Abstract
Objective:
A meta-analysis was carried out to further evaluate the relationship between ALDH2 Glu487Lys polymorphism and esophageal cancer risk.
Methods:
A total number of 15 studies that included 3812 cases and 7376 controls were identified for our meta-analysis.
Results:
Our findings indicated that individuals with the combination of Glu/Lys and Lys/Lys genotype had an increased risk of getting esophageal cancer (GA + AA vs. GG: odds ratio [OR] 1.36, 95% confidence interval [CI] 0.93–2.00, P = 0.113) with a shift pattern. Although Lys/Lys genotype carriers showed areduced esophageal cancer risk (AA vs. GA + GG: OR 0.41, 95% CI 0.23–0.72, P = 0.002). Similarly, a negative association was observed under homozygote comparison (AA vs. GG: OR 0.49, 95% CI 0.29–0.85, P = 0.011). In the China subgroup analysis, the similar results were found.
Conclusions:
This meta-analysis concluded that there was a strong association between ALDH2 Glu487Lys polymorphism and the risk of esophageal cancer. It further confirmed that ALDH2 Glu487Lys polymorphism was a high-risk factor for esophageal cancer.
Keywords: ALDH2 Glu487Lys polymorphism, genotype, meta-analysis, the risk of esophageal cancer
1. Introduction
Esophageal carcinoma (EC) is the 8th most common cause of cancer-related death worldwide. Esophageal squamous cell carcinoma (ESCC) and adenocarcinoma (EAC) are 2 major histological types.[1] It has been reported that the highest incidences occurred in Asian countries, such as China, Iran, and Japan.[2] Despite of the use of surgery in combination with radiotherapy and chemotherapy, the overall 5-year survival rate remains <40% in patients with advanced disease.[3] Therefore, risk evaluation and elucidation of molecular mechanisms are required to improve treatment efficacy for patients with EC.
EC is a complex disease, and it is well accepted that environment factors play an important role as genetic factors in the development of EC. Alcohol consumption has been considered as a high-risk factor for EC patients.[4] Alcohol itself is not a carcinogen, but its metabolite acetaldehyde has carcinogenic property. Alcohol in humans can be oxidized to acetaldehyde, which is further oxidized into harmless acetate by aldehyde dehydrogenases (ALDHs). ALDH2 is one of the major enzymes to eliminate acetaldehyde, and thus determines blood acetaldehyde concentrations after drinking. ALDH2 displays a polymorphism that may affect alcohol-oxidizing capacity. A single point mutation of a lysine amino acid has been found at residue 487 instead of glutamic acid in ALDH2 gene (ALDH2 Glu487Lys), resulting in a reduced enzyme activity. Individuals with the ALDH2 Lys allele have a high concentration of blood acetaldehyde after drinking alcohol, thus enhances the risk for esophageal cancer.[5] The polymorphisms of ALDH2 associated with the risk of esophageal cancer have been described in several studies, but the results are still inconsistent.
Meta-analysis provides an opportunity to aggregate information from multiple studies, improving statistical power by increasing the sample size to precisely evaluate genetic polymorphisms effects on disease susceptibility. To assess the association between ALDH2 Glu487Lys polymorphism and the risk of esophageal cancer, we performed a meta-analysis based on 3812 cases and 7376 controls. Multiple databases under NCBI global database and Google Scholar were searched for relative studies. Fifteen case–control studies related to ALDH2 Glu487Lys polymorphism and esophageal cancer risk were covered in this meta-analysis. The statistical analysis was performed with STATA 12 software.
