Abstract
Background:
Lynch syndrome (LS) due to an inherited damaging mutation in mismatch repair (MMR) genes comprises 3% of all incident colorectal cancer (CRC). Molecular testing using immunohistochemistry (IHC) for MMR proteins is a recommended screening tool to identify LS in incident CRC. This study assessed outcomes of population-based routine molecular screening for diagnosis of LS in a regional center.
Methods:
We conducted a prospective, consecutive case series study of universal IHC testing on cases of resected CRC from September 2004–December 2013. Referred cases with abnormal IHC results that attended a familial cancer clinic were assessed according to modified Bethesda criteria (until 2009) or molecular criteria (from 2009).
Results:
1612 individuals underwent resection for CRC in the study period and had MMR testing by IHC. Of these, 274 cases (16.9%) exhibited loss of expression of MMR genes. The mean age at CRC diagnosis was 68.1 years (± standard deviation 12.7) and the mean age of those with an IHC abnormality was 71.6 (± 11.8). A total of 82 (29.9%) patients with an abnormal result were seen in a subspecialty familial cancer clinic. Patients aged under 50 (p = 0.009) and those with loss of MSH6 staining (p = 0.027) were more likely to be referred and to attend. After germ-line sequencing, 0.6% (10 of 82) were identified as having a clinically significant abnormality. A further eight probands with pathogenic germ-line mutations were identified from other referrals to the service over the same time period.
Conclusions:
While technically accurate, the yield of ‘universal’ IHC in detecting new Lynch probands is limited by real-world factors that reduce referrals and genetic testing. We propose an alternative approach for universal, incident case detection of Lynch syndrome with ‘one-stop’ MMR testing and sequencing.
Keywords: colorectal neoplasms, DNA mismatch repair, immunohistochemistry, Lynch syndrome
Introduction
Lynch syndrome (LS) is the most common inherited colorectal cancer (CRC) syndrome, accounting for 3% of colorectal cancers.1,2 Affected pedigrees bear a deleterious mutation in one allele of a DNA mismatch repair (MMR) gene – MLH1, MSH2, MSH6 or PMS2, or a mutation of EPCAM, which leads to MSH2 silencing.3,4 These defects predispose affected individuals to CRC as well as endometrial, pancreatic, genitourinary and other cancers.5,6 These cancers arise in adult life after acquired (somatic) inactivation of the wild type MMR allele and loss of MMR function. Loss of MMR can be detected within tumor tissue as microsatellite instability (MSI)7 or by loss of MMR protein expression.8
The high penetrance of CRC, endometrial and other cancers in those with Lynch syndrome has driven efforts to diagnose the disorder at the preclinical stage. Confirmation of LS by genetic testing leads to enhanced surveillance and cancer prevention9,10,11 reduces anxiety,12 the need for surveillance in family members not bearing the familial mutation, and is cost effective.13,14 In 1991, clinical criteria for the definition of LS were published15 and were revised in 1999.16 While highly specific, these criteria lack sensitivity, leading to underdiagnosis of LS.17,18 This experience has led to proposals for routine molecular screening for diagnosis of LS in incident CRC17,8 and incident adenoma,19 as well as hybrid schemes utilizing clinical and molecular criteria.20 The aim of these schemes is to limit the number of cases of cancer in the affected family to the incident case. As these schemata rely on clinicians to recognize and apply the criteria, conduct genetic counselling, order genetic tests where indicated, interpret and communicate the results, and perform follow up and surveillance in probands and affected family members,21,22,23 all such schemes will underdiagnose LS. Those that live in regions lacking formal cancer genetic services may be particularly vulnerable to underdiagnosis.
After Lynch and de la Chappelle8 and without a dedicated cancer genetic service, we commenced molecular screening for LS in incident CRC cases undergoing resection in the Australian Capital Territory (ACT). As refinements of molecular screening strategies were developed, these were incorporated into our clinical practice. We present the outcome of 8 years of molecular screening for LS, show that significant underdiagnosis occurs, and propose a change in practice to address this underdiagnosis.
Methods
Population: The Australian Capital Territory (ACT) has a population of 390,000 but the hospitals draw additional patients from the surrounding New South Wales region to service approximately 550,000 people.
