Abstract
Corynebacterium pseudotuberculosis is a Gram-positive, pleomorphic, facultative intracellular pathogen that causes Oedematous Skin Disease (OSD) in buffalo. To better understand the pathogenic mechanisms of OSD, we performed a comparative genomic analysis of 11 strains of C. pseudotuberculosis isolated from different buffalo found to be infected in Egypt during an outbreak that occurred in 2008. Sixteen previously described pathogenicity islands (PiCp) were present in all of the new buffalo strains, but one of them, PiCp12, had an insertion that contained both a corynephage and a diphtheria toxin gene, both of which may play a role in the adaptation of C. pseudotuberculosis to this new host. Synteny analysis showed variations in the site of insertion of the corynephage during the same outbreak. A gene functional comparison showed the presence of a nitrate reductase operon that included genes involved in molybdenum cofactor biosynthesis, which is necessary for a positive nitrate reductase phenotype and is a possible adaptation for intracellular survival. Genomes from the buffalo strains also had fusions in minor pilin genes in the spaA and spaD gene cluster (spaCX and spaYEF), which could suggest either an adaptation to this particular host, or mutation events in the immediate ancestor before this particular epidemic. A phylogenomic analysis confirmed a clear separation between the Ovis and Equi biovars, but also showed what appears to be a clustering by host species within the Equi strains.
Introduction
Corynebacterium, Mycobacterium, Nocardia, and Rhodococcus, collectively known as the CMNR group, are all members of the order Corynebacteriales. This group contains species of medical, veterinary and biotechnological importance with shared common characteristics that include a high GC content and a cell wall composed mainly of peptidoglycans, arabinogalactans and mycolic acids [1]. Corynebacterium species are Gram-positive, rod-shaped, non-motile and non-spore forming bacteria [2], and include both pathogens like C. diphtheria, C. jeikeium and C. pseudotuberculosis, and non-pathogenic strains like C. glutamicum [3]. Corynebacterium pseudotuberculosis are facultative intracellular pathogens that are divided into two distinct biovars, each of which can infect a range of mammalian hosts. Nitrate reductase activity seems to correlate with the type of host that they infect, and thus the biovar that each strain belongs to. Isolates belonging to the Ovis biovar infect sheep and goats, and they are nitrate reductase negative. Members of the Equi biovar infect larger hosts like the horses, cattle, and buffalo [4], and they are have positive nitrate reductase activity [5,6]. The separation into two biovars is also supported by molecular markers [6,7] and by distinct differences in gene content that has observed across genomes [8].
C. pseudotuberculosis causes different diseases and symptoms in these various hosts. Infection in goats and sheep results in a manifestation called Caseous Lymphadenitis [9]. Infected cattle have ulcerative granulomatous lesions and mastitis [10,11], while horses are diagnosed with ulcerative lymphangitis, or with pigeon fever, so named due to a swelling in the chest of infected animals [12]. Oedematous Skin Disease (OSD) is the manifestation seen in buffalo. OSD is characterized by an extensive cutaneous, oedematous swelling in the dewlap, whole hind or forelimbs, and belly [13]. The disease causes economic loss due to reduced milk and meat production as well as reduced work efficiency of the animals. Moreover, the treatment is expensive and can last for months [13,14]. OSD is endemic in regions of lower Egypt and the areas surrounding Cairo, with sporadic cases throughout the year and periodic epidemics seen in the summer [13]. Unlike some of the other hosts that are infected by C. pseudotuberculosis, there is no documented transmission directly between infected buffalo. Instead, it has been suggested that insects are involved with transmitting the disease between these animals. Specifically, biting flies of the genus Hippobosca have been associated with transmission, the suggestion being that they inject the bacteria intradermally with each bite [15]. Further proof of involvement is the close correlation between the breeding season of these blood-sucking flies with outbreaks of OSD in buffalo [13].
All strains that have been isolated in Egypt from infected buffalo belonged to the Equi biovar, and all those sequenced genomes contain a gene that produces the diphtheria toxin [16]. This toxin is found in other Corynebacterium species, but within C. pseudotuberculosis it has only been found in isolates from buffalo to date. It is unclear if the presence of this gene indicates an adaptation that allows it to specifically infect buffalo, or if it is present due to a horizontal transfer event that occurred recently and spread across the specific geographic region where the animals were found [8].
To fully characterize these isolates and determine additional genomic features that not only distinguish them, but might explain the pathogenic mechanisms of OSD, a comparative analysis of 11 strains of C. pseudotuberculosis isolated from different buffalo in Egypt was conducted. These genomes were compared to other C. pseudotuberculosis strains from both Ovis and Equi biovars. Here we describe those differences, explore the species-wide pangenome, and define the differences related to not only the biovars, but also to the hosts they have been isolated from.
Materials and methods
Strains isolation
Each of the 11 C. pseudotuberculosis strains were isolated from individual water buffalo (Bubalus bubalis) in Egypt during an outbreak of OSD that occurred during the summer of 2008 (Table 1). The infected animals were all located in separate farms either in the Menofia (located 89 km north of Cairo) or El-Fayoum (50 km west Cairo) regions. The samples were collected following permission from the owners of the individual animals. No additional permission was required as Egyptian buffalo are not protected, and are considered to be domesticated livestock. Samples were collected from internal lesions that were located within the brisket, fore or hind limbs of these animals. The samples were plated on selective media (Brain Heart Agar with Fosfomycin). A single colony from each of the samples was selected for species identification, and identification of C. pseudotuberculosis was determined by API Coryne (BioMerieux, France) [17] according to the manufacturer’s instructions. An aliquot of each sample was lyophilized and sent to the Laboratory of Cellular and Molecular Genetics in Brazil where they were grown again, and a second confirmation showed them to be C. pseudotuberculosis. Each strain was demonstrated to produce diphtheria toxin by both API Coryne System and multiplex PCR tests [18]. Colonies from these pure cultures were plated on agar, and one colony from each of these was randomly selected for sequencing.
Table 1. Strains of Corynebacterium pseudotuberculosis isolated from buffalo diagnosed with Oedematous Skin Disease in Egypt during the summer of 2008.
| Strain | Genome size (bp) | Mean coverage depth | De novo assembler | CDS | tRNA | rRNA | Accession number |
|---|---|---|---|---|---|---|---|
| 31 [19] | 2,402,956 | 550x | MIRA 4.0.2; SPADES 3.1.1 | 2248 | 47 | 12 | CP003421.3 |
| 32 | 2,403,533 | 56.61x | Newbler 2.9 | 2272 | 52 | 12 | CP015183 |
| 33 | 2,403,550 | 104.11x | Newbler 2.9 | 2281 | 52 | 12 | CP015184 |
| 34 | 2,403,454 | 58.94x | MIRA 3.9.18 | 2286 | 49 | 12 | CP015192 |
| 35 | 2,403,502 | 112.68x | Newbler 2.9 | 2276 | 52 | 12 | CP015185 |
| 36 | 2,403,412 | 172.02x | SPAdes 3.6.0 | 2279 | 49 | 12 | CP015186 |
| 38 | 2,403,515 | 160.27x | SPAdes 3.6.0 | 2281 | 49 | 12 | CP015187 |
| 39 | 2,403,579 | 241.79x | SPAdes 3.6.0 | 2281 | 49 | 12 | CP015188 |
| 43 | 2,365,075 | 218.07x | Newbler 2.9 | 2218 | 51 | 12 | CP015189 |
| 46 | 2,366,565 | 249.57x | SPAdes 3.6.0 | 2218 | 48 | 12 | CP015190 |
| 48 | 2,403,301 | 306.54x | SPAdes 3.6.0 | 2281 | 49 | 12 | CP015191 |
Genome sequencing and assembly
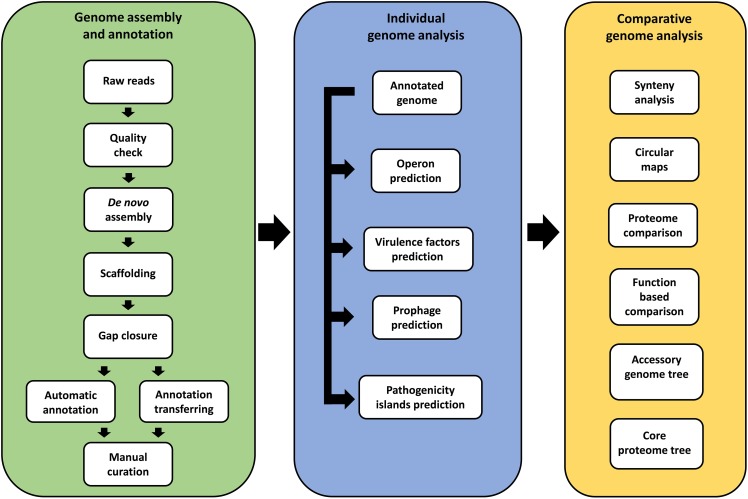
The in silico analysis workflow is presented in Fig 1. Sequencing, assembly and annotation of individual genomes was conducted at one of three laboratories. Two of these (Laboratory of Cellular and Molecular Genetics and the National Reference Laboratory for Aquatic Animal Diseases) are part of the Federal University of Minas Gerais, Belo Horizonte, Minas Gerais, Brazil. The third (Center of Genomic and System Biology) is located at the Federal University of Pará in Belém, Pará, Brazil. All genomes were sequenced by Ion Personal Genome Machine (PGM) with the Ion 318TM chip, using Ion PGM Template OT2 400 Kit and Ion PGM Hi-Q Sequencing Kit. The quality of the reads for each genome was examined using FastQC 0.10.1 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/).
Fig 1. Workflow of the genome assembly and annotation of 11 Corynebacterium pseudotuberculosis strains isolated from buffalo, and comparative genomics analysis with other strains.
