Abstract
One of the defining characteristics of human and animal viruses is their ability to suppress host antiviral responses. Viruses express proteins that impair the detection of viral nucleic acids by host pattern-recognition receptors, block signaling pathways that lead to the synthesis of type I interferons (IFN) and other cytokines, or prevent the activation of virus-induced genes. We have identified a novel mechanism of virus-mediated suppression of antiviral gene expression that relies on the presence of histone-like sequences (histone mimics) in viral proteins. We describe how viral histone mimics can interfere with key regulators of gene expression and contribute to the suppression of antiviral responses. We will also describe how viral histone mimics can facilitate the identification of novel mechanisms of antiviral gene regulation and lead to the development of drugs that employ histone mimicry for interference with gene expression during diseases.
Introduction
Viral infection leads to a rapid onset of two major mutually antagonistic processes. Encounter of human or mouse cells with virus results in rapid activation of intracellular signaling networks that trigger expression of genes responsible for the elimination of the virus and the establishment of lasting antiviral immunity (Katze et al. 2002; Garcia-Sastre and Biron 2006; Kawai and Akira 2006). In turn, viruses can target the antiviral signaling events for the benefit of virus replication and long-term virus propagation (Katze et al. 2002; Garcia-Sastre and Biron 2006; Kawai and Akira 2006).
In the nucleus, the virus-host antagonism evolves around key regulatory processes that govern gene transcription and DNA replication. The structural and non-structural proteins of numerous viruses accumulate in the nuclei of infected cells, where they interact with the distinct chromatin proteins, including histones, transcription factors as well as regulators of DNA repair, cell cycle transition, mRNA splicing, export and catabolism (Nemeroff et al. 1998; Ballestas et al. 1999; Friborg et al. 1999; Fujimuro et al. 2003; Krug et al. 2003; You et al. 2004; Zhao and Elder 2005; Satterly et al. 2007; Ashour et al. 2009; Colpitts et al. 2011; Fonseca et al. 2012; Banerjee et al. 2013).
One of the most common anti-host viral strategies involves the disruption of the host protein-protein interactions by viral mimics of cellular proteins (Elde and Malik 2009; Davey et al. 2011; Tarakhovsky 2013). In particular, the inherent functions and properties of host signaling and regulatory networks make them extremely attractive targets for viral mimic-derived interference. For one, such networks typically rely on highly specific and dynamic protein-protein interactions to be able to quickly transmit information within the cell (Smock and Gierasch 2009; Stein et al. 2009). This is in part supported by the utilization of low affinity protein-protein interactions that involve compact (3–20 amino acid long), degenerate and evolvable modules that have been defined as Short Linear Motifs (SLIMs) (Diella et al. 2008; Davey et al. 2012; Edwards et al. 2012; Van Roey et al. 2012; Weatheritt et al. 2012a; Weatheritt et al. 2012b; Van Roey et al. 2013). Most of the binding specificity and affinity of a SLiM is embedded within a 2–5 residue-long core of amino acids. This paucity of amino acids limits the strength of SLiM-mediated interactions to a relatively low affinity range (1–150 μM) that supports transiency and reversibility of SLiM mediated protein-protein binding (Diella et al. 2008; Davey et al. 2012; Edwards et al. 2012; Van Roey et al. 2012; Weatheritt et al. 2012a; Weatheritt et al. 2012b; Van Roey et al. 2013; Dinkel et al. 2014).
The described features of SLiMs match well the features of the short protein binding sequences within the amino-terminal (N-terminal) domains of histone proteins (histone tail). The histone tail could be viewed as a collection of multiple overlapping SLiMs, with each motif (and its modification state) functioning as a discrete unit of information for histone-binding proteins (Fischle et al. 2003; Ruthenburg et al. 2007; Taverna et al. 2007). This feature of the histone tail may contribute to its unique capability to facilitate binding of numerous non-histone proteins. However, the minimal size of the interacting histone sequences can potentially compromise the specificity. Therefore, it is tempting to speculate that histone tail-dependent formation of the chromatin complexes relies on a kinetic proofreading mechanism that has been initially introduced by Hopfield (Hopfield 1974) and Ninio (Ninio 1975) as a mechanism for error correction in biochemical processes. The proofreading mechanism connects the small differences in binding energy of interacting partners and the amount of extra free energy required to generate the desirable reaction product. When undesirable reactants participate in a reaction, free energy consumption drives molecular circuits in cycles before completing the reaction (Hopfield 1974; Ninio 1975). It is plausible that the shortness of the histone tail and, even more, the minimal size of the functionally distinct histone tail sub-domains, e.g. H3K4 vs H3K9 vs H3K27, increases the length of time and amount of energy required for formation of functional transcription complexes. While it is important to keep the error rates of transcription complex assembly low, the potential multiplicity of the error-correcting cycles may increase the potentially damaging effect of “foreign” histone-like sequences that, by competing with cognate histone sequences, may prolong proofreading at the expense of cell fitness. In essence, while highly useful for the diversity of the chromatin complexes, the paucity of the amino acid sequence within the histone tail may create an opportunity for interference with the histone-based chromatin complexes by endogenous or exogenous histone-like SLiMs.
Here we provide evidence for the existence and functional relevance of viral histone mimics. We also show that synthetic compounds that resemble histone mimics are able to interfere with the formation of transcription protein complexes and gene expression in cells of various types. Our studies suggest that targets of pathogen-derived or synthetic histone mimics can lead to identification of critical regulators of gene expression associated with inflammatory and viral diseases.
Results and Discussion
The endogenous and exogenous histone mimics
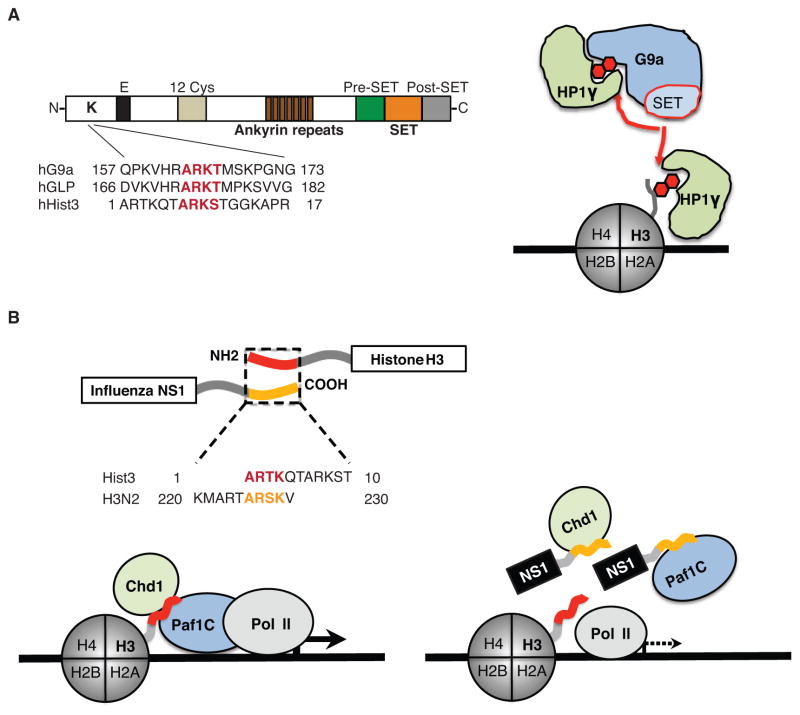
The existence of the histone–like SLiMs (histone mimics) in non-histone proteins has been initially demonstrated in our earlier findings that show the presence of the histone H3–like sequence within the histone methyltransferase G9a that catalyzes di-methylation at lysine 9 of histone H3 (Sampath et al. 2007). G9a bears a 163-ARKT-166 motif that strongly resembles the 7-ARKS-10 motif of its target H3 target residue (Figure 1A, left). Consistent with the presence of the histone mimic, G9a can auto-methylate itself on lysine 165 (Sampath et al. 2007) and this methylation facilitates the formation of the G9a bound repressor complex with chromodomain-containing protein, HP1γ (Figure 1A, right) (Sampath et al. 2007). The H3-like sequence in G9a is conserved in its homologue and hetero-dimerization partner GLP (Figure 1A, left) (Tachibana et al. 2005; Sampath et al. 2007), although the two proteins share relatively poor primary sequence conservation in their N-terminal domains. The initial discovery of the histone mimics in G9a/GLP led to identification of numerous histone mimics in nuclear and non-nuclear proteins, where these mimics contribute to protein-protein interaction or protein stability (Lee et al. 2010; Donlin et al. 2012; Lee et al. 2012; Badeaux and Shi 2013; Shi et al. 2014).