All CRC cases were drawn from three hospitals with pathology services provided by one public provider (ACT Pathology) and one private provider (Capital Pathology). For retrospective analysis, CRC cases were retrieved from the pathology providers’ databases. We included all resected colorectal adenocarcinomas identified in the ACT from September 2004 until December 2012. We also retrieved index cases of LS diagnosed in the ACT from September 2004 until December 2012 from the ACT Genetic Service Database. We identified the reason for referral and means of identifying the patients as having LS. The study was approved by the ACT Health Human Research Ethics Committee (ETH.10/02.376).
Screening strategy: The study aim was that all resected CRC in the study period would be prospectively screened for LS using immunohistochemistry (IHC) as a routine diagnostic test. Study investigators wrote to all treating surgeons at the study commencement providing an ‘opt out’ from universal testing. MMR IHC staining results were reported to requesting doctors in the body of the histopathology report, together with an advisory for referral to the sub-speciality clinic of the gastroenterology unit. This advisory recommended referral for cases with abnormal MMR, in a patient aged less than 50, or a prior history of any LS cancer, first-degree relative with any LS cancer, or synchronous cancers. There was no involvement of the study team in the decision to refer to the service.
Immunohistochemistry: MMR IHC was performed on relevant formalin-fixed, paraffin-embedded (FFPE) tumour blocks and reported in line with accepted criteria by pathologists expert in interpretation of MMR staining.24 From September 2004, specimens were analyzed using anti-MLH1 and anti-MSH2. Staining for MSH6 expression was introduced in 2005 and for PMS2 in 2008 in the public sector, while MSH6 and PMS2 tests were introduced in 2009 in the private sector. Expanded testing was initiated collaboratively between the investigators and providers. We retrospectively tested any cases that were not tested initially for MMR abnormality using the four MMR protein stains.
Clinical Assessment: Clinic attendance at a subspecialty gastroenterology clinic or to a general genetics service was recorded until 31 December 2013, allowing at least 1 year from cancer diagnosis for attendance to occur. For each patient, we recorded the specialty of the referring clinician. Patients who attended the service were offered genetic counseling and given the option for further genetic testing, when appropriate. In referred patients with loss of MLH1 staining, somatic loss was further assessed by V600E BRAF and MLH1 methylation assays from 2009. Prior to 2009, a detailed family history was taken and patients assessed as high risk using the modified Bethesda criteria16 were offered germ-line testing. Patients with loss of expression of MSH2, MSH6 and PMS2 were given genetic counseling before genetic testing and offered a consultation with a cancer geneticist by videoconference. Testing was conducted by an accredited diagnostic molecular service (Hunter Genetics Lab, New England, NSW). MMR alterations were classified according to the International Society for Gastroin-testinal Hereditary Tumors (InSIGHT) Variant Interpretation Committee Classification.25,26
Statistical analysis: Demographic data was recorded and summarised as mean (± SD). The effect of individual characteristics on clinic attendance was estimated with clinic attendance as the outcome and the factors as predictors. Fisher’s exact test was used to test the effect of the predictors. Age was analyzed as a categorized variable, grouping individuals by age (<50, 50–70, and >70). All statistical analyses were performed using SPSS (version 20; SPSS Chicago, IL). Statistical significance was defined as p < 0.05.
Results
Patient characteristics: A total of 1612 individuals underwent resection for CRC in the study period and had MMR testing by IHC. Of these, 274 individuals (17.0%) had loss of expression of at least one of the four mismatch repair genes. The mean age at CRC diagnosis was 68.0 (range 22–95) and 49% of individuals were female (Table 1). Most patients were aged over 70 (175, 68.9%). The mean age of those with an IHC abnormality was 71.6 (± 11.8) and 58.4% were female (Table 2).
Table 1.
Characteristics of individuals described in the study.
| Total number of individuals with resected CRC | 1612 |
| Average age (years ± SD) | 68.1 ± 12.7 |
| Males | 824 (51%) |
| Females | 788 (49%) |
CRC, colorectal cancer; SD, standard deviation.
Table 2.
Characteristics of the 274 individuals with abnormal mismatch repair testing.
| Age ± SD | 71.6 ± 11.8* |
| Sex (M:F) | 114:160 (41.6 : 58.4%) |
| Proximal § | 223 (81.3%) |
| Mucinous | 51 (18.6%) |
| Tumor stage | |
| I–III | 213 |
| IV | 61 |
| IHC staining pattern | |
| Absence MLH1 ± PMS2 | 211 |
| Absence MSH2 ± MSH6 | 18 |
| Absence PMS2 | 13 |
| Absence MSH6 | 19 |
| Absence all | 5 |
| Mispaired loss# | 8 |
Significantly different to group without abnormal MMR testing, p = 0.007.
