of transmission, they are also linked to faster progression to AIDS 40, 41 and are therefore more pathogenic. It has been calculated that the number of onward transmissions during the course of an infection is maximised when viral load, and by implication viral replicative capacity, is intermediate 42, 43. However, the predicted replicative capacity of HIV-1 in untreated patients tends to slowly increase during the course of infection [9], presumably a consequence of within-host competition. In other words, between-host evolution favours viruses with intermediate levels of virulence, but within-host evolution favours viruses with high levels of virulence. All else being equal, over the long term, within-host competition is expected to result in the evolution of highly pathogenic viruses at the host population level, even though these viruses will generate fewer onward transmissions than less pathogenic strains, a consequence of short-sighted evolution [43].
Life History Strategies That Mitigate Short-Sighted Evolution
The potential for short-sighted evolution will be greatest for chronic viruses with low fidelity replication and long transmission intervals (Figure 1). To persist at the epidemiological scale, we suggest that such viruses require life-history traits that avoid or reduce short-sighted evolution. These include traits resulting in a low rate of evolution across the whole within-host population, or a reduced rate in one or more subsets of the within-host viral population. We know very little about the within-host life history of most viruses, except those that infect humans, so it is not clear how many viruses fit into this category. It could be that chronic viruses with long transmission intervals are uncommon because few viruses are able to mitigate short-sighted evolution. Moreover, if life-history traits that ameliorate short-sighted evolution are partly under the control of the virus, then evolutionary theory predicts those traits should be under selection (see Box 1). Here, we describe potentially relevant life-history traits in four human chronic viruses with long transmission intervals [Human T-lymphotropic virus-1 (HTLV-1), HBV, HIV-1, and HCV].
Decreased Mutation Rate during Viral Replication
HTLV-1 is a human deltaretrovirus that causes adult T-cell lymphoma in some infected individuals. The majority of infections in endemic countries are through mother-to-child transmission via breast milk, resulting in a long transmission interval. As with all retroviruses, HTLV-1 uses an error-prone reverse transcriptase to generate complementary DNA (cDNA) from an RNA template, which is then integrated into the genome of the host cell, where it is referred to as provirus. Subsequently, however, the vast majority of viral reproduction is via mitotic division of proviral-containing host cells, leading to clonal expansion of the provirus. Since this uses the host cell polymerase to copy the provirus, error rates are extremely low, and within-host evolution comes to an almost standstill: the within-host rate of evolution of HIV-1, also a retrovirus, is in the order of 1.5×10−2 substitutions per site per year (s/s/yr) in the Env region of the genome 44, 45, 46, whereas it is probably four orders of magnitude lower for HTLV-1 [47]. As with other slowly evolving viruses, HTLV-1 does not try to outpace the host adaptive immune system but instead avoids it by inhibiting the transcription of viral genes and therefore reducing the immunogenicity of infected cells [48].
Increased Viral Generation Time
HBV is a hepadnavirus that typically causes acute infection in newly infected adults, but chronic infection in infants. In endemic areas most infections are acquired at birth or during infancy, and many of these are due to mother-to-child transmission, resulting in a long transmission interval. Hepadnaviruses are reverse-transcribing DNA viruses, and therefore have high mutation rates during replication. However, the unique life cycle of hepadnaviruses results in long viral (cell-to-cell) generation times even though chronic infection is productive, with infected cells producing 1 to 10 virions per day [49]. When a hepatocyte is infected with a hepadnavirus particle, viral relaxed circular DNA (RC-DNA) is transported to the nucleus of the cell where it is converted into covalently closed circular DNA (cccDNA). This cccDNA then acts as a template for the production of more RC-DNA, via an RNA intermediate, which is either packaged into virions that are released from the host cell or, during early infection of the hepatocyte, are transported back into the nucleus to form more cccDNA [50]. During chronic infection, cccDNA is incredibly stable, with an estimated half-life for duck hepatitis B virus (DHBV) cccDNA of 33–57 days (reviewed in [51]), giving a viral generation time around 20–40 days in ducks (and possibly much longer in humans). This leads to a rate of evolution 20–40 times slower than if the viral generation time were 1 day. As a result of this long generation time, and the compact nature of the HBV genome, which constrains its evolution, the within-host rate of HBV evolution is much slower (on the order 5×10−5 s/s/yr [52]) than would be expected given its high rate of spontaneous mutation. HCV, in comparison, has a within-host rate of evolution of about 1×10−2 s/s/yr in the E1/E2 gene region [53], with rates about five times lower in other gene regions [54]. Despite its slow rate of evolution, HBV is able to persist due to immunotolerance and/or immune exhaustion driven by excessive production of viral antigens (sAg and HBeAg) [55].