Simba 1.2.1 [20] was used to assemble the genomes, unite them into a single scaffold, and establish the start of the chromosome. Simba includes three different assembly packages: SPAdes 3.6.0 [21]; MIRA 3.9.18 (http://sourceforge.net/projects/mira-assembler/); and Newbler 2.9 (http://swes.cals.arizona.edu/maier_lab/kartchner/documentation/index.php/home/docs/newbler). A “best” assembly was selected for each isolate based on having the highest N50, the smallest number of contigs, and higher values of maximum and minimum contigs size [22]. The contigs of this “best” assembly were then united into a single scaffold using CONTIGuator [23], with C. pseudotuberculosis 31 (CP003421.2) [19] used as a reference. The overlap between contigs was verified by BLASTN, and an in-house script was used to establish the beginning of the chromosome at the dnaA gene. The gaps were closed using CLC Genomics Workbench 6.5 (http://www.clcbio.com/products/clc-main-workbench/) in the following way. First, the reads were mapped to the reference genome to establish a consensus sequence. Next, the sequences of the flanking regions around each gap were identified in the reference genome. Then, the consensus sequence that had been generated from the reads was used to fill the gap when it was inserted between the flanking sequences.
Genome annotation
All genomes, including strain 31 (CP003421.2) [24], were annotated consistently using the RASTtk (Rapid Annotation Using Subsystem Technology) [25] annotation service in PATRIC (Pathosystems Resource Integration Center) [26]. To verify the authenticity of the identified frameshifts, a specific curation process that involved manual examination of potential frameshifts was performed. Insertion/deletion (indel) errors are associated to a certain extent with any sequencing platform, and those associated with homopolymers in the Ion Torrent sequencing platform are well defined [27]. Our curation process involved the following steps. First, a manually curated annotation of the reference genome was transferred to each of the other 10 genomes using an in-house script to identify the pseudogenes. Frameshifts were examined in Artemis v16.0.0 [28]. To identify the frameshift location and see if the mutation was conserved across other genomes, the genes with potential frameshifts were compared to other complete genes by BLASTN against the NR database at NCBI (National Center for Biotechnology Information). Second, we checked for indels among the reads at the location of the frameshift by mapping them against the assembled genome using the CLC Genomics Workbench v6.5. When an examination of the reads showed that an identified frameshift was a sequencing artifact, the sequence was adjusted. The new sequence, translated into a protein sequence, was verified by BLASTP against the Uniprot database [29]. The genome with the corrected sequences, which included the fixed frameshifts, was then re-annotated using RASTtk.
Genome plasticity
Pathogenicity islands (PAIs) were predicted by GIPSy [30]. All 11 genomes were individually compared to the C. glutamicum ATCC1302 genome (NC_006958.1), a non-pathogenic strain. DoubleACT v2 (http://www.hpa-bioinfotools.org.uk/pise/double_act.html) was used to align the pairs of genomes, and ACT (Artemis Comparison Tool) v13.0.0 [28] was used to visualize the alignments and check if the sequence of a predicted island in one genome was present but not annotated in the second genome. Comparison maps of the genomes were generated using BRIG (Blast Ring Image Generator) v0.95 [31].
PHAST, which predicts and annotates prophages from raw DNA sequence data or Genbank files, was used to predict prophages in the genomes by BLASTing against the NCBI and the prophage databases [32]. Syntenic and unique regions were both verified in the genomes by the progressiveMauve algorithm [33].
Phylogenomics
Gegenees v2.2.1 [34] was used to create a matrix of similarity across the genomes. This matrix was exported as a nexus file and used to generate a phylogenomic tree using Splitstree v4.14.2 [35] with the UPGMA method.
The PEPR (Phylogenomic Estimation with Progressive Refinement) program (https://github.com/enordber/pepr.git) was used to generate a phylogenomic tree with the publicly available and complete C. pseudotuberculosis genomes, and also included the new buffalo isolates. This is an automated system for generation of phylogenomic trees from amino acid sequences by a maximum likelihood algorithm. It identifies the common orthologs among all genomes, filters out genes that have been transferred horizontally, aligns and concatenates sequences, and generates a tree. Subtrees with low bootstrap values are refined by subsequent steps of addition genes that are shared across smaller clusters. This method is appropriated for uneven sampling in databases, where taxons of interest have a denser sampling. The resulting Newick tree file was visualized using Mega 6 [36].
Pangenomics
We compared the 11 buffalo isolates with the 33 other genomes publicly available in NCBI (Table 2). The Protein Family Sorter [26] was used to examine the pan-, core- and accessory genomes using the Corynebacterium genus-specific protein families (PLfams) that are available in PATRIC [37]. In addition, PATRIC’s Proteome Comparison tool (https://www.patricbrc.org/app/SeqComparison), an adaptation of RAST’s Sequence Based Comparison Tool [38], was used to generate a matrix of the bidirectional BLASTP hits across the proteins annotated in the genomes, with each genome used in a separate comparison as the reference genome to which the other 10 were compared. RAST’s Function based Comparison tool (http://rast.nmpdr.org/seedviewer.cgi) was used to assess similarities and differences in the presence of functional roles among the genomes. FgenesB (http://www.softberry.com/berry.phtml?topic=fgenesb) was used to identify operons in regions of interest.
Table 2. List of the 33 strains of Corynebacterium pseudotuberculosis used in this study that were isolated from hosts other than buffalo.
| Strain | Biovar | Host | Country | Accession number |
|---|---|---|---|---|
| Cp162 | Equi | Camel | UK | CP003652.1 |
| 262 | Equi | Cow | Belgium | CP012022.1 |
| 258 | Equi | Horse | Belgium | CP003540.2 |
| E19 | Equi | Horse | Chile | CP012136.1 |
| CIP52.97 | Equi | Horse | Kenya | CP003061.1 |
| 1/06-A | Equi | Horse | USA | CP003082.1 |
| 316 | Equi | Horse | USA | CP003077.1 |
| MB11 | Equi | Horse | USA | CP013260.1 |
| MB14 | Equi | Horse | USA | CP013261.1 |
| MB30 | Equi | Horse | USA | CP013262.1 |
| MB66 | Equi | Horse | USA | CP013263.1 |
| 29156 | Ovis | Cow | Israel | CP010795.1 |
| I19 | Ovis | Cow | Israel | CP002251.1 |
| 1002B | Ovis | Goat | Brazil | CP012837.1 |
| VD57 | Ovis | Goat | Brazil | CP009927.1 |
| CS_10 | Ovis | Goat | Norway | CP008923.1 |
| Ft_2193/67 | Ovis | Goat | Norway | CP008924.1 |
| PO222/4-1 | Ovis | Goat | Portugal | CP013698.1 |
| PO269-5 | Ovis | Goat | Portugal | CP012695.1 |
| 226 | Ovis | Goat | USA | CP010889.1 |
| FRC41 | Ovis | Human | France | CP002097.1 |
| 48252 | Ovis | Human | Norway | CP008922.1 |
| 267 | Ovis | Llama | USA | CP003407.1 |
| N1 | Ovis | Sheep | Equatorial Guinea | CP013146.1 |
| PAT10 | Ovis | Sheep | Argentina | CP002924.1 |
| C231 | Ovis | Sheep | Australia | CP001829.1 |
| 42/02-A | Ovis | Sheep | Australia | CP003062.1 |
| 12C | Ovis | Sheep | Brazil | CP011474.1 |
| PA01 | Ovis | Sheep | Brazil | CP013327.1 |
| E56 | Ovis | Sheep | Egypt | CP013699.1 |
| MEX25 | Ovis | Sheep | Mexico | CP013697.1 |
| 3/99-5 | Ovis | Sheep | Scotland | CP003152.1 |
| P54B96 | Ovis | Wildebeest | South Africa | CP003385.1 |
Specialty genes search
PATRIC’s Specialty Genes Search tool [39] was used to identify some of the virulence genes that were found in the 11 genomes. This tool BLASTs all genes in the annotated genome against a virulence factor database that includes VFDB [40] and manually curated virulence genes [39].
Results and discussion
Genome sequencing, assembly and annotation
All the 10 new genomes were closed, with all reads united into a single chromosome. For each genome, the theoretical mean coverage depth varied between 56.72 and 307.02x, while mean coverage depth genome varied between 56.61 and 306.54x. The 11 strains (including strain 31) have a genome size of approximately 2.4 Mb, a GC content that ranges between 52.07 and 52.1%, 2218 to 2,286 coding sequences (CDSs), 44 to 52 tRNA and 12 rRNA genes (Table 1). These values are within the ranges seen among other strains of C. pseudotuberculosis, which is considered to be more clonal compared to its closest relatives C. diphtheriae and C. ulcerans [8].
Genome plasticity
Genome plasticity analysis can identify areas of the genome, known as PAIs, which have been acquired by horizontal transfer and frequently include virulence genes [30]. Sixteen PAIs (PiCp1-16) were previously described in C. pseudotuberculosis and are shared across the biovars [8,41–43]. The variability in the size, gene content and deletion patterns in these islands explain most of the genomic differences seen between the two biovars. The content of sixteen PAIs are highly conserved in all the genomes belonging to Ovis, while biovar Equi has a greater variability, specifically with regard to deletions within the pilus genes [8].
