Supplemental Digital Content is available in the text
Keywords: cadherins, CDH1, clinical significance, HCC, methylation
Abstract
Background:
Cadherins (CDHs) have been reported to be associated with cancer. However, the clinical significance of CDH gene methylation in hepatocellular carcinoma (HCC) remains unclear.
Methods:
Based on the preferred reporting items for systematic reviews and meta-analyses (PRISMA) statement criteria, available studies were identified from online electronic database. The overall odds ratio (OR) and the corresponding 95% confidence interval (95% CI) were calculated and analyzed.
Results:
A total of 29 eligible studies with 2562 HCC samples and 1685 controls were included. E-cadherin (CDH1) hypermethylation was observed to be significantly higher in HCC than in benign, adjacent, or normal samples. Moreover, CDH1 hypermethylation was not associated with gender, tumor grade, clinical stage, hepatitis B virus (HBV), or hepatitis C virus (HCV) infection in HCC patients. H-cadherin (CDH13), protocadherin-10 (PCDH10), P-cadherin (CDH3), and M-cadherin (CDH15) methylation may have an increased risk of HCC in fewer than 4 studies, and methylated cadherin 8, type 2 (CDH8) and OB-cadherin (CDH11) had a similar OR in HCC and adjacent samples. When HCC samples were compared with normal samples, the analysis of sample type revealed a significantly higher OR in normal blood samples than in normal tissues for hypermethylated CDH1 (50.82 vs 4.44).
Conclusion:
CDH1 hypermethylation may play a key role in the carcinogenesis of HCC. However, CDH1 hypermethylation was not correlated with clinicopathological features. Methylated CDH13, PCDH10, CDH3, and CDH15, but not methylated CDH8 or CDH11, may lead to an increased risk of HCC. Hypermethylated CDH1 may become a noninvasive blood biomarker. Further studies with more data are necessary.
1. Introduction
Worldwide, hepatocellular carcinoma (HCC) is the 5th most frequent malignant disease and the 2nd most common cause of cancer death, with China accounting for approximately half of HCC cases and deaths.[1] A low incidence of this disease has been recorded in Europe.[2] According to global cancer statistics, an estimated 782,500 new cases of HCC were diagnosed in 2012, with approximately 745,500 HCC-related deaths.[1] Several risk factors have been suggested to be associated with the majority of HCC cases, such as chronic liver diseases associated with hepatitis B virus (HBV) or hepatitis C virus (HCV) infection, liver cirrhosis, aflatoxin exposure, alcohol consumption, obesity, type 2 diabetes, and tobacco smoking, etc.[1,3] The majority of HCC patients are diagnosed at a late stage, and symptomatic HCC has a 5-year survival rate of 3%.[1,4]
Epigenetic events involving DNA methylation, histone modifications, nucleosome positioning, and noncoding RNAs have been reported to play key roles in the carcinogenesis and progression of cancers.[5,6] DNA methylation, a major mechanism of epigenetic alterations, occurs more often in human tumor cells than gene mutations.[7] Epigenomic regulation displays 2 essential molecular mechanisms: hypermethylation of tumor suppressor genes (TSGs) and hypomethylation of oncogenes.[8,9] The classical cadherins (CDHs) are a superfamily of transmembrane glycoproteins involved in calcium-dependent cell–cell adhesion in embryonic development and epithelial tissues.[10] CDHs are also associated with signaling, mechanotransduction, cancer progression, and tissue morphogenesis.[11–14] Many CDHs are related to cancer. For example, the expression of E-cadherin (CDH1), a TSG, is reduced through hypermethylation in several cancers, including in HCC.[15–17] In addition, aberrant methylation of the TSG H-cadherin (CDH13) associated with gene inactivation has been found in some primary tumors, including ovarian, renal cell, and breast cancers.[18] Reduction of protocadherin-10 (PCDH10) expression via methylation has also been reported in some cancers.[19,20] In contrast, increased expression of P-cadherin (CDH3) is clinically associated with several cancers, such as breast and colon tumors.[21,22]
In the present study, we first determined whether methylated CDH genes were significantly associated with the risk of HCC. In addition, we assessed the clinicopathological significance of CDH1 hypermethylation in HCC patients.
2. Materials and methods
2.1. Search strategy
We systemically performed literature searches in electronic databases (PubMed, EMBASE, Web of Science, EBSCO, and Cochrane Library) for eligible studies published in English prior to October 10th, 2016. The following keywords and search terms were used: (liver OR hepatocellular OR hepatic) AND (cancer OR tumor OR neoplasm OR carcinoma) AND (CDH∗ OR cadherin) AND (methylation OR epigene∗). Furthermore, the reference lists of the identified articles were manually searched to identify additional relevant studies.
2.2. Study criteria
The studies included in our analysis had to meet the following inclusion criteria: patients were limited to individuals with HCC based on histopathological examination, without restriction of sample type; studies reported that sufficient data were obtained to evaluate the association between CDH gene methylation and HCC in the cancer and control groups; and studies on CDH1 methylation were performed to estimate the clinicopathological significance of CDH1 methylation in HCC patients. The major exclusion criteria were as follows: studies using cell lines and animal studies; reviews, case reports, letters, and conference abstracts; or studies with insufficient data or duplicated data.
2.3. Ethical review from patients
The current study was a secondary analysis regarding human subject data published in the public domain.