Figure 1. The structure and function of histone mimics.
A. The methyltransferases G9a and GLP possess a functional histone mimic. The histone-like sequence (red letters) is localized within the N-terminal domain of the proteins (Sampath et al. 2007). The G9a histone mimic methylation (red hexagon) is mediated in cis by the catalytic SET domain (Sampath et al. 2007) that is flanked by pre- and post-SET domains. The ankyrin repeat domain is involved in G9a interaction with methylated histones (Collins et al. 2008), and the methylated histone mimic in G9a (red hexagon) binds to the chromodomain-containing protein HP1γ that can also interact with methylated histone H3 (red hexagon). B. The NS1 proteins of the influenza A H3N2 virus possess a functional histone H3K4-like sequence. The histone-like sequence of NS1 (yellow letters) is localized within the non-structured carboxy-terminus of the protein whereas the homologous H3 sequence (red letters) is localized within the amino-terminus of histone H3 (Marazzi et al. 2012). The NS1 histone mimic (yellow tail) is present in the nucleus (Greenspan et al. 1988) where it interacts with Paf1 and Chd1 proteins. Interaction with Chd1 depends on NS1 lysine methylation, whereas Paf1 can bind to the unmethylated or methylated NS1 histone mimic. The pattern of Paf1 and Chd1 binding to NS1 is similar to these protein interactions with histone H3. The schematic model describes a putative mechanism of NS1 interference with Paf1-mediated transcription of virus-induced genes.
The compactness and intrinsic evolutionary plasticity of SLiMs, including those that are present within histones, make them easy to imitate, thus enabling pathogen-derived SLiMs to compete with the host counterparts. The viral histone mimicry has been initially demonstrated by identification of the histone H3-like sequence within the carboxy-terminal (C-terminal) portion of the non-structural protein 1 (NS1) derived from the H3N2 subtype of influenza A virus (Marazzi et al. 2012). We found that the C-terminal domain of the NS1 protein carries the sequence 226-ARSK-229 (yellow) that resembles the first 4 amino acids of the histone H3 protein 1-ARTK-4 (red; Figure 1B, top). Strikingly, both of these sequences were localized to the unstructured terminal domains of their respective proteins, indicating that the NS1 histone-like sequence is present within a similarly disordered structural context as the histone H3 tail.
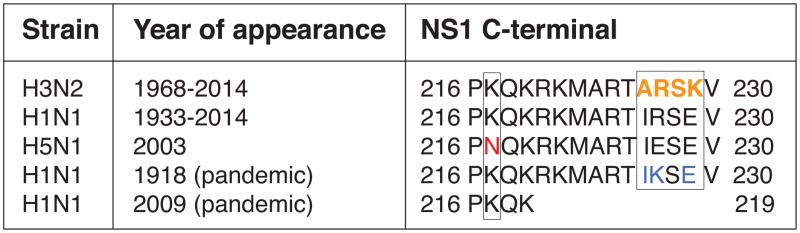
The NS1 protein plays a key role in virus propagation by suppressing the type I IFN response during influenza infection (Garcia-Sastre et al. 1998; Wang et al. 2000). Interestingly, amongst the 2753 unique full-length NS1 sequences obtained from the Influenza Virus Resource (Bao et al. 2008), the H3N2 subtype of viruses appears to be the predominant carrier of the bona fide histone H3K4-like sequence. In contrast, “tails” of NS1 proteins from other influenza subtypes vary significantly and display either no recognizable motifs or possess highly specific motifs such as PDZ ligand (PL) motifs (Obenauer et al. 2006; Jackson et al. 2008; Thomas et al. 2011) or the sumoylation sequence within the NS1 tail of the H1N1 (1918) influenza A virus (Table 1) (Santos et al. 2013).
Table 1. The heterogeneity of the NS1 histone-like tails in influenza A subtypes.
The NS1 tails of the displayed influenza A subtypes vary at amino acid positions 217 and 226–229 (boxed). The bona fide H3K4-like sequence in NS1 of the influenza A H3N2 virus (226–229) is shown in yellow. The sumoylation sequence within the NS1 tail of the influenza A H1N1 1918 virus is shown in blue.
This NS1 tail diversity may contribute to unique features of the individual virus subtypes. The NS1 C-terminal tail domain has previously been implicated as a virulence factor for some avian influenza A viral isolates (Jackson et al. 2008). The PL motifs ESEV and EPEV, found in the NS1 C-terminus from multiple avian-derived influenza strains, have been associated with several highly-pathogenic human isolates such as H5N1 1997 and H5N1 2003 respectively (Obenauer et al. 2006; Jackson et al. 2008; Thomas et al. 2011). These PL motifs enable viral interference with the apoptotic death of the infected cells, thus contributing to viral virulence (Golebiewski et al. 2011). Contrary to the avian viruses, the commonly circulating and relatively low-pathogenic human viruses predominantly contain PL motif sequences RSKV (H3N2 isolates) or RSEV (H1N1 isolates) that do not bind to known PDZ domain containing proteins (Obenauer et al. 2006; Thomas et al. 2011). Furthermore, the overall loss of the NS1 C-terminal tail from low-pathogenic viruses resulted in major attenuation of the virus (Jackson et al. 2008).
In the H3N2 strain, the presence of the histone mimic may contribute to the unique ability of the virus to compete with the cognate histone sequences for their common binding partners, including those that play an important role in regulation of antiviral gene expression. In support of this model, we found that both the histone H3 tail and the H3N2 NS1 tail bind in a sequence-dependent fashion to the Paf1 component of the polymerase-associated factor 1 complex (Paf1C) that contributes to RNA elongation, as well as other co-transcriptional processes (Marazzi et al. 2012). As expected, the NS1 tails from influenza H5N1 (Marazzi et al. 2012) and H1N1 (Marazzi and Tarakhovsky, unpublished data) that do not carry the histone mimic did not bind to Paf1.