Establishing Slowly Evolving ‘Germline’ Lineages
Unlike HTLV-1 and HBV, HIV-1 and HCV have very high rates of within-host evolution. However, accumulating evidence suggests that minority viral populations persist within HIV-1- and HCV-infected hosts that have much lower rates of evolution. HIV-1 replication requires integration of virus into the genome of newly infected CD4+ T cells. A small proportion of these provirus-containing cells enter a long-lived resting phase with estimated half-lives ranging between ∼0.75 and ∼3.6 years 56, 57, 58. It is these latently-infected resting CD4+ T cells that constitute the bulk of the HIV reservoir and which represent the major barrier to finding a true cure. The reservoir is established early in infection 59, 60, and then maintained by newly infected cells entering the reservoir, and by the high-fidelity proliferation of cells within the reservoir 61, 62. Consequently, a proportion of the provirus population in the reservoir is expected to be identical, or similar, to the virus(es) that initiated the infection 63, 64; a prediction supported by phylogenetic analysis 56, 65. The latent reservoir is not visible to the host immune system, and therefore proviral populations originating from early infection are expected to avoid immune-mediated deletion. This is supported by a recent study of antiretroviral-treated patients that documented the persistence of a minority population of provirus, which had not accumulated CTL escape mutations in the reservoir of most patients despite robust CTL responses [66]. If within-host selection pressures are not too strong, reactivation of latently-infected cells is also expected to result in a minority RNA viral population resembling TF virus years after initial infection 63, 64, 67. Variation in the rate of evolution along different branches of within-host HIV-1 phylogenies (which exclude viruses in the reservoir) provides support for this 44, 68, although the short sequence lengths used to generate these phylogenies currently makes it difficult assess the importance of this process.
Our current understanding of the HCV life cycle does not include direct mechanisms for establishing latent or dormant infections. However, phylogenetic analyses of longitudinally sampled HCV-infected patients has revealed the persistence of independently evolving viral lineages within individuals, with unusually high heterogeneity in evolutionary rates along different lineages – often higher than found for HIV-1 [53]. This is indicative of a complex within-host population that includes slowly evolving lineages [53].
The origin of these subpopulations is unclear, and might represent infection of long-lived hepatocytes [53], or infections of cells outside of the liver. Extra-hepatic replication of HCV remains controversial, not least because the HCV life cycle is seemingly intrinsically linked to liver biology [69]. Nonetheless, numerous studies suggest limited genome replication in neurological tissue, gastrointestinal cells, and B-lymphocytes 70, 71, 72, 73, 74. Moreover, genetic analysis of virus isolated from hepatocytes, plasma, and peripheral blood mononuclear cells (PBMCs) from the same patients has shown that virus in PBMCs typically represents a distinct subpopulation (discussed in [53]), and recent work has shown that HCV infecting B cells shows tropism for lymphocytes rather than hepatocytes, indicating the presence of subpopulations specialised on infecting B cells [73]. Among the PBMC types it has been suggested that long-lived memory B cells are primarily infected, with these infected cells avoiding host antiviral immune responses [74]. These factors will potentially increase the viral generation time of extra-hepatic lineages and provide a way of maintaining a compartment of slowly evolving viruses that do not undergo extensive within-host adaptation.
Transmission of Founder-like Lineages
We propose that chronic viral infections with long transmission intervals can only persist in a host population if viral populations that have undergone little within-host adaptation remain available for onward transmission. We call these ‘founder-like’ populations because they will be genetically similar to the TF virus(es). For chronic viruses with slow rates of within-host evolution, such as HTLV-1 and HBV, the entire within-host population will be founder-like, and therefore transmitted viruses will be representative of the viruses present in the donor at the time of transmission. However, for chronic viruses with fast rates of within-host evolution, such as HIV-1 and HCV, this leads to the expectation that viral subpopulations that have experienced low rates of within-host evolution will be more likely to be transmitted (Figure 2 ). We draw an analogy between these subpopulations and the germline in most animals, and possibly also in plants [75]. For both HIV-1 and HCV, transmission is relatively inefficient, with only one or a few viral strains transmitted between hosts 76, 77, 78, and studies of both HIV-1 and HCV have revealed that TF viruses are often under-represented in donor viral populations 1, 19, 79. This has led to the suggestion that, for these rapidly evolving viruses at least, the majority of virions circulating in an individual are adapted to the within-host environment, but are poorly adapted for transmission between hosts.