2.4. Data extraction
For eligible studies, the following data were extracted: surname of the first author, year of publication, country, race, methylation detection methodology, sample type, number of HCC cases, number of HCC controls, methylation rate, number of CDH gene methylations, and clinicopathological parameters (including gender status, tumor grade, clinical stage, HBV status, and HCV status). Benign samples included liver disease associated with chronic hepatitis or cirrhosis, normal samples were from normal healthy subjects without chronic hepatitis, cirrhosis, or other disease. The present meta-analysis met the guidelines of Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA).[23]
2.5. Data analysis
The pooled odds ratios (ORs) were calculated using STATA software (version 12.0, Stata Corporation, College Station, TX). The pooled ORs with the corresponding 95% confidence intervals (95% CIs) were calculated to assess the correlation between CDH gene methylation and HCC in the HCC and control groups. In addition, the correlation of the CDH1 hypermethylation status with gender status, tumor grade, tumor stage, HBV status, and HCV status in HCC patients was determined to estimate the clinicopathological significance of CDH1 hypermethylation in HCC patients. The heterogeneity among studies was examined based on the chi-square test and Q statistics.[24] The pooled OR was calculated and summarized under a random-effects model. If heterogeneity was significant (I2 ≥ 50%), meta-regression analyses were performed to assess the potential sources of heterogeneity. Moreover, a sensitivity analysis was conducted to evaluate the influence of an individual study on the results and the effect of omitting a single study on the stability of the results.[25,26] Possible publication bias was identified using Egger test.[27]
3. Results
3.1. General characteristics of the included studies
After being carefully retrieved from the above electronic databases according to our inclusion criteria (as shown in Fig. 1), 29 eligible studies[17,28–55] involving 2562 HCC specimens and 1685 controls were included in the final meta-analysis. Twenty-three of the studies analyzed the association between CDH1 hypermethylation and HCC in cancer and control groups.[17,28,29,31,32,34,36–42,46–55] Among these 23 studies, 12 studies that included a total of 594 HCC samples and 581 benign samples analyzed the association between CDH1 hypermethylation and HCC.[28,29,31,34,40,46,48–50,53–55] Twelve studies, involving 598 HCC tissue samples and 471 adjacent tissue samples, analyzed the association between CDH1 hypermethylation and HCC.[17,29,36–39,41,42,47,49,51,52] Nine studies, involving 494 HCC samples and 166 normal samples, analyzed the association between CDH1 hypermethylation and HCC.[28,29,32,38,40,47,48,50,51] Sixteen studies, involving 1110 HCC patients, assessed the correlation of CDH1 hypermethylation with clinicopathological parameters[17,28,32–34,36,38,41–44,46,49,50,52,53] in HCC. Three studies analyzed the association between CDH13 methylation and HCC in cancer and control groups.[35,39,51] Two studies evaluated the relationship between PCDH10 methylation and HCC in cancer and control groups.[30,45] One study evaluated the relationship of methylated CDH3, VE-cadherin (CDH5), cadherin 8, type 2 (CDH8), OB-cadherin (CDH11), and M-cadherin (CDH15) with HCC in cancer and control groups.[39] The basic characteristics of the included studies are summarized in supplemental Table S1.
Figure 1.
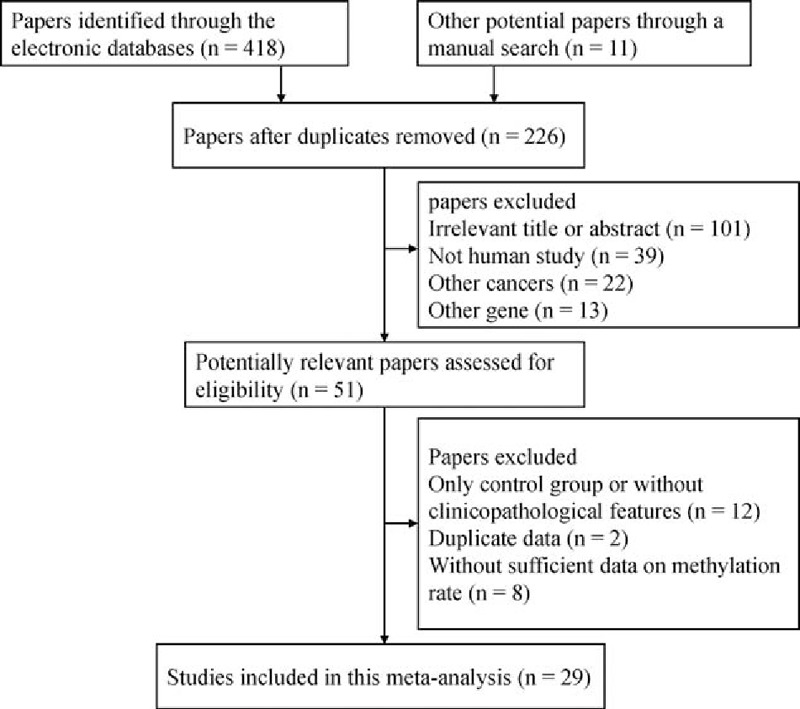
Flow chart of the literature search strategy.
3.2. Correlation between CDH1 hypermethylation and HCC
In a random-effects model (Figs. 2–4), the pooled OR of CDH1 hypermethylation showed that HCC exhibited a significantly different OR than the benign samples, adjacent samples, and normal samples (benign samples: OR = 3.40, 95% CI = 1.35–8.59, P = .01; adjacent samples: OR = 2.42, 95% CI = 1.01–5.83, P = .049; and normal samples: OR = 7.17, 95% CI = 2.80–18.35, P < .001), which demonstrated that CDH1 hypermethylation was significantly associated with an increased risk of HCC.
Figure 2.