Overall, the NS1 tail could be viewed as a highly interactive internally disorganized motif, where amino acid variations contribute to target specificity and hence to the unique features of individual virus subtypes. One of the extreme extensions of this model is that the diversity of the entire NS1 tails collection of the global influenza pool matches the diversity of the histone tails, as well as other “tail”–like disordered structures in other host proteins. This feature of NS1 may support the influenza subtypes ability to choose their optimal strategy for the host-pathogen interaction, based on the genetic and epigenetic state of the infected host during the emergence of infection. In turn, the counter-selection against certain viral subtypes, including some of the NS1 tail-less viruses, could be explained by the inability of NS1 to match the host factor(s), including those localized in the cell nucleus. Most provoking could be a consideration of the existence of the unique host-binding partner(s) for the sumoylated NS1 of the 1918 Spanish flu that was “removed” from the human population due to the exceptionally high morbidity of the Spanish influenza.
Viral “histone mimics” reveal novel regulators of antiviral response
NS1 binding to Paf1, while apparently specific for H3N2, may offer a clue for understanding of the general mechanism that governs antiviral transcriptional responses. Paf1 is an obligate member of the multi-protein Paf1 complex that contributes to RNA elongation (Kim et al. 2010) in addition to other co-transcriptional processes. The latter include transcription-coupled histone modifications (Wood et al. 2003; Kim and Roeder 2009; Hahn et al. 2012) and regulation of mRNA 3′ end processing (Mueller et al. 2004; Penheiter et al. 2005; Nordick et al. 2008; Rozenblatt-Rosen et al. 2009; Nagaike et al. 2011). In particular, the Paf1C has been shown to impact monoubiquitylation of histone H2BK120 in human cells, by promoting recruitment of the ubiquitin-conjugating enzyme Rad6 and the ubiquitin protein ligase Bre1 to chromatin (Wood et al. 2003; Kim and Roeder 2009). Rad6/Bre1 mediated monoubiquitylation of H2BK120 facilitates H3K4 tri-methylation and H3K79 di-methylation by the Set1/MLL-1 and Dot1 complexes respectively (Dover et al. 2002; Sun and Allis 2002; Kim and Roeder 2009; Nakanishi et al. 2009).
By binding to Paf1 the NS1 protein has the potential to interfere with multiple transcriptional processes, including those that support antiviral gene expression (Figure 1B, bottom). Indeed, truncation of the Paf1-binding NS1 sequence decreased virus ability to suppress antiviral gene expression and attenuated influenza infectivity in vitro (Marazzi et al. 2012). Conversely, the association with NS1 points to an important role of Paf1C in antiviral transcriptional response.
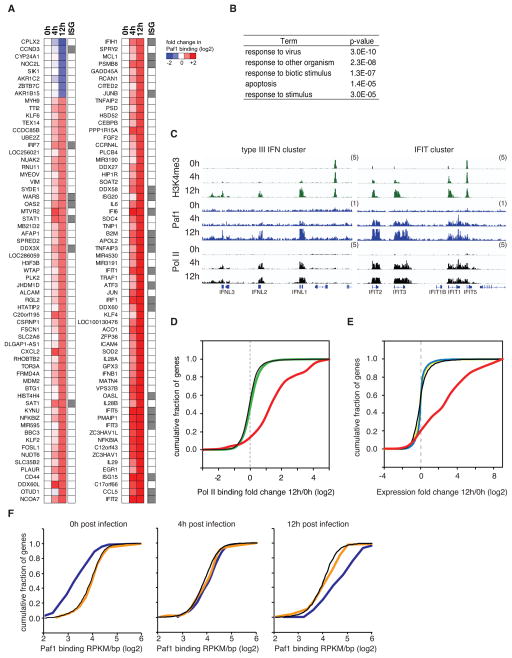
In a course of cell infection with the influenza virus that lacks NS1 and hence does not interfere with Paf1 function, Paf1 is recruited de novo to approximately 200 genes associated with antimicrobial and antiviral response, including numerous type I/III IFN stimulated genes (ISGs; Figures 2A–C). The virus-induced recruitment of Paf1 correlates with recruitment of RNA Poll II and an increase in Paf1 bound gene expression levels (Figures 2C–E). Accordingly, the siRNA mediated knock-down of Paf1 suppressed selectively the expression of genes that recruit Paf1 during infection, but did not affect the expression of the majority of the approximately 10,000 genes that bind Paf1 in a non-inducible fashion (Figure 2F).
Figure 2. Virus infection leads to Paf1 recruitment to the virus activated gene loci.
A. A549 lung epithelia cells were infected with PR8/ΔNS1 virus that due to the lack of NS1 induces strong antiviral responses. The virus infection resulted in time-dependent increase (red) or decrease (blue) in Paf1 binding. The heatmap shows relative abundance of Paf1 binding to the gene transcriptional start sites (TSS) in control or infected cells as determined by ChIP-sequencing. Grey boxes at the right of the heat map indicate known interferon stimulated genes (ISGs) (Schoggins et al. 2011). B. The table shows the top five functional categories associated with genes that display > 2-fold change in Paf1 binding at 12 hours (h) after infection. C. Virus infection increases Paf1 abundance at key antiviral genes. Binding of H3K4me3 (green), Paf1 (blue) and Pol II (black) at virus-induced genes in non-infected cells (0 hours) and PR8/ΔNS1-infected cells at 4 hours and 12 hours post infection is shown. The y-axes represent the average number of tags per gene per 25 base pairs per 1,000,000 mapped reads. Scale values are indicated in parentheses. D. Virus induced Paf1 binding correlates with recruitment of RNA Pol II to antiviral gene loci. The plot shows the cumulative distribution function (CDF) of changes in Pol II binding to different groups of Pol II or Paf1-bound genes at 12 hours post infection with influenza PR8/ΔNS1. Genes that change Paf1 binding upon infection: red line; all genes that bind Paf1: black line; all genes that bind Pol II: green line. The broken grey line indicates a fold change of 1. E. Virus induced Paf1 binding correlates with up-regulation of antiviral gene expression. The plot shows the CDF of changes in gene expression for different groups of Paf1-associated genes at 12 hours post infection with influenza PR8/ΔNS1. Genes that change Paf1 binding upon infection: red line; all genes that bind Paf1: black line; genes that do not bind to Paf1: blue line; all genes: yellow line. The broken grey line indicates a fold change of 1. F. Paf1 deficiency down-regulates selectively expression of genes that recruit Paf1 during infection. The plot shows the CDF of Paf1 binding levels (log2 RPKM/bp) in non-infected A549 cells (left), in A549 cells 4 hours post infection with influenza PR8/ΔNS1 (middle), or 12 hours post infection (right) for genes that were suppressed by Paf1 knockdown (dark blue line), up-regulated by Paf1-deficiency (orange line), or for all genes (black line).