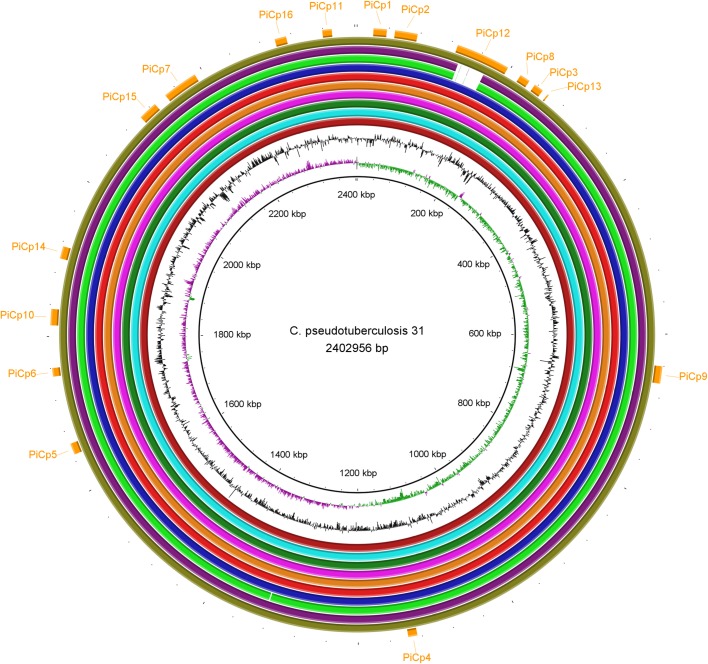
These same sixteen PAIs were found in all the genomes isolated from buffalo (Fig 2). Interestingly, we discovered three regions that were missing in the second assembly of strain 31 (CP003421.2, re-sequenced by Ion PGM) when it was compared to other buffalo strains, but found that these regions were present in the first assembly of the same genome (CP003421.1, sequenced by Solid v3) (S1 Fig, S1 File, S1 Table). To fix the second assembly, we mapped the reads of the Ion PGM to the first assembly to verify whether those regions were represented within the reads, and to generate a consensus sequence. The second assembly was updated to include these regions, verified using an optical map, and deposited at GenBank under the accession number CP003421.3.
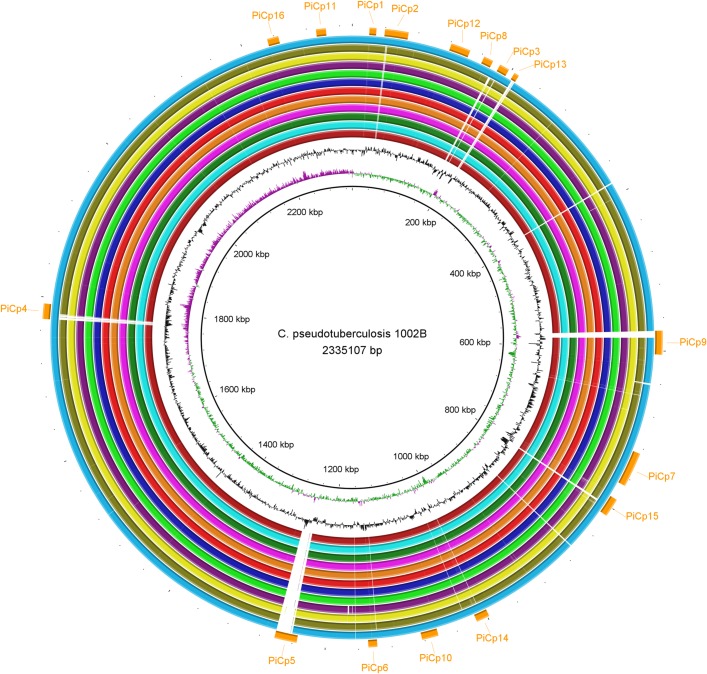
Fig 2. A circular genomic map that compares 11 Corynebacterium pseudotuberculosis strains isolated from Egyptian buffalo.
The rings, from the inner to outer circle, are: strain 31, GC skew, GC content, strains 32, 33, 34, 35, 36, 38, 39, 43, 46, and 48, and pathogenicity islands.
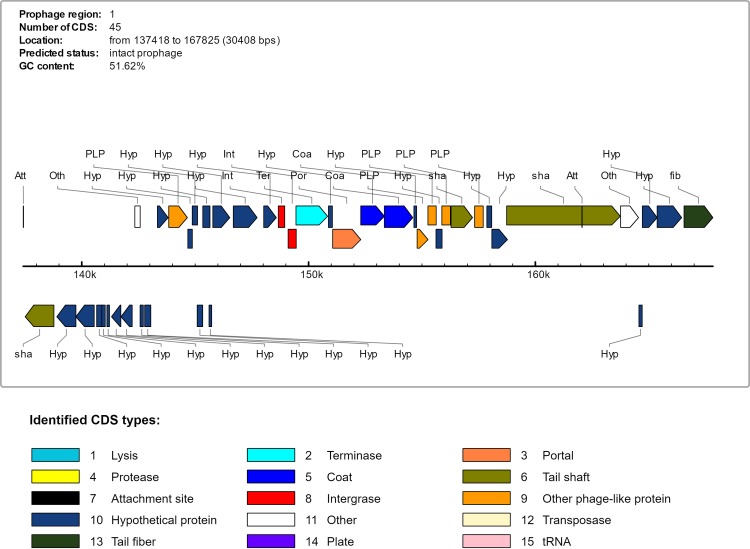
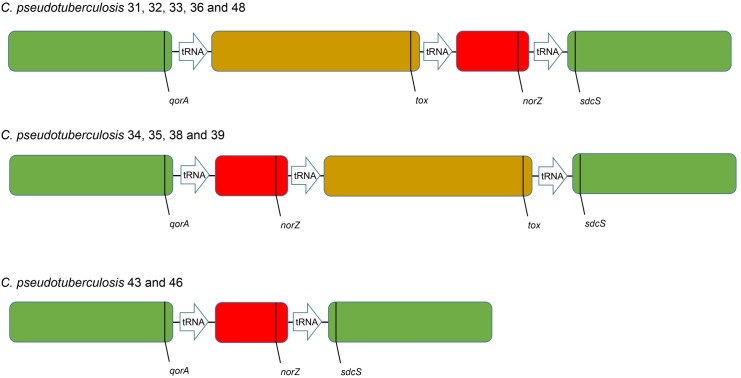
The Ovis and Equi strains share a genomic island known as PiCp12. All the buffalo isolates except strains 43 and 46 have a large insertion in PiCp12 that the other isolates from other hosts lack. This insertion is 36.6 Kb in length and includes 48 CDS and which are flanked by tRNA-Arg genes (Table 3). This insertion includes an intact β-corynephage, a prophage that was predicted by PHAST and estimated to be 30.4Kb and contains 39 CDSs (Fig 3) (Table 3, CDSs 3 to 41). A search of the NCBI database showed that the 36.6 Kb insertion sequence is unique and probably a new corynephage. This insertion also includes a tyrosine integrase gene, which codes for an enzyme known for site specific recombination [44]. tRNA-Arg genes, known as integration sites of β-corynephages [45], flank this region. This provides a possible explanation for the insertion of this unique 36.6 Kb region inside of the PiCp12 genomic island.
Table 3. Gene content of pathogenicity island PiCp12, a 36.6 Kb insertion sequence found in the buffalo isolates.
| CDS | Position | Strand | Product | Refseq locus tag |
|---|---|---|---|---|
| 1 | 136439..137053 | + | Hypothetical protein | CP31_RS00695 |
| 2 | 137233..137475 | + | Hypothetical protein | CP31_RS00700 |
| 137418..137430 | + | attL GCTAAAAAGGGGC | CP31_RS00715 | |
| 3 | 137506..138771 | - | PHAGE_Mycoba_Ariel_NC_028876: tyrosine integrase; phage(gi971751237) | CP31_RS00705 |
| 4 | 138901..139740 | - | Hypothetical protein | CP31_RS00710 |
| 5 | 139742..140548 | - | Hypothetical protein | CP31_RS00715 |
| 6 | 140658..140882 | - | PHAGE_Rhodoc_REQ2_NC_016652: Hypothetical protein; phage(gi372449852) | CP31_RS00720 |
| 7 | 140885..141031 | - | Hypothetical protein | None |
| 8 | 141106..141219 | - | Hypothetical protein | None |
| 9 | 141313..141720 | - | No significant database matches | CP31_RS00725 |
| 10 | 141730..141981 | - | No significant database matches | CP31_RS0073 |
| 11 | 142339..142575 | + | Transcriptional regulator | CP31_RS00735 |
| 12 | 142624..142884 | + | Hypothetical protein | None |
| 13 | 143334..143807 | + | Hypothetical protein | CP31_RS00745 |
| 14 | 143834..144655 | + | PHAGE_Bacter_Lily_NC_028841: antirepressor; phage(gi971748300) | CP31_RS00750 |
| 15 | 144870..145103 | + | Hypothetical protein | CP31_RS00760 |
| 16 | 145127..145351 | + | Hypothetical protein | CP31_RS00765 |
| 17 | 145612..145725 | - | Hypothetical protein | None |
| 18 | 145779..146525 | + | Hypothetical protein | CP31_RS00775 |
| 19 | 146683..147729 | + | Hypothetical protein | CP31_RS00780 |
| 20 | 148215..148577 | + | No significant database matches | CP31_RS00785 |
| 21 | 148665..148946 | + | PHAGE_Coryne_BFK20_NC_009799: gp55, HNH endonuclease; phage(gi157168428) | None |
| 22 | 149063..149455 | + | Hypothetical protein | CP31_RS00790 |
| 23 | 149445..150839 | + | PHAGE_Coryne_BFK20_NC_009799: gp2, terminase; phage(gi157168375) | CP31_RS00795 |
| 24 | 150875..151048 | + | Hypothetical protein | CP31_RS00795 |
| 25 | 151061..152302 | + | PHAGE_Coryne_BFK20_NC_009799: gp3, phage portal protein; phage(gi157168376) | CP31_RS00800 |
| 26 | 152299..153345 | + | PHAGE_Coryne_BFK20_NC_009799: gp5, head maturation protease; phage(gi157168378) | CP31_RS00805 |
| 27 | 153342..154592 | + | PHAGE_Coryne_BFK20_NC_009799: gp6, major capsid protein; phage(gi157168379) | CP31_RS00810 |
| 28 | 154592..154762 | + | No significant database matches | None |
| 29 | 154785..155267 | + | PHAGE_Coryne_BFK20_NC_009799: gp8; phage(gi157168381) | CP31_RS00815 |
| 30 | 155264..155626 | + | PHAGE_Coryne_BFK20_NC_009799: gp9; phage(gi157168382) | CP31_RS00820 |
| 31 | 155619..155885 | + | PHAGE_Coryne_BFK20_NC_009799: gp10; phage(gi157168383) | CP31_RS00825 |
| 32 | 155875..156252 | + | PHAGE_Coryne_BFK20_NC_009799: gp11; phage(gi157168384) | CP31_RS00830 |
| 33 | 156281..157228 | + | PHAGE_Coryne_BFK20_NC_009799: gp12, major tail protein; phage(gi157168385) | CP31_RS00835 |
| 34 | 157323..157700 | + | PHAGE_Coryne_BFK20_NC_009799: gp13; phage(gi157168386) | CP31_RS00840 |
| 35 | 157862..158071 | + | No significant database matches | CP31_RS00845 |
| 36 | 158081..158770 | + | PHAGE_Coryne_BFK20_NC_009799: gp15, minor tail protein; phage(gi157168388) | CP31_RS00850 |
| 37 | 158733..163745 | + | PHAGE_Coryne_P1201_NC_009816: putative tail measure protein; phage(gi157310951) | CP31_RS00850 |
| 162051..162063 | + | attR GCTAAAAAGGGGC | None | |
| 38 | 163755..164546 | + | Immunity-specific protein Beta201 | CP31_RS00855 |
| 39 | 164587..165369 | + | Immunity-specific protein Beta286 | CP31_RS00860 |
| 40 | 165370..166449 | + | Immunity-specific protein Beta371 | CP31_RS00865 |
| 41 | 166449..167825 | + | PHAGE_Salmon_Fels_1_NC_010391: putative bacteriophage tail fiber protein; Lambda gpN homolog; phage(gi169257204) | CP31_RS00870 |
| 42 | 167829..168005 | + | Hypothetical protein | None |
| 43 | 168307..168861 | + | Hypothetical protein | CP31_RS00875 |
| 44 | 168969..169706 | + | Teichoic acid phosphorylcholine esterase/choline binding protein E (cbpE) | CP31_RS00880 |
| 45 | 169703..169945 | + | Hypothetical protein | CP31_RS00885 |
| 46 | 169948..170412 | + | Putative membrane protein | CP31_RS00890 |
| 47 | 170409..170747 | + | Putative secreted protein | CP31_RS00895 |
| 48 | 171092..172774 | + | Diphtheria toxin | CP31_RS00900 |
Fig 3. Gene content of the intact prophage predicted by PHAST.