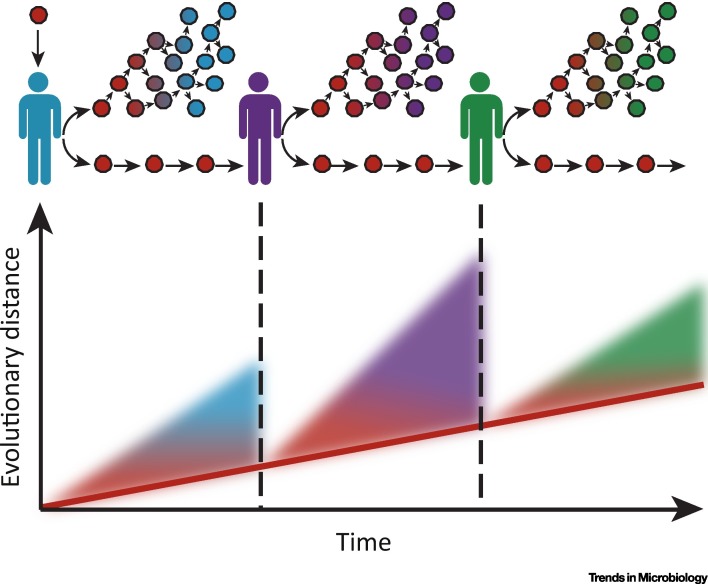
Figure 2.
The Germline Hypothesis of Chronic Viral Infections. Once a chronic virus has been transmitted to a new host (indicated by the vertical lines), within-host rates of viral evolution can be extremely rapid, indicated by increasing evolutionary distance as time progresses. If there is heterogeneity in the rates of evolution among different within-host viral lineages, and if more slowly evolving lineages are more likely to be transmitted because they contain fewer transmission-reducing mutations, the rate of evolution of the virus at the between-host level will be slower than the rate of evolution measured at the within-host level (indicated by the red line). These slowly evolving lineages can be considered the viral germline.
There is mounting evidence that less-evolved founder-like HIV-1 viruses are more likely to be successfully transmitted, probably because they have greater fitness in the new host. The original indication of this came from the observation that HIV evolves between two and four times faster within-hosts than at the epidemiological level 44, 45, 46, 80, as would be expected if founder-like viruses are transmitted and within-host evolution is bypassed 67, 81. Another mechanism likely to contribute to this mismatch in rates is ‘adapt-and-revert’, whereby host-specific adaptations accumulate in one host, only to be lost in the next host 82, 83. However, this is unlikely to be the primary mechanism since it will only result in a mismatch at sites under selection, whereas the mismatch in evolutionary rates is seen for both synonymous and nonsynonymous mutations 67, 84, 85, and across the whole genome [46]. Saturation effects might also contribute [86]. However, because the mismatch can be observed over short transmission chains [80], whereas saturation effects are expected to accumulate over longer timescales, the transmission of germline lineages remains the most likely primary mechanism.
A similar mismatch in evolutionary rates is seen for HCV. For E1/E2, the only genomic region for which corresponding within- and among-host rates exist, the rate of evolution is about fourfold higher at the within-host level (∼1×10−2 s/s/yr within hosts [53] compared to ∼2.5×10−3 s/s/yr among hosts [54]). Moreover, the mismatch is roughly similar for synonymous and nonsynonymous mutations. As with HIV-1, this implicates the transmission of less-evolved virus, rather than a process of adaptation and reversion, as the primary mechanism leading to the mismatch in evolutionary rates. Finally, a mismatch also appears to exist for HIV-2 87, 88, although the relative rates of synonymous and nonsynonymous mutations have not been measured. Interestingly, for HBV, the only other chronic virus for which comparable within- and between-host rates of evolution have been estimated, there is no mismatch in evolutionary rates between the two levels [52].
Additional support for the transmission of founder-like viruses comes from the analysis of HIV-1 transmission pairs. Two studies of HIV-1 discordant couples from Rakai, Uganda, found evidence for the preferential transmission of more founder-like virus 89, 90. Furthermore, a study of 137 linked transmission pairs from Zambia concluded that viruses within a donor are more likely to be transmitted if their genomes more closely resemble viral genomes circulating in the population as a whole, even if these viruses are minority variants in the donor [28]. This observation is expected if founder-like viruses are more likely to be transmitted.
In summary, there is indirect evidence that ‘germline viral lineages’ of some rapidly evolving chronic viruses are preferentially transmitted. However direct empirical evidence is lacking (see Outstanding Questions). For a study to demonstrate this it would need to (i) identify transmission pairs in which an untreated donor individual is longitudinally sampled since early infection, (ii) include samples from the donor and recipient taken around the time of transmission, and (iii) sequence large numbers of individual virus genomes from both donor and recipient. Since it is unethical to enrol patients into a study without offering treatment, suitable pairs must be searched for retrospectively in the sample archives of earlier studies.