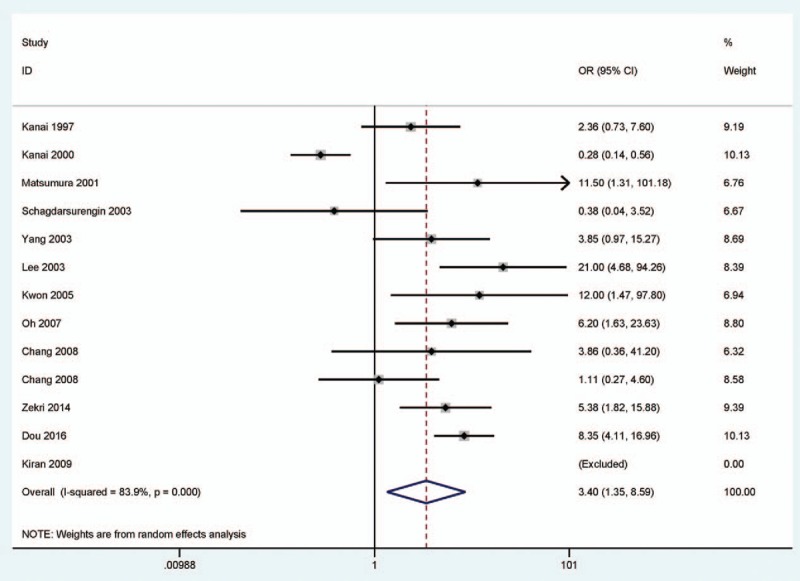
Forest plot for the correlation of CDH1 hypermethylation status from 12 publications showing the pooled OR under a random-effects model in 594 HCC versus 581 benign samples, OR = 3.40, 95% CI = 1.35 to 8.59. CDH1 = E-cadherin, CI = confidence interval, HCC = hepatocellular carcinoma, OR = odds ratio.
Figure 4.
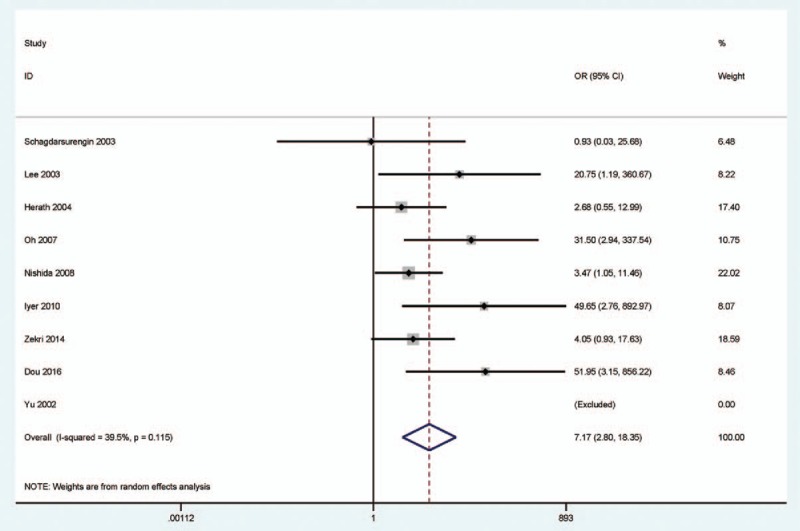
Forest plot for the relationship of CDH1 hypermethylation status from 9 studies showing the pooled OR under a random-effects model in 494 HCC versus 166 normal samples, OR = 7.17, 95% CI = 2.80 to 18.35. CDH1 = E-cadherin, CI = confidence interval, HCC = hepatocellular carcinoma, OR = odds ratio.
Figure 3.
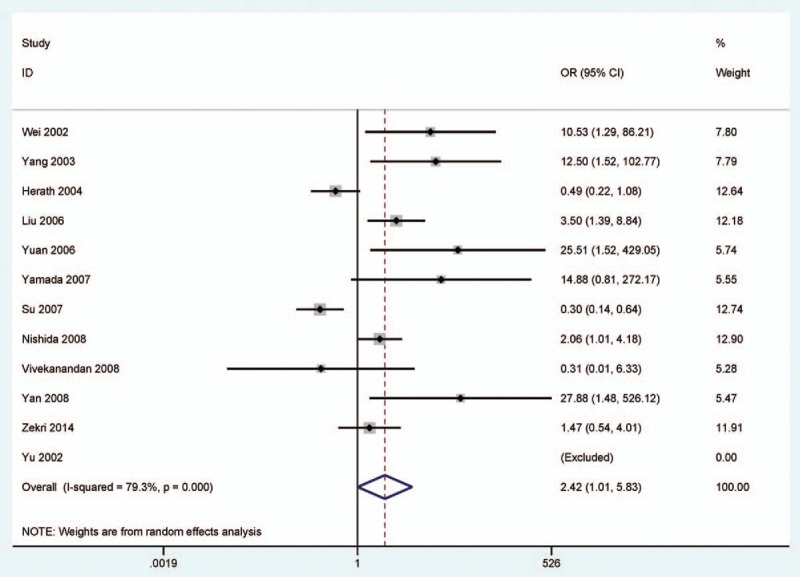
Forest plot for the association of CDH1 hypermethylation status from 12 studies showing the pooled OR under a random-effects model in 598 HCC versus 471 adjacent samples, OR = 2.42, 95% CI = 1.01 to 5.83. CDH1 = E-cadherin, CI = confidence interval, HCC = hepatocellular carcinoma, OR = odds ratio.
3.3. Correlation between the methylation of other CDH genes and HCC
As shown in Table 1, the overall OR from 3 studies involving 125 HCC samples and 125 adjacent tissue samples demonstrated that the level of CDH13 methylation was slightly higher in HCC than adjacent tissues (OR = 4.11, 95% CI = 1.04–16.23, P = .044).
Table 1.
The summary of the ORs for CDH gene methylation in HCC versus controls.