The prophage is inserted in pathogenicity island PiCp12 of the strains isolated from buffalo.
The sequence that is most similar to this new corynephage is BFK20 (NC_009799), a non-toxigenic dsDNA virus that infects Brevibacterium flavum. BFK20 has structural and lytic genes closely related to known phages found in the Corynebacterinae and to C. diphtheriae prophages [46]. The sequence of the new β-corynephage of strain 31 was deposited in Genbank under accession number KY566218. Additional accession numbers for the sequences of all of the buffalo strains are provided in S2 Table.
The 3’ end of the 36.6 Kb insertion into the PiCp12 island includes a gene of particular interest that produces the diphtheria toxin. This gene is only found in C. pseudotuberculosis strains that were isolated from buffalo, and is found 3,266 bp from the end of the predicted prophage in our isolates. The sequence is identical in all isolates, and differs from the toxin genes that are found in C. diphtheria and C. ulcerans (Fig 4). Maximescu et al. [47] have reported the presence of a diphtheria toxin from two separate strains isolated from Egyptian buffalo, indicating that this gene has been associated with infection in these animals since at least 1974. We cannot be certain that the entire insert, including the prophage, was also present in these previous infections as we do not have genomic sequences from these animals. However, previous studies have suggested that toxins can be acquired through a lysogenic conversion during an infection by a β-corynephage [2,48], and this gene in the buffalo isolates is downstream from a corynephage in the same insertion sequence. If these studies are correct, then the proximity of the corynephage and the toxin gene supports the premise that the acquisition of the two together is linked in buffalo, but it is certainly not definitive proof.
Fig 4. Phylogeny of diphtheria toxin gene (tox) from Corynebacterium pseudotuberculosis, C. diphtheriae and C. ulcerans inferred by using the maximum likelihood method based on the Tamura 3-parameter model, on Mega v6.
Toxins can cause disease. When injected intradermally into susceptible animals, the toxin from C. diptheriae resulted in erythema, induration, and dermonecrosis [48]. Parenteral injection caused myocarditis, polyneuritis, and focal necrosis in organs that included the adrenal glands, kidneys, and liver [48]. The toxin gene described in this study was detected in C. pseudotuberculosis isolated from buffalo with OSD [16]. The presence of this toxin may be responsible for the unique disease manifestations seen in the infected buffaloes. These animals have huge abscesses in the draining lymph nodes of dewlap, belly, and limbs that can extend across the whole limb, giving an aspect of elephantiasis. Other manifestations include skin eruptions and hair loss around the eruptions, extensive dermal necrosis, spontaneous bleeding from the skin, rapid respiration, coughing, dyspnea, and haemoglobinuria [13,16]. In contrast, horses infected by C. pseudotuberculosis (biovar Equi), which lack the toxin, have abscesses in different locations that include the pectoral or ventral abdomen, or in internal organs, or ulcerative lymphangitis of the limbs [12]. As buffalo are immune to any strains from the Ovis biovar [49], and there has never been a report of buffalo infected with any other strain within the Equi biovar, a possible preventative measure could be to use an inactivated form of the C. pseudotuberculosis diphtheria toxin as an antigen in a vaccine.
A synteny graph including all the C. pseudotuberculosis buffalo genomes showed two large regions (green blocks) that had the same relative position, and two smaller regions (brown and red blocks) that were close to each other, but had a variable position or were missing (Fig 5). These variable regions are found within the same pathogenicity island PiCp12 and are flanked by tRNA-Arg-ACG genes. The brown block shows the 36.6 kb insertion that harbors the prophage and the diphtheria toxin (tox), and is absent in strains 43 and 46. The red block is also present in the genomes of the Ovis and Equi strains, and contains the Nitric-oxide reductase (norZ) gene. This enzyme is a defense against the cytotoxic actions of Nitrous-oxide (NO) that the host uses as a defensive mechanism [50]. As stated previously, the tRNA-Arg genes are known to be a hotspot for phage integration [45] and the fact that we see these rearrangements within genomes isolated during the same outbreak, in the same region and from the same host species confirms that this is a volatile region in these genomes. As if in confirmation of this, strains 43 and 46 were found to be positive for the toxin gene by PCR when originally isolated from infected animals. By the time they were sequenced, this region was missing, indicating that there was an excision of the 36.6 Kb insert that occurred between isolation and sequencing. Another indication of volatility is the fact that we observed not only a prophage inserted in a copy of a tRNA-Arg gene, but also saw rearrangements within this same region in other genomes that were collected during a short period of time during the summer when this disease outbreak occurred.
Fig 5. Synteny graph of 11 Corynebacterium pseudotuberculosis strains isolated from Egyptian buffalo.
A previous study showed that all Equi strains shared the same deletion pattern when compared to strains in biovar Ovis [8]. When we compared the Equi strains with a single Ovis strain, 1002B, we, too, found that they all shared the same deletion pattern except for strain 262 (Fig 6). However, the comparison between Equi strain 31 and the other Equi strains showed that E19, 258, MB11, MB14, MB30 and MB66 had no gaps, besides the 36.6 Kb insertion exclusive of buffalo isolated strains. Also, nitrate reductase genes are missing in the strains that have gaps (Fig 7). This suggests that the gaps in those Equi strains are assembly issues instead of genetic differences.
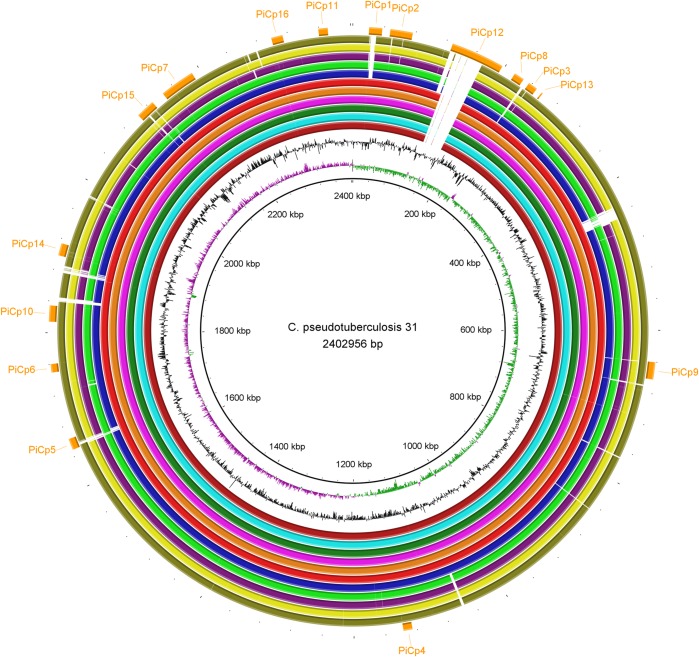
Fig 6. Circular genomic maps comparing Corynebacterium pseudotuberculosis 1002B (biovar Ovis) with other Equi strains.
The rings, from the inner to outer circle, are strain 1002B, CG Skew, CG Content, strains 31, 258, E19, MB11, MB14, MB30, MB66, 316, CIP52.97, 1/06-A, Cp162 and 262, and pathogenicity islands.
Fig 7. Circular genomic maps comparing Corynebacterium pseudotuberculosis 31 with other Equi strains.
