ABSTRACT
Development of the insect compound eye requires a highly controlled interplay between transcription factors. However, the genetic mechanisms that link early eye field specification to photoreceptor terminal differentiation and fate maintenance remain largely unknown. Here, we decipher the function of 2 transcription factors, Glass and Hazy, which play a central role during photoreceptor development. The regulatory interactions between Glass and Hazy suggest that they function together in a coherent feed-forward loop in all types of Drosophila photoreceptors. While the glass mutant eye lacks the expression of virtually all photoreceptor genes, young hazy mutants correctly express most phototransduction genes. Interestingly, the expression of these genes is drastically reduced in old hazy mutants. This age-dependent loss of the phototransduction cascade correlates with a loss of phototaxis in old hazy mutant flies. We conclude that Glass can either directly or indirectly initiate the expression of most phototransduction proteins in a Hazy-independent manner, and that Hazy is mainly required for the maintenance of functional photoreceptors in adult flies.
KEYWORDS: cell fate maintenance, Drosophila, eye development, Glass, Hazy, photoreceptor differentiation, phototransduction, Pph13
Introduction
Cell differentiation is typically controlled by networks of transcription factors, which gradually shift during development from specifying organ and cellular identity to activating terminal gene expression in mature cells. Such a network acts during the formation of photoreceptors (PRs) in Drosophila.1-3 During early eye development, an evolutionarily conserved set of transcription factors, called the retinal determination network (RDN), specifies eye field identity in the eye imaginal disc. RDN genes are both necessary and sufficient for eye formation, and thus can induce formation of ectopic eyes when misexpressed in other imaginal discs.4-8
Recently, we have shown that PR differentiation is critically regulated by the zinc finger transcription factor Glass. Glass provides a genetic link between the early-acting RDN, terminal differentiation transcription factors, and genes functioning in mature PRs, such as those involved in the phototransduction cascade (Bernardo-Garcia et al.9). In the absence of Glass, PR precursors retain a neuronal identity but fail to express any PR markers. Therefore, PR precursors require Glass for differentiating into functional light-sensing cells.
A direct target of Glass is the homeobox transcription factor Hazy. Ectopic expression of Glass is sufficient to induce expression of Hazy and some phototransduction proteins. Similarly, ectopic expression of Hazy can only induce a subset of phototransduction components. However, when co-expressed, Glass and Hazy can ectopically induce most of the phototransduction cascade, suggesting that both Glass and Hazy act synergistically during PR development.9 Thus, the combinatorial action of these 2 transcription factors appears to play a central role in directing PR precursors toward a terminal differentiation program.
Here, we investigate the regulatory interaction between Glass and Hazy. We found that, while Glass is able to activate its own promoter, Hazy does not appear to auto-activate its own expression nor that of Glass. Also, by analyzing hazy mutants we disentangle the individual roles of Glass and Hazy in regulating the expression of phototransduction genes. Interestingly, we found that Hazy is particularly relevant to ensure the continued expression of phototransduction proteins in adult PRs. Young hazy mutants correctly express most of the phototransduction components, and show a similar attraction to white light as wild-type flies. By contrast, the expression of most phototransduction proteins is reduced in old hazy mutants, and they fail to show phototaxis. Together with previous results, our data suggest that Glass and Hazy are required for different tasks and at different steps in PR development. During early eye development, Glass contributes to specifying PR identity.9-12 Later, during terminal differentiation, both Glass and Hazy activate genes that are required for the maturation of functional PRs.9,12,13 Finally, Hazy is required for maintaining expression of phototransduction genes, and thus ensures the continued functionality of adult PRs.
Results and discussion
Hazy is a direct target of Glass in all visual organs in the fly
PR development in the eye imaginal disc starts after the passage of the so-called morphogenetic furrow (MF), which sweeps across the disc from the posterior edge toward anterior, initiating the formation of ommatidia. RDN genes are present in the eye disc prior to the MF, while the proneural gene atonal is transiently expressed at the MF.1,2 Glass expression is initiated at the MF, and maintained in differentiating cells in the retina.14,15 In contrast, the expression of Hazy starts later during pupation, after all PRs have been specified.3,13 We and others have shown that the expression of a hazy(wt)-GFP reporter in compound eye PRs depends on Glass binding to 2 sites in the hazy promoter.9,12 In addition to the compound eye, flies also have PRs in the ocelli (3 separate eyes located at the top of the adult head) and in the larval eye, also termed Bolwig's organ. Since both Glass and Hazy also play a role in the development of these organs,9,11,14,16-19 we hypothesized that Glass might similarly activate hazy in PRs outside the compound eye.