Proximal CRC were defined as those up to but not including the splenic flexure.
Mispaired loss included MLH1/MSH2, MLH1/MSH2/MSH6, MLH1/MSH6/PMS2; all were BRAF V600E + or exhibited MLH1 hypermethylation.
SD, standard deviation; IHC, immunohistochemistry; CRC, colorectal cancer; MMR, mismatch repair.
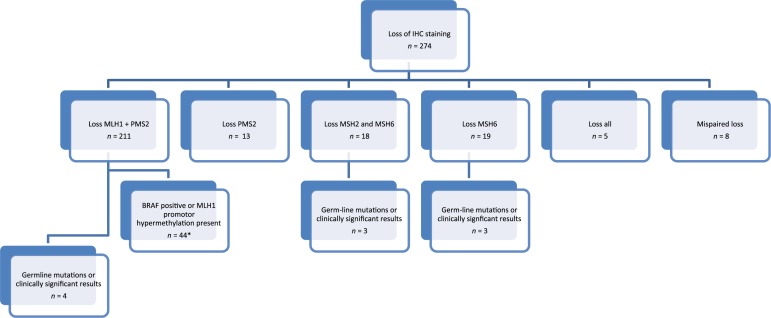
Result of immunohistochemistry: Of individuals with abnormal staining, the majority as expected (211) had loss of expression of MLH1 with or without PMS2 (77.0%). There were 18 (6.6%) with loss of MSH2 with or without MSH6 loss, 19 (6.9%) with MSH6 only loss, 13 (4.7%) with PMS2 loss, 5 (1.8%) with loss of all 4 MMR and 8 with atypical staining patterns or mispaired loss (e.g. MSH2/PMS2, MLH1/MSH6: Table 2). Of cancers with atypical staining patterns or mispaired loss, all were found to be either MLH1 negative or BRAF positive. 145 individuals (9%) did not have IHC performed at the time of resection. Of these, 18 (1.1% of CRC resections for the period) had abnormal staining on retrospective analysis and all of those were found to have loss of MLH1 staining.
Pathological findings: Of those with an abnormality on IHC, 24.3% were mucinous adenocarcinomas and 81% of the tumors were proximal (Table 2). Of tumors with MMR abnormality, 78% (213) were stage I–III while 61 (22%) were stage IV. Seven individuals had synchronous tumors at the time of resection.
Results of Screening Strategy: Figure 1 summarises the results of the screening strategy. A total of 145 (9.0%) did not have IHC staining done at the time of resection due to oversight or the preference of the reporting pathologist. A total of 82 (29.9%) patients with an abnormal result were seen in a familial cancer clinic; 77 in a subspecialty gastroenterology clinic and 5 in a collaborating general genetics service (Table 3). Factors predicting referral and attendance were age under 50 (p = 0.009) and individuals with tumors having a significant mucinous component (p = 0.012). Of all IHC abnormalities, individuals with loss of MSH6 staining were more likely to attend (p = 0.027). Rates of clinic attendance were not associated with gender, tumor site or place of surgery (public versus private hospital). Patients were most likely to be referred to the clinic by treating surgeons and oncologists (Table 4).
Figure 1.
Flow diagram of the mismatch repair screening results in the study, and results of further testing.
IHC, immunohistochemistry.
Table 3.
Factors associated with attendance at a familial cancer clinic.
| Attendance at familial cancer clinic |
p value (Fishers exact) | ||
|---|---|---|---|
| YES | NO | ||
| Sex (M:F) | 32.5: 28.1% | 67.5: 71.9% | 0.504 |
| Tumour site (proximal: distal) | 30.6: 27.3% | 69.4: 72.7% | 0.742 |
| Site of resection (public: private hospital) | 29.9: 30.0% | 70.1: 70.0% | 0.55 |
| Mucinous histology | 45.3: 26.2% | 54.7: 73.8% | 0.012 |
| Age < 50 | 62% | 39% | |
| Age ⩾ 50 < 70 | 36% | 64% | 0.009 |
| Age ⩾ 70 | 25% | 75% | |
| MMR staining pattern | |||
| Absence MLH1 ± PMS2 | 28.4% | 71.6% | 0.202 |
| Absence MSH2 ± MSH6 | 44.4% | 55.6% | 0.186 |
| Absence PMS2 | 15.4% | 84.6% | 0.356 |
| Absence MSH6 | 52.6% | 47.4% | 0.027 |
| Mispaired loss | 40.0% | 60.0% | 0.637 |
Table 4.