Concluding Remarks
We have proposed that chronic viruses with long transmission intervals require mechanisms to avoid or reduce ‘short-sighted’ evolution. For some viruses, these mechanisms lead to surprisingly low rates of within-host viral evolution, limiting the capacity for short-sighted evolution. For viruses exhibiting continual adaptive evolution during infection, we propose that the maintenance of a ‘germline’ viral lineage (one that has experienced comparatively little ‘short-sighted’ evolution) is required in order for the virus to be maintained in a host population (Figure 2). Moreover, we speculate that the mechanisms for maintaining proposed germline lineages in viral populations could themselves be under selection (Box 1).
Evidence for this argument is available for six of the eight chronic RNA viruses (including DNA viruses with RNA intermediates) that are known to be prevalent in humans; less is known about the natural history and evolution of the other two viruses, human pegiviruses 1 and 2 (HPgV, and HPgV-2) 91, 92, or for chronic RNA viruses infecting other animals. All six of these chronic viruses have long transmission intervals, ranging from years to decades, and three of them (HTLV-1, HTLV-2, and HBV) have life history traits that result in slower rates of within-host evolution than would be expected given their high mutation rates during replication. The other three (HIV-1, HIV-2, and HCV) have fast rates of within-host evolution, but may bypass short-sighted evolution through the maintenance and preferential transmission of a subpopulation of viruses that retain the transmissibility of the TF virus that initiated the infection (a ‘germline’ lineage).
An important factor that influences the potential impact of short-sighted evolution is the transmission interval, or more specifically, the number of viral generations between the time of infection and the average time of onward transmission. This will depend on a number of variables, including the mode of transmission, host behaviour and life-history traits, and viral generation time. The potential for short-sighted evolution could therefore explain why some families of chronic viruses persist in some host species, but not others. For example, arenaviruses and hantaviruses cause chronic infections in rodents and are endemic in these populations, but not in human populations. This might be because transmission intervals are likely to be shorter in rodents, limiting the amount of short-sighted evolution that can accrue between transmission events. Similarly, pestiviruses might be able to persist at the host population level in livestock (despite occasional host death due to within-host viral evolution) but not in humans, because the mother-to-child transmission interval in humans would be much longer. Whether this is a general pattern is unknown; longitudinal sampling of individual animals to determine whether infections are acute, persistent and/or chronic is extremely challenging, particularly in wild populations [93], and in itself should become a research priority, not least because many of these potentially zoonotic infections are highly pathogenic in humans.
The ability of viruses to evolve rapidly is one of the secrets of their success, allowing them to evade host immune responses, evolve novel functions and explore new niches. However, this genetic plasticity may, for a virus, represent a double-edged sword that needs to be controlled. Indeed, it could be that overcoming short-sighted evolution is a necessary condition for the success of some viruses. We have speculated on some of the mechanisms by which this may be achieved, but confirming and understanding these processes will require further investigation.
Outstanding Questions.
How many wild-animal chronic viral infections are there?
Are chronic viral infections with short transmission intervals more common than those with long transmission intervals?
What are the relative rates of within- and between-host evolution of chronic viral infections other than HIV, HCV, and HBV?
Can we find direct evidence that founder-like viruses are more likely to be transmitted?
Does the probability of transmission decrease as infections progress?
Can we conclusively show that HCV has ‘germline’ lineages, and what is the mechanism maintaining them?
Ackowledgments
We would like to thank Jayna Raghwani, Lenka Stejskal, Robin Thompson, Lucas Walker, Chris Wymant, and two anonymous referees for helpful comments and discussions. This work was funded by The Wellcome Trust and The Royal Society grant numbers wtvm055984 (KAL) and 107653/Z/15/Z (JG), The Natural Environment Research Council grant number NE/K009524/1 (AG), and The European Research Council under the European Union’s Seventh Framework Programme (FP7/2007-2013)/ERC grant number 614725-PATHPHYLODYN (OGP)
Glossary
- Acute viral infection
infection characterised by early high viral loads, followed by clearance by the host immune system within days or weeks of initial infection.
- Chronic viral infection
a persistent infection characterised by continual viral/proviral production and high viral/proviral loads.
- Founder strain(s)
the viral strain(s) that initiate an infection in a host. These are also referred to as transmitted/founder (TF) viruses.
- Germline lineage (population)
a within-host viral lineage (population) that has undergone less evolutionary change than the majority of viral lineages (populations) within the host.
- Mutation rate
the rate at which spontaneous mutations are generated, measured per viral replication.
- Persistent viral infection
an infection that is not cleared and lasts for the lifetime of the host.
- Provirus
a viral genome that has integrated into the DNA of a host cell.
- Short-sighted evolution
evolutionary change that is adaptive within a host, but limits the ability of the virus to transmit to new hosts.
- Transmission interval
the typical time between infection of a host and onward transmission to a new host.
- Viral generation time
the average time between two consecutive generations along a within-host viral lineage. This can be estimated as the average time for viruses/proviruses to complete a full replication cycle in a given population, and therefore can also be considered the cell-to-cell generation time.