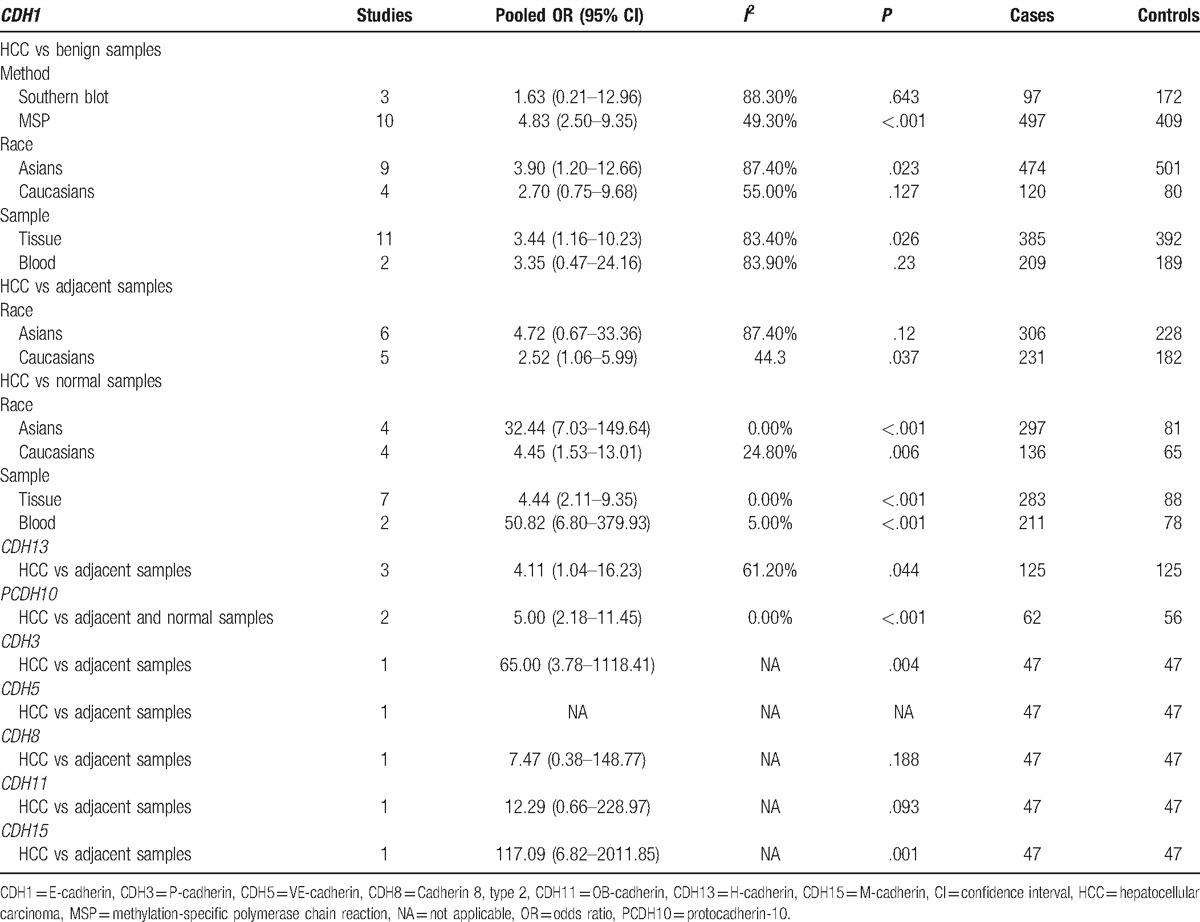
The pooled OR from 2 studies involving 62 HCC tissues and 56 adjacent and normal tissues showed that PCDH10 methylation exhibited a significantly higher OR in HCC than in adjacent and normal tissue samples (OR = 5.00, 95% CI = 2.18–11.45, P < .001).
The pooled OR from 1 study involving 47 HCC tissues and 47 adjacent tissues showed that CDH3 and CDH15 methylation exhibited a higher OR in HCC than in adjacent tissue samples (P < .01), while methylated CDH8 and CDH11 showed a similar OR between HCC and adjacent tissues (P > .05). The CDH5 gene exhibited no methylation in 47 HCC tissues and 47 adjacent tissues.
Our findings showed that methylated CDH13, PCDH10, CDH3, and CDH15 may be associated with an increased risk of HCC. However, the results regarding the methylation of other CDH genes should be carefully considered, as only small sample sizes were included in the current study.
3.4. Subgroup analyses of the CDH1 hypermethylation in cancer and control groups
Subgroup analyses based on ethnicity (Asians and Caucasians), sample types (tissue and blood), and detection methods (Southern blot and methylation-specific polymerase chain reaction [MSP]) were conducted to detect the various correlations with CDH1 hypermethylation under a random-effects model (Table 1).
When HCC samples were compared with benign samples, the results related to ethnicity showed that CDH1 hypermethylation was only significantly associated with HCC in the Asian population (P = .023), and not the Caucasian population (P = .127). The results regarding sample types showed that CDH1 hypermethylation was only significantly associated with HCC in the tissue subgroup (P = .026), and not in the blood subgroup (P = .23). Subgroup analysis based on the testing method demonstrated that CDH1 hypermethylation exhibited a significant association in the MSP subgroup (P < .001), but not in the Southern blot subgroup (P = .643).
When HCC samples were compared with adjacent tissue samples, subgroup analysis of ethnic populations showed that CDH1 hypermethylation was correlated with the Caucasian population subgroup (P = .037), but not with the Asian population subgroup (P = .12).
When HCC samples were compared with normal samples, subgroup analysis of ethnic populations showed that CDH1 hypermethylation presented a significant association in Asians and Caucasians (OR = 32.44, P < .001; OR = 4.45, P = .006, respectively). Subgroup analysis based on sample types showed that CDH1 hypermethylation exhibited a significant association in tissue and blood samples (OR = 4.44; P < .001; OR = 50.82, P < .001, respectively). Although the analysis of blood samples only involved in 2 studies, the obtained OR was significantly higher than that for tissue samples, indicating that CDH1 hypermethylation may be a potential noninvasive biomarker.
3.5. Meta-regression analysis of CDH1 hypermethylation in cancer and control groups
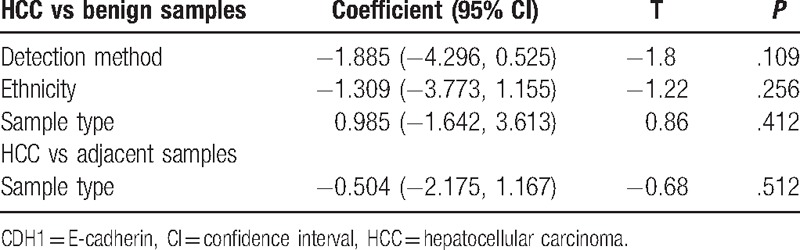
Meta-regression analysis was performed to explain the sources of heterogeneity (Table 2). In the comparison of HCC and benign samples, the results revealed no detectable heterogeneity related to ethnicity, sample type, or testing method (all P > .05). In the comparison of HCC and adjacent tissues, the result of the ethnicity failed to explain the source (P > .05). Therefore, sensitivity analyses were required.