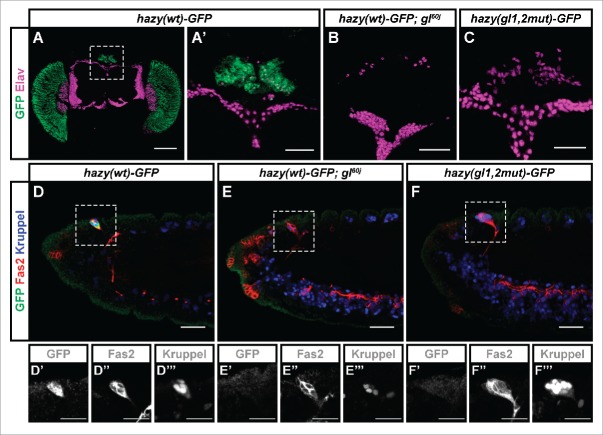
The hazy(wt)-GFP reporter, which we have previously used to study the expression of hazy in the compound eye, was also expressed in PRs of the ocelli and Bolwig's organ (Figs. 1A, A′, D–D‴), reflecting the expression pattern of the endogenous Hazy protein.13,17 When the hazy(wt)-GFP transgene was placed in glass mutant background, GFP expression was lost in the ocelli and the Bolwig's organ (Figs. 1B, E–E‴). The wild-type embryonic Bolwig's organ consists of 12 PRs, and can be identified in stage 14 embryos because of the co-expression of Kruppel and Fasciclin 2 (Fas2).17,20 Interestingly, in glass mutants we only found 4 Kruppel-positive cells in the Bolwig's organ (Fig. 1E‴), indicating an early defect in larval eye formation. In addition, a hazy(gl1,2mut)-GFP reporter in which the Glass binding sites were mutated was not expressed in the ocelli nor in the Bolwig's organ (Figs. 1C, F–F‴).
Figure 1.
Expression analysis of the hazy(wt)-GFP reporter in the ocelli and Bolwig's organ PRs. (A-C) In the case of the ocelli, these are 3 visual organs located dorsally on the head of adult flies (A). Samples were stained with antibodies against GFP (green), and Elav (used as a neuronal marker, magenta). The hazy(wt)-GFP reporter was expressed in the ocelli in wild-type (A, A'), but not glass mutant background (B). A hazy(gl1,2mut)-GFP reporter in which the 2 Glass binding sites were mutated was not expressed in the ocelli (C). (D-F) In the case of the Bolwig's organ, this is a larval eye that develops from the optic placode during embryogenesis. Stage 14 embryos were stained with antibodies against GFP (green), Fas2 (red) and Kruppel (blue). At this stage, the developing Bolwig's organ is located dorsally, still in contact with the surface, and can be identified both because of its position and the co-expression of Fas2 and Kruppel.,17,20,36 Similar to the ocelli, the hazy(wt)-GFP reporter was expressed in the Bolwig's organ in wild-type (D), but not glass mutant background (E). Also, hazy(gl1,2mut)-GFP was not expressed in the Bolwig's organ (F). For each image, the 3 channels from a close-up of the Bolwig's organ were separated and are shown below in grayscale (D′-F‴). Scale bars represent 20 μm in D′-F‴; 30 μm in A′, B-F; and 100 μm in A.
Together, these results corroborate our findings for the compound eye.9 We conclude that the hazy promoter is directly bound and activated by Glass in all PRs in the 3 visual organs of Drosophila.
Glass can auto-activate its own expression
We have previously shown that ectopically expressing Glass or Hazy induces the expression of some phototransduction proteins in the central nervous system (CNS). Co-expressing Glass and Hazy displays a synergistic effect on the induction of phototransduction components. Not only the genes that are activated by either Glass or Hazy alone become ectopically expressed, but also additional phototransduction proteins are induced, suggesting that Glass and Hazy function together in a coherent feed-forward loop. Glass activates the expression of Hazy and together they are able to activate the expression of more target genes than either Glass or Hazy could activate on their own.9 Here we tested additional regulatory interactions between Glass and Hazy.
For this, we used a glass-DsRed and a hazy(wt)-GFP reporter.9,21 glass-DsRed larvae expressed nuclear DsRed in the Bolwig's organ and in Glass-expressing cells in the brain (Figs. 2A–A‴), while hazy(wt)-GFP was expressed exclusively in the Bolwig's organ PRs (Figs. 1D, 2B), whose axons could be seen projecting into the brain (Fig. 2B′). Thus, both reporters faithfully reflect the expression patterns of Glass and Hazy.13,14 We performed flip-out experiments in which we ectopically induced either Glass or Hazy in the CNS of third instar larvae, and found that Glass was able to activate the glass-DsRed reporter in a subset of cells (Figs. 2C–C‴). Therefore, Glass may be able to maintain its own expression by auto-regulation. In contrast, ectopic expression of Hazy did not induce the expression of the hazy(wt)-GFP reporter (Figs. 2D, D′), suggesting that Hazy cannot activate its own expression. Similarly, we did not observe ectopic expression of Glass in the CNS after ectopically expressing Hazy (Figs. 2E, E′), suggesting that Glass expression is not activated by Hazy.