Source of referrals to familial cancer clinic for the 82 individuals seen with an immunohistochemistry abnormality.
| Specialist group | Percentage (%) |
|---|---|
| Surgeon | 52.4 |
| Medical Oncologist | 23.2 |
| Gastroenterologist | 15.9 |
| General practitioner | 7.3 |
| General physician | 1.2 |
Outcomes of germ-line testing: A total of 10 individuals of the 82 with abnormal MMR IHC and who underwent MMR germ-line testing were considered likely to have LS by combined molecular and clinical criteria (Table 5). The mean age of these 10 cases was 52.1 years (range 19–80). Only 7 had any germ-line alteration identified; of these, 3 had a pathogenic (Class 5) mutation using InSIGHT criteria allowing for predictive testing in the proband’s family. One had a variant of uncertain significance, one had compound MSH6 variants (Class 3/Class 1), two had variants that are novel at the time of submission and one had a benign (intronic, single nucleotide missense) variant. A further three cases did not have a germ-line mutation confirmed. One was not tested due to the restricted pedigree and patient preference, and two cases had no mutation identified after sequencing but were considered as likely LS because of compelling clinical features.
Table 5.
Characteristics of individuals considered Lynch syndrome on combined clinical and molecular criteria.
| Patient no. | Sex | Age at first appointment | CRC site | IHC Result | Gene | Exon | Mutation | Variant ID | Class |
|---|---|---|---|---|---|---|---|---|---|
| 1 | F | 19 | Synchronous ascending colon | MLH1 negative | MLH1 | 17 | 1975_1976 delCG | MLH1_00694 | 5 |
| 2 | M | 54 | Ascending colon | MSH2/MSH6 negative | MSH6 | No mutation identified* | |||
| 3 | M | 80 | Rectal | MSH6 negative | MSH6 | 7 | c.3583A > G | Novel | N/A |
| 4 | F | 59 | Transverse colon | MSH2/MSH6 negative | MSH2 | 7 | c.1216C > T | MSH2_00312 | 5 |
| 5 | M | 33 | Splenic flexure | MLH1/PMS2 negative | MLH1 | No mutation identified§ | |||
| 6 | M | 56 | Rectal | MSH6 negative | MSH6 | 4 | 1186 C > G | MSH6_00118 | 1 |
| 8 | 3694_369delGTT | MSH6_00721 | 3 | ||||||
| 7 | M | 51 | Rectal | MHS6 negative | MSH6 | 5 | c.3261dup | MSH6_00201 | 5 |
| 8 | M | 33 | Caecum | MSH2 negative | MSH2 | 6 | c.1042C > T | Novel | N/A |
| 9 | M | 67 | Sigmoid | MLH1/PMS2 negative | MLH1 | 12 | c.1039-8T > A | Benign | |
| 10 | M | 69 | Transverse colon | MLH1 and PMS2 negative | MLH1 | Not tested# |
No mutation in MSH2 and MSH6 or EPCAM on sequencing or MSH2 rearrangement by MLPA. Family history of CRC and uterine cancer.
MLH1 – BRAF negative, MLH1 methylation neg, no mutation on MLH1 sequencing.
Not tested due to restricted pedigree and patient preference.
CRC, colorectal cancer, IHC, immunohistochemistry.
Alternate Index case identification: Eight probands with pathogenic (Class 5) germ line mutations were identified after referral to the familial cancer service (n = 4) or a general genetics service (n = 4, Table 6). These patients were referred for previous history of other cancer, family history of CRC, or a resected colorectal cancer that fell outside the study period. MMR IHC was used retrospectively in these eight patients and led to identification of the pathogenic mutation (Table 6).
Table 6.