Table 2.
Meta-regression analysis of CDH1 hypermethylation in HCC versus controls.
3.6. Sensitivity analysis of the CDH1 hypermethylation in cancer and controls
Next, a sensitivity analysis was performed to assess the influence of an individual study on the pooled OR and the effect of omitting a single study on its stability. In the comparison of HCC and benign samples, when we removed 2 studies (Kanai et al 2000, Japan;[54] and Schagdarsurengin et al 2003, Germany[50]) and recalculated the pooled OR from the remaining studies (OR = 5.41, 95% CI = 3.25–9.01, P < .001), a very low heterogeneity was observed (I2 = 29.4%). In the comparison of HCC and adjacent tissues, when we deleted 3 studies (Su et al 2007, China[41]; Herath et al 2004, Australia[47]; and Yuan et al 2006, China [42]) and recalculated the overall OR (OR = 3.31, 95% CI = 1.64–6.67, P = .001), a decreased heterogeneity was observed (I2 = 41.8%). The results of the sensitivity analyses showed that the pooled OR for CDH1 hypermethylation was not significantly altered, indicating stability of our results.
3.7. Correlation of CDH1 hypermethylation with clinicopathological features of HCC patients
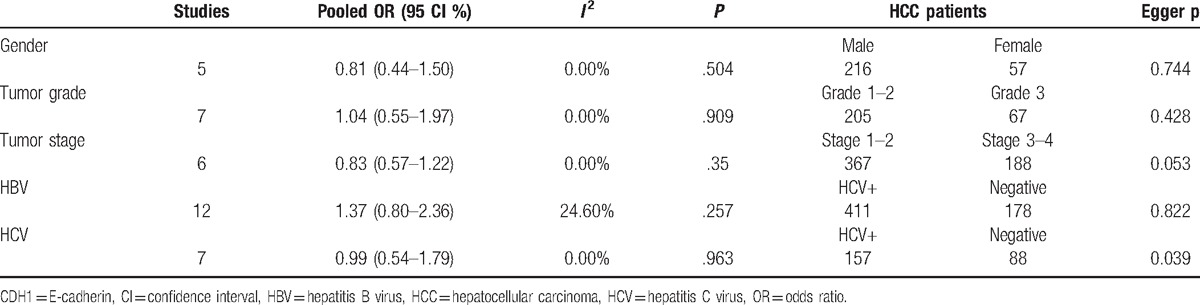
We further determined whether CDH1 hypermethylation was associated with clinicopathological characteristics in HCC patients. Our findings revealed that a hypermethylated CDH1 gene did not present a significant association in relation to gender status, tumor grade, clinical stage, HBV status, or HCV status in HCC (all P > .1) (Table 3).
Table 3.
The association of CDH1 hypermethylation with clinicopathological features in HCC patients.
3.8. Publication bias
Possible publication bias was detected using Egger test (Figure S1 and Table 3). A slight publication bias was only found in relation to HCV status in HCC (P = .039). However, there was no obvious publication bias in the cancer or control groups in relation to gender status, tumor grade, pathological stage, or HBV status in cancer (all P > .05).
4. Discussion
The hypermethylation of TSGs and hypomethylation of oncogenes play crucial roles in the initiation and development of HCC.[56,57] DNA methylation of CpG islands is one of the important causes of the downregulation of gene expression in cancer.[58] Reduction of the expression of the TSG CDH1 may be an important event in cancer invasion and metastasis.[12,59] CDH13 expression in human cancer cells can inhibit their invasion and markedly reduce their proliferation.[60,61]PCDH10 has been demonstrated to act as a TSG in some cancers and plays a key role in proliferation inhibition, apoptosis induction, and invasion repression.[62,63]
However, the results are still inconsistent and controversial. Different methylation rates of the CDH1 gene are reported in different studies, ranging from 0%[51] to 100%.[31] Yu et al[51] reported that the CDH13 gene displayed a similar methylation rate in HCC tissues and adjacent tissues. Two studies reported that the CDH13 methylation rate in HCC was significantly higher than in adjacent tissue samples.[35,39] In addition, we found that the CDH1 gene showed a significantly different methylation frequency in HCC samples than in benign and adjacent samples. For example, 2 studies reported that the CDH1 gene showed a significantly lower methylation rate in HCC than in benign samples.[50,54] Su et al and Herath et al reported that adjacent tissues exhibited a higher methylation frequency of the CDH1 gene than HCC tissue samples.[41,47] Therefore, our study is the first to evaluate the role of CDH gene methylation in HCC risk. Moreover, we are the first to determine whether CDH1 hypermethylation is associated with clinicopathological characteristics in HCC.
The results regarding CDH1 hypermethylation demonstrated that HCC showed a significantly higher OR than that benign, adjacent, and normal samples, suggesting that CDH1 hypermethylation may play a crucial role in the tumorigenesis of HCC. Regarding our finding that the OR of CDH1 hypermethylation was lower in the comparison of HCC samples and adjacent samples (adjacent samples: OR = 2.42; benign samples: OR = 3.40; and normal samples: OR = 7.17), the possible reason may have been impure adjacent specimens contaminated by HCC cells. In addition, methylated CDH13, PCDH10, CDH3, and CDH15 were associated with an increased risk of HCC, while no significant association was found between methylated CDH8 and CDH11 and HCC. However, the results regarding methylated CDH13, PCDH10, CDH3, CDH15, CDH8, and CDH11 should be considered carefully because of the small number of subjects included in this analysis.