The ability of the NS1 histone mimic to interact with host nuclear proteins could be expanded by its post-translational modification. Similar to histone H3, the lysine residue within the NS1 histone mimic could be acetylated or methylated in influenza infected cells (Marazzi et al. 2012). While this methylation appears to have no obvious impact on the NS1–Paf1 interaction, the lysine methylation of NS1 supported its interaction with the chromatin remodeling protein Chd1 (Marazzi et al. 2012). Moreover, the affinity of the methylated NS1 interaction with Chd1 is nearly equal to the affinity of Chd1 binding to the methylated lysine 4 of histone H3. Despite tight association of Chd1 with NS1, the siRNA-mediated deficiency of Chd1 had no effect on virus-induced gene expression. However, combined Paf1 and Chd1 deficiencies resulted in a nearly complete abrogation of antiviral gene expression and 50-fold increase in influenza virus replication in vitro (data not shown). The major negative and cumulative effect of Chd1 and Paf1 deficiencies on antiviral defense may reflect the Chd1 and Paf1C interaction (Simic et al. 2003) that has been implicated in control of pre-mRNA splicing (Sims et al. 2007). NS1 interaction with Chd1 may provide the virus with the access to the host splicing machinery and contribute to viral mRNA splicing and hence to viral replication (Chua et al. 2013).
The Paf1C and associated proteins contribute to replication of the human adenovirus (HAdV). During HAdV infection, E1A recruits the Paf1C to the viral genome via Bre1, thus promoting viral early gene expression (Fonseca et al. 2013). The interaction of E1A with Bre1 was also shown to inhibit the expression of IFN and ISGs, suggesting that Rad6/Bre1 mediated ubiquitylation of H2BK120 was important for the expression of these genes (Fonseca et al. 2012). Given that the Paf1 complex also facilitates Rad6/Bre1 activity, these data support the notion that the Paf1C and its related activities are key regulators of inflammatory response. Binding of the HIV protein TAT to Paf1C has also been implicated in generating the permissive transcription environment for HIV replication (Sobhian et al. 2010). Reduced levels of HIV pro-viral integration in cells that over-express Paf1 highlighted the importance of Paf1 expression in the HIV viral life cycle (Liu et al. 2011).
Functional similarity between synthetic and natural histone mimics
The important role of Paf1C in up-regulation of virus-inducible gene expression corroborates well with earlier data that showed the key role of transcriptional elongation in inflammatory gene expression. RNA elongation depends greatly on the signal-induced lysine acetylation of histone H3 and H4 at gene promoters followed by the acetylated histone association with bromodomain and ET domain (BET) family of proteins (Hargreaves et al. 2009). The interaction between BET and acetylated histones is mediated by the evolutionary conserved ~ 110aa long bromodomain, which is present in each of the two tandem arranged modules (BDI and BDII) in BRD4, as well as related BRDT, BRD2 and BRD3 proteins of the BET family (Dey et al. 2000; Dey et al. 2003; Kanno et al. 2004; Huang et al. 2007). The mechanism by which individual BETs contribute to gene regulation is not fully understood. BRD4 is the only BET protein that links acetylated chromatin to the P-TEFb complex that mediates the signal-induced phosphorylation and activation of RNA Pol II (Jang et al. 2005; Yang et al. 2005; Hargreaves et al. 2009; Zhou et al. 2012). The BET proteins can also contribute to RNA synthesis by a bromodomain independent association with Paf1C as well as with numerous effector proteins such as NSD3, a SET domain-containing histone methyltransferase; JMJD6, a histone arginine demethylase; or Chd4, a catalytic component of the NuRD nucleosome remodeling complex (Rahman et al. 2011; Liu et al. 2013).
We were the first to demonstrate the powerful anti-inflammatory effect of synthetic antagonists of BET bromodomains (Nicodeme et al. 2010). The pharmacological inhibitor of BET proteins that we named I-BET, binds to the acetylated lysine-binding pockets of BET proteins and prevents these proteins from interacting with their cognate acetyl-lysine ligands (Nicodeme et al. 2010). In addition to suppression of inflammatory genes, the inhibitor of BET proteins (I-BET) possess a remarkable capacity to inhibit expression of numerous inducible genes, including those that are driven by transcription factors such as c-Myc in tumor cells (Dawson et al. 2011; Delmore et al. 2011; Zuber et al. 2011; Loven et al. 2013), NFkB in macrophages (Nicodeme et al. 2010; Belkina et al. 2013) or OCA-B in B cells (Chapuy et al. 2013). In a certain sense, I-BET appears as a chemical equivalent of a histone SLiM and hence could be qualified as a synthetic histone mimic.
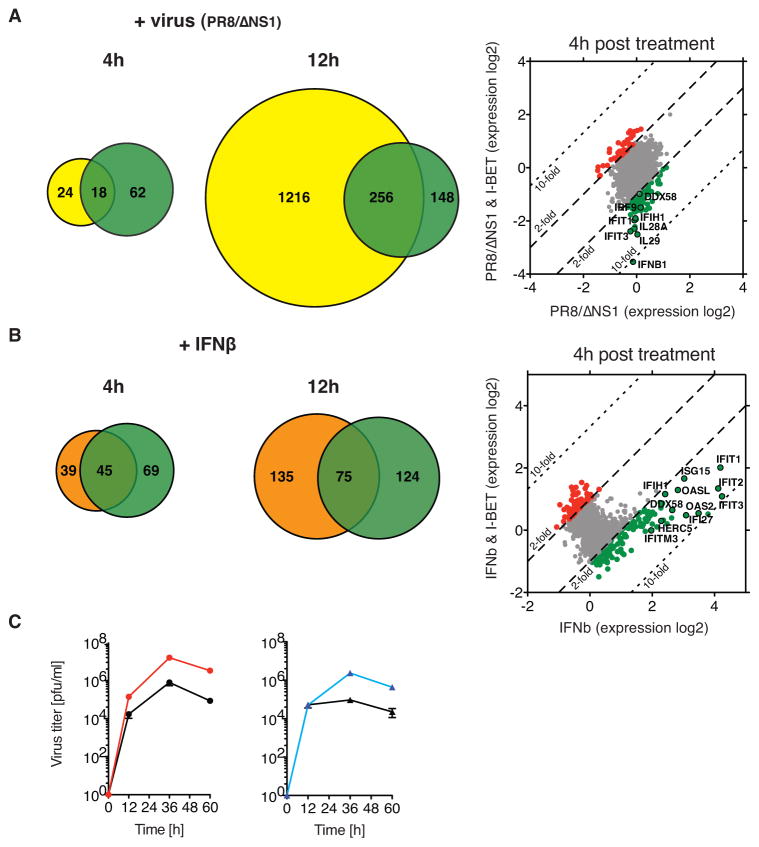
We found that similar to Paf1C, the BET proteins play an important role in regulation of antiviral gene expression and antiviral defense. Incubation of A549 cells with I-BET resulted in greatly attenuated expression of 18 and 256 genes induced by infection with influenza A PR8/ΔNS1 at 4 hours and 12 hours post infection, respectively (Figure 3A). The impact of I-BET on antiviral gene expression reflects the ability of the inhibitor to suppress expression of the virus-induced type I/III IFN genes as well as multiple ISGs. Incubation with I-BET led to down- regulation of 45 and 75 ISGs in A549 cells treated with IFNβ at 4 hours and 12 hours post stimulation, respectively (Figure 3B). The suppressive effect of I-BET on antiviral gene expression was associated with greatly increased cell susceptibility to viral infection. Similar to Paf1-deficient cells, the I-BET-treated A549 cells display a nearly 10-fold increase in levels in replication of NS1-deficient influenza A PR8/ΔNS1 as compared to non-treated cells (Figure 3C).
