Abstract
While detailed pictures are emerging of the structures of ribosomes, little is known at the atomic level about the structural and co-translational folding properties of nascent polypeptide chains. Here we have used solution-state NMR spectroscopy to define a structural ensemble of a ribosome-nascent chain complex (RNC) formed during biosynthesis in E. coli, where a pair of immunoglobulin-like domains adopts a folded N-terminal domain (FLN5) and a disordered but compact C-terminal domain (FLN6). To study how FLN5 acquires its native structure co-translationally, we progressively shortened the RNC constructs. We find that the ribosome modulates the folding process, as the complete sequence of FLN5 emerges well beyond the tunnel before acquiring native structure, while in isolation it folds spontaneously, even when truncated. This finding suggests that regulating structure acquisition during biosynthesis can reduce the probability of misfolding, particularly of homologous domains.
The manner by which a protein acquires its correct tertiary structure, whilst avoiding alternative pathways which lead to aberrant folding, is a fundamental processes and one which underpins the biological activity of all living systems1. Our mechanistic understanding of the inherent nature of protein folding has come predominantly from extensive studies of isolated polypeptides renatured in dilute aqueous solutions, and where the folding process can be elegantly described using energy landscapes2,3 but the extent to which such characteristics are shared during folding within the cell is a prominent question in contemporary biology4. For the vast majority of proteins, folding processes can begin in a co-translational manner during biosynthesis on the ribosome5-7, which leads to constant remodelling of the energy landscape as translation proceeds8. Co-translational folding is thought to be a vital means by which the cell can promote successful folding, particularly for polypeptide chains that would otherwise readily misfold7,9,10.
A mechanistic understanding of protein biosynthesis is emerging through detailed structures of the functional ribosome11, but there is little structural understanding of the emerging nascent chain as its inherent dynamics has eluded most high-resolution techniques. During biosynthesis, the nascent chain is synthesized at a rate of ca. 10-20 amino acids per second in prokaryotes12, and its folding is at least under some form of translational control; thus, for example, the presence of synonymous codons within mRNA sequences has been observed to affect adversely the folding efficiency6,10 of nascent chains. As the nascent chain elongates, it emerges in a vectorial manner from the restricted environment of the ribosomal exit tunnel, enters the crowded cellular milieu and begins to explore conformational space and acquire its complex tertiary structure. A range of ancillary proteins such as molecular chaperones13 and those mediating processing and translocation14 are present, and the ribosome is a central hub for many of these proteins, which compete for the nascent chain15. Most notable of these proteins, is the ribosome-associated molecular chaperone, trigger factor16,17, which can bind to emerging polypeptide chains at the ribosomal exit tunnel18. In addition, the ribosomal surface itself has been suggested to influence this process through transient electrostatic interactions between the emerging nascent chain and the ribosomal surface9,19, that in some cases appear to alter the rate and efficiency of folding9.
The manner by which nascent chains sample structural conformations has been investigated largely using translationally-arrested RNCs, and local compaction in nascent chains observed using FRET probes on the nascent chain has been used to propose structure formation10. Putative co-translational protein folding intermediates20 have also been identified using fluorescence measurements20 and biochemical studies21, suggesting that structural conformations formed on the ribosome may differ from those populated in vitro. Cryo EM analysis of RNCs22 shows that nascent chains remain largely extended as they are extruded through the ribosomal exit tunnel, with additional structural23 and biochemical evidence24,25 indicating that some amino acid sequences can promote the formation of incipient structure, such as α-helices, as well as a simple tertiary motif26 in distinct regions of the tunnel. Although it has been shown more recently that a simple tertiary motif can form within the exit tunnel26, higher-order structure appears to be formed only when a nascent chain has emerged. A detailed understanding of the progressive acquisition of the tertiary structure of the nascent chain outside the ribosome is absent. In this study, therefore, we set out to utilize the ability of NMR spectroscopy to report upon both structure and dynamics during folding at a residue specific level27,28, and produced a structural ensemble of a highly dynamic nascent chain of a pair of immunoglobulin-like domains emerging during biosynthesis. In addition, we have characterized in solution, a set of RNCs generated in vivo in E. coli, to produce a series of high-resolution snapshots that reveals structural details of co-translational protein folding.
Results
Isotopically-labelled RNCs produced in vivo within E. coli
To explore the structure and dynamics of nascent chains as they emerge from the ribosome, we studied a polypeptide chain whose sequence is based upon a pair of immunoglobulin-like proteins, FLN5646-750 and FLN6751-857, the fifth and sixth filamin domains of the Dictyostelium discoideum gelation factor (FLN)27, respectively. We initially designed a FLN5-6 ribosome-nascent chain complex (RNC), FLN5+11027, in which the C-terminus of the 105 residue FLN5 domain is separated from the peptidyl transferase centre (PTC) by 110 residues, comprised of a folding-incompetent FLN6 domain (with an 18 amino acid truncation at its C-terminus (residues 840-857)28, referred to here as FLN6∆18), and the 17 amino acid SecM translation-arrest motif29. Supplementary Fig. 1 shows all RNC and isolated protein designations for FLN5 and FLN6 variants used in this study. The RNCs were all generated in E. coli27, where folding takes place within the cellular milieu, and the intact RNCs were purified in high yield as previously described27 (Fig. 1a,).
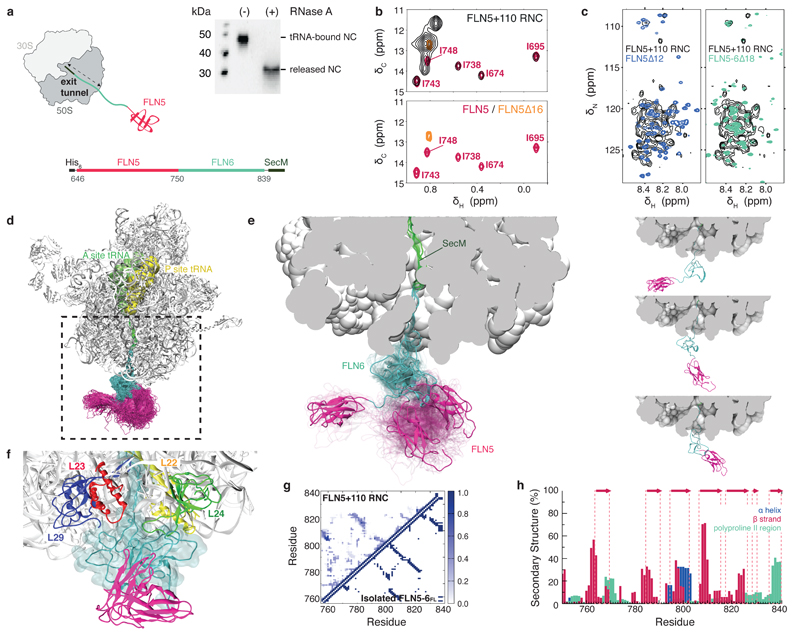
Figure 1. Structural ensemble of a ribosome-bound nascent chain.
(a) Schematic of the FLN5+110 RNC used for the ensemble calculations. The FLN5 sequence is tethered to the ribosome by a C-terminally truncated FLN6751-839 sequence and stalled using the SecM translational-arrest motif27,29. Anti-His western blots of purified FLN5+110 RNC, ribosome-attached with bound prolyl P-site tRNA, and also in its released form. (b) Overlay of 1H-13C correlation spectra of [U-2H; Ileδ1-13CH3 labeled] FLN5+110 RNC (black) with isolated, natively folded FLN5 (pink) and isolated unfolded FLN5∆16 (orange). (c) Overlay of 1H-15N correlation spectra of U-15N-labelled FLN5+110 RNC with isolated FLN5∆12 (blue), and unfolded FLN5-6∆18 (green). (d) NMR chemical shift restrained structural ensemble of FLN5+110 RNC, showing the disordered FLN6 linker (cyan) and the native fold acquired by FLN5 (pink). (Accession codes: PDB ID 2N62; BMRB ID 25748). (e) Close-up view of the ribosomal exit tunnel, highlighting three representative conformations of the nascent chain ensemble (left); the three representative conformations are also shown separately (right). (f) Transient interactions made between the disordered FLN6 linker in close proximity with the ribosomal proteins at the surface. (g) Probability of the formation of inter-residue contacts in the FLN5+110 RNC (shown above diagonal) and in the native state of full length (FL), isolated FLN5-6 (below diagonal) (h) Secondary structure populations of the RNC depicting β-strands (red), α-helices (blue) and polyproline II regions (green); native β-strands are indicated (red arrows).