Subgroup analyses of CDH1 hypermethylation were performed to identify various associations in the current study. When HCC samples were compared with benign samples, the results related to ethnicity suggested that only Asians were susceptible to CDH1 hypermethylation. Subgroup analysis of sample types showed that the CDH1 gene only exhibited a significant methylation frequency in the tissue subgroup. Subgroup analysis of detection methods revealed that only MSP was a sensitive method of methylation detection for identifying CDH1 hypermethylation. When HCC samples were compared with adjacent tissue samples, an analysis of ethnic populations showed that hypermethylated CDH1 was a susceptibility gene for the Caucasian population. When HCC samples were compared with normal samples, the results regarding race showed that the CDH1 gene was a susceptibility gene for Asians and Caucasians, with the Asian population (OR = 32.44) showing a higher OR than the Caucasian population subgroup (OR = 4.45), suggesting that hypermethylated CDH1 may be a stronger susceptibility gene for Asians. According to subgroup analysis of sample types, there was a significant correlation between hypermethylated CDH1 and HCC in both tissue and blood samples, and the pooled OR for blood samples (OR = 50.82) was significantly higher than that for tissue samples (OR = 4.44), which suggested that hypermethylated CDH1 may be a useful noninvasive biomarker for blood detection. Due to the limitations of the available studies, we could not further evaluate the diagnostic capacity of CDH1 hypermethylation as a promising biomarker for HCC diagnosis. The results should be interpreted with caution, as only small sample sizes were involved, especially in the analysis of the Southern blot subgroup.
Additionally, CDH1 hypermethylation exhibited substantial heterogeneity between the cancer and benign or adjacent samples. Next, a meta-regression analysis was performed to identify the sources of heterogeneity. Our results showed that the meta-regression analysis could not reveal the sources of heterogeneity. Subsequently, sensitivity analyses were performed to evaluate the changes in the pooled OR and heterogeneity by omitting 2 studies[50,54] comparing cancer and benign samples or by deleting 3 studies[41,42,47] comparing cancer and adjacent samples. The results showed that the overall OR of hypermethylated CDH1 was not significantly altered, and no significant heterogeneity was observed, suggesting that the results regarding CDH1 hypermethylation were stable and reliable.
Finally, the relationship of CDH1 hypermethylation with clinicopathological parameters was determined in the present study. Our findings showed that CDH1 hypermethylation was not correlated with gender status, tumor grade, tumor stage, HBV status, or HCV status in HCC.
Several limitations of the present meta-analysis should be acknowledged. First, a slight publication bias was detected in relation to HCV status in cancer, with papers with positive results being more frequently published than papers with negative results. Second, only studies published in English were included in our analyses; potentially high-quality articles published in other languages were excluded because of the limitations of our language abilities, which may have led to selection bias. Third, results for methylated CDH13, PCDH10, CDH3, CDH15, CDH8, and CDH11 that were based on smaller sample sizes were analyzed in the current study; thus, additional studies with larger sample sizes are essential to further validate the obtained results.
In conclusion, our findings suggested that CDH1 hypermethylation is involved in HCC hepatocarcinogenesis. However, CDH1 hypermethylation was not found to be correlated with clinicopathological features. Moreover, methylated CDH13, PCDH10, CDH3, and CDH15 are associated with an increased risk of HCC, while methylated CDH8 and CDH11 were not observed to be associated with a risk of HCC. Hypermethylated CDH1 may become a potential noninvasive biomarker based on blood sample analysis. Further large-scale studies are required to strengthen the findings of our study in the future.
Supplementary Material
Footnotes
Abbreviations: 95% CI = 95% confidence interval, CDH = cadherin, CDH1 = E-cadherin, CDH13 = H-cadherin, CDH3 = P-cadherin, HBV = hepatitis B virus, HCC = hepatocellular carcinoma, HCV = hepatitis C virus, MSP = methylation-specific polymerase chain reaction, OR = odds ratio, PCDH10 = protocadherin-10, TSG = tumor suppressor genes.
Authorship: Conceptualization: TH and GY; methodology: CZ and XF; software: CZ and XF; validation: CZ, XF, and TH; formal analysis: XF; writing-original draft preparation: XF and SZ; writing-review and editing: TH and GY; and revised manuscript: XF.
The authors have no funding and conflicts of interest to disclose.
Supplemental Digital Content is available for this article.