The rings, from the inner to outer circle, are strain 31, CG Skew, CG Content, strains 258, E19, MB11, MB14, MB30, MB66, 316, CIP52.97, 1/06-A, Cp162 and 262, and pathogenicity islands.
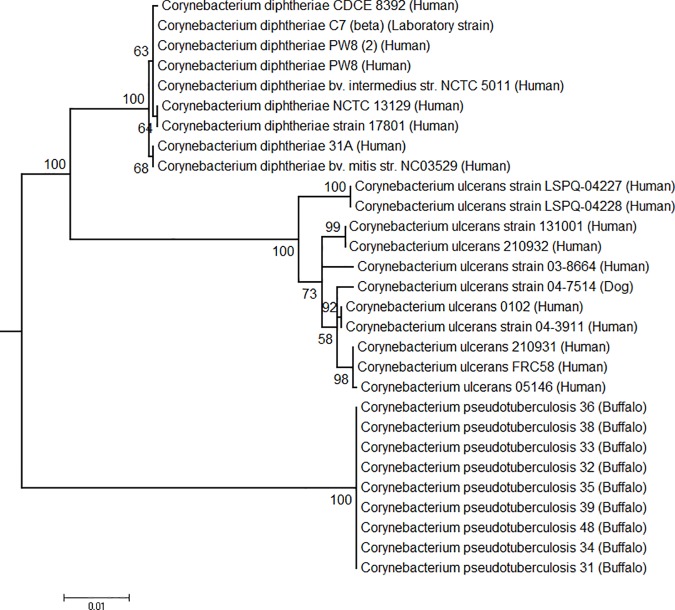
Phylogenomics
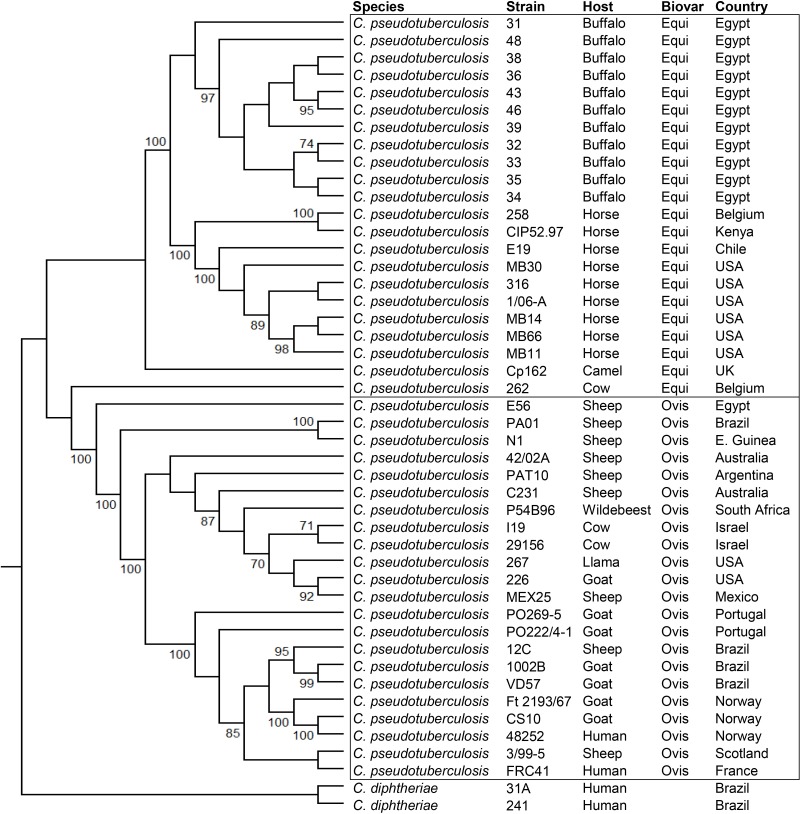
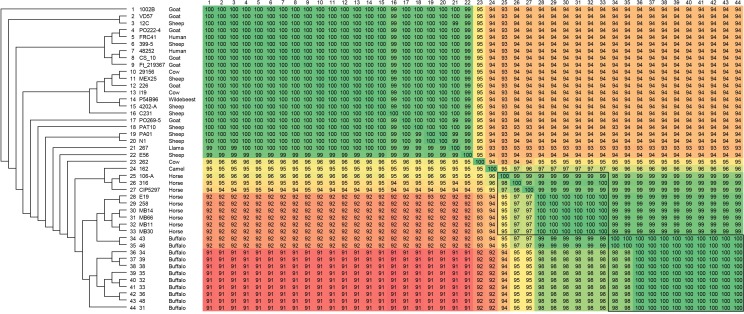
The phylogenomic tree generated by PEPR has two clusters representing the two biovars, with support values of 100 (Fig 8). In the Equi cluster, the buffalo were clearly separated from the horse isolates, which were collected in different countries. In the Ovis cluster, a group containing most of the goat isolates were separated from other hosts. These results suggest that C. pseudotuberculosis strains are grouped by host, at least in the Equi biovar. The fact that Equi strains 262, isolated from a cow (Bos taurus), and Cp162, isolated from a camel are outside of the two biovar clusters, with support values of 100, corroborates this hypothesis. Also, strain 262 was closer to biovar Ovis, as suggested by the circular map (Fig 6). It is possible that isolations from additional hosts like camels will create additional subclusters based on these hosts.
Fig 8. Phylogenomic tree of Corynebacterium pseudotuberculosis genomes based on the proteome of 44 complete genomes, generated by PEPR.
The core proteins were used to produce a tree, and additional protein families were added to refine subtrees with low bootstrap values.
The phylogenomic tree produced by Gegenees showed two clusters representing the two biovars, with similarity percentages within Equi strains (92–99%) being lower than within Ovis strains (99–100%) (Fig 9), a result similar to what was found previously [8]. Here, the clustering of strains by host is also apparent. Buffalo are grouped with the horse and camel isolates, with strains 43 and 46 separated from the rest of the buffalo isolates, probably due to the absence of the 36.6 Kb insertion in PiCp12. The heatmap shows higher values of similarity within isolates of the same type of host, with 262 (the cow isolate) and 162 (the camel isolate) having the lowest values of similarity when compared to the other Equi strains.
Fig 9. Phylogenomic tree of Corynebacterium pseudotuberculosis genomes based on the variable content of 44 complete genomes.
The percentages of similarity were plotted on a heatmap generated by Gegenees 2.2.1, and then used to produce a phylogenomic tree using Splitstree v4.14.2 with the UPGMA method.
Pangenomics
The “pangenome” is the complete gene inventory of a species. The “core genome” is the subset of orthologous genes present in all genomes, the “accessory genome” is a subset present in more than one genome, but not all genomes. “Singletons” are genes that are present in only one genome [51]. A previous C. pseudotuberculosis study identified 1,504 genes in the core genome and a pangenome with 2,782 genes. The pangenome was characterized as open [8], meaning that sequencing new genomes should significantly contribute to the identification of new genes and thus better characterize the genetic repertoire of the species [52].
Using PATRIC’s Protein Family Sorter, we identified a pangenome that included 2,172 protein families among those genomes isolated from buffalo. Most of these same protein families were also conserved as part of the core genome, which included 2,058 families. The accessory genome was limited to 91 protein families and 13 singletons, indicating that these genomes are all very similar. Analyzing the 11 buffalo with the other 33 strains public available showed that the C. pseudotuberculosis pangenome has 3,067 genes, and 1,541 in its core genome. In addition, we used the Proteome Comparison tool, a bidirectional BLASTP analysis, to examine the genomes and found 48 genes that were unique to the buffalo isolates. All of these were part of the 36.6 kb prophage that is located in the PiCp12 island.
When C. pseudotuberculosis strain 31 was used as the reference to compare all the strains isolated from buffalo, a strong homology was shared across the genes within these genomes. All genes had a highly conserved sequence identity of at least 90.2%. Also, we identified regions absent in the Ovis biovar that were present in most of the genomes of the Equi strains (S2 Table). A comparison of the functionality of these genes was conducted to look for metabolic and functional changes across the different groups. As expected, the buffalo genomes were all consistently similar, but when compared to genomes from the Ovis biovar, we saw that the buffalo genomes had some unique functionality. The buffalo genomes contained CRISPR and phage genes that were absent in Ovis. They also included the genes involved in nitrate reduction, and additional genes that are part of molybdenum cofactor biosynthesis. The presence of nitrate reductase is one characteristic that differentiates the biovar Equi from Ovis [5,8].
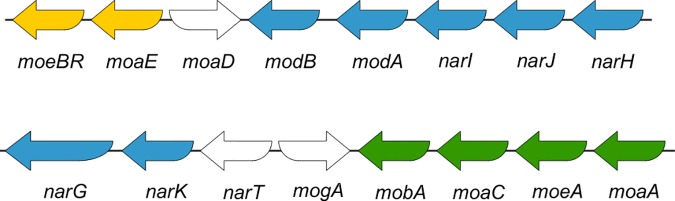
The use of nitrogen oxides as alternative electron acceptors, providing energy in an anaerobic environment and contributing to the persistence of the bacteria in their host, has been suggested as an advantage in adaptation to an intracellular lifestyle [50]. When we examined the region containing nitrate reduction and molybdenum cofactor biosynthesis genes, we found 13 of 16 genes were in operons: moeBR-moaE, narKGHJI-modAB, and mobA-moaC-moeA-moaA (Fig 10). The respiratory nitrate reductase enzyme has three subunits that are the product of the genes narGHI. NarI, the gamma subunit, is a transmembrane peptide that oxidizes quinol, liberates protons in the periplasm, and transports the electrons to NarH. NarH is the beta subunit, and it transports electrons from NarI to NarG. The alpha subunit NarG contains the catalytic domain that requires a molybdenum cofactor Mo-(bis-MGD) for activity. This subunit uses nitrate as the electron acceptor, reducing it to nitrite [53]. NarJ assists in the insertion of molybdenum cofactor in NarG, which must happen before the interaction between NarGH and NarI [54]. NarK is a transporter that performs nitrate/nitrite exchange [55]. NarT is probably involved in nitrate/nitrite transport as it has 80% identity with narK, and also includes the same Major Facilitator Superfamily (MFS) domain. MFS transporters are single-polypeptide secondary carriers that transport small solutes in response to a chemiosmotic ion gradients [56]. The nitrate reductase has been suggested as a drug target in Mycobacterium tuberculosis [53], and could be used in a similar fashion in C. pseudotuberculosis.