2. Materials and Methods
2.1. Selection criteria and data Collection
Only case–control studies that investigated the relationship between ALDH2 polymorphisms and EC were included in the meta-analysis. For the first-round exclusion, articles were searched with NCBI Global Cross-database, including PubMed, PMC, Gene, PubChem, among others, as well as Google Scholar by using "ALDH2 polymorphism? "ALDH2 Glu487Lys polymorphism? "ALDH2 rs671 polymorphism? and "esophageal cancer?as key words. There were 189 results after searching. We excluded books and other literatures that were not related with case–control studies, as well as literatures that were published before January 1st, 2000, and then 122 articles were obtained. For the second-round selection, we excluded the articles that were not aimed at investigating the association between ALDH2 Glu487Lys polymorphisms and esophageal cancer risk, and then 61 articles were identified. Articles that did not contain control group information or retrieve the original data were excluded. When the studies covered in each article were overlapped, we only kept the ones that showed the most extensive results. As a result, 15 case–control studies were covered in the final meta-analysis. The data collection flow chart was shown in Figure 1.
Figure 1.

Flow chart of selection of studies and specific reasons for exclusion from our meta-analysis.
2.2. Statistical analysis
Owing to the relatively larger database formed by studies performed in China (9 studies), we created a subgroup that covered all studies from China. To get a more reasonable result, we used 3 different methods: dominant model (AA + GA vs. GG), recessive model (AA vs. GA + GG), and homozygote comparison (AA vs. GG). In dominant model, we investigated the distribution of AA + GA genotype referred to GG genotype. In recessive model, we investigated the distribution of AA genotype referred to GA + GG genotype. In homozygote comparison, we used GG as reference genotype, and investigated the distribution of AA genotype. For each study, numbers of 3 genotypes in case and control group were used as pooled data. When dealing with large samples, Peto method might be misleading. However, inverse variance method only works on continuous data. Another key factor for choosing analysis model was to test the heterogeneity involved in the studies. Mantel-Haenszel (M-H) fixed-effect model should be applied to analyze datasets without significant heterogeneity, and DerSimonian and Laird (D-L) random-effected model should be applied for datasets that showed obvious heterogeneity. In our analysis, the heterogeneity among studies was tested by using I2 index, with the equation of, where Q is statistical data and df is its freedom. Higher I2 represents more significant heterogeneity. Values of I2 = 25%, 50%, and 75% represented low, medium, and high heterogeneity, respectively. When I2 ≤50%, there was no significant heterogeneity between pooled data. In this meta-analysis, 15 studies were included in the final analysis for ALDH2 Glu487Lys polymorphism. For each analysis, we used M-H fixed-effect model to test the heterogeneity first, and then chose different model based on the testing results. To get a reasonable statistical conclusion, association between ALDH2 Glu487Lys polymorphism and the risk of EC was evaluated using odds ratio (OR) derived from different analysis models. ORs were calculated with each model within 95% confidence intervals (CIs). The available polymorphism data were analyzed with the STATA 12 (Stata Statistical Software, Release 12; StataCorp LP, College Station, TX). Forest plots were generated to summarize the results. To evaluate publication bias, Begg funnel plots were generated based on the analysis results and database size. The more asymmetric the funnel plot looked, the more publication biases were introduced. Meanwhile, Egger test was also performed for furtherinvestigation.
2.3. Ethics statement
In our study, we just utilized previous articles to review the association between ALDH2 Glu487Lys polymorphism and the risk of esophageal cancer, and it did not include clinical trials and animal experiments. So we state that there is no ethical problem in our study.
3. Results
3.1. Characteristics of studies
A total of 15 publications (Figure 1) were included in the final meta-analysis by using the search method as described above.[6–20] All the data in these studies were related to association between ALDH2 Glu487Lys polymorphism and human esophageal cancer risk. The qualities of the studies were considered acceptable for our further analysis. The studies have been carried out in China (n = 9), Japan (n = 3), Africa (n = 1), Thailand (n = 1), and mixed (n = 1). Hardy–Weinberg equilibrium (HWE) was calculated for all 15 publications and P < 0.05 was considered as a departure from HWE. We found that Yokoyama et al's study was inconsistent with HWE (P < 0.0001). Study characteristics and the gene distribution of Glu/Glu (GG), Glu/Lys (GA), and Lys/Lys (AA) of all studies were included in Table 1.[6–20]
Table 1.