FLN5 acquires native-like structure near the exit tunnel
To use NMR spectroscopy to probe folded and unfolded conformations of FLN5+110 RNC, a dual isotopic labeling scheme was developed, using both selective protonation and uniform labelling approaches. As methyl group resonances are highly sensitive reporters of changes in protein tertiary structure, we generated selectively-labeled RNCs on a perdeuterated (2H) background in which only the Ile-δ1 side chain of the nascent chain was labeled as 13CH3. The replacement of all surrounding 1H by 2H nuclei results in longer relaxation times and more intense signals30. Samples of the uniform (U)-2H; Ileδ1-13CH3 labeled RNC were then examined via 1H-13C correlation spectra (using methyl TROSY NMR methods30). Resonances from all five FLN5 isoleucine residues could be identified in 1H-13C correlation spectra of FLN5+110 RNC, and these were found to overlay closely (1H and 13C chemical shift changes < 0.01 and 0.1 ppm respectively) with those of isolated FLN5 (Fig. 1b), indicating that in this nascent chain the FLN5 domain had folded into a native conformation. In parallel, we produced uniformly (U-) 15N-labeled FLN5+110 RNCs, in which the peptide backbone was isotopically labeled and we recorded 1H-15N correlation spectra via rapid acquisition SOFAST-HMQCs31. Examination of the 1H-15N correlation spectra of the U-15N-labelled FLN5+110 RNC showed nascent chain resonances within a narrow window of 1H chemical shifts, indicative of disordered structure. The chemical shifts of the nascent chain resonances corresponded closely to those observed of unfolded FLN6 (in spectra of isolated FLN5-6∆1828), rather than unfolded FLN5 (in spectra of a 12-residue C-terminal truncation, FLN5∆12) (Fig. 1c).
These combined NMR data are exquisite probes for both the folded and unfolded structural preferences of FLN5 and FLN6 tethered to the ribosome, and enabled us to use chemical shifts measured for FLN5+110 RNC as replica-averaged structural restraints in molecular dynamics simulations32 to determine a structural ensemble of the RNC (Fig. 1d,e and Supplementary Video 1). This ensemble showed folded FLN5 tethered to the ribosome by a disordered FLN6. Despite lacking persistent structure, FLN6 was compact and exhibits transient populations (of about 20% on average) of native-like secondary structure elements and inter-residue contacts (Fig. 1d-h). The ensemble also illustrated that FLN5 had substantial access to a broad region of the ribosomal surface, including forming transient contacts with both 23S rRNA and ribosomal protein L29, as a result of its tethering to the disordered FLN6 (Fig. 1d,e). We analyzed the regions of the ribosome in close proximity to FLN6, and observed transient interactions being made with both 23S RNA and several ribosomal proteins associated with the exit port (Fig. 1f). The most frequent degrees of contact are made with L24 (55%), specifically with a prominent loop in close proximity to the exterior of the exit port; such contact is further supported by nascent chain crosslinking studies33 and a cryo EM structure of a ribosome-SecYE complex34 which shows that this loop can undergo marked conformational changes in the presence of a nascent chain derived from the periplasmic protein, FtsQ. In addition, FLN6 made transient yet substantial contacts with L23 (30%), whose position on the surface near to the exit tunnel is shown in structural studies15 to be an adapter site for ancillary proteins including the molecular chaperone trigger factor.
These RNC ensemble structures suggested that the conformational freedom of the nascent chain was likely to be tempered by its interactions with the ribosomal surface (Fig. 1d) and that these interactions had both structural and dynamical implications for the processes by which a vectorially emerging nascent chain sequence forms its complex tertiary structure beyond the ribosomal tunnel. Previous studies35 show that isolated FLN5 folds highly co-operatively via a low population of a folding intermediate, raising the question of a possible role for the ribosome itself modulating the folding of FLN5 nascent chains as they emerge during biosynthesis.
Structural evidence for co-translational folding of FLN5
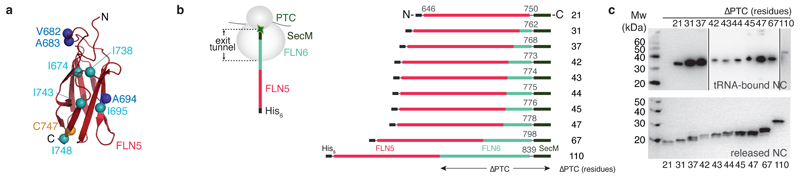
In order to probe how FLN5 acquires its structure during biosynthesis, we extended our NMR approach to analyze a series of twelve RNCs, in which the FLN6 linker was progressively shortened (Fig. 2a,b); each of these NMR spectra represented then a unique snapshot during biosynthesis that reported on co-translational protein folding at equilibrium. The series of SecM-arrested nascent chains consisted of FLN5 with decreasing numbers of residues of the FLN6 sequence, ranging from 21 to 110 residues (Fig. 2b). The RNCs, denoted FLN5+L (with L = 21 to 110), were purified from E. coli cells in similar yields to that of the FLN5+110 RNC, and a series of biochemical and biophysical analyses showed that all were completely intact (Fig. 2c), and free of any extraneous proteins including, notably, the ribosome-associated molecular chaperone trigger factor, as well as DnaK (Supplementary Fig. 1). The continuous cycling of these ubiquitous cytosolic chaperones17,36 and others with the ribosome and nascent chains alike meant that the RNCs had considerable access to these during co-translational folding within the cell; but their absence following purification (< 1.5% occupancy, Supplementary Fig. 1) indicated, however, that FLN5 RNCs are relatively poor substrates16 for these particular species. Each of the FLN5 RNCs samples was isotopically-labeled as U-2H; Ileδ1-13CH3 or U-15N-labeled in the peptide backbone and we acquired 1H-13C and 1H-15N correlation spectra, respectively. For all samples, the acquisition of these spectra was accompanied by rigorous control experiments including interleaved NMR diffusion and cross peak intensity measurements, in conjunction with western blots (Fig. 2c and Supplementary Fig. 2), to ensure that the data used for structural analysis were derived exclusively from intact RNCs.
Figure 2. Design and in vivo production of FLN5 ribosome-nascent chain complexes in E. coli.
(a) Structure of isolated, natively folded FLN5 (PDB: 1QFH). Mapped onto the FLN5 structure are the five isoleucines (∂ 1 methyl groups) of FLN5 (Ile674, 695, 738, 743, 748, cyan) used as probes of native structure acquisition, and the amide groups of three residues (Val682, Ala683 & Ala694, blue) selected for analysis of unfolded conformations (see text). (b) Design of the translationally-arrested RNCs27 to monitor nascent chain emergence and folding, in which the FLN5 sequence is tethered to the PTC via increasing lengths of the FLN6 sequence and the SecM translational arrest motif, (c) Anti-His western blots of the library of purified FLN5 RNCs shown in ribosome-bound (upper panel, see also Supplementary Fig. 1) and released (lower panel) forms.
As observed in 1H-13C correlation spectra of FLN5+110 RNC (Fig. 1b), resonances from all five FLN5 isoleucine residues could be similarly identified in [1H-13C correlation spectra of FLN5+67 and FLN5+47 samples, indicating that in these nascent chains the FLN5 domain had also folded into a native or near-native conformation (Fig. 3a). The intensities of the dispersed resonances in FLN5+47 were, however, only 30% (±12%) of those within the corresponding spectra of FLN5+67 and FLN5+110, a feature that we discuss below. Moreover, in spectra of FLN5+45, only three of the five native-like isoleucine resonances are visible, all with very low intensity (Fig. 3a), and no such resonances at all could be detected in spectra of the RNC with the shortest linker length, FLN5+21.
