Abstract
Aging is characterized by progressive loss of physiological and cellular functions, but the molecular basis of this decline remains unclear. We explored how aging impacts transcriptional dynamics using single-cell RNA-sequencing of unstimulated and stimulated naive and effector memory CD4+ T cells from young and old mice from two divergent species. In young animals, immunological activation drives a conserved transcriptomic switch resulting in tightly regulated gene expression, characterized by a strong up-regulation of a core activation program, coupled with a decrease in cell-to-cell variability. Aging perturbed the activation of this core program, and increased expression heterogeneity across populations of cells in both species. These discoveries suggest that increased cell-to-cell transcriptional variability will be a hallmark feature of aging across most, if not all, mammalian tissues.
The progressive decline of physiological and cellular functions (1) caused by aging is associated with complex effects on tissue-specific and species-specific gene expression levels (2, 3). For instance, aging affects distinct functional pathways even in closely related CD4+ and CD8+ T cells (4). Approaches analyzing the expression of sets of genes in single-cells have suggested that aging alters cell-to-cell transcriptional variability (5), though this may not be a universal attribute of aging (6). Whether cell-to-cell gene expression variability increases during aging on a genome-wide basis, particularly for dynamic activation programs, remains largely unexplored.
Naive CD4+ T cells are an excellent model system to evaluate the impact of aging on gene expression levels and cell-to-cell transcriptional variability. They are readily isolated as single, phenotypically homogeneous cells and can be stimulated into a physiologically-relevant activated transcriptional state in vitro. Naive CD4+ T cells are maintained in a quiescent state, and are essential for lifelong maintenance of adaptive immune function against infection and cancer (7). Comparing gene expression levels in matched tissues from different mammalian species is a powerful strategy to reveal conserved cell-type-specific regulatory programs (8) (9, 10), and has been successfully employed to identify a conserved set of immune activation genes in CD4+ T cells (11). However, it is not known whether conservation of gene expression levels is also reflected in cell-to-cell variability.
Here, we dissect the activation dynamics of naive CD4+ T cells at the single cell level during aging in two inbred mouse sub-species separated by 1 million years of divergence: the reference C57BL/6J, Mus musculus domesticus (B6), and Mus musculus castaneus (CAST). These mice have similar lifespans and aging phenotypes (12). We isolated naive CD4+ T cells and characterized their gene expression programs by single-cell RNA-sequencing (scRNA-seq) during aging in young (~3 months) and old (~21 months) individuals of each strain (Fig. 1A, (13)).
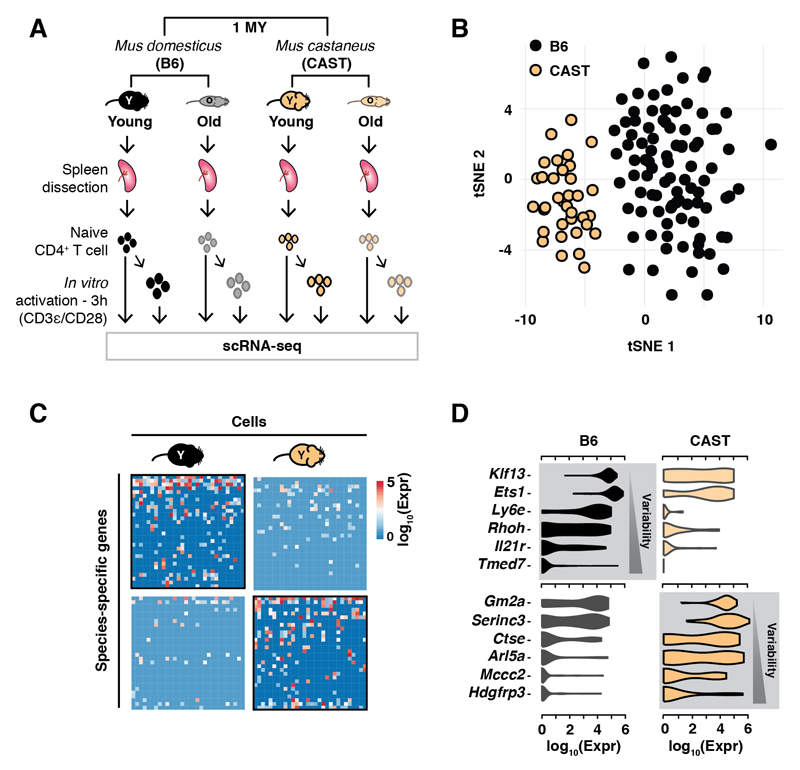
Fig. 1. Divergence in gene expression levels and cell-to-cell variability among naive CD4+ T cells isolated from two species of mice.