Pooled data for ALDH2 Glu487Lys polymorphism analysis.

3.2. Evaluation of ALDH2 Glu487Lys polymorphisms and EC risk
First of all, we performed the analysis for the entire database. The M-H fixed-effect model was applied on the subgroup dataset with 3 different analysis models (dominant, recessive, and homozygote) to test the heterogeneity. Two different methods (M-H fixed effect model and D-L random effect model) were used according to different analysis results. By definition, with I2 <25% we should definitely apply M-H fixed model, while with I2 >75% we should apply D-L random effect model due to the significant heterogeneity. However, for database <10 studies, it was more reasonable to apply fixed-effect model on the analysis. Odds ratio (OR) was derived based on the analysis and corresponding P value was acquired as well. To test publication bias, both Begg and Egger tests were performed. Forest plot and Funnel plot were also generated. Final results were shown in Table 2. Forest plots and Funnel plots for each model were shown as Figure 2. An OR <1 indicated a reduced risk of esophageal cancer in case group. For dominant model, the overall OR was 1.36 (95% CI 0.93–2.00, P = 0.113), indicating that an increased EC risk was associated with ALDH2 Glu487Lys polymorphisms in the dominant model (GA+AA vs GG, Figure 2A) even though a shift pattern was detected (P > 0.05). Random-effect model was applied because of the significant heterogeneity (I2 = 94.3%). The overall OR for recessive model (AA vs. GA + GG) was 0.41 (95% CI 0.23–0.72, P = 0.002), suggesting that AA genotype of ALDH2 Glu487Lys polymorphisms had the association with the reduced EC risk (Fig. 2C). Random-effect model was applied (I2 = 81.1%). Significant difference was observed between control group and case group. A similar association was observed under homozygote comparison (AA vs. GG)based on the fact that the overall OR was 0.49 (95% CI 0.29–0.85, P = 0.011, Fig. 2E). Random-effect model was also applied (I2 = 77.8%). Funnel plot did not show significant publication bias in all 3 methods.
Table 2.
Meta-analysis for entire database with dominant model (GA + AA vs. GG), recessive model (AA vs. GA + GG), and homozygote comparison (AA vs. GG).
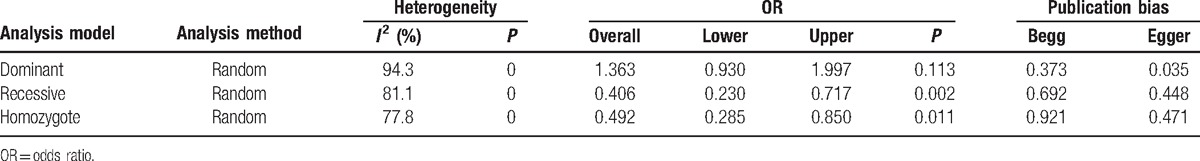
Figure 2.