Figure 3. Nascent chains of FLN5 emerging from the ribosome monitored by NMR spectroscopy.
(a) 1H-13C correlation spectra of [U-2H; Ileδ1-13CH3 labeled] FLN5 RNCs (black), isolated, natively folded FLN5 (cyan) and isolated unfolded FLN5∆16 (orange). Resonances marked “R” arise from background labeling of 70S ribosomal proteins27. (b) 1H-15N correlation spectra of U-15N-labelled FLN5 RNCs, isolated FLN5∆12 (blue), and unfolded FLN5-6∆18 (green). Resonances used for the analysis of unfolded conformations are labeled in the FLN5+21 RNC spectrum.
Using each of the FLN5 RNCs, 1H-15N correlation spectra were recorded to monitor in each sample the presence or absence of resonances of the unfolded form of the FLN5 domain. Examination of the spectra (Fig. 3b) revealed that when L is between 21 and 44 residues, all the resonances of the nascent chain appeared within a narrow window of 1H chemical shifts, indicative of disordered structure. The chemical shifts of the nascent chain resonances corresponded closely to those observed in spectra of unfolded forms of isolated FLN5 generated by a C-terminal truncation, FLN5∆12 (Fig. 3b and Supplementary Fig. 3), or by a destabilizing mutation in the FLN5 variant, Y719E (Supplementary Fig. 3). The average intensities of these RNC cross-peaks were, however, found to be reduced substantially in spectra of FLN5+43 and FLN5+44 RNCs, and no comparable unfolded FLN5 resonances were visible in spectra of FLN5+45 to FLN5+110 RNCs. In addition, cross-peaks attributable to the emerging FLN6 sequence in an unfolded state could be identified (Supplementary Fig. 3) in spectra of FLN5+67 (Fig. 3b), as in the FLN5+110 RNC (Fig. 1c). These NMR data clearly showed the increasing population of the folded state of FLN5 relative to its unfolded state as the length of the sequence joining it to the PTC increased, and also the concomitant appearance of peaks from disordered residues from FLN6.
To evaluate further the transition from the unfolded to the folded state as FLN5 emerged from the tunnel, three amide resonances of FLN5 were selected from the spectra of the U-15N-labelled RNCs, that were particularly well resolved and not overlapping with other resonances (Fig. 3b, Supplementary Fig. 3). These resonances had comparable 1H linewidths (20 ± 3 Hz) in all RNCs from FLN5+21 to FLN5+42 (Supplementary Fig. 3), indicating that, for these residues at least, any differences in intensity associated with the nascent chain length could be attributed to changes in the population of the unfolded form of the nascent chain rather than to changes in relaxation behavior. Indeed analysis of the signal intensities indicated that the population of the unfolded state of FLN5 decreased substantially in samples for which L = 42 to 45, and the length-dependent changes in the amide resonance intensities of the disordered FLN5 nascent chain were consistent with an unfolded-to-folded transition with a mid-point between L = 42 and 45 (Fig. 4a). Also consistent with this conclusion, native-like resonances of the isoleucine methyl groups of FLN5 were observable in 1H-13C correlation spectra starting from FLN5+45 through to FLN5+110 RNCs. We attributed the weak intensity of the methyl resonances in nascent chains with L = 45 and 47 to the low mobility of the folded FLN5 domain as a result of its proximity to the slowly tumbling ribosome, rather than to a reduction in the population of the folded state. In support of this conclusion, the increases in the intensity of these resonances, evident for nascent chains with L = 67 and 110, reflected the gain in mobility of the folded FLN5 domain as the length of the chain linking it to the PTC increased (Fig. 3a).
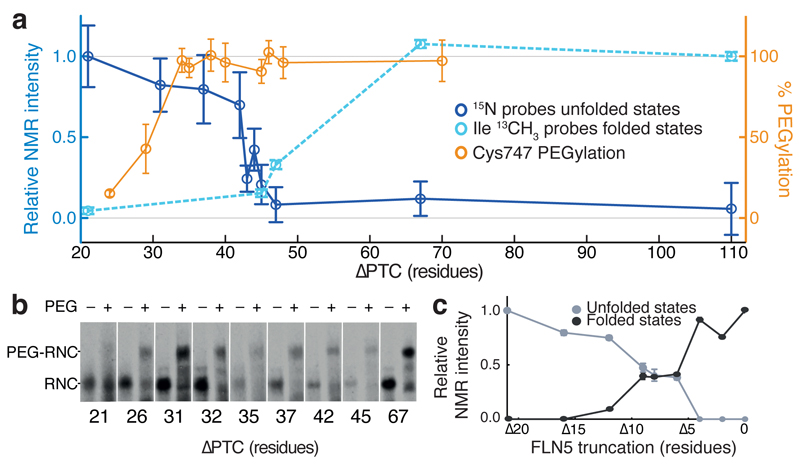
Figure 4. Folding of FLN5 on the ribosome monitored by NMR spectroscopy and PEGylation.
(a) FLN5 nascent chain folding as measured by intensity changes of 15N amide resonances (blue) arising from the unfolded FLN5 domain (mean ± s.d. for n=4 (n=3 for +45 and n=2 +47); nascent chain concentration from western blot replicates), and intensity changes in Ile 13CH3 resonances (cyan) arising from native FLN5 structure (mean ± s.d. of spectral noise, n=1). Intensities are normalized and scaled relative to L = 21 (unfolded) or L = 110 (folded). The solvent accessibility of the FLN5 domain from the ribosomal exit tunnel was probed using PEGylation (orange) (mean ± s.d) of folding-incompetent FLN5 Y719E RNCs, where the native Cys747 is close to the FLN5 and FLN6 boundary. (b) Cys747 PEGylation of FLN5 Y719E RNCs results in a band shift (PEG-RNC). (c) C-terminal truncations of isolated FLN5 as measured by NMR. Averaged cross-peak intensities of folded (black) and unfolded (grey) states of FLN5 (see Supplementary Fig. 5) are mapped against truncation length.
Folding of FLN5 RNCs is offset relative to isolated FLN5
To rationalize these spectral observations of folding as the nascent chain elongates, the accessibility to solvent of the emerging FLN5 domain was examined by probing the native cysteine residue (C747 in orange, Fig. 2a), which is located close to the FLN5-FLN6 boundary, for its susceptibility to modification by methoxypolyethylene glycol maleimide (PEG-Mal) (Fig. 4a,b). Under the experimental conditions used, the 5 kDa moiety, as shown previously24, can only substantially (> 80%) modify a cysteine in a nascent chain if it is beyond the exit vestibule, i.e. more than ca. 100 Å from the PTC37. We used a series of RNCs of the folding incompetent variant, FLN5 Y719E (Supplementary Fig. 4), to monitor the emergence of Cys747 from the ribosomal exit tunnel, without the complication of the cysteine residue being shielded from solvent as a result of structure acquisition in the FLN5 domain. Under conditions analogous to those of the NMR experiments (and adapted from those well established24 to achieve PEGylation of a nascent chain entirely emerged from the vestibule), we observed complete PEGylation for L ≥ 31, i.e., when Cys747 was ≥ 34 residues from the PTC (Fig. 4a,b and Supplementary Fig. 4). This result showed that at these nascent chain lengths the entire FLN5 sequence had emerged from the tunnel to an extent that enabled it to be accessible by PEG-Mal, but well before the folding of the domain could be observed (L > 44) by NMR spectroscopy as discussed above.
We next generated a series of C-terminal truncations of the isolated FLN5646-750 domain so as to examine the length-dependence of folding of this domain in the absence of the ribosome. We analyzed 1H-15N correlation spectra of nine C-terminal truncations ranging from FLN5∆2 to FLN5∆21. The spectra indicated that FLN5∆12 and its shorter variants, in which the C-terminal β-strand G and its adjacent loop in the native structure were absent, were fully unfolded under the conditions used in this study (Fig. 4c and Supplementary Fig. 5). By contrast, the longer variants FLN5∆2 and FLN5∆4 were fully natively folded, while sequences of intermediate lengths between FLN5∆6 and FLN5∆9 populated both folded and unfolded states at equilibrium. From these results, we concluded that the isolated FLN5 domain in bulk solution could tolerate truncation of up to nine residues and still populate a folded state to a very significant degree.