- Viral lineage
the line of descent connecting a contemporary virus to a virus that founded the infection.
References
- 1.Foster T.L. Resistance of transmitted founder HIV-1 to IFITM-mediated restriction. Cell Host Microbe. 2016;4:429–442. doi: 10.1016/j.chom.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murphy M.K. Viral escape from neutralizing antibodies in early subtype A HIV-1 infection drives an increase in autologous neutralization breadth. PLoS Pathog. 2013;9:e1003173. doi: 10.1371/journal.ppat.1003173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asquith B. Inefficient cytotoxic T lymphocyte-mediated killing of HIV-1-infected cells in vivo. PLoS Biol. 2006;4:e90. doi: 10.1371/journal.pbio.0040090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goonetilleke N. The first T cell response to transmitted/founder virus contributes to the control of acute viremia in HIV-1 infection. J. Exp. Med. 2009;206:1253–1272. doi: 10.1084/jem.20090365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herbeck J.T. Demographic processes affect HIV-1 evolution in primary infection before the onset of selective processes. J. Virol. 2011;85:7523–7534. doi: 10.1128/JVI.02697-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leviyang S., Ganusov V.V. Broad CTL response in early HIV infection drives multiple concurrent CTL escapes. PLoS Comput. Biol. 2015;11:1–21. doi: 10.1371/journal.pcbi.1004492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petrovic D. Hepatitis C virus – T-cell responses and viral escape mutations. Eur. J. Immunol. 2012;42:17–26. doi: 10.1002/eji.201141593. [DOI] [PubMed] [Google Scholar]
- 8.Bull R.A. Transmitted/founder viruses rapidly escape from CD8(+) T cell responses in acute hepatitis C virus infection. J. Virol. 2015;89:5478–5490. doi: 10.1128/JVI.03717-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kouyos R.D. Assessing predicted HIV-1 replicative capacity in a clinical setting. PLoS Pathog. 2011;7:e1002321. doi: 10.1371/journal.ppat.1002321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deymier M.J. Heterosexual transmission of subtype C HIV-1 selects consensus-like variants without increased replicative capacity or interferon-α resistance. PLoS Pathog. 2015;11:1–22. doi: 10.1371/journal.ppat.1005154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Honegger J.R. Loss of immune escape mutations during persistent HCV infection in pregnancy enhances replication of vertically transmitted viruses. Nat. Med. 2013;19:1529–1533. doi: 10.1038/nm.3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levin B.R., Bull J.J. Short-sighted evolution and the virulence of pathogenic microorganisms. Trends Microbiol. 1994;2:76–81. doi: 10.1016/0966-842x(94)90538-x. [DOI] [PubMed] [Google Scholar]
- 13.Duffy S. Rates of evolutionary change in viruses: patterns and determinants. Nat. Rev. Genet. 2008;9:267–276. doi: 10.1038/nrg2323. [DOI] [PubMed] [Google Scholar]
- 14.Swanstrom R., Coffin J. HIV-1 pathogenesis: the virus. Cold Spring Harb. Perspect. Med. 2012;2:a007443. doi: 10.1101/cshperspect.a007443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chalmet K. Presence of CXCR4-using HIV-1 in patients with recently diagnosed infection: correlates and evidence for transmission. J. Infect. Dis. 2012;205:174–184. doi: 10.1093/infdis/jir714. [DOI] [PubMed] [Google Scholar]
- 16.Frange P. Sexually-transmitted/founder HIV-1 cannot be directly predicted from plasma or PBMC-derived viral quasispecies in the transmitting partner. PLoS One. 2013;8:e69144. doi: 10.1371/journal.pone.0069144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joseph S.B. Bottlenecks in HIV-1 transmission: insights from the study of founder viruses. Nat. Rev. Microbiol. 2015;13:414–425. doi: 10.1038/nrmicro3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei X. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 19.Iyer S.S. Resistance to type 1 interferons is a major determinant of HIV-1 transmission fitness. Proc. Natl. Acad. Sci. U.S.A. 2017;114:E590–E599. doi: 10.1073/pnas.1620144114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mason W.S. Immune selection during chronic hepadnavirus infection. Hepatol. Int. 2008;2:3–16. doi: 10.1007/s12072-007-9024-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frelin L. A mechanism to explain the selection of the hepatitis e antigen-negative mutant during chronic hepatitis B virus infection. J. Virol. 2009;83:1379–1392. doi: 10.1128/JVI.01902-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kramvis A. The clinical implications of hepatitis B virus genotypes and HBeAg in pediatrics. Rev. Med. Virol. 2016;26:285–303. doi: 10.1002/rmv.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen H. Woodchuck hepatitis. J. Virol. 1992;66:5682–5684. doi: 10.1128/jvi.66.9.5682-5684.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tong S., Revill P. Overview of hepatitis B viral replication and genetic variability. J. Hepatol. 