Fig 10. Organization of nitrate reductase and molybdenum cofactor biosynthesis genes in Corynebacterium pseudotuberculosis 31.
Genes with the same color are transcribed in the same operon.
Several studies have attempted to define the roles that the nar genes play in survival within the host. NarGH nitrate reductase provides resistance from acid stress and reactive nitrogen species in mycobacteria [57]. A narG mutant of M. tuberculosis was unable to persist in the lung, kidney and liver of immunocompetent mice, but it was able to grow and persist in the spleen as the wild-type strain, suggesting that the role of nitrate reduction in virulence is tissue specific [58]. However, another study showed that there no difference in persistence in the mice lungs between a narG mutant and wild-type M. tuberculosis, probably because mouse granulomas are not sufficiently hypoxic to affect the growth and survival of these bacteria [59]. The benefit, if any, that these genes give to the Equi strains has not been determined, but could be a target of further experimentation to see if they are important in either host specificity or disease manifestation. A genetic manipulation of narG in C. pseudotuberculosis could shed light on the different disease manifestations seen between hosts infected with either of the two biovars.
NarG requires molybdenum to function, and, in the buffalo genomes, the molybdenum transport genes are adjacent to the ones needed for nitrate reduction. The molybdenum cofactor biosynthesis operon modABC encodes a molybdate transmembrane transporter. ModA binds molybdate, ModB is a transmembrane subunit, and ModC provides the energy required for transportation by ATPase activity [60]. MoaA and MoaC convert a guanine nucleotide (GTP) to cyclic pyranopterin monophosphate (cPMP). MoaD and MoaE are subunits of the molybdopterin (MPT) synthase, which converts cPMP to MPT. MoeBR (a sulfurtransferase) activates MPT synthase by transferring sulfur groups to MoaD subunit. MogA (an adenylyltransferase) adenylates MPT, resulting in MPT-AMP. Molybdate is added by MoeA (MPT Mo-transferase), resulting in Mo-MPT (molybdenum cofactor). MobA (guanylyltransferase) converts Mo-MPT to a bis-Mo-MPT intermediary and attaches two guanines (GMP) to its phosphate groups, converting it to bis-MGD (molybdopterin guanine dinucleotide), which is the required cofactor for dimethylsulfoxide reductase (DMSO) enzymes, including nitrate reductase [60,61]. The insertion of the molybdenum cofactor in nitrate reductase NarGHI is performed by NarJ [62]. We found that the genes coding the transmembrane and ATP binding subunits of molybdate transporter (modB and modC) are fused in C. pseudotuberculosis and are transcribed with the nitrate reductase genes narKGHJI in the same operon, as predicted by FgenesB. The fusion between modB and modC does not appear to affect the phenotype, as all Equi strains that have been sequenced are positive for nitrate reduction, and they all share this fusion.
Specialty genes search
Specific genes that function as virulence factors can determine bacterial adhesion, invasion, colonization, dissemination within the host and evasion of the immune system [63]. The diphtheria toxin gene was the only virulence factor found that was unique to the buffalo isolates. An additional virulence factor, inositol-1-phosphate synthase, a product of the ino1 gene, was found in the buffalo strains. This gene is broadly shared across the Corynebacterium. Ino1 catalyzes the first step in the synthesis of inositol [64], a compound required for the production of essential cell wall lipoglycans in Mycobacterium [65], and is a major thiol that plays a role in protection from oxidative stress [66,67]. In an experiment using M. tuberculosis, the role that ino1 plays in virulence was demonstrated when the CFUs of ino1 mutants fell sharply, and the bacteria were virtually cleared in seven days by infected macrophages, while they remained constant in wild type strains [68]. Furthermore, mice infected with wild type ino1 strains died in 38 days, while mice infected by the ino1 mutant were alive and healthy when the experiment concluded at 56 days post infection [68]. The role, if any, that this gene plays in the virulence of C. pseudotuberculosis has not yet been determined.
Another virulence factor identified was the exotoxin phospholipase D (pld gene), which promotes bacterial dissemination by degradation of sphingomyelin in endothelial cell membranes, and also plays a role in macrophage death [69,70]. In strain 31, pld was found to have a frameshift mutation near the 3’, and it was suggested that this could decrease the ability of this strain to spread throughout the host, and this, along with the tox gene, have been proposed as either a requirement for infection or a geographic variant [8]. This mutation was not seen after resequencing strain 31, however, nor did we find it any other genomes used in this study. This suggests that the original finding was probably a sequencing artifact.
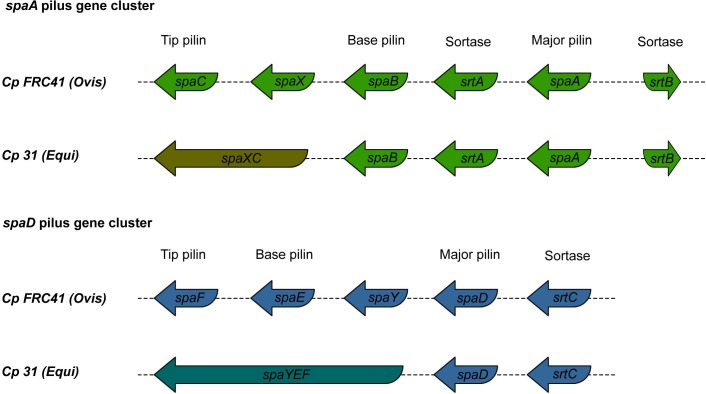
Pili are structures responsible for bacterial adhesion and play a major role in the initiation of extracellular and intracellular invasion and proliferation [71]. Pathogenicity islands PiCp7 and PiCp15 harbor the pilus gene cluster spaA (srtB-spaA-srtA-spaB-spaX-spaC) and spaD (srtC-spaD-spaY-spaE-spaF), respectively [8]. The genes srtABC are specific pilus sortases, while spaAD, spaBE and spaCF are major, base, and tip proteins, respectively. The genes spaX and spaY have unknown functions [72]. Specific sortases cleave the LPTxG motif of the pilin proteins and polymerize them to assembly the pilus, while the housekeeping sortase incorporates the final structure to the cell wall [73]. When we compared the Ovis and Equi strains, we saw some unique differences in these regions.
A comparison of pilus gene clusters spaD and spaA showed conservation among buffalo isolates (Fig 11) and polymorphisms when compared to other Equi strains. The spaD pilus genes from the buffalo isolates had a fusion between three genes, including the base and tip pilin (genes spaE-spaF-spaY), which is also seen in the Equi strains 258, E19 (isolated from a horse) and Cp162 (camel isolate). In all of the buffalo isolates except for strain 31, the major pilin gene spaA has a frameshift. A similar frameshift is also seen in all the other Equi isolates except for strains 316 (isolated from a horse), and 262 (isolated from a cow). Fusion is also seen between with the spaC (tip pilin) and spaX genes in the buffalo isolates. Not all members of Equi share this fusion, but it is also found in 262 (cow), Cp162 (camel), E19 and 258 (horse isolates). The major pilin (spaA) gene is frameshifted in most of the Equi strains.
Fig 11. Comparison of pilus gene cluster spaA and spaD in Corynebacterium pseudotuberculosis isolated from buffalo, represented by strain 31, and biovar Ovis, represented by strain FRC41.
In C. diphtheria, the three pili structures SpaA, SpaD and SpaH have been found to be necessary for adhesion to pharyngeal, laryngeal and lung human epithelial cells, respectively. In this same species, a deletion of the major pilin gene showed that the tip and base pilin were the most important proteins for adhesion on the host cell wall [74]. The frameshift in spaA (major pilin) found in C. pseudotuberculosis may be not critical to the adhesion. Although, given the importance of base and tip pilin, the fusions of these genes and the conservation within strains may represent an adaptation to the buffalo host, or to mutation events in the immediate ancestor this particular outbreak. More data, perhaps from isolates of future outbreaks of OSD before could determine if this an anomaly or a true adaptation. The polymorphisms in the Equi pilus genes could be due to the higher variability of host species in this biovar, leading to specificity for tissues of different host species [74].
The cow isolate 262 is classified as Equi and has the same nitrate reductase genes, but the sequences of two pilus genes (srtB and spaA) are identical to those found in the Ovis biovar. Also, the phylogenetic tree (Fig 8) and the circular map (Fig 7) showed the similarity of this strain with the Ovis isolates, yet it could be viewed as ancestral to that clade. Perhaps other Equi isolates, including those from B. taurus cattle, will help determine if this is an anomaly, or perhaps the first of a new biovar of C. pseudotuberculosis.
Conclusion
C. pseudotuberculosis strains isolated from buffalo showed an overall synteny, conservation in pilus genes, and a unique insertion that contained both a corynephage and the diphtheria toxin gene. This insertion could explain the expansion of the known host range for C. pseudotuberculosis to include buffalo, as these genes may play a role in adaptation to this new host. The tRNA-Arg gene was identified as a hotspot of phage insertion and rearrangements, events observed during in this outbreak of OSD. By comparison with Ovis strains, we identified and described the known nitrate reductase (narGHI) genes and identified genes involved molybdenum cofactor biosynthesis, which are necessary for the action of the nar genes. A phylogenomic tree confirmed a clear separation between the Ovis and Equi biovars, and indicated that Equi strains are clustered depending on the host they infected.