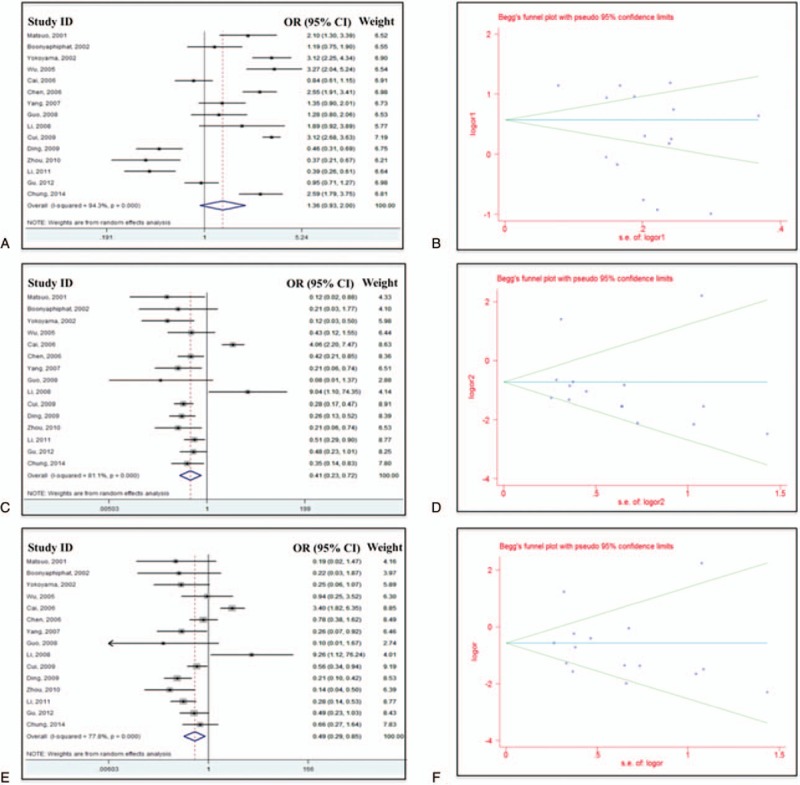
Forest plots (A, C, and E) and Begg funnel plot (B, D, and F) for all studies under dominant model (A and B), recessive model (C and D), and homozygote model (E and F).
3.3. The association between ALDH2 Glu487Lys polymorphisms and EC risk in China subgroup
Similarly, we used all 3 analysis methods (dominant, recessive, and homozygote) to analyze the China subgroup. Analysis results were included in Table 3. Significant difference was observed between case group and control group (with P < 0.05) with recessive model and homozygote model. Shift pattern was observed with dominant model. Forrest plots and funnel plots for China subgroup were summarized in Figure 3. For dominant model, overall OR was 0.98 (95% CI 0.92–1.56, P = 0.931, heterogeneity index I2 = 91.8%, Fig. 3A). For recessive model, the overall OR was 0.44 (95% CI 0.20–0.94, P = 0.034, heterogeneity index I2 = 85%, Fig. 3C). For homozygote comparison, the overall OR was 0.44 (95% CI 0.20–0.98, P = 0.043, heterogeneity index I2 = 84.7%, Fig. 3E). Publication bias might exist because of the size of database.
Table 3.
Meta-analysis for China subgroup with dominant model (GA + AA vs. GG), recessive model (AA vs. GA + GG), and homozygote comparison (AA vs. GG).
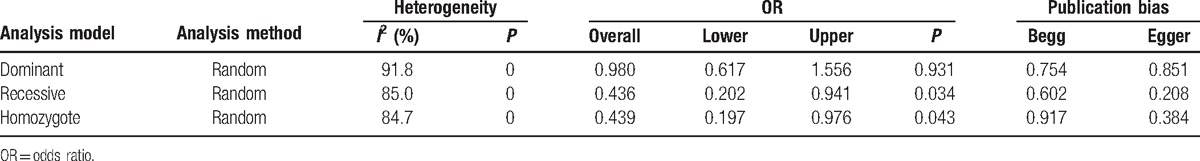
Figure 3.