Characteristics of the eight individuals diagnosed with Lynch syndrome but not presenting with colorectal cancer in the study period.
| Patient no. | Sex | Age at first appointment | Reason for assessment | Use of IHC | Gene | Exon | Mutation | Variant ID | Class |
|---|---|---|---|---|---|---|---|---|---|
| 1 | M | 21 | Family history | Loss of PMS2 restrospectively analyzed cancer tissue from a relative | PMS2 | 1 | C1A>g | PMS2_00130 | 5 |
| 3 | M | 53 | Age at CRC diagnosis, family history | IHC on CRC specimen diagnosed before universal screening | MSH2 | 8 | del1277-1386 | MSH2_01582 | 5 |
| 4 | F | 37 | Age at CRC diagnosis | IHC on endometrial cancer specimen | MSH2 | 1 – 16 | Gene deletion | MSH2_00043 | 5 |
| 5 | M | 59 | Multiple cancers | IHC on sebaceous adenoma and renal tract tumour specimen | MSH2 | 2, 6 | Exon 2 – 6 inversion | MSH2_01579 | 5 |
| 6 | F | 55 | Multiple cancers | IHC on small intestinal cancer | MSH2 | 14 | c2228c>G | MSH2_00646 | 5 |
| 7 | F | 38 | Family history | IHC on adenomatous polyp | MSH2 | 3 | c484G>A | MSH2_00152 | 5 |
| 8 | F | 66 | Multiple cancers | IHC on ovarian cancer specimen | MSH2 | 4 – 16 | DEL 646-2674 | MSH2_01255 | 5 |
| 9 | F | 57 | Age at CRC diagnosis, family history | IHC on CRC specimen diagnosed before universal screening | MLH1 | 16 | In-frame deletion 618 | MLH1_00652 | 5 |
IHC, immunohistochemistry;CRC, colorectal cancer.
Discussion
The yield of universal IHC in detecting new Lynch probands may be limited by factors that reduce referral for genetic assessment and testing.27,28 We detected an abnormal IHC result in 16.9% a population-based sample of incident CRC of all ages undergoing resection during the study period. This result is similar to other screening studies with differing methodologies and populations.29,3,30,31,24,6 Only 10/274, or 0.6% of our study population, were considered likely to have a germ-line MMR mutation, a figure similar to that from another Australian genetic service,28 although a germ-line alteration was only detected in 7 of these 10 cases. Other studies with universal MMR analysis have detected LS in between 0.7% and 3.1% of individuals with newly diagnosed CRC and at a higher rate, 4.5%, when only those under 70 are included for routine testing.6,29,24,32
Our study confirms that implementation of universal IHC/MSI screening does not on its own lead to diagnosis of the majority of LS, which accounts for 3% of all CRC.3,6 We identified the steps at which cases escaped detection. We identified 145 cases where the intended IHC screening was not performed at the time of resection, due to an oversight of the reporting pathologist. All of these cases were in the public sector, and on retrospective analysis, 18 patients cases exhibited loss of MLH1 expression. The majority of these cases were considered by the investigators to have somatic loss of MLH1; however, it is possible that some of these cases are LS.
Of patients who had contemporaneous MMR testing and returned an abnormal result, 30% attended further assessment at either specialty service. Referral to appropriate genetics review is highly variable, both locally and internationally,33,27,28,34 and is the subject of a current research intervention.35 In our study, one factor may have been the advisory attached to the MMR result, which may have created a referral bias; patients attending the services were more likely to be under the age of 50. Prior to 2009, we also used the modified Bethesda criteria16 in ordering mutation testing after abnormal MMR immunohistochemistry. In addition, some patients may be reluctant to undergo further assessment. A review of screening programmes in the Lynch Syndrome Screening Network found that the rate that patients proceed to further testing following a positive screen ranges from 10–85%.22 The yield of universal screening was enhanced when automatic testing of BRAF and MLH1 methylation was done in MLH1-negative cases, a measure we adopted in referred cases during the study and have now also automated.
The availability of expert cancer geneticists is a barrier to case detection. In Australia, expert cancer geneticists are located only in major population centers. While patients living in remote and regional Australia may access services remotely, distance, and the lack of genetic workforce, is a proven barrier to genetic testing.36 In response to this, we operate a subspecialty gastroenterology service, which the majority of new probands in this study attended. Patients were given informal counseling and those with abnormal sequencing were offered consultation with a cancer geneticist by videoconference. Our approach yielded a similar case detection rate for LS as a program from a major genetics service,28 noting, however, the same barriers to case detection. We conclude that in centers where cancer geneticists do not practice, that assessment for highly penetrant cancer syndromes can be performed adequately by nongeneticists.