References
- [1].Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87–108. [DOI] [PubMed] [Google Scholar]
- [2].Turdean S, Gurzu S, Turcu M, et al. Current data in clinicopathological characteristics of primary hepatic tumors. Rom J Morphol Embryol 2012;53(3 Suppl):719–24. [PubMed] [Google Scholar]
- [3].El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology 2012;142:1264–73. e1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Chen X, Liu HP, Li M, et al. Advances in non-surgical management of primary liver cancer. World J Gastroenterol 2014;20:16630–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Khan SA, Reddy D, Gupta S. Global histone post-translational modifications and cancer: biomarkers for diagnosis, prognosis and treatment? World J Biol Chem 2015;6:333–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 2012;13:484–92. [DOI] [PubMed] [Google Scholar]
- [7].Stewart DJ. Wnt signaling pathway in non-small cell lung cancer. J Natl Cancer Inst 2014;106: djt356. [DOI] [PubMed] [Google Scholar]
- [8].Ko M, An J, Pastor WA, et al. TET proteins and 5-methylcytosine oxidation in hematological cancers. Immunol Rev 2015;263:6–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Franco R, Schoneveld O, Georgakilas AG, et al. Oxidative stress, DNA methylation and carcinogenesis. Cancer Lett 2008;266:6–11. [DOI] [PubMed] [Google Scholar]
- [10].Bryan RT. Cell adhesion and urothelial bladder cancer: the role of cadherin switching and related phenomena. Philos Trans R Soc Lond Ser B, Biol Sci 2015;370: 20140042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Rajwar YC, Jain N, Bhatia G, et al. Expression and significance of cadherins and its subtypes in development and progression of oral cancers: a review. J Clin Diagn Res 2015;9:ZE05–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].van Roy F. Beyond E-cadherin: roles of other cadherin superfamily members in cancer. Nat Rev Cancer 2014;14:121–34. [DOI] [PubMed] [Google Scholar]
- [13].Takamura M, Yamagiwa S, Matsuda Y, et al. Involvement of liver-intestine cadherin in cancer progression. Med Mol Morphol 2013;46:1–7. [DOI] [PubMed] [Google Scholar]
- [14].Andrews JL, Kim AC, Hens JR. The role and function of cadherins in the mammary gland. Breast Cancer Res 2012;14:203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Tagde A, Rajabi H, Stroopinsky D, et al. MUC1-C induces DNA methyltransferase 1 and represses tumor suppressor genes in acute myeloid leukemia. Oncotarget 2016;7:38974–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Liu X, Chu KM. E-cadherin and gastric cancer: cause, consequence, and applications. Biomed Res Int 2014;2014:637308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Liu J, Lian Z, Han S, et al. Downregulation of E-cadherin by hepatitis B virus X antigen in hepatocellullar carcinoma. Oncogene 2006;25:1008–17. [DOI] [PubMed] [Google Scholar]
- [18].Andreeva AV, Kutuzov MA. Cadherin 13 in cancer. Genes Chromosomes Cancer 2010;49:775–90. [DOI] [PubMed] [Google Scholar]
- [19].Deng J, Liang H, Ying G, et al. Clinical significance of the methylated cytosine-phosphate-guanine sites of protocadherin-10 promoter for evaluating the prognosis of gastric cancer. J Am Coll Surg 2014;219:904–13. [DOI] [PubMed] [Google Scholar]
- [20].Narayan G, Xie D, Freddy AJ, et al. PCDH10 promoter hypermethylation is frequent in most histologic subtypes of mature lymphoid malignancies and occurs early in lymphomagenesis. Genes Chromosomes Cancer 2013;52:1030–41. [DOI] [PubMed] [Google Scholar]
- [21].Broussard EK, Kim R, Wiley JC, et al. Identification of putative immunologic targets for colon cancer prevention based on conserved gene upregulation from preinvasive to malignant lesions. Cancer Prev Res (Phila) 2013;6:666–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Paredes J, Albergaria A, Oliveira JT, et al. P-cadherin overexpression is an indicator of clinical outcome in invasive breast carcinomas and is associated with CDH3 promoter hypomethylation. Clin Cancer Res 2005;11:5869–77. [DOI] [PubMed] [Google Scholar]
- [23].Moher D, Liberati A, Tetzlaff J, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med 2009;6:e1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zintzaras E, Ioannidis JP. HEGESMA: genome search meta-analysis and heterogeneity testing. Bioinformatics 2005;21:3672–3. [DOI] [PubMed] [Google Scholar]
- [25].Higgins JP, Thompson SG, Deeks JJ, et al. Measuring inconsistency in meta-analyses. BMJ 2003;327:557–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Lau J, Ioannidis JP, Schmid CH. Quantitative synthesis in systematic reviews. Ann Intern Med 1997;127:820–6. [DOI] [PubMed] [Google Scholar]
- [27].Egger M, Davey Smith G, Schneider M, et al. Bias in meta-analysis detected by a simple, graphical test. BMJ 1997;315:629–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Dou CY, Fan YC, Cao CJ, et al. Sera DNA methylation of CDH1, DNMT3b and ESR1 promoters as biomarker for the early diagnosis of hepatitis B virus-related hepatocellular carcinoma. Dig Dis Sci 2016;61:1130–8. [DOI] [PubMed] [Google Scholar]
- [29].Zekri AR, Bahnasy AA, Shoeab FE, et al. Methylation of multiple genes in hepatitis C virus associated hepatocellular carcinoma. J Adv Res 2014;5:27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Fang S, Huang SF, Cao J, et al. Silencing of PCDH10 in hepatocellular carcinoma via de novo DNA methylation independent of HBV infection or HBX expression. Clin Exp Med 2013;13:127–34. [DOI] [PubMed] [Google Scholar]
- [31].Kiran M, Chawla YK, Kaur J. Methylation profiling of tumor suppressor genes and oncogenes in hepatitis virus-related hepatocellular carcinoma in northern India. Cancer Genet Cytogenet 2009;195:112–9. [DOI] [PubMed] [Google Scholar]
- [32].Iyer P, Zekri AR, Hung CW, et al. Concordance of DNA methylation pattern in plasma and tumor DNA of Egyptian hepatocellular carcinoma patients. Exp Mol Pathol 2010;88:107–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Ko E, Kim Y, Kim SJ, et al. Promoter hypermethylation of the p16 gene is associated with poor prognosis in recurrent early-stage hepatocellular carcinoma. Cancer Epidemiol Biomarkers Prev 2008;17:2260–7. [DOI] [PubMed] [Google Scholar]