Figure 3. I-BET suppresses the antiviral response.
A. Venn diagrams display the number of virus-induced genes (> 2-fold, yellow circle) that were suppressed (> 2-fold, green circle) by I-BET treatment of A549 cells at 4 hours or 12 hours after infection with influenza PR8/ΔNS1. The scatter plot shows gene expression levels in control (x-axis) versus I-BET-treated cells (y-axis) at 4 hours after infection. The down- or up-regulated genes are shown in green or red, respectively. A black circle highlights genes that play an important role in antiviral response. The broken black line indicates a 2-fold change in expression; the dotted black line indicates a 10-fold change in expression. B. Venn diagrams display the number of IFNβ-induced genes (> 2-fold, orange circle) that were suppressed (> 2-fold, green circle) by I-BET treatment of A549 cells at 4 hours or 12 hours after incubation with IFNβ. The scatter plot shows gene expression levels in control (x-axis) versus I-BET-treated cells (y-axis) at 4 hours after IFNβ treatment. The down- or up-regulated genes are shown in green or red, respectively. A black circle highlights examples of genes that play an important role for the cellular antiviral response. The broken black line indicates a 2-fold change in expression; the dotted black line indicates a 10-fold change in expression. C. I-BET increases virus growth in vitro. The kinetics of influenza PR8/ΔNS1 virus replication in control (black circle) or I-BET-treated A549 cells (red circle) has been measured as described (Marazzi et al. 2012). The viral replication in I-BET treated cells was compared to viral replication in Paf1-deficient A549 cells (blue triangle) that display significant increase in viral replication as compared to control cells (black triangle).
The profound and selective impact of I-BET on antiviral gene expression in infected cells is similar to a negative effect of NS1 on the antiviral state of the infected cells. Assuming that the histone mimic mediates a significant part of the NS1 effect on gene expression, our finding shows a remarkable similarity between natural, e.g. the NS1 tail and synthetic, e.g. I-BET, histone mimics.
Conclusion
Our finding identified the ability of viral and synthetic histone mimics to interfere with the formation of transcriptional complexes and gene expression. We show that the immunosuppressive NS1 protein of influenza A virus employs histone mimicry for suppression of antiviral gene expression. We also demonstrate the potent role of synthetic histone mimics in suppression of antiviral response.
The presence of histone mimics in structurally distinct viral proteins (Table 2) underscores the ability of viruses to employ highly evolvable host proteins sequences for developing the optimal strategy for pathogen-host interaction. By imitating histones, the viruses can challenge the adaptive capacity of the host that cannot modify the histone primary sequences without endangering the very foundation of eukaryotic cell organization. Therefore, it’s plausible that many of the histone modifying enzymes that operate in the nucleus have a dual function, both as regulator of host gene expression as well as possible modifiers and attenuators of the viral histone mimic function. In support of this model, acetylation of the histone mimic within NS1, that prevents its binding to Paf1C (Marazzi et al. 2012), could be seen as a part of the host defense against the virus. Accordingly, identification of enzymes that modify viral mimics may lead to the development of novel antiviral drugs.
Table 2. A variety of human pathogens bear putative histone-like sequences.
The viral proteins that are homologous to the H3K4- or H3K9/H3K27-sequences in histone H3 are shown. The position of the putative viral histone mimics, with respect to the carboxy- or amino-termini, as well as the cellular localization of the viral proteins is indicated.
| Viral protein with H3K4-like sequence | Accession | Motif | Distance from terminus | Terminus | Localization |
|---|---|---|---|---|---|
|
| |||||
| Non structural protein 1 [Influenza A virus (A/New York/392/2004(H3N2))] | YP_308845.1 | ARSK | 4 | C | Nuclear |
| DNA polymerase catalytic subunit [Human herpesvirus 6A] | NP_042931.1 | ARSK | 72 | N | Nuclear |
| E2 protein [Semliki forest virus] | NP_819006.1 | ARSK | 31 | C | Membrane |
| Attachment glycoprotein G [Human metapneumovirus] | YP_012612.1 | ARSK | 23 | N | Membrane/Cytoplasm |
| Truncated structural polyprotein [Semliki forest virus] | YP_006390078.1 | ARSK | 106 | C | Membrane/Cytoplasm |
| NS5b [Hepatitis C virus genotype 3] | YP_001491557.1 | ARSK | 97 | N | Perinuclear |
| Envelope glycoprotein B [Human herpesvirus 5] | YP_081514.1 | ARSK | 257 | N | Membrane/Perinuclear |
| HCV polyprotein [Hepatitic C virus genotype 4] | YP_001469632.1 | ARSK | 494 | C | Varied |
| Polyprotein [Hepatitis C virus genotype 5] | YP_001469633.1 | ARSK | 494 | C | Varied |
| Polyprotein [Hepatitis C virus genotype 2] | YP_001469630.1 | ARSK | 494 | C | Varied |
| Viral protein with H3K9/H3K27-like sequence | Accession | Motif | Distance from terminus | Terminus | Localization |
|---|---|---|---|---|---|
|
| |||||
| Pol [Human adenovirus 35] | AP_000576.1 | ARKS | 28 | N | Nuclear |
| DNA polymerase [Human adenovirus B] | YP_002213842.1 | ARKS | 28 | N | Nuclear |
| Encapsidation protein IVa2 [Human adenovirus C] | NP_040515.1 | ARKT | 5 | C | Nuclear |
| IVa2 [Human adenovirus 2] | AP_000165.1 | ARKT | 5 | C | Nuclear |
| IVa2 [Human adenovirus 5] | AP_000201.1 | ARKT | 5 | C | Nuclear |
| IVa2 [Human adenovirus 1] | AP_000502.1 | ARKT | 5 | C | Nuclear |
| E2 protein [Human papillomavirus type 88] | YP_001672011.1 | ARKS | 52 | N | Nuclear |
| Single-stranded DNA-binding protein [Human adenovirus D] | YP_001974427.1 | ARKT | 72 | N | Nuclear |
| ORF29 [Human herpesvirus 8] | YP_001129382.1 | ARKT | 126 | N | Nuclear |
| BALF5 [Human herpesvirus 4] | YP_401712.1 | ARKT | 60 | C | Nuclear |
The histone mimics may not only potentiate the ability of the virus to interfere with the host, but also increase virus dependence on host transcription. It is tempting to speculate, that by using histone mimicry the viruses may become more dependent on the host. As a consequence, the histone mimic-bearing viruses or other pathogens may evolve eventually as symbiotic pathogens in a fashion similar to the commensal organisms in humans and animals.