The fact that FLN5∆4 (residues 646 to 746 of FLN5) was fully folded in its isolated state was highly significant as the next residue, Cys747, in the RNC FLN5+31, was solvent accessible and hence clear of the exit vestibule as shown by the PEGylation experiments discussed above (Fig. 4a,b). Acquisition of native structure would therefore, in principle, be possible even in the case of FLN5+31, i.e. when L = 31 and where Cys747 has emerged from the tunnel, as indicated by its accessibility to PEGylation. The NMR data, however, showed that folding of the FLN5 domain takes place only when a further 11 to 14 residues of FLN6 have been added to the sequence (Fig. 4a). There is thus a substantial difference between the length of the FLN5 polypeptide sequence required for the acquisition of native structure by the isolated domain in bulk solution, and by the domain when attached to the ribosome; indeed, the folding transition on the ribosome required, remarkably, the availability of an additional 17 residues compared to that observed for the isolated protein (Fig. 5).
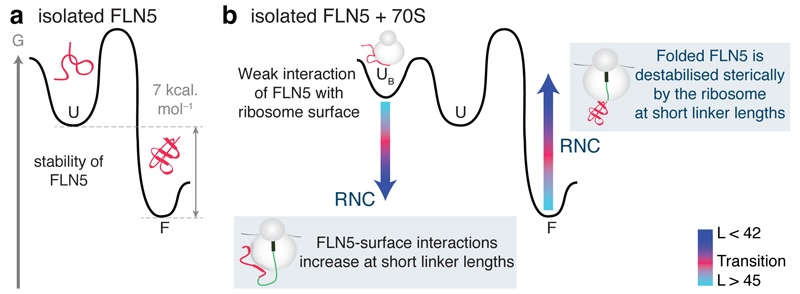
Figure 5. FLN5 folding is offset on the ribosome.
A comparison of FLN5 folding on the ribosome and in isolation as depicted by the conformational states observed for FLN5 RNCs with different linker lengths, L, from the PTC, and those of C-terminal FLN5 truncations: unfolded (blue), folded (cyan) and folding transition (pink). The sequence of the FLN5 nascent chains is solvent exposed, as monitored by PEGylation, at L ≥ 31 residues where Cys747 is 34 residues from the PTC yet the domain only acquires native-like structure upon addition of a further 11-14 residues, at 42 ≤ L ≤ 45 as shown by NMR spectroscopy. Isolated FLN5 truncations are shown alongside the RNC lengths with FLN5∆4 a reference for L = 31, at which point complete PEGylation is observed. A folding offset (pink dotted arrow) is the difference observed between the initiation of FLN5 structure acquisition on the ribosome compared to that of the protein in isolation.
The origins of the offset between the solvent accessibility of the complete FLN5 domain and its folding during biosynthesis were explored, initially by examining the inter-domain interactions between the emerged FLN5 and sections of the successive FLN6 linking sequence (Fig. 1d,g) by substituting the FLN6 residues with a poly glycine-serine linker (LGS) to generate a series of FLN5+LGS RNCs (Supplementary Fig. 6). Using identical conditions to those used for the FLN5 RNCs, complete PEGylation of the FLN5+LGS RNCs occurred at LGS ≥ 35 (Cys751, i.e. 34 residues from the PTC). NMR observations of LGS=31, 37 and 42 showed only disordered FLN5 resonances, suggesting that FLN5 folded independently, regardless of the linking sequence, and it is unlikely that inter-domain interactions alone were the cause of the offset observed for folding (Supplementary Fig. 6).
The ribosome surface modulates the energy landscape of FLN5
Our structural ensemble of the FLN5+110 RNC revealed that the emerging nascent chain interacted transiently with ribosomal surface proteins (Fig. 1d,f), and we assessed this issue further using high-resolution 2D 1H-15N correlation spectra (Fig. 6a). We reasoned that such interactions might influence the capacity for a nascent chain to acquire structure. Addition of an equimolar concentration of 70S ribosomes to samples of the isolated unfolded variants FLN5 Y719E (Fig. 6b) and FLN5∆12 (Supplementary Fig. 7) resulted in moderate (ca. 30%) reductions in the intensities of the resonances of Lys663 to Val677 and Gly713 to Gly750. Analogous intensity changes were not, however, observed following addition of 70S ribosomes to a sample of full-length, folded FLN5 (Fig. 6b and Supplementary Fig. 7), indicating that the intensity changes were the result of broadening attributable to the binding of unfolded FLN5 to the slowly tumbling ribosome particle. Analysis of the spectra of the various FLN5 RNCs showed that when L = 21 to 42, where the domain is unfolded, resonances of Phe665 to Val677 and Gly713 to Gly750 were similarly reduced in intensity. The effects were, however, much more substantial than those observed for the isolated domain, with the resonances of Phe665 to Val667 losing more than 70% of their intensities, and those of Gly713 to Gly750 became completely undetectable (Fig. 6a,c).
Figure 6. Residue-specific mapping of RNC interactions.
(a) An overlay of 1H-15N correlation spectra (recorded at a 1H frequency of 950 MHz) of FLN5+31 RNC (black) and unfolded, isolated FLN5∆12 (red) highlighting resonances that are significantly broadened in the RNC. (b) Relative intensities of 1H-15N resonances of folded FLN5 (5 µM) and unfolded FLN5 Y719E (8 µM) in the presence of 1 molar equivalent of 70S ribosomes. (c) Relative intensities of FLN5+21, FLN5+31, FLN5+42, FLN5+67 and FLN5+110 RNCs as compared to a reference, made up of a composite consisting of FLN5∆12 and FLN5 Y719E, to monitor unfolded FLN5 (red), and FLN5-6∆18 to monitor unfolded FLN6 (green), and folded FLN5 from 1H-13C correlation spectra (cyan). A 5-point moving average is plotted as a guide; errors derived from spectral noise, n=1. The grey shaded area denotes occluded residues inaccessible to PEGylation.
The same FLN5 residues showed similar reductions in intensity in analogous spectra of LGS= 31 and 42 in the FLN5+LGS RNCs (Supplementary Fig. 6). These data indicated, therefore, that the specific stretches of sequence identified from the spectra of unfolded FLN5 interact with the ribosomal surface9,19. The greater extent of ribosome surface interactions of FLN5 in the RNCs, relative to the isolated FLN5 domain in the presence of the ribosome, can be attributed to a higher effective concentration of the ribosome as a result of its anchoring to the PTC. Such an effect will be most pronounced at short linker lengths (L = 21 to 42); indeed the effective concentration was estimated to be 20 mM for a C-terminal residue located 10 residues beyond the exit tunnel (see Online Methods). This effect will increase the magnitude of the interaction between unfolded FLN5 and the ribosomal surface, particularly at the C-terminus, which includes residues Gly713 to Gly750, a result consistent with our observations. As a consequence, the unfolded state will be stabilized relative to the native state8 at short RNC linker lengths, 21 < L < 44 (Fig. 7b), and will therefore inhibit folding of the domain when attached to the ribosome relative to its isolated state. As the nascent chain elongates, the interactions with the ribosome surface are progressively reduced, and only at L > 42 do they become insufficient to overcome the 7 kcal mol-1 free energy of folding measured for isolated FLN5 in bulk solution35 (Fig. 7a,b).
Figure 7. The ribosome modulates the folding landscape of FLN5 nascent chains.
(a) Schematic of a free energy diagram for isolated FLN5, showing the difference in free energy of 7 kcal mol-1 between the folded state, F, and the unfolded state, U. (b) Schematic free energy diagram for isolated FLN5 in the presence of ribosomes shows a ribosome-bound state, UB, accessible from the unfolded state. A model for how the ribosome could alter this landscape and inhibit nascent chain folding is indicated (arrows): At short linker lengths, the tethered nascent chain is subject to high effective ribosome concentrations, favoring a ribosome-bound state UB. The native state, F, is also likely to be energetically unfavorable, due to steric interactions with the ribosome. As the nascent chain increases in length, the steric effects and ribosome- associated interactions experienced by the tethered nascent chain are overcome by the stability of the folded FLN5 domain.