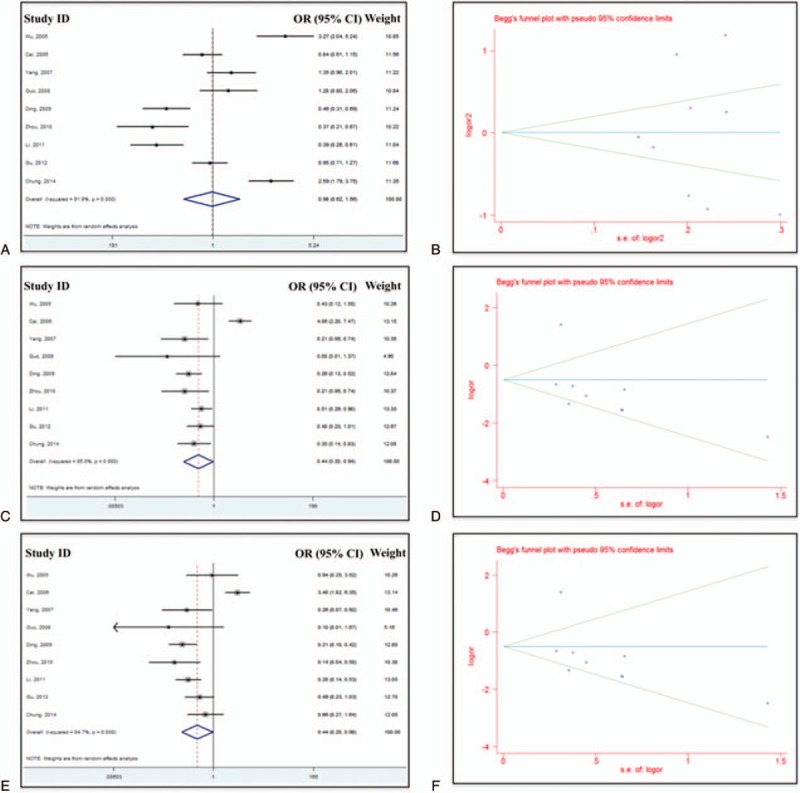
Forest plots (A, C, and E) and Begg funnel plot (B, D, and F) for China subgroup under dominant model (A and B), recessive model (C and D), and homozygote model (E and F).
4. Discussion
In this article, we presented a meta-analysis to investigate the association between ALDH2 Glu487Lys polymorphisms and the risk of human esophageal cancer. A total of 15 studies wasincluded in our analysis. According to our knowledge, this is the first meta-analysis generated to summarize the effects of the specific single-nucleotide polymorphism (SNP) (ALDH2 Glu487Lys) on human esophageal cancer. Meanwhile, the databases used in our analysis were large enough to generate a comprehensive and convincing conclusion. Totally 3812 cases and 7376 controls were included in our meta-analysis, which highly increased the statistical power. Several studies have been performed to investigate the association between this SNP and the risk of esophageal cancer, but the results were not consistent. The data from this meta-analysis showed that there was a significant association between ALDH2 Glu487Lys polymorphism and the risk of esophageal cancer. The combination of Glu/Lys and Lys/Lys genotype showed an increased risk of esophageal cancer even though a shift pattern was detected (P > 0.05). However, compared to Glu/Glu genotype, we observed that individuals with Lys/Lys genotype presented a decreased risk of esophageal cancer. The overall OR for homozygote comparison (AA vs. GG) was estimated to be 0.49 (95% CI 0.29–0.85, P = 0.011). Similar result was acquired with recessive model (AA vs. GA + GG). The overall OR was 0.41 (95% CI 0.23–0.72, P = 0.002). Random-effect method was used for both these 3 models because of the high heterogeneity (I2 >75%). Similar results were found in China subgroup. The study by Zhang et al[21] also confirmed that ALDH2 Glu/Lys had statistically significant effects on ESCC, which is consistent with our study.
Previous studies have shown that ADH1B and ALDH2 polymorphism was one of risk-conferring factors for alcohol dependence.[22] ALDH 487Lys allele encodes an inactive subunit of ALDH2, leading to large amounts of acetaldehyde accumulation after alcohol consumption. The concentration of blood acetaldehyde was 6-fold in individuals with inactive ALDH2 than those with active ALDH2.[23] Our dominant model results in this study suggest the hypothesis that ALDH2 Lys allele may increase the susceptibility to EC because of longer exposure to alcohol and high concentration of acetaldehyde. Thus, this metabolite of alcohol is a strong inducer for carcinogenesis of EC. It may be very important to avoid alcohol to prevent EC for those carriers with ALDH2 Lys allele.