Figure 2.
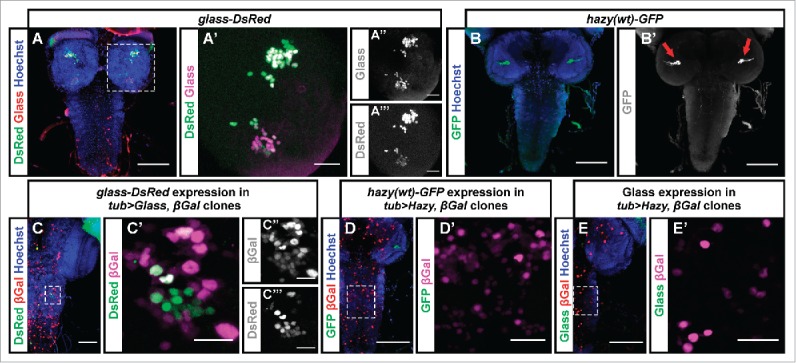
Test for additional regulatory interactions between Glass and Hazy. (A, B) We used Hoechst (blue), which labels cell nuclei, as a counterstain to analyze the expression pattern of the glass-DsRed and hazy(wt)-GFP reporters in the CNS of third instar larvae. The glass-DsRed reporter was expressed the nuclei of some cells in the brain (green, A). A close-up to the right shows that those neurons endogenously expressing Glass (red/magenta) also co-express the reporter (green, A′). These two channels are shown separately to the right in grayscale (A,″ A‴). The hazy(wt)-GFP reporter is exclusively expressed in PRs (green, B). A grayscale image to the right shows GFP labeling the axonal projections of the PRs in the brain (arrows, B′). (C-E) In flip-out experiments we ectopically induced either Glass or Hazy expression in clones labeled with nuclear β-galactosidase (βGal). We stained the CNS of third instar larvae with antibodies against βGal (red/magenta); either DsRed, GFP or Glass (green) and with Hoechst (blue). We found that Glass ectopically induced the expression of the glass-DsRed reporter in the ventral nerve cord (C, C′; channels are also shown separately in grayscale in C,″ C‴). By contrast, Hazy did not ectopically induce the hazy(wt)-GFP reporter (D, D′) nor Glass (E, E′). Scale bars represent 20 μm in A′, C′- E′; and 80 μm in A-E.
Glass can initiate the expression of most phototransduction proteins independently of Hazy
Both Glass and Hazy are required for the expression of PR genes.9,13 Since we have shown that Glass directly activates hazy, and that inducing the expression of Hazy partly rescues the glass mutant phenotype,9 it would be possible that Glass mainly relies on Hazy for activating the expression of phototransduction proteins. To test this, we examined the individual role of Hazy.
We found that young hazy mutant flies—less than one day old—failed to express Rhodopsin 6 (Rh6) and Transient receptor potential-like (Trpl), but did express correctly most of the phototransduction proteins that we tested: Rhodopsin 1 (Rh1), G protein α q subunit (Gαq), No receptor potential A (NorpA), Transient receptor potential (Trp), Inactivation no afterpotential D (InaD) and Arrestin 1 (Arr1) (Figs. 3A–P).22-30 Of these, after 10 d the levels of Rh1, Gαq, NorpA, Trp and InaD were strongly reduced, and only Arr1 expression appeared unchanged (Figs. 3a–p). Thus, our results indicate that Hazy is not required for initiating the expression of most phototransduction proteins, but that it plays an important role in maintaining the differentiated state of PRs. These results contrast with an earlier description of the hazy mutant phenotype.13 The differences between our findings and this report may be explained because we analyzed young and old flies separately.
Figure 3.