- [34].Chang H, Yi B, Li L, et al. Methylation of tumor associated genes in tissue and plasma samples from liver disease patients. Exp Mol Pathol 2008;85:96–100. [DOI] [PubMed] [Google Scholar]
- [35].Chan DW, Lee JM, Chan PC, et al. Genetic and epigenetic inactivation of T-cadherin in human hepatocellular carcinoma cells. Int J Cancer 2008;123:1043–52. [DOI] [PubMed] [Google Scholar]
- [36].Yan Q, Zhang ZF, Chen XP, et al. Reduced T-cadherin expression and promoter methylation are associated with the development and progression of hepatocellular carcinoma. Int J Oncol 2008;32:1057–63. [PubMed] [Google Scholar]
- [37].Vivekanandan P, Torbenson M. Epigenetic instability is rare in fibrolamellar carcinomas but common in viral-associated hepatocellular carcinomas. Mod Pathol 2008;21:670–5. [DOI] [PubMed] [Google Scholar]
- [38].Nishida N, Nagasaka T, Nishimura T, et al. Aberrant methylation of multiple tumor suppressor genes in aging liver, chronic hepatitis, and hepatocellular carcinoma. Hepatology 2008;47:908–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Yamada S, Nomoto S, Fujii T, et al. Frequent promoter methylation of M-cadherin in hepatocellular carcinoma is associated with poor prognosis. Anticancer Res 2007;27(4B):2269–74. [PubMed] [Google Scholar]
- [40].Oh BK, Kim H, Park HJ, et al. DNA methyltransferase expression and DNA methylation in human hepatocellular carcinoma and their clinicopathological correlation. Int J Mol Med 2007;20:65–73. [PubMed] [Google Scholar]
- [41].Su PF, Lee TC, Lin PJ, et al. Differential DNA methylation associated with hepatitis B virus infection in hepatocellular carcinoma. Int J Cancer 2007;121:1257–64. [DOI] [PubMed] [Google Scholar]
- [42].Yuan Y, Wang J, Li J, et al. Frequent epigenetic inactivation of spleen tyrosine kinase gene in human hepatocellular carcinoma. Clin Cancer Res 2006;12:6687–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Cui X, Wakai T, Shirai Y, et al. Arsenic trioxide inhibits DNA methyltransferase and restores methylation-silenced genes in human liver cancer cells. Hum Pathol 2006;37:298–311. [DOI] [PubMed] [Google Scholar]
- [44].Katoh H, Shibata T, Kokubu A, et al. Epigenetic instability and chromosomal instability in hepatocellular carcinoma. Am J Pathol 2006;168:1375–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Ying J, Li H, Seng TJ, et al. Functional epigenetics identifies a protocadherin PCDH10 as a candidate tumor suppressor for nasopharyngeal, esophageal and multiple other carcinomas with frequent methylation. Oncogene 2006;25:1070–80. [DOI] [PubMed] [Google Scholar]
- [46].Kwon GY, Yoo BC, Koh KC, et al. Promoter methylation of E-cadherin in hepatocellular carcinomas and dysplastic nodules. J Korean Med Sci 2005;20:242–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Herath NI, Walsh MD, Kew MC, et al. Cadherin/catenin complex appears to be intact in hepatocellular carcinomas from Australia and South Africa. J Gastroenterol Hepatol 2004;19:676–82. [DOI] [PubMed] [Google Scholar]
- [48].Lee S, Lee HJ, Kim JH, et al. Aberrant CpG island hypermethylation along multistep hepatocarcinogenesis. Am J Pathol 2003;163:1371–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Yang B, Guo M, Herman JG, et al. Aberrant promoter methylation profiles of tumor suppressor genes in hepatocellular carcinoma. Am J Pathol 2003;163:1101–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Schagdarsurengin U, Wilkens L, Steinemann D, et al. Frequent epigenetic inactivation of the RASSF1A gene in hepatocellular carcinoma. Oncogene 2003;22:1866–71. [DOI] [PubMed] [Google Scholar]
- [51].Yu J, Ni M, Xu J, et al. Methylation profiling of twenty promoter-CpG islands of genes which may contribute to hepatocellular carcinogenesis. BMC Cancer 2002;2:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Wei Y, Van Nhieu JT, Prigent S, et al. Altered expression of E-cadherin in hepatocellular carcinoma: correlations with genetic alterations, beta-catenin expression, and clinical features. Hepatology 2002;36:692–701. [DOI] [PubMed] [Google Scholar]
- [53].Matsumura T, Makino R, Mitamura K. Frequent down-regulation of E-cadherin by genetic and epigenetic changes in the malignant progression of hepatocellular carcinomas. Clin Cancer Res 2001;7:594–9. [PubMed] [Google Scholar]
- [54].Kanai Y, Ushijima S, Tsuda H, et al. Aberrant DNA methylation precedes loss of heterozygosity on chromosome 16 in chronic hepatitis and liver cirrhosis. Cancer Lett 2000;148:73–80. [DOI] [PubMed] [Google Scholar]
- [55].Kanai Y, Ushijima S, Hui AM, et al. The E-cadherin gene is silenced by CpG methylation in human hepatocellular carcinomas. Int J Cancer 1997;71:355–9. [DOI] [PubMed] [Google Scholar]
- [56].Nishida N, Kudo M. Oxidative stress and epigenetic instability in human hepatocarcinogenesis. Digest Dis 2013;31:447–53. [DOI] [PubMed] [Google Scholar]
- [57].Tischoff I, Tannapfe A. DNA methylation in hepatocellular carcinoma. World J Gastroenterol 2008;14:1741–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Yoshiura K, Kanai Y, Ochiai A, et al. Silencing of the E-cadherin invasion-suppressor gene by CpG methylation in human carcinomas. Proc Natl Acad Sci U S A 1995;92:7416–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Oka H, Shiozaki H, Kobayashi K, et al. Expression of E-cadherin cell adhesion molecules in human breast cancer tissues and its relationship to metastasis. Cancer Res 1993;53:1696–701. [PubMed] [Google Scholar]
- [60].Kuphal S, Martyn AC, Pedley J, et al. H-cadherin expression reduces invasion of malignant melanoma. Pigment Cell Melanoma Res 2009;22:296–306. [DOI] [PubMed] [Google Scholar]
- [61].Lee SW, Reimer CL, Campbell DB, et al. H-cadherin expression inhibits in vitro invasiveness and tumor formation in vivo. Carcinogenesis 1998;19:1157–9. [DOI] [PubMed] [Google Scholar]
- [62].Yu J, Cheng YY, Tao Q, et al. Methylation of protocadherin 10, a novel tumor suppressor, is associated with poor prognosis in patients with gastric cancer. Gastroenterology 2009;136:640–51. e641. [DOI] [PubMed] [Google Scholar]
- [63].Ying J, Gao Z, Li H, et al. Frequent epigenetic silencing of protocadherin 10 by methylation in multiple haematologic malignancies. Br J Haematol 2007;136:829–32. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