The degree of heterogeneity is one of the major concerns in meta-analysis because nonhomogeneous study has high possibility to mislead results. In the present study, I2 index was carried out to test the significance of heterogeneity. For 3 analysis models, there was significant heterogeneity in all studies, as well as china subgroup. Publication bias is another key factor that might affect the quality of meta-analysis. Both Begg and Egger test were used to assess publication bias in this study. No significant publication bias was observed when all studies were included. However, there was publication bias for china subgroup. One of explanations may because of insufficient size of database.
There are still several limitations in this study, although our primary results are suggestive. First, in 15 studies included in this meta-analysis, 9 studies were conducted in China. There are no white samples in our analysis, we thus miss the cases occurred in these high-risk areas. Larger samples are needed to assess the effect among different ethnicities and to validate our results. Second, the samples from different countries and controls were not uniform, results should be interpreted with caution. Third, there was heterogeneity between studies of ALDH2 polymorphisms. Most studies performed analysis focusing on drinking status. Therefore, other risk factors are not well considered into analysis, such as smoking. It has been accepted that drinkers tend to smoke, which may affect the risk of EC.
5. Conclusions
Based on the results of our meta-analysis, we concluded that a strong association was existed between ALDH2 Glu487Lys polymorphism and the risk of esophageal cancer. The combination of ALDH2 Glu/Lys and Lys/Lys genotypes was associated with the increased risk of esophageal cancer with a shift pattern. When compared to Glu/Lys and Glu/Glu or Glu/Glu genotypes, individuals with Lys/Lys genotype had a reduced risk of getting esophageal cancer. These findings confirmed a significant interaction between gene and environment for risk of EC, and may provide a new strategic method to prevent the EC.
Footnotes
Abbreviations: ALDHs = aldehyde dehydrogenases, EC = esophageal carcinoma, ESCC = esophageal squamous cell carcinoma, HWE = Hardy–Weinberg equilibrium, OR = odds ratio.
KL and GS contributed equally to this article.
The authors report no conflicts of interest.
References
- [1].Rice TW, Rusch VW, Apperson-Hansen C, et al. Worldwide esophageal cancer collaboration. Dis Esophagus 2009;22:1–8. [DOI] [PubMed] [Google Scholar]
- [2].Su M, Liu M, Tian D-P, et al. Temporal trends of esophageal cancer during1995-2004inNanao Island, an extremely high-risk area in China. Eur J Epidemiol 2007;22:43–8. [DOI] [PubMed] [Google Scholar]
- [3].Uemura N, Kondo T. Current advances in esophageal cancer proteomics. Biochim Biophys Acta 2015;1854:687–95. [DOI] [PubMed] [Google Scholar]
- [4].Hanaoka T, Tsugane S, Ando N, et al. Alcohol consumption and risk of esophageal cancer in japan: a case-control study in seven hospitals. Jpn J Clin Oncol 1994;24:241–6. [PubMed] [Google Scholar]
- [5].Crabb DW, Edenberg HJ, Bosron WF, Li TK. Genotypes for aldehyde dehydrogenase deficiency and alcohol sensitivity. The inactive ALDH2(2) allele is dominant. J Clin Invest 1989;83:314–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Boonyaphiphat P, Thongsuksai P, Sriplung H, Puttawibul P. Lifestyle habits and genetic susceptibility and the risk of esophageal cancer in the Thai population. Cancer Lett 2002;186:193–9. [DOI] [PubMed] [Google Scholar]
- [7].Cai L, You NC, Lu H, et al. Dietary selenium intake, aldehyde dehydrogenase-2 and X-ray repair cross-complementing 1 genetic polymorphisms, and the risk of esophageal squamous cell carcinoma. Cancer 2006;106:2345–54. [DOI] [PubMed] [Google Scholar]