The mechanism by which the ribosome-nascent chain interactions specifically acquire the capacity to modulate nascent chain folding and achieve the observed folding offset, is likely to be related to the effects of steric occlusion (particularly at short linker lengths) (Fig. 7b), as well as being directed by the sequence determinants inherent to the nascent chain. In the latter case, the close homology between FLN5 and FLN6 suggests that the two domains are likely to form similar transient interactions with the ribosome in their disordered states (Fig. 1). As replacing FLN6 with a poly (GS) linker did not abrogate the folding offset (Supplementary Fig. 6), this result suggested that the transient interactions with the ribosome act independently on each emerging domain, rather than requiring a preceding domain to interact with a specific ribosomal protein or RNA at the ribosomal exit, and be responsible for transmitting a “folding trigger”. Therefore, for a multi-domain protein such as FLN, comprised of homologous domains, ribosome-nascent chain interactions to produce a folding offset may occur as each domain emerges sequentially, rather than in a coordinated intra-domain manner.
Discussion
In summary, we have used NMR spectroscopy to determine the structural ensemble of a folded multi-domain nascent chain on its parent ribosome. The ensemble provides clear insights into the dynamic process of co-translational folding: the globular FLN5 domain possessed a high degree of conformational freedom resulting from the presence of a compact, disordered FLN6 domain, the latter showed transient yet significant interactions with both ribosomal RNA and the ribosomal proteins surrounding the exit site, in particular L24. Our studies of the changes to the structural ensembles formed by shortened RNCs further revealed a residue-specific understanding of how a nascent chain acquires native-like structure during its progressive emergence from the ribosomal exit tunnel. Indeed, in the case of the protein domain studied here, the folding of the tethered nascent chain did not take place as soon as a sequence of polypeptide chain that is capable of folding in bulk solution, emerges from the ribosomal tunnel.
It appeared instead that a certain degree of compaction of the nascent chain along with contributions from specific interactions of the disordered state of FLN5 (that are likely to be analogous to those observed for FLN6 in the structural ensemble) with the ribosomal surface permitting persistent folding to occur only after an additional segment, here consisting of 11 to 14 residues of the subsequent FLN6 sequence, has also emerged. Within living cells, however, co-translational folding is not an equilibrium process but occurs in parallel with the process of translation (10-20 a.a. s-1)12, which can result in an offset between the point at which folding occurs on actively translating ribosomes, compared to those that are stalled38. Thus, the continuous translation process would indicate that the folding of FLN5 may be completed at longer linker lengths than at the point at which we observed FLN5 folding to occur in stalled RNCs; folding in vitro on a timescale of ca. 1 s-1, typical of immunoglobulin domains39 could produce an offset of 10-20 a.a. between the polypeptide chain length at which FLN5 folding becomes thermodynamically favorable, and the point at which folded populations can form kinetically.
At least for this system, therefore, interactions with the ribosome during emergence from the tunnel inhibit the acquisition of stable structure by a nascent chain, rather than promote nativelike contacts in a progressive manner as suggested for other systems5,7,9,10. This phenomenon has apparent similarities to the behavior of some molecular chaperones described as holdases40 that inhibit the formation of misfolded and potentially toxic aggregates by stabilizing more highly unfolded states41. We suggest that regulating the acquisition of partially folded structures within a nascent chain during co-translational folding of a protein may act in a similar manner to ensure efficient generation of functional proteins within living systems by reducing the probability of misfolding, particularly of multi-domain proteins with high sequence identities between domains42. Indeed, such a mechanism suggests that cotranslational folding of neighboring individual domains may be remarkably similar to the cooperative folding in vitro3, rather than the gradual acquisition of native-like structure during the process of biosynthesis.
Online Methods
Generation of ribosome-nascent chain complexes (RNCs) and isolated C-terminal truncations
DNA constructs of RNCs of tandem domains FLN5-6 were derived from a FLN5+110 RNC construct described previously27. Site-directed mutagenesis was used to manipulate the length of the 110 amino acid FLN6 linker to generate a set of SecM-stalled FLN5 RNCs with linker lengths L, ranging from 21 to 110 residues (L = 21, 26, 31, 35, 37, 42, 43, 44, 45, 47, 67, 110). Selectively isotopically-labeled, His-tagged RNCs were generated in BL21(DE3) E. coli using an in vivo procedure described previously27 with modifications. Following growth in an unlabeled MDG medium at 37°C, the cells were washed and resuspended in an M9-based expression medium (“EM9”, adapted from43) enriched with the relevant isotopes. RNC expression was induced with 1 mM IPTG, and after 10 min, 150 mg/mL rifampicin44 was added and the cells were harvested 35 min later. Uniform 15N labeling was performed as described previously27. The production of 2H,13CH3-Ile-δ1 methyl-labeled perdeuterated (U-2H; Ileδ1-13CH3) RNCs, in which the δ-CH3 group of the isoleucine side-chain was selectively protonated, was achieved by using perdeuterated conditions, employing the isoleucine precursor 2-ketobutyric-4-13C,3,3-d2 acid in a procedure adapted from that described previously for U-15N-labelled RNCs27, in which the cells were progressively adapted into the deuterated isotopes and precursors. Rifampicin was omitted during the induction period, and cells were harvested after 1.5 h. The purification of RNCs from E. coli was performed as described previously27, except the ribosomal material was recovered from the lysate using a 35% (w/v) high salt sucrose cushion prior to purification using a Ni-IDA column followed by a 10-35% w/v sucrose gradient. Site-directed mutagenesis was used to introduce the Y719E point mutation into FLN5 RNCs and into isolated FLN5, as well as the substitution of the FLN6 linker for a glycine-serine repeat sequence (poly (GS)). Isolated C-terminal truncations of FLN5 (residues 646 to 750) were generated by removing between 2 and 21 amino acids (FLN5∆2 ∆4, ∆6, ∆8, ∆9, ∆12, ∆16, ∆21), using mutagenesis and each of the FLN5 variants was expressed and purified as previously described for full-length FLN528.
RNC integrity and the determination of nascent chain occupancy
For evaluating RNC integrity, samples were run on denaturing 12% (w/v) polyacrylamide bistris gels at neutral pH and using a sample dye at pH 5.7 to maintain the ester bond between the tRNA and the nascent chain. Released forms of the nascent chain were obtained by treatment of the RNC samples with RNase A. For determination of nascent chain occupancy, RNase A treated RNCs were run alongside isolated protein concentration standards and anti-His western blots analyzed using ImageJ (Rasband, W.S., U.S National Institutes of Health) software.
RNC integrity over the time course of NMR experiments as monitored by western blot
Purified RNCs were incubated at 25°C and 5 pmol aliquots were collected periodically to examine the integrity of the tRNA-bound form of the nascent chain over time, in conjunction with NMR experiments being recorded on an identical sample. All samples were analyzed by western blotting and the intensity of the band corresponding to tRNA-bound nascent chain was assessed by densitometry.
Trigger factor and DnaK detection and quantification within RNC samples
Purified RNC samples were treated with RNase A and then were assessed by western blotting for the presence of trigger factor using a rabbit polyclonal anti-trigger factor antibody (Cat No. A01329, GenScript, UK). A similar procedure was employed for the detection of DnaK using an anti-DnaK antibody (Cat No. LS-C63274-50, Source Bioscience, UK). The residual amount of both trigger factor and DnaK present within the RNCs was determined using densitometry analysis using purified trigger factor and DnaK proteins as standards, as described for RNC integrity and the determination of nascent chain occupancy.