We detected an abnormal IHC result in 16.9% of a population-based sample of incident CRC of all ages undergoing resection during the study period. This result is similar to other screening studies with differing methodologies and populations. The negative predictive value of MMR IHC has not been assessed in any population-based prospective series against a standard of germ-line testing, to the best of our knowledge, although smaller series indicate a relative lack of sensitivity for MSH6 in late-onset cancers (reviewed in Resnick37).
Our study evolved due to expansion of MMR tests available, and this may have limited the study by failing to detect LS in unscreened individuals. Offsetting this, an alternate referral pathway based on clinical criteria was available, in line with previous practice.
The detection rate of LS from MMR screening is unsatisfactory, considering the potential cancer burden that flows from failure to detect LS. This detection rate could greatly increase if the analysis were rapid, decisive and required less of the patients and their referring doctors. A ‘one-stop’ approach of initial MMR screening by immunohistochemistry, complemented by sequencing of blood for MMR genes at diagnosis is feasible and affordable.38 The use of parallel sequencing of cancer susceptibility genes through multigene panels has become widespread. Its value is great if there are clear indications, a focused genomic panel and appropriate follow up.39 One challenge these panels pose to clinicians is the interpretation of unexpected findings in low-penetrance genes40 but this issue does not arise in MMR mutations, which are both penetrant and well curated. The ability to gain truly informed consent for these tests is acknowledged.41 The broad ethical principles governing point-of-care sequencing in cancers have been developed in this country and elsewhere.42,43 We propose that such an approach be adopted in a pilot, clinical setting.
Acknowledgments
The authors thank Ms Linda Warwick, ACT Genetic Service, Dr Kathy Tucker, Prince of Wales Hospital Hereditary Cancer Clinic, and Dr James Riddell, Gastroenterology and Hepatology Unit, The Canberra Hospital, for their contributions to results in this study.
Footnotes
Funding: This work was supported by a grant from the Canberra Hospital Private Practice Fund (to DT).
Conflict of interest statement: The authors declare that there is no conflict of interest.
Contributor Information
Beatrice Brennan, Gastroenterology and Hepatology Unit, The Canberra Hospital, Garran, ACT, Australia.
Christine T. Hemmings, ACT Pathology, The Canberra Hospital, Garran, ACT, Australia Current address: Anatomic Pathology (WA) & Head of Molecular Oncology, St John of God Pathology, Subiaco, WA, Australia.
Ian Clark, Capital Pathology, Deakin, ACT, Australia; Current address: Australian Pathology at Sonic Healthcare, Macquarie Park, New South Wales, Australia.
Desmond Yip, Department of Medical Oncology, Canberra and Calvary Hospitals, Garran, Australian Capital Territory, Australia.
Mitali Fadia, ACT Pathology, The Canberra Hospital, Garran, ACT, Australia.
Douglas R. Taupin, Cancer Research, B10 L5, The Canberra Hospital, Yamba Drive, Garran, ACT 2605, Australia.
References
- 1. Lynch HT, Lynch PM, Lanspa SJ, et al. Review of the Lynch syndrome: history, molecular genetics, screening, differential diagnosis, and medicolegal ramifications. Clin Genet 2009; 76: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vasen HF, Blanco I, Aktan-Collan K, et al. Revised guidelines for the clinical management of Lynch syndrome (HNPCC): recommendations by a group of European experts. Gut 2013; 62: 812–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hampel H, Frankel WL, Martin E, et al. Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer). N Engl J Med 2005; 352: 1851–1860. [DOI] [PubMed] [Google Scholar]
- 4. Lung MS, Trainer AH, Campbell I, et al. Familial colorectal cancer. Intern Med J 2015; 45: 482–491. [DOI] [PubMed] [Google Scholar]
- 5. Bonadona V, Bonaïti B, Olschwang S, et al. Cancer risks associated with germline mutations in MLH1, MSH2, and MSH6 genes in Lynch syndrome. JAMA 2011; 305: 2304–2310. [DOI] [PubMed] [Google Scholar]