Supporting information
The rings, from the inner to outer circle, are strain 31 v3 (CP003421.3), GC skew, GC content, strains 31 v1 (CP003421.1), 31 v2 (CP003421.2), and 32 (CP015183.1), and pathogenicity islands.
(TIF)
(DOCX)
(DOCX)
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by: Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Ministério da Pesca e Agricultura. A.R. Wattam was supported in part by federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under contract no. HHSN272201400027C.
References
- 1.Dorella FA, Carvalho Pacheco L, Oliveira SC, Miyoshi A, Azevedo V. Corynebacterium pseudotuberculosis: microbiology, biochemical properties, pathogenesis and molecular studies of virulence. Vet Res. 2006;37: 201–218. doi: 10.1051/vetres:2005056 [DOI] [PubMed] [Google Scholar]
- 2.Guaraldi AL de M, Júnior RH, Azevedo CVA de. Corynebacterium diphtheriae, Corynebacterium ulcerans and Corynebacterium pseudotuberculosis—General Aspects. In: Burkovski A, editor. Corynebacterium diphtheriae and Related Toxigenic Species. London, New York; 2014. pp. 15–37.
- 3.Bernard K. The genus Corynebacterium and other medically relevant coryneform-like bacteria. J Clin Microbiol. 2012;50: 3152–8. doi: 10.1128/JCM.00796-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oliveira A, Teixeira P, Azevedo M, Jamal SB, Tiwari S, Almeida S, et al. Corynebacterium pseudotuberculosis may be under anagenesis and biovar Equi forms biovar Ovis: a phylogenic inference from sequence and structural analysis. BMC Microbiol. BMC Microbiology; 2016;16: 100 doi: 10.1186/s12866-016-0717-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biberstein EL, Knight HD, Jang S. Two biotypes of Corynebacterium pseudotuberculosis. Vet Rec. 1971;89: 691–692. [DOI] [PubMed] [Google Scholar]
- 6.Songer JG, Beckenbach K, Marshall MM, Olson GB, Kelley L. Biochemical and genetic characterization of Corynebacterium pseudotuberculosis. Am J Vet Res. 1988;49: 223–226. [PubMed] [Google Scholar]
- 7.Sutherland SS, Hart RA, Buller NB. Genetic differences between nitrate-negative and nitrate-positive C. pseudotuberculosis strains using restriction fragment length polymorphisms. Vet Microbiol. 1996;49: 1–9. [DOI] [PubMed] [Google Scholar]
- 8.Soares SC, Silva A, Trost E, Blom J, Ramos R, Carneiro A, et al. The Pan-Genome of the Animal Pathogen Corynebacterium pseudotuberculosis Reveals Differences in Genome Plasticity between the Biovar ovis and equi Strains. PLoS One. 2013;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Windsor PA, Bush RD. Caseous lymphadenitis: Present and near forgotten from persistent vaccination? Small Rumin Res. 2016;142: 6–10. [Google Scholar]
- 10.Silva A, Schneider MPC, Cerdeira L, Barbosa MS, Ramos RTJ, Carneiro AR, et al. Complete genome sequence of Corynebacterium pseudotuberculosis I19, a strain isolated from a cow in Israel with bovine mastitis. J Bacteriol. 2011;193: 323–324. doi: 10.1128/JB.01211-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yeruham I, Friedman S, Perl S, Elad D, Berkovich Y, Kalgard Y. A herd level analysis of a Corynebacterium pseudotuberculosis outbreak in a dairy cattle herd. Vet Dermatol. 2004;15: 315–320. doi: 10.1111/j.1365-3164.2004.00388.x [DOI] [PubMed] [Google Scholar]
- 12.Spier SJ, Azevedo V. Corynebacterium pseudotuberculosis infection in horses: Increasing frequency and spread to new regions of North America. Equine Vet Educ. 2016; [Google Scholar]
- 13.Selim SA. Oedematous skin disease of buffalo in Egypt. Journal of Veterinary Medicine, Series B. 2001. pp. 241–258. [DOI] [PubMed] [Google Scholar]
- 14.Ahmed IM, El-Tahawy AS. Prevalence of So-called Oedematous Skin Disease in Egyptian buffaloes with particular study on its economic influence. Alexandria J Vet Sci. 2012;37: 129–133. [Google Scholar]
- 15.Ghoneim MA, Mousa AW, Ibrahim AK, Amin AS, Khafagy A, Selim SA. Role of Hippobosca equina as a transmitter of C. pseudotuberculosis among buffaloes as revealed by PCR and dot blot hybridization. J Egypt Vet Med Assoc. 2001;61: 165–176. [Google Scholar]
- 16.Selim SA, Mohamed FH, Hessain AM, Moussa IM. Immunological characterization of diphtheria toxin recovered from Corynebacterium pseudotuberculosis. Saudi J Biol Sci. King Saud University; 2015; 0–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soto a, Zapardiel J, Soriano F. Evaluation of API Coryne system for identifying coryneform bacteria. J Clin Pathol. 1994;47: 756–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Costa Torres L de F, Ribeiro D, Hirata R, Pacheco LGC, Souza MC, dos Santos LS, et al. Multiplex polymerase chain reaction to identify and determine the toxigenicity of Corynebacterium spp with zoonotic potential and an overview of human and animal infections. Mem Inst Oswaldo Cruz. 2013;108: 272–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silva A, Ramos RTJ, Carneiro AR, Pinto AC, Soares SDC, Santos AR, et al. Complete genome sequence of Corynebacterium pseudotuberculosis Cp31, isolated from an Egyptian buffalo. J Bacteriol. 2012;194: 6663–6664. doi: 10.1128/JB.01782-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mariano DCB, Pereira FL, Aguiar EL, Oliveira LC, Benevides L, Guimarães LC, et al. SIMBA: a web tool for managing bacterial genome assembly generated by Ion PGM sequencing technology. BMC Bioinformatics. 2016;17: 456 doi: 10.1186/s12859-016-1344-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J Comput Biol. 2012;19: 455–477. doi: 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics. 2013;29: 1072–1075. doi: 10.1093/bioinformatics/btt086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galardini M, Biondi EG, Bazzicalupo M, Mengoni A. CONTIGuator: a bacterial genomes finishing tool for structural insights on draft genomes. Source Code Biol Med. 2011;6: 11 doi: 10.1186/1751-0473-6-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramos RTJ, Carneiro AR, de Castro Soares S, Barbosa S, Varuzza L, Orabona G, et al. High efficiency application of a mate-paired library from next-generation sequencing to postlight sequencing: Corynebacterium pseudotuberculosis as a case study for microbial de novo genome assembly. J Microbiol Methods. 2013;95: 441–447. doi: 10.1016/j.mimet.2013.06.006 [DOI] [PubMed] [Google Scholar]
- 25.Brettin T, Davis JJ, Disz T, Edwards R a, Gerdes S, Olsen GJ, et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep. 2015;5: 8365 doi: 10.1038/srep08365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wattam AR, Abraham D, Dalay O, Disz TL, Driscoll T, Gabbard JL, et al. PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res. 2014;42: D581–91. doi: 10.1093/nar/gkt1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bragg LM, Stone G, Butler MK, Hugenholtz P, Tyson GW. Shining a Light on Dark Sequencing: Characterising Errors in Ion Torrent PGM Data. PLoS Comput Biol. 2013;9: e1003031 doi: 10.1371/journal.pcbi.1003031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carver T, Berriman M, Tivey A, Patel C, Böhme U, Barrell BG, et al. Artemis and ACT: Viewing, annotating and comparing sequences stored in a relational database. Bioinformatics. 2008;24: 2672–2676. doi: 10.1093/bioinformatics/btn529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wasmuth E V, Lima CD. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017;45: D158–D169. doi: 10.1093/nar/gkw1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soares SC, Geyik H, Ramos RTJ, de Sá PHCG, Barbosa EGV, Baumbach J, et al. GIPSy: Genomic island prediction software. J Biotechnol. 2016;232: 2–11. doi: 10.1016/j.jbiotec.2015.09.008 [DOI] [PubMed] [Google Scholar]
- 31.Alikhan N- F, Petty NK, Ben Zakour NL, Beatson SA. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 2011;12: 402 doi: 10.1186/1471-2164-12-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. PHAST: a fast phage search tool. Nucleic Acids Res. 2011;39: W347–52. doi: 10.1093/nar/gkr485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Darling AE, Mau B, Perna NT. Progressivemauve: Multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 2010;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ågren J, Sundström A, Håfström T, Segerman B. Gegenees: Fragmented Alignment of Multiple Genomes for Determining Phylogenomic Distances and Genetic Signatures Unique for Specified Target Groups. PLoS One. 2012;7: e39107 doi: 10.1371/journal.pone.0039107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huson DH. Application of Phylogenetic Networks in Evolutionary Studies. Mol Biol Evol. 2005;23: 254–267. doi: 10.1093/molbev/msj030 [DOI] [PubMed] [Google Scholar]