Coupled transcription-translation of RNCs in vitro
An E. coli S30 cell extract was prepared as described elsewhere25. A pair of primers: 5’ primer upstream of the T7 promoter (5’- CTCGATCCCGCGAAATTAATACG-3’) and a 3’ primer partially overlapping the SecM-stalling sequence (5’- AGGTCCATGGTTAAGGGCCAG-3’), was used to produce linear templates encoding SecM-stalled RNCs from the relevant plasmids. Reactions were performed in 25 µL volumes using an S30 extract and a translation premix25, containing 1.5 µg linear DNA,, 0.04 mM L-amino acids, 20 µL, 10 units of T7 RNA polymerase, 5 µCi [35S]-methionine and 200 ng/µL anti-sense ssrA oligonucleotide. Transcription-translation reactions were incubated at 37°C for 30 min and the RNCs isolated from a 30% (w/v) sucrose cushions centrifuged at 100,000 rpm for 1 h.
PEGylation gel shift assay of RNCs
Pelleted in vitro derived RNCs, corresponding to approximately 6 pmol of 70S ribosomes were resuspended in buffer A (20 mM Hepes (pH 7.2), 100 mM NaCl, 5 mM MgCl2). Samples were divided, and in which the PEGylation reaction set were incubated in buffer A containing 1 mM methoxypolyethylene glycol maleimide (5 kDa). Samples were then incubated at 25°C for 1 h. Following PEGylation reactions, the samples were run using PAGE conditions as described above for RNC integrity determination. The gel was exposed to film and the extent of PEGylation in each RNC was evaluated by densitometry using ImageJ software, in which the intensity of the PEGylated, tRNA-bound form was evaluated relative to the unPEGylated tRNA-bound form within the same sample. The PEGylation data reported are the average of at least six independent experiments.
NMR spectroscopy of RNCs
Prior to NMR spectroscopy, each sample was buffer-exchanged into Tico buffer27 at pH 7.5 (containing d8-HEPES for 13CH3-labelled RNCs), supplemented with 1 mM EDTA and protease inhibitors. The samples also contained 7% (v/v) D2O (U-15N samples) or 100% D2O ([U-2H; Ileδ1-13CH3] samples) as a lock signal and 0.001% (w/v) DSS as an internal reference. Sample concentrations were based upon the nascent chain content and ranged from 2 to 12 µM. NMR data were acquired on a 700 MHz Bruker Avance III spectrometer (University College London) equipped with a TXI cryoprobe, and in specific cases using 800 and 950 MHz Bruker Avance III HD spectrometers (NMR Centre, Crick Institute) and all spectra were recorded at 298 K unless otherwise stated and using an interleaved manner27. For samples of U-15N-labelled RNCs, 1H-15N SOFAST-HMQC spectra at 700 MHz were recorded with 1024 points in the direct (1H) dimension (Taq=46 ms) and 64 points (128 points for poly (GS) linker RNCs) in the indirect (15N) dimension (Taq=14.1 ms) and using a recycling delay of 50 ms. 1H-13C HMQC spectra of [U-2H; Ileδ1-13CH3]-labeled RNCs at 700 MHz were recorded with 3072 points in the direct (1H) dimension (Taq=137.6 ms) and 128 points in the indirect (13C) dimension (Taq=12.1 ms). For all RNCs recorded at 700 MHz, either 15N XSTE45 or 1H STE-1H,13C-HMQC46 diffusion measurements were acquired using a diffusion delay of 100 ms and bipolar trapezoidal gradient pulses (total length 4 ms, shape factor 0.9) with strengths of 0.028 and 0.529 T m-1. Spectra recorded at 800 and 950 MHz were recorded with a non-uniform weighted sampling scheme, a 50 ms acquisition time in the direct (1H) dimension (spectral width 16 ppm), 160 points in the indirect (15N) dimension (spectral width 22 ppm), and a recycling time of 50 ms. The indirect dimension was acquired using a cosine non-uniform weighted scheme, providing an 11% increase in intensity47. These data were interleaved with SORDID diffusion measurements48 using a diffusion delay of 190 ms and trapezoidal gradient pulses (total length 4 ms, shape factor 0.9) with strengths of 0.058 and 0.387 T m-1. All data were processed and analyzed using NMRPipe49 and Sparky50.
RNC labeling efficiency and selectivity as assessed by 15N filtered/edited difference spectroscopy
Isotopic labeling of the 70S ribosome particle was monitored in U-15N-labelled RNCs: A 15N-edited 1D experiment was recorded using modified 15N-SOFAST-HMQC sequences with 500 ms pre-saturation of water for suppression of the disordered nascent chain resonances that exchange rapidly with the solvent. The observed signals therefore arise predominantly from non-labile amides of folded domain of ribosomal protein L7/L1251. A 15N-filtered experiment was run identically, except with the phase-cycle of the receiver inverted to reject 15N-labelled magnetization (14N-bound 1H). The intensity of the 1H envelope of 70S ribosomal resonances bound to 14N (15N-filtered 1D) was matched by scaling to that of 15N-bound 1H (15N-edited 1D) in order to quantify the ratio of unlabeled to labeled ribosomal protons. From these measurements, the extent of background labeling arising from the ribosomal proteins was determined to range between 1 and 15% across all samples (ca. 50 samples) of 15N-labelled RNCs. An analogous approach was applied to purified, released nascent chains and the extent of nascent chain labeling was determined to be > 90%.
Co-translational folding as monitored by 1H-15N correlation spectra
Three well-resolved resonances with signal-to-noise ratio of 12 ± 2 (corresponding to residues Ala683, Ala694, Val682) within 1H-15N correlation spectra of RNCs L = 21 to 42 were used for lineshape analysis. Spectra were processed with exponential window functions and 1D cross-sections were fitted to Lorentzian lineshapes. The averaged calculated linewidths of the resonances of these residues were 12 ± 1 Hz for FLN5∆16 and 20 ± 3 Hz for FLN5 RNCs (error taken as the standard deviation). The similar linewidths measured for Ala683, Ala694 and Val682 in disordered FLN5 RNCs (L = 21 to 42) indicated that these resonances have uniform relaxation properties and could therefore be used to monitor the populations of the disordered state. Peak intensities of FLN5+L RNCs (L = 21, 31, 37, 42, 43, 44, 45, 47, 67 and 110) were determined using Sparky50, scaled for the number of scans and relative nascent chain concentrations, and averaged across the 3 peaks. Peak height errors were calculated as the standard deviation of 100 points randomly picked in the baseline of these RNC spectra. Nascent chain concentrations were determined using anti-His western blot analysis of the RNC (taken at t = 0 h), as described in RNC integrity and the determination of nascent chain occupancy. At least two independent experiments were performed for each RNC sample, and the error was determined from the standard deviation of these experiments.
Assignment of FLN5 Y719E and disordered FLN6
FLN5 Y719E amide chemical shifts were assigned on the basis of an assigned FLN5∆12 spectrum and using 15N-NOESY-HSQC (200 ms mixing time) and 15N-TOCSY-HSQC (70 ms mixing time) experiments recorded at 277 K. Unfolded FLN6 1HN and 15N chemical shifts were assigned (except for residues 810-832), using a FLN5-6∆18 construct28, which gives rise to resonances that closely overlay with those of natively folded FLN5 and with additional resonances of characteristic disordered chemical shifts of unfolded FLN6. FLN5-6∆18 amide chemical shifts were assigned at 283 K using uniformly 15N,13C-labelled samples via standard triple resonance experiments (HNCO, HN(CA)CO, HNCACB and HN(CO)CACB).
Estimation of the effective ribosome concentration experienced by a nascent chain
The effective local concentration of a binding site near the exit tunnel on the ribosome surface as experienced by a residue in a nascent chain, can be estimated using previously described methods52. Treating the unfolded polypeptide outside the exit tunnel using a random flight model, the mean distance from a residue at the end of the exit tunnel (taken here to be 31 residues from the PTC based on PEGylation measurements, Fig. 4a,b) to a point N residues along the chain (i.e. N+31 residues from the PTC) is approximately <r2>=CNl2, where the Cα-Cα distance l=3.8 Å and the characteristic ratio C=9 accounts for the stiffness of a typical polypeptide chain52. By modeling the ribosome surface as an infinite plane, the effective local concentration of a binding site situated close to the exit tunnel can be determined to be cL=2(3/2π<r2>)3/2/1000NA (in mol L-1), where NA is Avogadro's number52. For residues 10 to 20 amino acids beyond the exit tunnel (i.e. linker lengths L = 41-51), this corresponds to effective concentrations of between 8 and 23 mM.