- 6. Moreira L, Balaguer F, Lindor N, et al. EPICOLON Consortium. Identification of Lynch syndrome among patients with colorectal cancer. JAMA 2012; 308: 1555–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Peltomäki P, Lothe RA, Aaltonen LA, et al. Microsatellite instability is associated with tumors that characterize the hereditary non-polyposis colorectal carcinoma syndrome. Cancer Res 1993; 53: 5853–5855. [PubMed] [Google Scholar]
- 8. Lynch and de la, Chapelle A. Hereditary colorectal cancer. N Engl J Med 2003; 348: 919–932. [DOI] [PubMed] [Google Scholar]
- 9. Järvinen HJ, Mecklin JP, Sistonen P. Screening reduces colorectal cancer rate in families with hereditary nonpolyposis colorectal cancer. Gastroenterology 1995; 108: 1405–1411. [DOI] [PubMed] [Google Scholar]
- 10. Järvinen HJ, Renkonen-Sinisalo L, Aktán-Collán K, et al. Ten years after mutation testing for Lynch syndrome: cancer incidence and outcome in mutation-positive and mutation-negative family members. J Clin Oncol 2009; 27: 4793–4797. [DOI] [PubMed] [Google Scholar]
- 11. Palomaki GE, McClain MR, Melillo S, et al. EGAPP supplementary evidence review: DNA testing strategies aimed at reducing morbidity and mortality from Lynch syndrome. Genet Med 2009; 11: 42–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hunter JE, Zepp JM, Gilmore MJ, et al. Universal tumor screening for Lynch syndrome: assessment of the perspectives of patients with colorectal cancer regarding benefits and barriers. Cancer 2015; 121: 3281–3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Severin F, Stollenwerk B, Holinski-Feder E, et al. Economic evaluation of genetic screening for Lynch syndrome in Germany. Genet Med 2015; 17: 765–773. [DOI] [PubMed] [Google Scholar]
- 14. Snowsill T, Huxley N, Hoyle M, et al. A model-based assessment of the cost-utility of strategies to identify Lynch syndrome in early-onset colorectal cancer patients. BMC Cancer 2015; 15: 313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vasen HF, Mecklin JP, Khan PM, et al. The International Collaborative Group on Hereditary Non-Polyposis Colorectal Cancer (ICG-HNPCC). Dis Colon Rectum 1991; 34: 424–425. [DOI] [PubMed] [Google Scholar]
- 16. Vasen HF, Watson P, Mecklin JP, et al. New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative group on HNPCC. Gastroenterology 1999; 116: 1453–1456. [DOI] [PubMed] [Google Scholar]
- 17. Salovaara R, Loukola A, Kristo P, et al. Population-based molecular detection of hereditary nonpolyposis colorectal cancer. J Clin Oncol 2000; 18: 2193–2200. [DOI] [PubMed] [Google Scholar]
- 18. Balmaña J, Balaguer F, Castellví-Bel S, et al. Gastrointestinal Oncology Group of the Spanish Gastroenterological Association. Comparison of predictive models, clinical criteria and molecular tumour screening for the identification of patients with Lynch syndrome in a population-based cohort of colorectal cancer patients. J Med Genet 2008; 45: 557–563. [DOI] [PubMed] [Google Scholar]
- 19. Loukola A, de la Chapelle A, Aaltonen LA. Strategies for screening for hereditary non-polyposis colorectal cancer. J Med Genet 1999; 36: 819–822. [PMC free article] [PubMed] [Google Scholar]
- 20. Umar A. Lynch syndrome (HNPCC) and microsatellite instability. Dis Markers 2004; 20: 179–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ahnen DJ, Wade SW, Jones WF, et al. The increasing incidence of young-onset colorectal cancer: a call to action. Mayo Clin Proc 2014; 89: 216–224. [DOI] [PubMed] [Google Scholar]
- 22. Cragun D, DeBate RD, Vadaparampil ST, et al. Comparing universal Lynch syndrome tumor-screening programs to evaluate associations between implementation strategies and patient follow-through. Genet Med 2014; 16: 773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mange S, Bellcross C, Cragun D, et al. Creation of a network to promote universal screening for Lynch syndrome: the LynchSyndrome Screening Network. J Genet Couns 2015; 24: 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. van Lier MG, Leenen CH, Wagner A, et al. Yield of routine molecular analyses in colorectal cancer patients ⩽70 years to detect underlying Lynch syndrome. J Pathol 2012; 226: 764–774. [DOI] [PubMed] [Google Scholar]