- 36.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30: 2725–9. doi: 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davis JJ, Gerdes S, Olsen GJ, Olson R, Pusch GD, Shukla M, et al. PATtyFams: Protein Families for the Microbial Genomes in the PATRIC Database. Front Microbiol. 2016;7: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014;42: D206–14. doi: 10.1093/nar/gkt1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mao C, Abraham D, Wattam AR, Wilson MJC, Shukla M, Yoo HS, et al. Curation, integration and visualization of bacterial virulence factors in PATRIC. Bioinformatics. 2015;31: 252–258. doi: 10.1093/bioinformatics/btu631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen F, Ding X, Ding Y, Xiang Z, Li X, Ghosh D, et al. Proinflammatory caspase-2-mediated macrophage cell death induced by a rough attenuated Brucella suis strain. Infect Immun. 2011;79: 2460–2469. doi: 10.1128/IAI.00050-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruiz JC, D’Afonseca V, Silva A, Ali A, Pinto AC, Santos AR, et al. Evidence for reductive genome evolution and lateral acquisition of virulence functions in two Corynebacterium pseudotuberculosis strains. PLoS One. 2011;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramos RTJ, Carneiro AR, Soares SDC, Santos AR Dos, Almeida S, Guimarães L, et al. Tips and tricks for the assembly of a Corynebacterium pseudotuberculosis genome using a semiconductor sequencer. Microb Biotechnol. 2013;6: 150–156. doi: 10.1111/1751-7915.12006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soares SC, Trost E, Ramos RTJ, Carneiro AR, Santos AR, Pinto AC, et al. Genome sequence of Corynebacterium pseudotuberculosis biovar equi strain 258 and prediction of antigenic targets to improve biotechnological vaccine production. J Biotechnol. 2013;167: 135–141. doi: 10.1016/j.jbiotec.2012.11.003 [DOI] [PubMed] [Google Scholar]
- 44.Fogg PCM, Colloms S, Rosser S, Stark M, Smith MCM. New applications for phage integrases. Journal of Molecular Biology. 2014. pp. 2703–2716. doi: 10.1016/j.jmb.2014.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sekizuka T, Yamamoto A, Komiya T, Kenri T, Takeuchi F, Shibayama K, et al. Corynebacterium ulcerans 0102 carries the gene encoding diphtheria toxin on a prophage different from the C. diphtheriae NCTC 13129 prophage. BMC Microbiol. 2012;12: 72 doi: 10.1186/1471-2180-12-72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bukovska G, Klucar L, Vlcek C, Adamovic J, Turna J, Timko J. Complete nucleotide sequence and genome analysis of bacteriophage BFK20—a lytic phage of the industrial producer Brevibacterium flavum. Virology. 2006;348: 57–71. doi: 10.1016/j.virol.2005.12.010 [DOI] [PubMed] [Google Scholar]
- 47.Maximescu P, Oprişan A, Pop A, Potorac E. Further studies on Corynebacterium species capable of producing diphtheria toxin (C. diphtheriae, C. ulcerans, C. ovis). J Gen Microbiol. 1974;82: 49–56. doi: 10.1099/00221287-82-1-49 [DOI] [PubMed] [Google Scholar]
- 48.Holmes RK. Biology and molecular epidemiology of diphtheria toxin and the tox gene. J Infect Dis. 2000;181: S156—S167. doi: 10.1086/315554 [DOI] [PubMed] [Google Scholar]
- 49.Moussa IM, Ali MS, Hessain AM, Kabli SA, Hemeg HA, Selim SA. Vaccination against Corynebacterium pseudotuberculosis infections controlling caseous lymphadenitis (CLA) and oedematousskin disease. Saudi J Biol Sci. 2016;23: 718–723. doi: 10.1016/j.sjbs.2016.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vázquez-Torres A, Baumler AJ. Nitrate, nitrite and nitric oxide reductases: From the last universal common ancestor to modern bacterial pathogens. Curr Opin Microbiol. 2016;29: 1–8. doi: 10.1016/j.mib.2015.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rouli L, Merhej V, Fournier P-E, Raoult D. The bacterial pangenome as a new tool for analysing pathogenic bacteria. New Microbes New Infect. 2015;7: 72–85. doi: 10.1016/j.nmni.2015.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Medini D, Donati C, Tettelin H, Masignani V, Rappuoli R. The microbial pan-genome. Curr Opin Genet Dev. 2005;15: 589–594. doi: 10.1016/j.gde.2005.09.006 [DOI] [PubMed] [Google Scholar]
- 53.Huang Q, Abdalla AE, Xie J. Phylogenomics of Mycobacterium Nitrate Reductase Operon. Curr Microbiol. 2015;71: 121–128. doi: 10.1007/s00284-015-0838-2 [DOI] [PubMed] [Google Scholar]
- 54.Zakian S, Lafitte D, Vergnes A, Pimentel C, Sebban-Kreuzer C, Toci R, et al. Basis of recognition between the NarJ chaperone and the N-terminus of the NarG subunit from Escherichia coli nitrate reductase. FEBS J. 2010;277: 1886–1895. doi: 10.1111/j.1742-4658.2010.07611.x [DOI] [PubMed] [Google Scholar]
- 55.Zheng H, Wisedchaisri G, Gonen T. Crystal structure of a nitrate/nitrite exchanger. Nature. 2013;497: 647–651. doi: 10.1038/nature12139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yan N. Structural Biology of the Major Facilitator Superfamily Transporters. Annu Rev Biophys. 2015;44: 257–283. doi: 10.1146/annurev-biophys-060414-033901 [DOI] [PubMed] [Google Scholar]
- 57.Tan MP, Sequeira P, Lin WW, Phong WY, Cliff P, Ng SH, et al. Nitrate respiration protects hypoxic Mycobacterium tuberculosis against acid- and reactive nitrogen species stresses. PLoS One. 2010;5: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fritz C, Maass S, Kreft A, Bange F. Dependence of Mycobacterium bovis BCG on Anaerobic Nitrate Reductase for Persistence Is Tissue Specific. Infect Immun. 2002;70: 286–291. doi: 10.1128/IAI.70.1.286-291.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aly S, Wagner K, Keller C, Malm S, Malzan A, Brandau S, et al. Oxygen status of lung granulomas in Mycobacterium tuberculosis-infected mice. J Pathol. 2006;210: 298–305. doi: 10.1002/path.2055 [DOI] [PubMed] [Google Scholar]
- 60.Williams M, Mizrahi V, Kana BD. Molybdenum cofactor: A key component of Mycobacterium tuberculosis pathogenesis? Crit Rev Microbiol. 2014;40: 18–29. doi: 10.3109/1040841X.2012.749211 [DOI] [PubMed] [Google Scholar]
- 61.Leimkühler S. The Biosynthesis of the Molybdenum Cofactor in Escherichia coli and Its Connection to FeS Cluster Assembly and the Thiolation of tRNA. Adv Biol. 2014;2014: 1–21. [Google Scholar]
- 62.Iobbi-Nivol C, Leimkühler S. Molybdenum enzymes, their maturation and molybdenum cofactor biosynthesis in Escherichia coli. Biochim Biophys Acta—Bioenerg. 2013;1827: 1086–1101. [DOI] [PubMed] [Google Scholar]
- 63.Tauch A, Burkovski A. Molecular armory or niche factors: virulence determinants of Corynebacterium species. FEMS Microbiol Lett. 2015;67: fnv185. [DOI] [PubMed] [Google Scholar]
- 64.Bachhawat N, Mande SC. Identification of the INO1 gene of Mycobacterium tuberculosis H37Rv reveals a novel class of inositol-1-phosphate synthase enzyme. J Mol Biol. 1999;291: 531–536. doi: 10.1006/jmbi.1999.2980 [DOI] [PubMed] [Google Scholar]
- 65.Jackson M, Crick DC, Brennan PJ. Phosphatidylinositol is an essential phospholipid of mycobacteria. J Biol Chem. 2000;275: 30092–30099. doi: 10.1074/jbc.M004658200 [DOI] [PubMed] [Google Scholar]
- 66.Fahey RC. Novel thiols of prokaryotes. Annu Rev Microbiol. 2001;55: 333–356. doi: 10.1146/annurev.micro.55.1.333 [DOI] [PubMed] [Google Scholar]
- 67.Newton GL, Buchmeier N, Fahey RC. Biosynthesis and functions of mycothiol, the unique protective thiol of Actinobacteria. Microbiol Mol Biol Rev. 2008;72: 471–94. doi: 10.1128/MMBR.00008-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Movahedzadeh F, Smith D a., Norman RA, Dinadayala P, Murray-Rust J, Russell DG, et al. The Mycobacterium tuberculosis ino1 gene is essential for growth and virulence. Mol Microbiol. 2004;51: 1003–14. [DOI] [PubMed] [Google Scholar]
- 69.McKean SC, Davies JK, Moore RJ. Expression of phospholipase D, the major virulence factor of Corynebacterium pseudotuberculosis is regulated by multiple environmental factors and plays a role in macrophage death. Microbiology. 2007;153: 2203–2211. doi: 10.1099/mic.0.2007/005926-0 [DOI] [PubMed] [Google Scholar]
- 70.D’Afonseca V, Moraes PM, Dorella FA, Pacheco LGC, Meyer R, Portela RW, et al. A description of genes of Corynebacterium pseudotuberculosis useful in diagnostics and vaccine applications. Genet Mol Res. 2008;7: 252–260. [DOI] [PubMed] [Google Scholar]
- 71.Rogers EA, Das A, Ton-That H. Adhesion by Pathogenic Corynebacteria. 2011. pp. 91–103. [DOI] [PubMed]
- 72.Trost E, Ott L, Schneider J, Schröder J, Jaenicke S, Goesmann A, et al. The complete genome sequence of Corynebacterium pseudotuberculosis FRC41 isolated from a 12-year-old girl with necrotizing lymphadenitis reveals insights into gene-regulatory networks contributing to virulence. BMC Genomics. BioMed Central Ltd; 2010;11: 728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mandlik A, Swierczynski A, Das A, Ton-That H. Pili in Gram-positive bacteria: assembly, involvement in colonization and biofilm development. Trends Microbiol. 2008;16: 33–40. doi: 10.1016/j.tim.2007.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mandlik A, Swierczynski A, Das A, Ton-That H. Corynebacterium diphtheriae employs specific minor pilins to target human pharyngeal epithelial cells. Mol Microbiol. 2007;64: 111–124. doi: 10.1111/j.1365-2958.2007.05630.x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The rings, from the inner to outer circle, are strain 31 v3 (CP003421.3), GC skew, GC content, strains 31 v1 (CP003421.1), 31 v2 (CP003421.2), and 32 (CP015183.1), and pathogenicity islands.
(TIF)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.