Structure calculations using chemical shift restrained molecular dynamics simulations
Structural ensemble calculations of the FLN5+110 RNC were performed using the replica-averaged metadynamics (RAM) method described32. In these calculations, chemical shifts are used as replica-averaged structural restraints in molecular dynamics simulations using GROMACS53 together with PLUMED254. We used the CHARMM22* force field55 with TIP3P water molecules56. A time step of 2 fs was used together with LINCS constraints57. The van der Waals and Coulomb interactions were cut-off at 0.9 nm, while long-range electrostatic effects were treated with the particle mesh Ewald method. All simulations were carried out in the canonical ensemble by keeping the volume fixed and by thermosetting the system at 300 K with the Bussi-Donadio-Parrinello thermostat58.
Supplementary Material
Acknowledgements
We thank John Kirkpatrick for his NMR technical assistance and useful discussions and Bernd Bukau (Ruprecht-Karls-Universität Heidelberg) for the kind gift of the anti-SecM antibody. JC acknowledges the use of the Biomolecular NMR Facility, University College London, and thanks the MRC Biomedical NMR Centre at the Crick Institute, London. JC and TW acknowledge the use of the ARCHER UK National supercomputing service (http://www.archer.ac.uk) A.L.R is a National Health and Medical Research Council (Australia) C.J Martin Fellow. The work of CMD and MV is supported by a Wellcome Trust Programme Grant (094425/Z/10/Z to C.M.D and M.V). This work was supported by a Biotechnology and Biochemical Sciences Research Council New Investigators Award (BBG0156511 to J.C.) and Wellcome Trust Investigator Award (097806/Z/11/Z to J.C.).
Footnotes
Accession numbers
Co-ordinates for the structural ensemble have been deposited in the RCSB Protein Data Bank (PDB) with accession ID 2N62 and NMR chemical shift restraints have been deposited in the Biological Magnetic Resonance Data Bank (BMRB) with accession ID 25748.
Author contributions
L.D.C., C.M.D., and J.C. designed the research. J.C. supervised the overall project. L.D.C., A.C., H.L., C.A.W., M.E.K., A.S.W. and X.W. performed the research. A.L.R., and L.D.C. performed the PEGylation experiments and C.A.W. (Glasgow) contributed valuable technical experience in in vitro translation experiments. C.C., T.W., L.G. and M.V. performed the NMR-restrained molecular dynamics simulations. L.D.C., A.C., A.L.R., C.A.W., and J.C. analyzed the data. L.D.C., A.C., C.A.W., M.V., C.M.D. and J.C. wrote the paper. All authors discussed the results and contributed to the final version of the manuscript.
Author Information
The authors declare no competing financial interests.
References
- 1.Dobson CM. Protein folding and misfolding. Nature. 2003;426:884–90. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 2.Onuchic JN, Luthey-Schulten Z, Wolynes PG. Theory of protein folding: the energy landscape perspective. Annu Rev Phys Chem. 1997;48:545–600. doi: 10.1146/annurev.physchem.48.1.545. [DOI] [PubMed] [Google Scholar]
- 3.Dobson CM, Sali A, Karplus M. Protein Folding: A perspective from theory and experiment. Angewandte Chemie-International Edition. 1998;37:868–93. doi: 10.1002/(SICI)1521-3773(19980420)37:7<868::AID-ANIE868>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 4.Cabrita LD, Dobson CM, Christodoulou J. Protein folding on the ribosome. Curr Opin Struct Biol. 2010;20:33–45. doi: 10.1016/j.sbi.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 5.Nicola AV, Chen W, Helenius A. Co-translational folding of an alphavirus capsid protein in the cytosol of living cells. Nat Cell Biol. 1999;1:341–5. doi: 10.1038/14032. [DOI] [PubMed] [Google Scholar]
- 6.Zhang G, Hubalewska M, Ignatova Z. Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nat Struct Mol Biol. 2009;16:274–80. doi: 10.1038/nsmb.1554. [DOI] [PubMed] [Google Scholar]
- 7.Frydman J, Erdjument-Bromage H, Tempst P, Hartl FU. Co-translational domain folding as the structural basis for the rapid de novo folding of firefly luciferase. Nat Struct Mol Biol. 1999;6:697–705. doi: 10.1038/10754. [DOI] [PubMed] [Google Scholar]
- 8.O’Brien EP, Christodoulou J, Vendruscolo M, Dobson CM. New Scenarios of Protein Folding Can Occur on the Ribosome. J Am Chem Soc. 2011;133:513–26. doi: 10.1021/ja107863z. [DOI] [PubMed] [Google Scholar]
- 9.Kaiser CM, Goldman DH, Chodera JD, Tinoco I, Jr, Bustamante C. The ribosome modulates nascent protein folding. Science. 2011;334:1723–7. doi: 10.1126/science.1209740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim SJ, et al. Protein folding. Translational tuning optimizes nascent protein folding in cells. Science. 2015;348:444–8. doi: 10.1126/science.aaa3974. [DOI] [PubMed] [Google Scholar]
- 11.Schmeing TM, Ramakrishnan V. What recent ribosome structures have revealed about the mechanism of translation. Nature. 2009;461:1234–42. doi: 10.1038/nature08403. [DOI] [PubMed] [Google Scholar]
- 12.Siller E, DeZwaan DC, Anderson JF, Freeman BC, Barral JM. Slowing bacterial translation speed enhances eukaryotic protein folding efficiency. J Mol Biol. 2010;396:1310–8. doi: 10.1016/j.jmb.2009.12.042. [DOI] [PubMed] [Google Scholar]
- 13.Deuerling E, Schulze-Specking A, Tomoyasu T, Mogk A, Bukau B. Trigger factor and DnaK cooperate in folding of newly synthesized proteins. Nature. 1999;400:693–6. doi: 10.1038/23301. [DOI] [PubMed] [Google Scholar]
- 14.Schaffitzel C, et al. Structure of the E. coli signal recognition particle bound to a translating ribosome. Nature. 2006;444:503–6. doi: 10.1038/nature05182. [DOI] [PubMed] [Google Scholar]
- 15.Kramer G, Boehringer D, Ban N, Bukau B. The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nat Struct Mol Biol. 2009;16:589–97. doi: 10.1038/nsmb.1614. [DOI] [PubMed] [Google Scholar]
- 16.Rutkowska A, et al. Dynamics of trigger factor interaction with translating ribosomes. J Biol Chem. 2008;283:4124–32. doi: 10.1074/jbc.M708294200. [DOI] [PubMed] [Google Scholar]
- 17.Kaiser CM, et al. Real-time observation of trigger factor function on translating ribosomes. Nature. 2006;444:455–60. doi: 10.1038/nature05225. [DOI] [PubMed] [Google Scholar]
- 18.Ferbitz L, et al. Trigger factor in complex with the ribosome forms a molecular cradle for nascent proteins. Nature. 2004;431:590–6. doi: 10.1038/nature02899. [DOI] [PubMed] [Google Scholar]
- 19.Knight AM, et al. Electrostatic effect of the ribosomal surface on nascent polypeptide dynamics. ACS Chem Biol. 2013;8:1195–204. doi: 10.1021/cb400030n. [DOI] [PubMed] [Google Scholar]
- 20.Kelkar DA, Khushoo A, Yang Z, Skach WR. Kinetic analysis of ribosome-bound fluorescent proteins reveals an early, stable, cotranslational folding intermediate. J Biol Chem. 2012;287:2568–78. doi: 10.1074/jbc.M111.318766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clark PL, King J. A newly synthesized, ribosome-bound polypeptide chain adopts conformations dissimilar from early in vitro refolding intermediates. J Biol Chem. 2001;276:25411–20. doi: 10.1074/jbc.M008490200. [DOI] [PubMed] [Google Scholar]
- 22.Bhushan S, et al. SecM-stalled ribosomes adopt an altered geometry at the peptidyl transferase center. PLoS Biol. 2011;9:e1000581. doi: 10.1371/journal.pbio.1000581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhushan S, et al. alpha-Helical nascent polypeptide chains visualized within distinct regions of the ribosomal exit tunnel. Nat Struct Mol Biol. 2010;17:313–7. doi: 10.1038/nsmb.1756. [DOI] [PubMed] [Google Scholar]