2016;64:S4–S16. doi: 10.1016/j.jhep.2016.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seki S., Matano T. CTL escape and viral fitness in HIV/SIV infection. Front. Microbiol. 2012;2:1–5. doi: 10.3389/fmicb.2011.00267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwei K. Impaired virion secretion by hepatitis B virus immune escape mutants and its rescue by wild-type envelope proteins or a second-site mutation. J. Virol. 2013;87:2352–2357. doi: 10.1128/JVI.02701-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uebelhoer L. Stable cytotoxic T cell escape mutation in hepatitis C virus is linked to maintenance of viral fitness. PLoS Pathog. 2008;4:1–15. doi: 10.1371/journal.ppat.1000143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carlson J.M. Selection bias at the heterosexual HIV-1 transmission bottleneck. Science. 2014;345:1254031. doi: 10.1126/science.1254031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leslie A.J. HIV evolution: CTL escape mutation and reversion after transmission. Nat. Med. 2004;10:282–289. doi: 10.1038/nm992. [DOI] [PubMed] [Google Scholar]
- 30.Kawashima Y. Adaptation of HIV-1 to human leukocyte antigen class I. Nature. 2009;458:641–645. doi: 10.1038/nature07746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bedford T., Malik H.S. Did a single amino acid change make Ebola virus more virulent? Cell. 2016;167:892–894. doi: 10.1016/j.cell.2016.10.032. [DOI] [PubMed] [Google Scholar]
- 32.Lackner T. Persistence of bovine viral diarrhea virus is determined by a cellular cofactor of a viral autoprotease. J. Virol. 2005;79:9746–9755. doi: 10.1128/JVI.79.15.9746-9755.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lackner T. Dissection of a viral autoprotease elucidates a function of a cellular chaperone in proteolysis. Proc. Natl. Acad. Sci. U. S. A. 2006;103:1510–1515. doi: 10.1073/pnas.0508247103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peterhans E. Cytopathic bovine viral diarrhea viruses (BVDV): emerging pestiviruses doomed to extinction. Vet. Res. 2010;41:41–44. doi: 10.1051/vetres/2010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bank-Wolf B.R. Mutations of 3c and spike protein genes correlate with the occurrence of feline infectious peritonitis. Vet. Microbiol. 2014;173:177–188. doi: 10.1016/j.vetmic.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Borschensky C.M.M., Reinacher M. Mutations in the 3c and 7b genes of feline coronavirus in spontaneously affected FIP cats. Res. Vet. Sci. 2014;97:333–340. doi: 10.1016/j.rvsc.2014.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kipar A., Meli M.L. Feline infectious peritonitis: still an enigma? Vet. Pathol. 2014;51:505–526. doi: 10.1177/0300985814522077. [DOI] [PubMed] [Google Scholar]
- 38.Quinones-Mateu M.E. A dual infection/competition assay shows a correlation between ex vivo human immunodeficiency virus type 1 fitness and disease progression. J. Virol. 2000;74:9222–9233. doi: 10.1128/jvi.74.19.9222-9233.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prince J.L. Role of transmitted Gag CTL polymorphisms in defining replicative capacity and early HIV-1 pathogenesis. PLoS Pathog. 2012;8:e1003041. doi: 10.1371/journal.ppat.1003041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mellors J.W. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science. 1996;272:1167–1170. doi: 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- 41.de Wolf F. AIDS prognosis based on HIV-1 RNA, CD4+ T-cell count and function: markers with reciprocal predictive value over time after seroconversion. AIDS. 1997;11:1799–1806. doi: 10.1097/00002030-199715000-00003. [DOI] [PubMed] [Google Scholar]
- 42.Fraser C. Variation in HIV-1 set-point viral load: Epidemiological analysis and an evolutionary hypothesis. Proc. Natl. Acad. Sci. U. S. A. 2007;104:17441–17446. doi: 10.1073/pnas.0708559104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lythgoe, K.A. et al. (2013) Is HIV short-sighted? Insights from a multistrain nested model. Evolution (N.Y.) 67, 2769–2782 [DOI] [PMC free article] [PubMed]
- 44.Lemey P. HIV evolutionary dynamics within and among hosts. AIDS Rev. 2006;8:125–140. [PubMed] [Google Scholar]
- 45.Pybus O.G., Rambaut A. Evolutionary analysis of the dynamics of viral infectious disease. Nat. Rev. Genet. 2009;10:540–550. doi: 10.1038/nrg2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alizon S., Fraser C. Within-host and between-host evolutionary rates across the HIV-1 genome. Retrovirology. 2013;10:49–58. doi: 10.1186/1742-4690-10-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Dooren S. The low evolutionary rate of human T-cell lymphotropic virus type-1 confirmed by analysis of vertical transmission chains. Mol. Biol. Evol. 2004;21:603–611. doi: 10.1093/molbev/msh053. [DOI] [PubMed] [Google Scholar]
- 48.Cook L.B. HTLV-1: persistence and pathogenesis. Virology. 2013;435:131–140. doi: 10.1016/j.virol.2012.09.028. [DOI] [PubMed] [Google Scholar]