(A) Single cells were isolated from spleens of young (~3 month) and old (~21 month) individuals of two mouse sub-species, and sequenced before or after activation (13) ; (B) Species-specific tSNE clustering of naive CD4+ T cells from young animals; (C) Representative heatmap of 30 genes and 30 cells randomly selected from young animals from both species shows species-specific variation; (D) Violin plots show distribution of expression levels of selected, species-specifically transcribed genes (grey background).
Purified naive CD4+ T cells were loaded directly into the Fluidigm C1, or were loaded three hours after stimulation in vitro with plate-bound anti-CD3ε/anti-CD28 antibodies (13). After extensive quality control and filtering (Fig. S1 and S2, (13)), a total of 1514 high-quality CD4+ T cell transcriptomes were analyzed with BASiCS (14) to identify differences in levels of gene expression and variability of gene expression. Naive CD4+ T cells cluster by species (Figs. 1B; S3A) with 15% of genes differentially transcribed between the two species (Figs. 1C; S3A, table S1). Species-specific genes were not enriched for any functional ontology and were not the result of errors in genome assembly or read mapping (Fig. S3A-D). Species-specific transcribed genes are more variable on a cell-to-cell basis than genes expressed in both species, consistent with neutral drift (Fig. 1D, Fig. S3E).
Functional CD4+ T cell transcriptional responses start with an early, targeted activation of translational machinery and cytokine networks, followed by large-scale transcriptional changes associated with lineage commitment (11, 15). We stimulated naive CD4+ T cells for three hours with plate-bound anti-CD3ε/anti-CD28 antibodies (Figs. 2A, S4A).
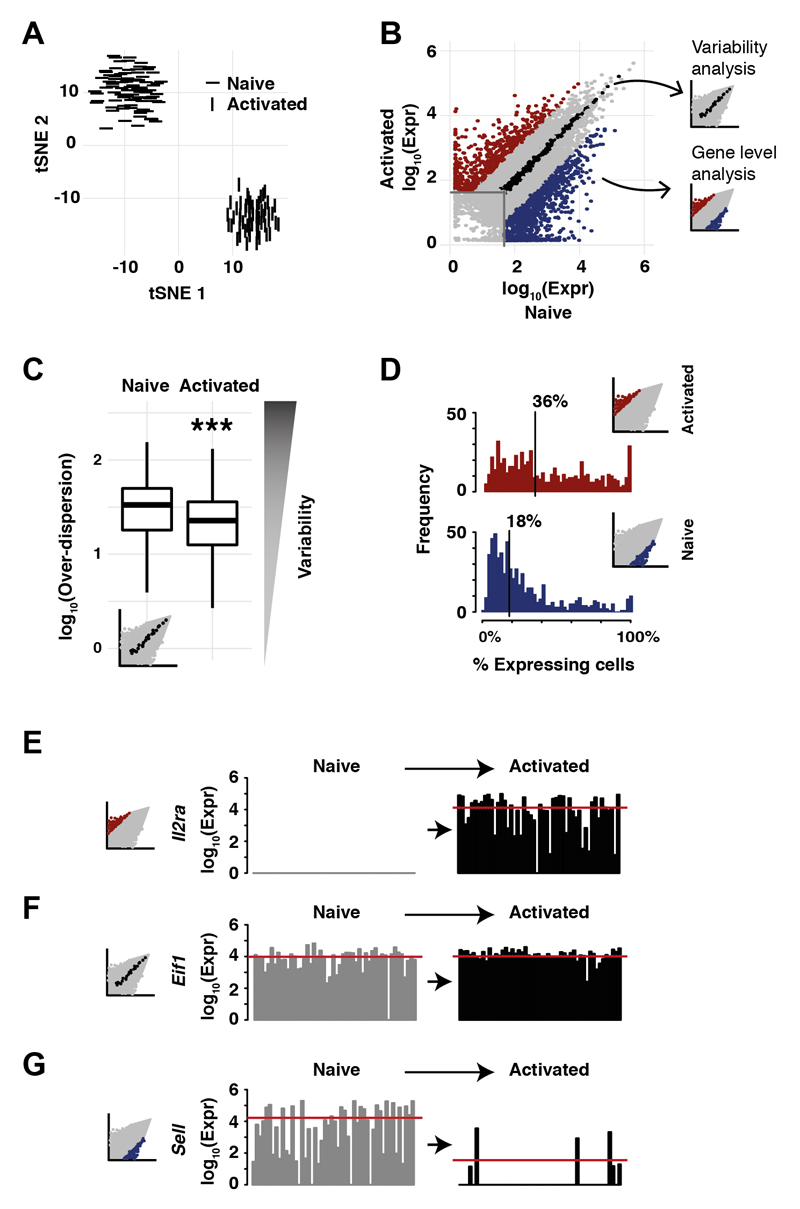
Fig. 2. Activation of naive CD4+ T cells induces a transcriptomic switch.