The synthetic histone mimics such as the BET inhibitor I-BET or, similar to it, JQ1 (Filippakopoulos et al. 2010) can interfere with a significant number of genes largely overlapping but subtly different, including those triggered by viruses. This effect of BET inhibitors relies on a highly specific interaction with the bromodomains of the BET family of proteins. By using the analogy to SLiMs, it is possible to speculate that minor changes in the structure of BET inhibitors, while altering the inhibitor binding to BET proteins, may reveal therapeutically relevant drug targets that could not have been predicted based on the rational design that commonly ignores the founding role of SLiM in protein-protein interaction.
Acknowledgments
We would like to thank Adolfo Garcia-Sastre for providing the PR8/ΔNS1 virus; Raphael Cohn for performing ChIP-sequencing experiments; the Rockefeller University Genomics Resource Center, Jonas Marcello and Ryan Kim for assistance with the data analysis. This work has been supported by the Lupus Foundation Grant (A.T.), the Rockefeller University and the Epinova DPU, Immuno-Inflammation Therapy Area, GlaxoSmithKline (A.T., U.S., and R.P.).
References
- Ashour J, Laurent-Rolle M, Shi PY, Garcia-Sastre A. NS5 of dengue virus mediates STAT2 binding and degradation. J Virol. 2009;83:5408–5418. doi: 10.1128/JVI.02188-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badeaux AI, Shi Y. Emerging roles for chromatin as a signal integration and storage platform. Nat Rev Mol Cell Biol. 2013;14:211–224. doi: 10.1038/nrm3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballestas ME, Chatis PA, Kaye KM. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science. 1999;284:641–644. doi: 10.1126/science.284.5414.641. [DOI] [PubMed] [Google Scholar]
- Banerjee P, DeJesus R, Gjoerup O, Schaffhausen BS. Viral interference with DNA repair by targeting of the single-stranded DNA binding protein RPA. PLoS Pathog. 2013;9:e1003725. doi: 10.1371/journal.ppat.1003725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y, Bolotov P, Dernovoy D, Kiryutin B, Zaslavsky L, Tatusova T, Ostell J, Lipman D. The influenza virus resource at the National Center for Biotechnology Information. J Virol. 2008;82:596–601. doi: 10.1128/JVI.02005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkina AC, Nikolajczyk BS, Denis GV. BET protein function is required for inflammation: Brd2 genetic disruption and BET inhibitor JQ1 impair mouse macrophage inflammatory responses. J Immunol. 2013;190:3670–3678. doi: 10.4049/jimmunol.1202838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapuy B, McKeown MR, Lin CY, Monti S, Roemer MG, Qi J, Rahl PB, Sun HH, Yeda KT, Doench JG, et al. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer Cell. 2013;24:777–790. doi: 10.1016/j.ccr.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua MA, Schmid S, Perez JT, Langlois RA, Tenoever BR. Influenza A virus utilizes suboptimal splicing to coordinate the timing of infection. Cell Rep. 2013;3:23–29. doi: 10.1016/j.celrep.2012.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins RE, Northrop JP, Horton JR, Lee DY, Zhang X, Stallcup MR, Cheng X. The ankyrin repeats of G9a and GLP histone methyltransferases are mono- and dimethyllysine binding modules. Nat Struct Mol Biol. 2008;15:245–250. doi: 10.1038/nsmb.1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colpitts TM, Barthel S, Wang P, Fikrig E. Dengue virus capsid protein binds core histones and inhibits nucleosome formation in human liver cells. PLoS One. 2011;6:e24365. doi: 10.1371/journal.pone.0024365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey NE, Trave G, Gibson TJ. How viruses hijack cell regulation. Trends Biochem Sci. 2011;36:159–169. doi: 10.1016/j.tibs.2010.10.002. [DOI] [PubMed] [Google Scholar]
- Davey NE, Van Roey K, Weatheritt RJ, Toedt G, Uyar B, Altenberg B, Budd A, Diella F, Dinkel H, Gibson TJ. Attributes of short linear motifs. Mol Biosyst. 2012;8:268–281. doi: 10.1039/c1mb05231d. [DOI] [PubMed] [Google Scholar]
- Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, Robson SC, Chung CW, Hopf C, Savitski MM, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478:529–533. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey A, Chitsaz F, Abbasi A, Misteli T, Ozato K. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc Natl Acad Sci U S A. 2003;100:8758–8763. doi: 10.1073/pnas.1433065100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey A, Ellenberg J, Farina A, Coleman AE, Maruyama T, Sciortino S, Lippincott-Schwartz J, Ozato K. A bromodomain protein, MCAP, associates with mitotic chromosomes and affects G(2)-to-M transition. Mol Cell Biol. 2000;20:6537–6549. doi: 10.1128/mcb.20.17.6537-6549.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diella F, Haslam N, Chica C, Budd A, Michael S, Brown NP, Trave G, Gibson TJ. Understanding eukaryotic linear motifs and their role in cell signaling and regulation. Front Biosci. 2008;13:6580–6603. doi: 10.2741/3175. [DOI] [PubMed] [Google Scholar]
- Dinkel H, Van Roey K, Michael S, Davey NE, Weatheritt RJ, Born D, Speck T, Kruger D, Grebnev G, Kuban M, et al. The eukaryotic linear motif resource ELM: 10 years and counting. Nucleic Acids Res. 2014;42:D259–266. doi: 10.1093/nar/gkt1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlin LT, Andresen C, Just S, Rudensky E, Pappas CT, Kruger M, Jacobs EY, Unger A, Zieseniss A, Dobenecker MW, et al. Smyd2 controls cytoplasmic lysine methylation of Hsp90 and myofilament organization. Genes Dev. 2012;26:114–119. doi: 10.1101/gad.177758.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dover J, Schneider J, Tawiah-Boateng MA, Wood A, Dean K, Johnston M, Shilatifard A. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J Biol Chem. 2002;277:28368–28371. doi: 10.1074/jbc.C200348200. [DOI] [PubMed] [Google Scholar]
- Edwards RJ, Davey NE, O’Brien K, Shields DC. Interactome-wide prediction of short, disordered protein interaction motifs in humans. Mol Biosyst. 2012;8:282–295. doi: 10.1039/c1mb05212h. [DOI] [PubMed] [Google Scholar]
- Elde NC, Malik HS. The evolutionary conundrum of pathogen mimicry. Nat Rev Microbiol. 2009;7:787–797. doi: 10.1038/nrmicro2222. [DOI] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Allis CD. Histone and chromatin cross-talk. Curr Opin Cell Biol. 2003;15:172–183. doi: 10.1016/s0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- Fonseca GJ, Cohen MJ, Nichols AC, Barrett JW, Mymryk JS. Viral retasking of hBre1/RNF20 to recruit hPaf1 for transcriptional activation. PLoS Pathog. 2013;9:e1003411. doi: 10.1371/journal.ppat.1003411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca GJ, Thillainadesan G, Yousef AF, Ablack JN, Mossman KL, Torchia J, Mymryk JS. Adenovirus evasion of interferon-mediated innate immunity by direct antagonism of a cellular histone posttranslational modification. Cell Host Microbe. 2012;11:597–606. doi: 10.1016/j.chom.2012.05.005. [DOI] [PubMed] [Google Scholar]