- 49.Nowak M.A. Viral dynamics in hepatitis B virus infection. Proc. Natl. Acad. Sci. U. S. A. 1996;93:4398–4402. doi: 10.1073/pnas.93.9.4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Summers J. Hepadnavirus envelope proteins regulate covalently closed circular DNA amplification. J. Virol. 1990;64:2819–2824. doi: 10.1128/jvi.64.6.2819-2824.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nassal M. HBV cccDNA: viral persistence reservoir and key obstacle for a cure of chronic hepatitis B. Gut. 2015;64:1972–1984. doi: 10.1136/gutjnl-2015-309809. [DOI] [PubMed] [Google Scholar]
- 52.Harrison A. Genomic analysis of hepatitis B virus reveals antigen state and genotype as sources of evolutionary rate variation. Viruses. 2011;3:83–101. doi: 10.3390/v3020083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Raghwani J. Exceptional heterogeneity in viral evolutionary dynamics characterises chronic hepatitis C virus infection. PLoS Pathog. 2016;12:e1005894. doi: 10.1371/journal.ppat.1005894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gray R.R. The mode and tempo of hepatitis C virus evolution within and among hosts. BMC Evol. Biol. 2011;11:131. doi: 10.1186/1471-2148-11-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nebbia G. Hepatitis B infection: current concepts and future challenges. Q. J. Med. 2012;105:109–113. doi: 10.1093/qjmed/hcr270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brodin J. Establishment and stability of the latent HIV-1 DNA reservoir. eLife. 2016;5:e18889. doi: 10.7554/eLife.18889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Finzi D. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat. Med. 1999;5:512–517. doi: 10.1038/8394. [DOI] [PubMed] [Google Scholar]
- 58.Crooks A.M. Precise quantitation of the latent HIV-1 reservoir: implications for eradication strategies. J. Infect. Dis. 2015;212:1361–1365. doi: 10.1093/infdis/jiv218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chun T.-W. Early establishment of a pool of latently infected, resting CD4+ T cells during primary HIV-1 infection. Proc. Natl. Acad. Sci. 1998;95:8869–8873. doi: 10.1073/pnas.95.15.8869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Whitney J.B. Rapid seeding of the viral reservoir prior to SIV viraemia in rhesus monkeys. Nature. 2014;512:74–77. doi: 10.1038/nature13594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Simonetti F.R. Clonally expanded CD4+ T cells can produce infectious HIV-1 in vivo. Proc. Natl. Acad. Sci. 2016;113:1883–1888. doi: 10.1073/pnas.1522675113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim M., Siliciano R.F. Reservoir expansion by T-cell proliferation may be another barrier to curing HIV infection. Proc. Natl. Acad. Sci. U. S. A. 2016;113:201600097. doi: 10.1073/pnas.1600097113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Doekes H.M.M. Effect of the latent reservoir on the evolution of HIV at the within- and between-host levels. PLoS Comput. Biol. 2017;13:e1005228. doi: 10.1371/journal.pcbi.1005228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Immonen T.T. Recombination enhances HIV-1 envelope diversity by facilitating the survival of latent genomic fragments in the plasma virus population. PLoS Comput. Biol. 2015;11:1–26. doi: 10.1371/journal.pcbi.1004625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Frenkel L.M. Multiple viral genetic analyses detect low-level human immunodeficiency virus type 1 replication during effective highly active antiretroviral therapy. J. Virol. 2003;77:5721–5730. doi: 10.1128/JVI.77.10.5721-5730.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Deng K. Broad CTL response is required to clear latent HIV-1 due to dominance of escape mutations. Nature. 2015;517:381–385. doi: 10.1038/nature14053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lythgoe K.A., Fraser C. New insights into the evolutionary rate of HIV-1 at the within-host and epidemiological levels. Proc. R. Soc. B. 2012;279:3367–3375. doi: 10.1098/rspb.2012.0595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Immonen T.T., Leitner T. Reduced evolutionary rates in HIV-1 reveal extensive latency periods among replicating lineages. Retrovirology. 2014;11:81. doi: 10.1186/s12977-014-0081-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Scheel T.K.H., Rice C.M. Understanding the hepatitis C virus life cycle paves the way for highly effective therapies. Nat. Med. 2013;19:837–849. doi: 10.1038/nm.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fletcher N.F. Hepatitis C virus infection of neuroepithelioma cell lines. Gastroenterology. 2010;139 doi: 10.1053/j.gastro.2010.06.008. 1365–1374.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mee C.J. Effect of cell polarization on hepatitis C virus entry. J. Virol. 2008;82:461–470. doi: 10.1128/JVI.01894-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Durand T. Occult infection of peripheral B cells by hepatitis C variants which have low translational efficiency in cultured hepatocytes. Gut. 2010;59:934–942. doi: 10.1136/gut.2009.192088. [DOI] [PubMed] [Google Scholar]