(A) Activation of CD4+ T cells from young B6 mice induces large-scale transcriptional changes (see S4 for CAST CD4+ T cells); (B) Genes up-regulated (red) and down-regulated (blue) by immune stimulation in young B6 mice (13); (C) Genes with no overall gene expression differences (black inset, (B)) during activation show decreased cell-to-cell variability in transcription (Mann-Whitney-Wilcoxon test, ***: p< 10 -10); (D) Up-regulated genes were expressed in a large fraction of activated CD4+ T cells after stimulation (median 36%); down-regulated genes were expressed in a smaller fraction of naive CD4+ T cells (median 18%); (E-G) Example genes showing transcriptional changes in unstimulated and activated CD4+ T cells: (E) Il2ra (Cd25); (F) Eif1; (G) Sell (Cd62l).
The resulting transcriptional response was highly coordinated, with 2063 genes (log2FC>2) differentially expressed in CD4+ T cells upon activation in vitro (Figs. 2B, S4B). We observed a reduction in cell-to-cell transcriptional variability among genes whose transcription levels do not change after activation (Mann-Whitney-Wilcoxon test, p<10-10) (Fig. 2C). Transcriptionally variable genes in the unstimulated condition showed no population substructure (Fig. S4C, (13)). Genes up-regulated after stimulation were expressed in more cells than downregulated genes (Figs. 2D; S4D).
The transcriptional switch driven by TCR engagement and co-stimulation included activation markers and translational machinery (11, 15) (Figs. 2E; S4E; table S2). Even genes not up-regulated after three hours of immune stimulation showed a marked decrease in their cell-to-cell variability, consistent with increased regulatory coordination (Fig. 2F). As expected, we observed the coordinated suppression of Sell (Cd62l) (Figs. 2G; S3D; table S2). CD4+ T cells isolated from CAST showed similar molecular phenotypes (Fig. S5).