- Friborg J, Jr, Kong W, Hottiger MO, Nabel GJ. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature. 1999;402:889–894. doi: 10.1038/47266. [DOI] [PubMed] [Google Scholar]
- Fujimuro M, Wu FY, ApRhys C, Kajumbula H, Young DB, Hayward GS, Hayward SD. A novel viral mechanism for dysregulation of beta-catenin in Kaposi’s sarcoma-associated herpesvirus latency. Nat Med. 2003;9:300–306. doi: 10.1038/nm829. [DOI] [PubMed] [Google Scholar]
- Garcia-Sastre A, Biron CA. Type 1 interferons and the virus-host relationship: a lesson in detente. Science. 2006;312:879–882. doi: 10.1126/science.1125676. [DOI] [PubMed] [Google Scholar]
- Garcia-Sastre A, Egorov A, Matassov D, Brandt S, Levy DE, Durbin JE, Palese P, Muster T. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology. 1998;252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- Golebiewski L, Liu H, Javier RT, Rice AP. The avian influenza virus NS1 ESEV PDZ binding motif associates with Dlg1 and Scribble to disrupt cellular tight junctions. J Virol. 2011;85:10639–10648. doi: 10.1128/JVI.05070-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenspan D, Palese P, Krystal M. Two nuclear location signals in the influenza virus NS1 nonstructural protein. J Virol. 1988;62:3020–3026. doi: 10.1128/jvi.62.8.3020-3026.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MA, Dickson KA, Jackson S, Clarkson A, Gill AJ, Marsh DJ. The tumor suppressor CDC73 interacts with the ring finger proteins RNF20 and RNF40 and is required for the maintenance of histone 2B monoubiquitination. Hum Mol Genet. 2012;21:559–568. doi: 10.1093/hmg/ddr490. [DOI] [PubMed] [Google Scholar]
- Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009;138:129–145. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfield JJ. Kinetic proofreading: a new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc Natl Acad Sci U S A. 1974;71:4135–4139. doi: 10.1073/pnas.71.10.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Zhang J, Shen W, Wang X, Wu J, Shi Y. Solution structure of the second bromodomain of Brd2 and its specific interaction with acetylated histone tails. BMC Struct Biol. 2007;7:57. doi: 10.1186/1472-6807-7-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D, Hossain MJ, Hickman D, Perez DR, Lamb RA. A new influenza virus virulence determinant: the NS1 protein four C-terminal residues modulate pathogenicity. Proc Natl Acad Sci U S A. 2008;105:4381–4386. doi: 10.1073/pnas.0800482105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- Kanno T, Kanno Y, Siegel RM, Jang MK, Lenardo MJ, Ozato K. Selective recognition of acetylated histones by bromodomain proteins visualized in living cells. Mol Cell. 2004;13:33–43. doi: 10.1016/s1097-2765(03)00482-9. [DOI] [PubMed] [Google Scholar]
- Katze MG, He Y, Gale M., Jr Viruses and interferon: a fight for supremacy. Nat Rev Immunol. 2002;2:675–687. doi: 10.1038/nri888. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S. Innate immune recognition of viral infection. Nat Immunol. 2006;7:131–137. doi: 10.1038/ni1303. [DOI] [PubMed] [Google Scholar]
- Kim J, Guermah M, Roeder RG. The human PAF1 complex acts in chromatin transcription elongation both independently and cooperatively with SII/TFIIS. Cell. 2010;140:491–503. doi: 10.1016/j.cell.2009.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Roeder RG. Direct Bre1-Paf1 complex interactions and RING finger-independent Bre1-Rad6 interactions mediate histone H2B ubiquitylation in yeast. J Biol Chem. 2009;284:20582–20592. doi: 10.1074/jbc.M109.017442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug RM, Yuan W, Noah DL, Latham AG. Intracellular warfare between human influenza viruses and human cells: the roles of the viral NS1 protein. Virology. 2003;309:181–189. doi: 10.1016/s0042-6822(03)00119-3. [DOI] [PubMed] [Google Scholar]
- Lee JM, Lee JS, Kim H, Kim K, Park H, Kim JY, Lee SH, Kim IS, Kim J, Lee M, et al. EZH2 generates a methyl degron that is recognized by the DCAF1/DDB1/CUL4 E3 ubiquitin ligase complex. Mol Cell. 2012;48:572–586. doi: 10.1016/j.molcel.2012.09.004. [DOI] [PubMed] [Google Scholar]
- Lee JS, Kim Y, Kim IS, Kim B, Choi HJ, Lee JM, Shin HJ, Kim JH, Kim JY, Seo SB, et al. Negative regulation of hypoxic responses via induced Reptin methylation. Mol Cell. 2010;39:71–85. doi: 10.1016/j.molcel.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Oliveira NM, Cheney KM, Pade C, Dreja H, Bergin AM, Borgdorff V, Beach DH, Bishop CL, Dittmar MT, et al. A whole genome screen for HIV restriction factors. Retrovirology. 2011;8:94. doi: 10.1186/1742-4690-8-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Ma Q, Wong K, Li W, Ohgi K, Zhang J, Aggarwal AK, Rosenfeld MG. Brd4 and JMJD6-associated anti-pause enhancers in regulation of transcriptional pause release. Cell. 2013;155:1581–1595. doi: 10.1016/j.cell.2013.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marazzi I, Ho JS, Kim J, Manicassamy B, Dewell S, Albrecht RA, Seibert CW, Schaefer U, Jeffrey KL, Prinjha RK, et al. Suppression of the antiviral response by an influenza histone mimic. Nature. 2012;483:428–433. doi: 10.1038/nature10892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller CL, Porter SE, Hoffman MG, Jaehning JA. The Paf1 complex has functions independent of actively transcribing RNA polymerase II. Mol Cell. 2004;14:447–456. doi: 10.1016/s1097-2765(04)00257-6. [DOI] [PubMed] [Google Scholar]
- Nagaike T, Logan C, Hotta I, Rozenblatt-Rosen O, Meyerson M, Manley JL. Transcriptional activators enhance polyadenylation of mRNA precursors. Mol Cell. 2011;41:409–418. doi: 10.1016/j.molcel.2011.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi S, Lee JS, Gardner KE, Gardner JM, Takahashi YH, Chandrasekharan MB, Sun ZW, Osley MA, Strahl BD, Jaspersen SL, et al. Histone H2BK123 monoubiquitination is the critical determinant for H3K4 and H3K79 trimethylation by COMPASS and Dot1. J Cell Biol. 2009;186:371–377. doi: 10.1083/jcb.200906005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemeroff ME, Barabino SM, Li Y, Keller W, Krug RM. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′end formation of cellular pre-mRNAs. Mol Cell. 1998;1:991–1000. doi: 10.1016/s1097-2765(00)80099-4. [DOI] [PubMed] [Google Scholar]
- Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung CW, Chandwani R, Marazzi I, Wilson P, Coste H, et al. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468:1119–1123. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninio J. Kinetic amplification of enzyme discrimination. Biochimie. 1975;57:587–595. doi: 10.1016/s0300-9084(75)80139-8. [DOI] [PubMed] [Google Scholar]