- 25. Plazzer JP, Sijmons RH, Woods MO, et al. The InSiGHT database: utilizing 100 years of insights into Lynch syndrome. Fam Cancer 2013; 12: 175–180. [DOI] [PubMed] [Google Scholar]
- 26. Thompson BA, Spurdle AB, Plazzer JP, et al. Application of a five-tiered scheme for standardized classification of 2,360 unique mismatch repair gene variants lodged on the InSiGHT locus-specific database. Nat Genet, 2014; 46: 107–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tan YY, McGaughran J, Ferguson K, et al. Improving identification of Lynch syndrome patients: a comparison of research data with clinical records. Int J Cancer 2013; 132: 2876–2883. [DOI] [PubMed] [Google Scholar]
- 28. Ward RL, Hicks S, Hawkins NJ. Population-based molecular screening for Lynch syndrome: implications for personalized medicine. J Clin Oncol 2013; 31: 2554–2562. [DOI] [PubMed] [Google Scholar]
- 29. Canard G, Lefevre JH, Colas C, et al. Screening for Lynch syndrome in colorectal cancer: are we doing enough? Ann Surg Oncol 2012; 19: 809–816. [DOI] [PubMed] [Google Scholar]
- 30. Hampel H, Frankel WL, Martin E, et al. Feasibility of screening for Lynch syndrome among patients with colorectal cancer. J Clin Oncol 2008; 26: 5783–5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Julié C, Trésallet C, Brouquet A, et al. Identification in daily practice of patients with Lynch syndrome (hereditary nonpolyposis colorectal cancer): revised Bethesda guidelines-based approach versus molecular screening. Am J Gastroenterol 2008; 103: 2825–2835. [DOI] [PubMed] [Google Scholar]
- 32. Pérez-Carbonell L, Ruiz-Ponte C, Guarinos C, et al. Comparison between universal molecular screening for Lynch syndrome and revised Bethesda guidelines in a large population-based cohort of patients with colorectal cancer. Gut 2012; 61: 865–872. [DOI] [PubMed] [Google Scholar]
- 33. Pujol P, Lyonnet DS, Frebourg T, et al. Lack of referral for genetic counseling and testing in BRCA1/2 and Lynch syndromes: a nationwide study based on 240,134 consultations and 134,652 genetic tests. Breast Cancer Res Treat 2013; 141: 135–144. [DOI] [PubMed] [Google Scholar]
- 34. Wong C, Gibbs P, Johns J, et al. Value of database linkage: are patients at risk of familial colorectal cancer being referred for genetic counselling and testing? Intern Med J 2008; 38: 328–333. [DOI] [PubMed] [Google Scholar]
- 35. Taylor N, Long JC, Debono D, et al. Achieving behaviour change for detection of Lynch syndrome using the Theoretical Domains Framework Implementation (TDFI) approach: a study protocol. BMC Health Serv Res 2016; 16: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Delikurt T, Williamson GR, Anastasiadou V, et al. A systematic review of factors that act as barriers to patient referral to genetic services. Eur J Hum Genet 2015; 23: 739–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Resnick KE, Hampel H, Fishel R, et al. Current and emerging trends in Lynch syndrome identification in women with endometrial cancer. Gynecol Oncol 2009; 114: 128–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. LaDuca H, Stuenkel AJ, Dolinsky JS, et al. Utilization of multigene panels in hereditary cancer predisposition testing: analysis of more than 2,000 patients. Genet Med 2014; 16: 830–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kohane IS, Masys D, Altman R. The incidentalome, a threat to genomic medicine. JAMA 2006; 296: 212–216. [DOI] [PubMed] [Google Scholar]
- 40. Slavin TP, Niell-Swiller M, Solomon I, et al. Clinical application of multigene panels: challenges of next-generation counseling and cancer risk management. Front Oncol 2015; 5: 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Winkler EC, Wiemann S. Findings made in gene panel to whole genome sequencing: data, knowledge, ethics – and consequences? Expert Rev Mol Diagn 2016; 16: 1259–1270. [DOI] [PubMed] [Google Scholar]
- 42. The NCCN Clinical Practice Guidelines in OncologyTM Genetic/Familial High-Risk Assessment: Colorectal National Comprehensive Cancer Network, http://www.nccn.org/ (2016, accessed 31 July 2016).
- 43. National Health and Medical Research Council. Principles for the translation of ‘omics’– based tests from discovery to health care. National Health and Medical Research Council, Canberra, Australia, 2015. [Google Scholar]