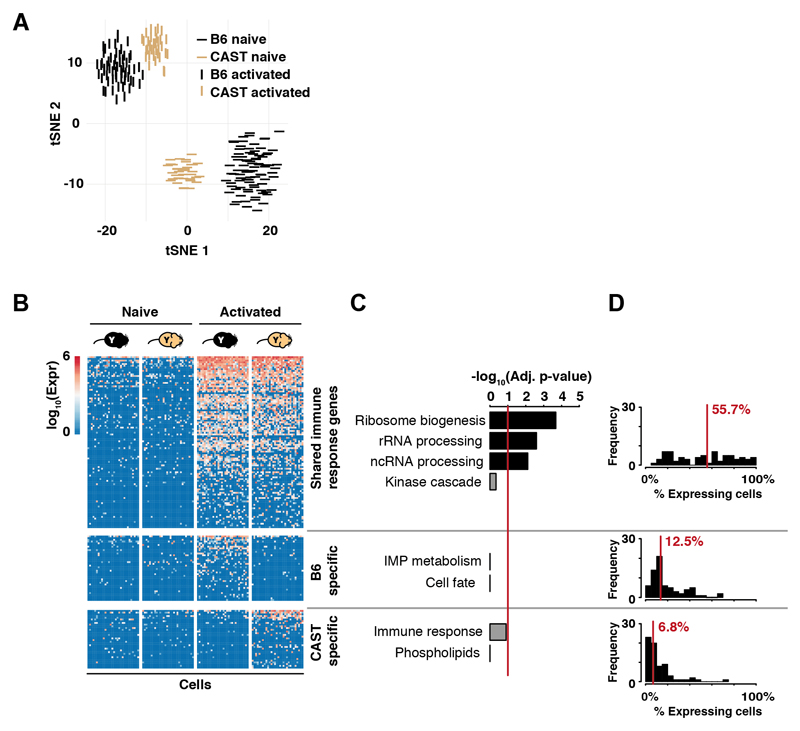
We identified functional targets activated upon immune stimulation by searching for gene expression levels conserved between species and expressed in most cells (11). Therefore, we stimulated naive CD4+ T cells isolated from young CAST males (13). The 225 up-regulated genes shared between B6 and CAST were enriched for cellular processes activated by immune stimulation (Fig. 3, table S3) and were expressed across most CD4+ T cells. Species-specific genes (96 for B6, 75 for CAST) showed no enrichment for biological function and were more sporadically expressed. Thus, target genes involved in translational control and immune function represent the conserved early activation response signature.
Fig. 3. Interspecies comparison of CD4+ T cell activation.
(A) CD4+ T cells isolated from B6 (black) and CAST (gold) show similar large-scale transcriptional changes upon immune stimulation; (B) Immune activation of CD4+ T cells triggers up-regulation of conserved and species-specific transcriptional programs. For visualization purposes, genes in shared and species-specific categories were proportionately and randomly selected (Material and Methods). Thirty cells were randomly selected for each condition/species (13); (C) Only genes up-regulated in both B6 and CAST highly enrich for known T cell functionality (Bonferroni multiple testing corrected p-values, red line is 0.1); (D) Fractions of cells in which a gene is detected are displayed as histograms; 70 genes were randomly selected from each gene set.
Aging can cause a shift in the functional balance between self-renewal and differentiation in hematopoietic stem cells (16). We considered whether aging might similarly perturb the transcriptional response of CD4+ T cells to immune stimulation.
The global expression profiles of naive or activated CD4+ T cells did not change due to aging (Fig. S6A, B). Nevertheless, approximately 10% of genes are differentially expressed between cells from young and old mice (Fig. S6C, D). These genes are only expressed in a small subset of cells, and are not conserved between B6 and CAST (Fig. S6E, F). Most genes in the core-conserved program responded even after three hours of stimulation, irrespective of age (Fig. S7). However, the magnitude of the response was consistently reduced in older mice (Fig 4A).
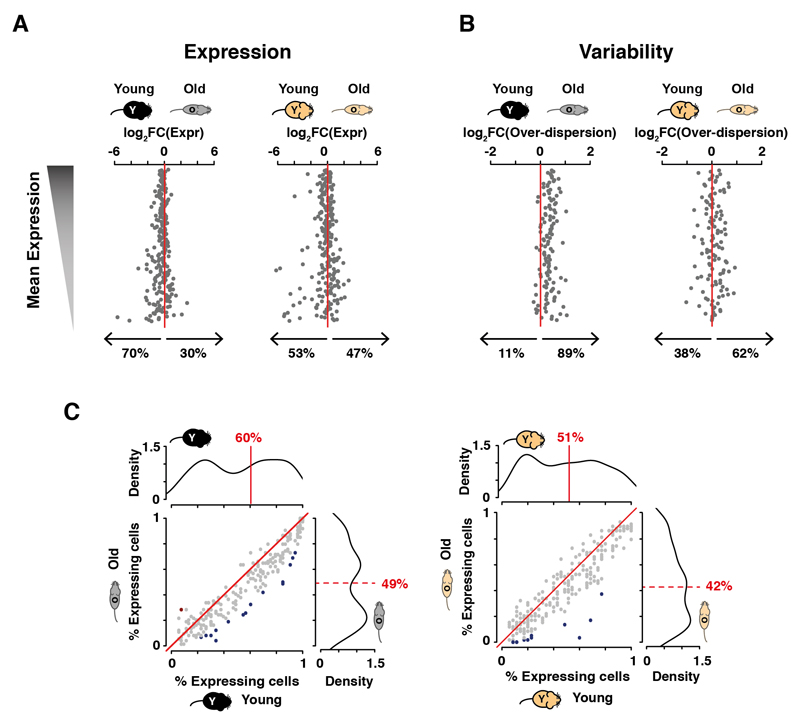
Fig. 4. Aging increases the cell-to-cell variability during early activation of CD4+ T cells.