- [8].Chen YJ, Chen C, Wu DC, et al. Interactive effects of lifetime alcohol consumption and alcohol and aldehyde dehydrogenase polymorphisms on esophageal cancer risks. Int J Cancer 2006;119:2827–31. [DOI] [PubMed] [Google Scholar]
- [9].Chung CS, Lee YC, Liou JM, et al. Tag single nucleotide polymorphisms of alcohol-metabolizing enzymes modify the risk of upper aerodigestive tract cancers: HapMap database analysis. Dis Esophagus 2014;27:493–503. [DOI] [PubMed] [Google Scholar]
- [10].Cui R, Kamatani Y, Takahashi A, et al. Functional variants in ADH1B and ALDH2 coupled with alcohol and smoking synergistically enhance esophageal cancer risk. Gastroenterology 2009;137:1768–75. [DOI] [PubMed] [Google Scholar]
- [11].Ding J-H, Li S-P, Cao H-X, et al. Polymorphisms of alcohol dehydrogenase-2 and aldehyde dehydrogenase-2 and esophageal cancer risk in Southeast Chinese males. World J Gastroenterol 2009;15:2395–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Gu H, Gong D, Ding G, et al. A variant allele of ADH1B and ALDH2, is associated with the risk of esophageal cancer. Exp Ther Med 2012;4:135–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Li D-P, Dandara C, Walther G, Parker MI. Genetic polymorphisms of alcohol metabolising enzymes: their role in susceptibility to oesophageal cancer. Clin Chem Lab Med 2008;46:323–8. [DOI] [PubMed] [Google Scholar]
- [14].Guo Y-M, Wang Q, Liu Y-Z, et al. Genetic polymorphisms in cytochrome P4502E1, alcohol and aldehyde dehydrogenases and the risk of esophageal squamous cell carcinoma in Gansu Chinese males. World J Gastroenterol 2008;14:1444–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Li QD, Li H, Wang MS, et al. Multi-susceptibility genes associated with the risk of the development stages of esophageal squamous cell cancer in Feicheng County. BMC Gastroenterol 2011;11:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Matsuo K, Hamajima N, Shinoda M, et al. Gene–environment interaction between an aldehyde dehydrogenase-2 (ALDH2) polymorphism and alcohol consumption for the risk of esophageal cancer. Carcinogenesis 2001;22:913–6. [DOI] [PubMed] [Google Scholar]
- [17].Wu C, Wu D, Hsu H, et al. Relationship between genetic polymorphisms of alcohol and aldehyde dehydrogenases and esophageal squamous cell carcinoma risk in males. World J Gastroenterol 2005;11:5103–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Yang S-J, Wang H-Y, Li X-Q, et al. Genetic polymorphisms of ADH2 and ALDH2 association with esophageal cancer risk in southwest China. World J Gastroenterol 2007;13:5760–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Yokoyama A, Kato H, Yokoyama T, et al. Genetic polymorphisms of alcohol and aldehyde dehydrogenases and glutathione S-transferase M1 and drinking, smoking, and diet in Japanese men with esophageal squamous cell carcinoma. Carcinogenesis 2002;23:1851–9. [DOI] [PubMed] [Google Scholar]
- [20].Zhou Y-Z, Diao Y-T, Li H, et al. Association of genetic polymorphisms of aldehyde dehydrogenase-2 with esophageal squamous cell dysplasia. World J Gastroenterol 2010;16:3445–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Zhang G-H, Mai R-Q, Huang B. Meta-analysis of ADH1B and ALDH2 polymorphisms and esophageal cancer risk in China. World J Gastroenterol 2010;16:6020–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Luczak SE, Glatt SJ, Wall TJ. Meta-analyses of ALDH2 and ADH1B with alcohol dependence in Asians. Psychol Bull 2006;132:607–21. [DOI] [PubMed] [Google Scholar]
- [23].Mizoi Y, Yamamoto K, Ueno Y, et al. Involvement of genetic polymorphism of alcohol and aldehyde dehydrogenases in individual variation of alcohol metabolism. Alcohol Alcohol 1994;29:707–10. [PubMed] [Google Scholar]