- Nordick K, Hoffman MG, Betz JL, Jaehning JA. Direct interactions between the Paf1 complex and a cleavage and polyadenylation factor are revealed by dissociation of Paf1 from RNA polymerase II. Eukaryot Cell. 2008;7:1158–1167. doi: 10.1128/EC.00434-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obenauer JC, Denson J, Mehta PK, Su X, Mukatira S, Finkelstein DB, Xu X, Wang J, Ma J, Fan Y, et al. Large-scale sequence analysis of avian influenza isolates. Science. 2006;311:1576–1580. doi: 10.1126/science.1121586. [DOI] [PubMed] [Google Scholar]
- Penheiter KL, Washburn TM, Porter SE, Hoffman MG, Jaehning JA. A posttranscriptional role for the yeast Paf1-RNA polymerase II complex is revealed by identification of primary targets. Mol Cell. 2005;20:213–223. doi: 10.1016/j.molcel.2005.08.023. [DOI] [PubMed] [Google Scholar]
- Rahman S, Sowa ME, Ottinger M, Smith JA, Shi Y, Harper JW, Howley PM. The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3. Mol Cell Biol. 2011;31:2641–2652. doi: 10.1128/MCB.01341-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenblatt-Rosen O, Nagaike T, Francis JM, Kaneko S, Glatt KA, Hughes CM, LaFramboise T, Manley JL, Meyerson M. The tumor suppressor Cdc73 functionally associates with CPSF and CstF 3′ mRNA processing factors. Proc Natl Acad Sci U S A. 2009;106:755–760. doi: 10.1073/pnas.0812023106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath SC, Marazzi I, Yap KL, Krutchinsky AN, Mecklenbrauker I, Viale A, Rudensky E, Zhou MM, Chait BT, Tarakhovsky A. Methylation of a histone mimic within the histone methyltransferase G9a regulates protein complex assembly. Mol Cell. 2007;27:596–608. doi: 10.1016/j.molcel.2007.06.026. [DOI] [PubMed] [Google Scholar]
- Santos A, Pal S, Chacon J, Meraz K, Gonzalez J, Prieto K, Rosas-Acosta G. SUMOylation affects the interferon blocking activity of the influenza A nonstructural protein NS1 without affecting its stability or cellular localization. J Virol. 2013;87:5602–5620. doi: 10.1128/JVI.02063-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterly N, Tsai PL, van Deursen J, Nussenzveig DR, Wang Y, Faria PA, Levay A, Levy DE, Fontoura BM. Influenza virus targets the mRNA export machinery and the nuclear pore complex. Proc Natl Acad Sci U S A. 2007;104:1853–1858. doi: 10.1073/pnas.0610977104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Wang Y, Zeng L, Wu Y, Deng J, Zhang Q, Lin Y, Li J, Kang T, Tao M, et al. Disrupting the Interaction of BRD4 with Diacetylated Twist Suppresses Tumorigenesis in Basal-like Breast Cancer. Cancer Cell. 2014;25:210–225. doi: 10.1016/j.ccr.2014.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simic R, Lindstrom DL, Tran HG, Roinick KL, Costa PJ, Johnson AD, Hartzog GA, Arndt KM. Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J. 2003;22:1846–1856. doi: 10.1093/emboj/cdg179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ, 3rd, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, Tempst P, Manley JL, Reinberg D. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell. 2007;28:665–676. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smock RG, Gierasch LM. Sending signals dynamically. Science. 2009;324:198–203. doi: 10.1126/science.1169377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobhian B, Laguette N, Yatim A, Nakamura M, Levy Y, Kiernan R, Benkirane M. HIV-1 Tat assembles a multifunctional transcription elongation complex and stably associates with the 7SK snRNP. Mol Cell. 2010;38:439–451. doi: 10.1016/j.molcel.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein A, Pache RA, Bernado P, Pons M, Aloy P. Dynamic interactions of proteins in complex networks: a more structured view. FEBS J. 2009;276:5390–5405. doi: 10.1111/j.1742-4658.2009.07251.x. [DOI] [PubMed] [Google Scholar]
- Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- Tachibana M, Ueda J, Fukuda M, Takeda N, Ohta T, Iwanari H, Sakihama T, Kodama T, Hamakubo T, Shinkai Y. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev. 2005;19:815–826. doi: 10.1101/gad.1284005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarakhovsky A. Logic of the inflammation-associated transcriptional response. Adv Immunol. 2013;119:107–133. doi: 10.1016/B978-0-12-407707-2.00004-7. [DOI] [PubMed] [Google Scholar]
- Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M, Kranjec C, Nagasaka K, Matlashewski G, Banks L. Analysis of the PDZ binding specificities of Influenza A virus NS1 proteins. Virol J. 2011;8:25. doi: 10.1186/1743-422X-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Roey K, Dinkel H, Weatheritt RJ, Gibson TJ, Davey NE. The switches. ELM resource: a compendium of conditional regulatory interaction interfaces. Sci Signal. 2013;6:rs7. doi: 10.1126/scisignal.2003345. [DOI] [PubMed] [Google Scholar]
- Van Roey K, Gibson TJ, Davey NE. Motif switches: decision-making in cell regulation. Curr Opin Struct Biol. 2012;22:378–385. doi: 10.1016/j.sbi.2012.03.004. [DOI] [PubMed] [Google Scholar]
- Wang X, Li M, Zheng H, Muster T, Palese P, Beg AA, Garcia-Sastre A. Influenza A virus NS1 protein prevents activation of NF-kappaB and induction of alpha/beta interferon. J Virol. 2000;74:11566–11573. doi: 10.1128/jvi.74.24.11566-11573.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weatheritt RJ, Davey NE, Gibson TJ. Linear motifs confer functional diversity onto splice variants. Nucleic Acids Res. 2012a;40:7123–7131. doi: 10.1093/nar/gks442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weatheritt RJ, Luck K, Petsalaki E, Davey NE, Gibson TJ. The identification of short linear motif-mediated interfaces within the human interactome. Bioinformatics. 2012b;28:976–982. doi: 10.1093/bioinformatics/bts072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood A, Schneider J, Dover J, Johnston M, Shilatifard A. The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J Biol Chem. 2003;278:34739–34742. doi: 10.1074/jbc.C300269200. [DOI] [PubMed] [Google Scholar]
- Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- You J, Croyle JL, Nishimura A, Ozato K, Howley PM. Interaction of the bovine papillomavirus E2 protein with Brd4 tethers the viral DNA to host mitotic chromosomes. Cell. 2004;117:349–360. doi: 10.1016/s0092-8674(04)00402-7. [DOI] [PubMed] [Google Scholar]
- Zhao RY, Elder RT. Viral infections and cell cycle G2/M regulation. Cell Res. 2005;15:143–149. doi: 10.1038/sj.cr.7290279. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Li T, Price DH. RNA polymerase II elongation control. Annu Rev Biochem. 2012;81:119–143. doi: 10.1146/annurev-biochem-052610-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]