(A) Upon activation, changes in expression of the core immune activation program are greater in younger mice than in older mice; (B) Among the genes from (A) that do not change their averaged gene expression, transcriptional variability increases during aging (13); (C) The fraction of cells in which genes of the shared activation process are expressed is reduced in CD4+ T cells from old animals. The distribution of fraction values is plotted on each corresponding axis (medians of fraction values are indicated in red); changes in fraction values were tested using a binomial test (significant changes are shaded in red or blue where multiple testing corrected p-values <0.1).
The cell-to-cell transcriptional variability of the core activation program was higher in older animals (Mann-Whitney-Wilcoxon test; p<0.05, Fig. 4B, Fig. S7), independent of changes in gene expression levels. In both B6 and CAST mice, genes involved in the activation process are typically expressed in fewer cells in old mice (Fig. 4C).
Finally, we asked whether additional cellular components of the immune system also show increased transcriptional variability upon aging. We used a stringent gating strategy to isolate effector memory CD4+ T cells in young and old B6 mice (17) (Fig. S8, (13)). scRNA-seq experiments revealed that immune activation genes showed an increase in variability in effector CD4+ T cells from older animals, similar to naïve CD4+ T cells (Fig. S9). These results indicated that aging reduces the fraction of cells in which immune activation genes are up-regulated, thus increasing cell-to-cell heterogeneity and attenuating the response to stimulation across multiple CD4+ T cell subsets.
How cell-type-specific gene expression programs change during organismal lifespan has long been debated (5, 6) and what cell-to-cell transcriptome-wide differences accumulate during aging are relatively unexplored (16). By activating naive CD4+ T cells and quantifying the transcriptional responses of hundreds of single-cells using scRNA-seq, we confirmed that translation processes and immune response genes are rapidly up-regulated (11) and discovered a reduction in cell-to-cell variability across thousands of transcripts. Thus, immune activation rapidly reduces transcriptional heterogeneity across the population of CD4+ T cells, revealing a regulatory strategy comparable to induced pluripotent stem cell reprogramming (18, 19).
Many attempts have been made to identify transcriptional signatures associated with aging (16, 20). Aging appears to have modest effects on mean expression levels in unstimulated and stimulated CD4+ T cells. However, regardless of species or analyzed cell-type, older mice showed substantially greater transcriptional heterogeneity; in simpler terms, older mice have more sporadic transcriptional responses. The discovery that CD4+ T cells from aged mice are less able to robustly up-regulate a core activation program may in part explain the aging associated decrease of immune function observed across mammals (21).
Our results indicate that in addition to transcriptional dysregulation and chromatin destabilization (1), increased cell-to-cell transcriptional variability is a major hallmark of aging.
Supplementary Material
One Sentence Summary.
Single-cell sequencing of unstimulated and stimulated CD4+ T cells reveals how aging destabilizes a conserved transcriptional activation program.
Acknowledgments
Thanks to R Miragaia, A Lun, A Scialdone and A Richard for input; A Mowbray, N Jacobs, M Clayton, M Mitchell for animal husbandry; S Lorenz and the Single Cell Genomics Core Facility at WTSI and the Genomics, Flow Cytometry, and BRU Cores at CRUK CI for technical support. Funded by the European Research Council (FC, TFR, DTO, SAT, MJTS), EMBO Young Investigators Programme (DTO), Cancer Research UK (HCC, MdLR, DTO, JCM), Janet Thornton Fellowship (WT098051) (CPJM), Sir Henry Dale Fellowship (WT107609, MdLR), EMBL (NE, AAK, MJTS, SAT, JCM), MRC Biostatistics Unit (CAV), Wellcome Trust Sanger Institute (CPMJ, SAT, JCM, DTO), BBSRC CASE Studentship with Abcam plc (AAK).