- 73.Douam F. Specialization of hepatitis C virus envelope glycoproteins for B lymphocytes in chronically infected patients. J. Virol. 2015;90:992–1008. doi: 10.1128/JVI.02516-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ito M. Peripheral B cells as reservoirs of persistent HCV infection. Front. Microbiol. 2011;2:1–3. doi: 10.3389/fmicb.2011.00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Watson J.M. Germline replications and somatic mutation accumulation are independent of vegetative life span in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 2016;113:12226–12231. doi: 10.1073/pnas.1609686113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Keele B.F. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A. 2008;105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bull R.A. Sequential bottlenecks drive viral evolution in early acute hepatitis C virus infection. PLoS Pathog. 2011;7:e1002243. doi: 10.1371/journal.ppat.1002243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang H.-Y. Distinct hepatitis B virus dynamics in the immunotolerant and early immunoclearance phases. J. Virol. 2010;84:3454–3463. doi: 10.1128/JVI.02164-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Brown R.J.P. Hepatitis C Virus envelope glycoprotein fitness defines virus population composition following transmission to a new host. J. Virol. 2012;86:11956–11966. doi: 10.1128/JVI.01079-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vrancken B. The genealogical population dynamics of HIV-1 in a large transmission chain: bridging within and among host evolutionary rates. PLoS Comput. Biol. 2014;10:e1003505. doi: 10.1371/journal.pcbi.1003505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fraser C. Virulence and pathogenesis of HIV-1 infection: an evolutionary perspective. Science. 2014;343:1243727. doi: 10.1126/science.1243727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Herbeck J.T. Human immunodeficiency virus type 1 env evolves toward ancestral states upon transmission to a new host. J. Virol. 2006;80:1637–1644. doi: 10.1128/JVI.80.4.1637-1644.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zanini F. Population genomics of intrapatient HIV-1 evolution. eLife. 2015;4:e11282. doi: 10.7554/eLife.11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lemey P. Synonymous substitution rates predict HIV disease progression as a result of underlying replication dynamics. PLoS Comput. Biol. 2007;3:e29. doi: 10.1371/journal.pcbi.0030029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Abecasis A.B. Quantifying differences in the tempo of human immunodeficiency virus type 1 subtype evolution. J. Virol. 2009;83:12917–12924. doi: 10.1128/JVI.01022-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Belshaw R. Pacing a small cage: mutation and RNA viruses. Trends Ecol. Evol. 2008;23:188–193. doi: 10.1016/j.tree.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rocha C. Evolution of the human immunodeficiency virus type 2 envelope in the first years of infection is associated with the dynamics of the neutralizing antibody response. Retrovirology. 2013;10:110. doi: 10.1186/1742-4690-10-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lemey P. Tracing the origin and history of the HIV-2 epidemic. Proc. Natl. Acad. Sci. U. S. A. 2003;100:6588–6592. doi: 10.1073/pnas.0936469100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sagar M. Selection of HIV variants with signature genotypic characteristics during heterosexual transmission. J. Infect. Dis. 2009;199:580–589. doi: 10.1086/596557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Redd A.D. Previously transmitted HIV-1 strains are preferentially selected during subsequent sexual transmissions. J. Infect. Dis. 2012;206:1433–1442. doi: 10.1093/infdis/jis503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Stapleton J.T. The GB viruses: a review and proposed classification of GBV-A, GBV-C (HGV), and GBV-D in genus Pegivirus within the family Flaviviridae. J. Gen. Virol. 2011;92:233–246. doi: 10.1099/vir.0.027490-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Berg M.G. Discovery of a novel human Pegivirus in blood associated with hepatitis C virus co-infection. PLoS Pathog. 2015;11:e1005325. doi: 10.1371/journal.ppat.1005325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Plowright R.K. Transmission or within-host dynamics driving pulses of zoonotic viruses in reservoir host populations. PLoS Negl. Trop. Dis. 2016;10:1–21. doi: 10.1371/journal.pntd.0004796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Otto S.P., Day T. Princeton University Press; 2007. A Biologist’s Guide to Mathematical Modelling in Ecology and Evolution. [Google Scholar]
- 95.Fisher R.A. Clarendon Press; 1930. The Genetical Theory of Natural Selection. [Google Scholar]