- 24.Lu J, Deutsch C. Folding zones inside the ribosomal exit tunnel. Nat Struct Mol Biol. 2005;12:1123–9. doi: 10.1038/nsmb1021. [DOI] [PubMed] [Google Scholar]
- 25.Woolhead CA, Johnson AE, Bernstein HD. Translation arrest requires two-way communication between a nascent polypeptide and the ribosome. Mol Cell. 2006;22:587–98. doi: 10.1016/j.molcel.2006.05.021. [DOI] [PubMed] [Google Scholar]
- 26.Nilsson OB, et al. Cotranslational Protein Folding inside the Ribosome Exit Tunnel. Cell Rep. 2015;12:1533–40. doi: 10.1016/j.celrep.2015.07.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cabrita LD, Hsu ST, Launay H, Dobson CM, Christodoulou J. Probing ribosome-nascent chain complexes produced in vivo by NMR spectroscopy. Proc Natl Acad Sci U S A. 2009;106:22239–44. doi: 10.1073/pnas.0903750106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsu ST, et al. Structure and dynamics of a ribosome-bound nascent chain by NMR spectroscopy. Proc Natl Acad Sci U S A. 2007;104:16516–21. doi: 10.1073/pnas.0704664104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakatogawa H, Ito K. The ribosomal exit tunnel functions as a discriminating gate. Cell. 2002;108:629–36. doi: 10.1016/s0092-8674(02)00649-9. [DOI] [PubMed] [Google Scholar]
- 30.Rosenzweig R, Kay LE. Bringing dynamic molecular machines into focus by methyl-TROSY NMR. Annu Rev Biochem. 2014;83:291–315. doi: 10.1146/annurev-biochem-060713-035829. [DOI] [PubMed] [Google Scholar]
- 31.Schanda P, Kupce E, Brutscher B. SOFAST-HMQC experiments for recording two-dimensional heteronuclear correlation spectra of proteins within a few seconds. J Biomol NMR. 2005;33:199–211. doi: 10.1007/s10858-005-4425-x. [DOI] [PubMed] [Google Scholar]
- 32.Camilloni C, Cavalli A, Vendruscolo M. Replica-averaged metadynamics. J Chem Theory Comput. 2013;9:5610–5617. doi: 10.1021/ct4006272. [DOI] [PubMed] [Google Scholar]
- 33.Peterson JH, Woolhead CA, Bernstein HD. The conformation of a nascent polypeptide inside the ribosome tunnel affects protein targeting and protein folding. Mol Microbiol. 2010;78:203–17. doi: 10.1111/j.1365-2958.2010.07325.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frauenfeld J, et al. Cryo-EM structure of the ribosome-SecYE complex in the membrane environment. Nat Struct Mol Biol. 2011;18:614–21. doi: 10.1038/nsmb.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsu ST, Cabrita LD, Fucini P, Dobson CM, Christodoulou J. Structure, dynamics and folding of an immunoglobulin domain of the gelation factor (ABP-120) from Dictyostelium discoideum. J Mol Biol. 2009;388:865–79. doi: 10.1016/j.jmb.2009.02.063. [DOI] [PubMed] [Google Scholar]
- 36.Deuerling E, et al. Trigger Factor and DnaK possess overlapping substrate pools and binding specificities. Mol Microbiol. 2003;47:1317–28. doi: 10.1046/j.1365-2958.2003.03370.x. [DOI] [PubMed] [Google Scholar]
- 37.Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–30. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 38.O’Brien EP, Ciryam P, Vendruscolo M, Dobson CM. Understanding the influence of codon translation rates on cotranslational protein folding. Acc Chem Res. 2014;47:1536–44. doi: 10.1021/ar5000117. [DOI] [PubMed] [Google Scholar]
- 39.Clarke J, Cota E, Fowler SB, Hamill SJ. Folding studies of immunoglobulin-like beta-sandwich proteins suggest that they share a common folding pathway. Structure. 1999;7:1145–53. doi: 10.1016/s0969-2126(99)80181-6. [DOI] [PubMed] [Google Scholar]
- 40.Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Hartl FU. Molecular chaperone functions in protein folding and proteostasis. Annu Rev Biochem. 2013;82:323–55. doi: 10.1146/annurev-biochem-060208-092442. [DOI] [PubMed] [Google Scholar]
- 41.Hartl FU, Hayer-Hartl M. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science. 2002;295:1852–8. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- 42.Wright CF, Teichmann SA, Clarke J, Dobson CM. The importance of sequence diversity in the aggregation and evolution of proteins. Nature. 2005;438:878–81. doi: 10.1038/nature04195. [DOI] [PubMed] [Google Scholar]
- 43.Sivashanmugam A, et al. Practical protocols for production of very high yields of recombinant proteins using Escherichia coli. Protein Sci. 2009;18:936–48. doi: 10.1002/pro.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rutkowska A, et al. Large-scale purification of ribosome-nascent chain complexes for biochemical and structural studies. FEBS Lett. 2009;583:2407–13. doi: 10.1016/j.febslet.2009.06.041. [DOI] [PubMed] [Google Scholar]
- 45.Ferrage F, Zoonens M, Warschawski DE, Popot JL, Bodenhausen G. Slow diffusion of macromolecular assemblies by a new pulsed field gradient NMR method. J Am Chem Soc. 2003;125:2541–5. doi: 10.1021/ja0211407. [DOI] [PubMed] [Google Scholar]
- 46.Didenko T, Boelens R, Rudiger SG. 3D DOSY-TROSY to determine the translational diffusion coefficient of large protein complexes. Protein Eng Des Sel. 2011;24:99–103. doi: 10.1093/protein/gzq091. [DOI] [PubMed] [Google Scholar]
- 47.Waudby CA, Christodoulou J. An analysis of NMR sensitivity enhancements obtained using non-uniform weighted sampling, and the application to protein NMR. J Magn Reson. 2012;219:46–52. doi: 10.1016/j.jmr.2012.04.013. [DOI] [PubMed] [Google Scholar]
- 48.Augustyniak R, Ferrage F, Damblon C, Bodenhausen G, Pelupessy P. Efficient determination of diffusion coefficients by monitoring transport during recovery delays in NMR. Chem Commun (Camb) 2012;48:5307–9. doi: 10.1039/c2cc30578j. [DOI] [PubMed] [Google Scholar]
- 49.Delaglio F, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 50.Goddard TD, Kneller DG. Sparky 3. University of California; San Francisco: [Google Scholar]
- 51.Christodoulou J, et al. Heteronuclear NMR investigations of dynamic regions of intact Escherichia coli ribosomes. Proc Natl Acad Sci U S A. 2004;101:10949–10954. doi: 10.1073/pnas.0400928101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Timpe LC, Peller L. A random flight chain model for the tether of the Shaker K+ channel inactivation domain. Biophys J. 1995;69:2415–8. doi: 10.1016/S0006-3495(95)80111-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pronk S, et al. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tribello GA, Bonomi M, Branduardi D, Camilloni C, Bussi G. PLUMED 2: New feathers for an old bird. Comp Phys Comm. 2014;185:604–613. [Google Scholar]
- 55.Piana S, Lindorff-Larsen K, Shaw DE. How robust are protein folding simulations with respect to force field parameterization? Biophys J. 2011;100:L47–9. doi: 10.1016/j.bpj.2011.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jorgensen WL, Chandrasekhar J, Madura JD, Impey ML, Klein L. Comparison of simple potential functions for stimulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 57.Hess B, Bekker H, Berendsen HJC, Fraaije J. LINCS: A linear constraint solver for molecular simulations. Journal of Computational Chemistry. 1997;18:1463–1472. [Google Scholar]
- 58.Bussi G, Donadio D, Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126:014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.