Footnotes
Author Contributions
CPMJ, NE, AAK, SAT, HCC, MdLR, JCM, DTO designed experiments; CPMJ performed experimental analyses; NE performed computational analyses; HCC, AAK, CAV, FC, TFR, MJTS, MdLR provided technical assistance/support; CPMJ, NE, HCC, MdLR, JCM, DTO wrote the manuscript. All authors commented on and approved the manuscript.
Accession Numbers The ArrayExpress accession number for all reported sequencing data is E-MTAB-4888. The R code for analysis is available at http://github.com/MarioniLab/ImmuneAging2017.
References
- 1.Booth LN, Brunet A. The Aging Epigenome. Mol Cell. 2016;62:728–744. doi: 10.1016/j.molcel.2016.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zahn JM, et al. AGEMAP: a gene expression database for aging in mice. PLoS Genet. 2007;3:e201. doi: 10.1371/journal.pgen.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McCarroll SA, et al. Comparing genomic expression patterns across species identifies shared transcriptional profile in aging. Nat Genet. 2004;36:197–204. doi: 10.1038/ng1291. [DOI] [PubMed] [Google Scholar]
- 4.Mirza N, Pollock K, Hoelzinger DB, Dominguez AL, Lustgarten J. Comparative kinetic analyses of gene profiles of naive CD4+ and CD8+ T cells from young and old animals reveal novel age-related alterations. Aging Cell. 2011;10:853–867. doi: 10.1111/j.1474-9726.2011.00730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bahar R, et al. Increased cell-to-cell variation in gene expression in ageing mouse heart. Nature. 2006;441:1011–1014. doi: 10.1038/nature04844. [DOI] [PubMed] [Google Scholar]
- 6.Warren LA, et al. Transcriptional instability is not a universal attribute of aging. Aging Cell. 2007;6:775–782. doi: 10.1111/j.1474-9726.2007.00337.x. [DOI] [PubMed] [Google Scholar]
- 7.Kim HJ, Cantor H. CD4 T-cell subsets and tumor immunity: the helpful and the not-so-helpful. Cancer Immunol Res. 2014;2:91–98. doi: 10.1158/2326-6066.CIR-13-0216. [DOI] [PubMed] [Google Scholar]
- 8.Brawand D, et al. The evolution of gene expression levels in mammalian organs. Nature. 2011;478:343–348. doi: 10.1038/nature10532. [DOI] [PubMed] [Google Scholar]
- 9.Romero IG, Ruvinsky I, Gilad Y. Comparative studies of gene expression and the evolution of gene regulation. Nat Rev Genet. 2012;13:505–516. doi: 10.1038/nrg3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perry GH, et al. Comparative RNA sequencing reveals substantial genetic variation in endangered primates. Genome Res. 2012;22:602–610. doi: 10.1101/gr.130468.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shay T, et al. Conservation and divergence in the transcriptional programs of the human and mouse immune systems. Proc Natl Acad Sci U S A. 2013;110:2946–2951. doi: 10.1073/pnas.1222738110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuan R, Peters LL, Paigen B. Mice as a mammalian model for research on the genetics of aging. ILAR J. 2011;52:4–15. doi: 10.1093/ilar.52.1.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Materials and methods are available as supplementary materials on Science Online.
- 14.Vallejos CA, Richardson S, Marioni JC. Beyond comparisons of means: understanding changes in gene expression at the single-cell level. Genome Biol. 2016;17:70. doi: 10.1186/s13059-016-0930-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Asmal M, et al. Production of ribosome components in effector CD4+ T cells is accelerated by TCR stimulation and coordinated by ERK-MAPK. Immunity. 2003;19:535–548. doi: 10.1016/s1074-7613(03)00268-1. [DOI] [PubMed] [Google Scholar]
- 16.Kowalczyk MS, et al. Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of hematopoietic stem cells. Genome Res. 2015;25:1860–1872. doi: 10.1101/gr.192237.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.den Braber I, et al. Maintenance of peripheral naive T cells is sustained by thymus output in mice but not humans. Immunity. 2012;36:288–297. doi: 10.1016/j.immuni.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 18.Buganim Y, et al. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell. 2012;150:1209–1222. doi: 10.1016/j.cell.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martinez-Jimenez CP, Odom DT. The mechanisms shaping the single-cell transcriptional landscape. Curr Opin Genet Dev. 2016;37:27–35. doi: 10.1016/j.gde.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 20.de Magalhaes JP, Curado J, Church GM. Meta-analysis of age-related gene expression profiles identifies common signatures of aging. Bioinformatics. 2009;25:875–881. doi: 10.1093/bioinformatics/btp073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goronzy JJ, Weyand CM. Understanding immunosenescence to improve responses to vaccines. Nat Immunol. 2013;14:428–436. doi: 10.1038/ni.2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations. Annu Rev Immunol. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramskold D, et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol. 2012;30:777–782. doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ilicic T, et al. Classification of low quality cells from single-cell RNA-seq data. Genome Biol. 2016;17:29. doi: 10.1186/s13059-016-0888-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brennecke P, et al. Accounting for technical noise in single-cell RNA-seq experiments. Nat Methods. 2013;10:1093–1095. doi: 10.1038/nmeth.2645. [DOI] [PubMed] [Google Scholar]
- 26.Buettner F, et al. Computational analysis of cell-to-cell heterogeneity in single-cell RNA-sequencing data reveals hidden subpopulations of cells. Nat Biotechnol. 2015;33:155–160. doi: 10.1038/nbt.3102. [DOI] [PubMed] [Google Scholar]
- 27.Scialdone A, et al. Computational assignment of cell-cycle stage from single-cell transcriptome data. Methods. 2015;85:54–61. doi: 10.1016/j.ymeth.2015.06.021. [DOI] [PubMed] [Google Scholar]
- 28.Vallejos CA, Marioni JC, Richardson S. BASiCS: Bayesian Analysis of Single-Cell Sequencing Data. PLoS Comput Biol. 2015;11:e1004333. doi: 10.1371/journal.pcbi.1004333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stubbington MJ, et al. T cell fate and clonality inference from single-cell transcriptomes. Nat Methods. 2016 doi: 10.1038/nmeth.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 31.Swain SL, McKinstry KK, Strutt TM. Expanding roles for CD4(+) T cells in immunity to viruses. Nat Rev Immunol. 2012;12:136–148. doi: 10.1038/nri3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ho IC, Tai TS, Pai SY. GATA3 and the T-cell lineage: essential functions before and after T-helper-2-cell differentiation. Nat Rev Immunol. 2009;9:125–135. doi: 10.1038/nri2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang W, Brahmakshatriya V, Swain SL. CD4 T cell defects in the aged: causes, consequences and strategies to circumvent. Exp Gerontol. 2014;54:67–70. doi: 10.1016/j.exger.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sakata-Kaneko S, Wakatsuki Y, Matsunaga Y, Usui T, Kita T. Altered Th1/Th2 commitment in human CD4+ T cells with ageing. Clin Exp Immunol. 2000;120:267–273. doi: 10.1046/j.1365-2249.2000.01224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wherry EJ. T cell exhaustion. Nat Immunol. 2011;12:492–499. doi: 10.1038/ni.2035. [DOI] [PubMed] [Google Scholar]
- 36.Goncalves A, et al. Extensive compensatory cis-trans regulation in the evolution of mouse gene expression. Genome Res. 2012;22:2376–2384. doi: 10.1101/gr.142281.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stegle O, Teichmann SA, Marioni JC. Computational and analytical challenges in single-cell transcriptomics. Nat Rev Genet. 2015;16:133–145. doi: 10.1038/nrg3833. [DOI] [PubMed] [Google Scholar]
- 38.Boursalian TE, Golob J, Soper DM, Cooper CJ, Fink PJ. Continued maturation of thymic emigrants in the periphery. Nat Immunol. 2004;5:418–425. doi: 10.1038/ni1049. [DOI] [PubMed] [Google Scholar]
- 39.Hale JS, Boursalian TE, Turk GL, Fink PJ. Thymic output in aged mice. Proc Natl Acad Sci U S A. 2006;103:8447–8452. doi: 10.1073/pnas.0601040103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fink PJ. The biology of recent thymic emigrants. Annu Rev Immunol. 2013;31:31–50. doi: 10.1146/annurev-immunol-032712-100010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.