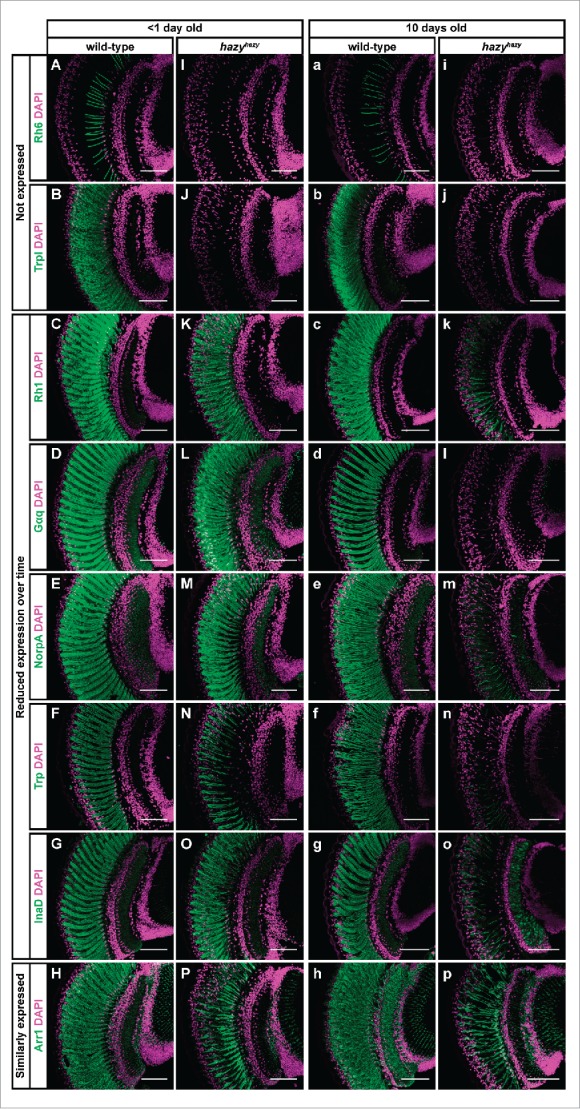
Expression of phototransduction proteins in the hazy mutant retina. Head sections were taken of control and hazyhazy flies, and stained with antibodies against different phototransduction proteins (green) and with Hoechst (used to label cell nuclei, magenta). (A-P) One group of flies was dissected on the day they eclosed. At this age we did not detect neither Rh6 (I) nor Trpl (J) in the retina of hazy mutants, but most of the phototransduction proteins that we tested were correctly expressed: Rh1 (K), Gαq (L), NorpA (M), Trp (N), InaD (O) and Arr1 (P). (a-p) A second group of flies were dissected 10 d after eclosion. Neither Rh6 (i) nor Trpl (j) were expressed in the retina of these older hazy mutants, and most phototransduction proteins showed decreased expression levels: Rh1 (k), Gαq (l), NorpA (m), Trp (n) and InaD (o). Only Arr1 (p) expression did not seem reduced over time in the hazy mutant retina. Scale bars represent 50 µm.
Since most phototransduction proteins are expressed in the retina of young hazy mutants, we infer that Glass does not mainly act via Hazy for initiating the expression of most phototransduction proteins. In the case of Rh6, Trpl, Rh1, Gαq, NorpA, Trp, InaD and Arr1, we found that all these genes contain putative Glass binding sites in their regulatory sequences (the GAARCC motif, which is present in either their promoter or their introns).31 Therefore, it would be possible that Glass either directly or indirectly activates the expression of these phototransduction components.
Hazy is not required for white light detection in young flies
To further assess the impact of their age-dependent loss of phototransduction genes we analyzed the phototactic behavior of young versus old hazy mutant flies. It has been previously reported that hazy mutants fail to detect light due to the absence of many phototransduction proteins.13,16 However, our finding suggests that young hazy mutant flies express a set of genes sufficient for the phototransduction machinery to detect white light.
Adult wild-type flies display a positive phototactic behavior. In a 2-choice assay, they move toward the light. This preference for light is very high in newly eclosed flies and shows some reduction when the flies get older.32 10 day old flies are still able to distinguish between light and darkness (Fig. 4). In contrast, glass mutant flies do not show phototaxis from the day they eclose (Fig. 4).33 This is in agreement with our previous finding that glass mutants do not express any of the proteins in the phototransduction cascade.9 In the case of hazy mutants, we observed normal phototactic behavior in young flies, comparable with that of wild-type. However, at the age of 10 d they differed from wild-type and displayed the same disability to distinguish between light and darkness as glass mutants (Fig. 4). These experiments are in agreement with our antibody analysis above, and show that young hazy mutants are able to detect white light, but lose this ability over time. Thus, Hazy is required for the maintenance of PR functionality.
Figure 4.

Age-related changes in the phototaxis of wild-type, glass and hazy mutant flies. Box plots show the light preference indices (PIs) of wild-type (yellow), glass (cyan) and hazy mutants (pink) of different ages. Bold lines represent medians. The upper and lower quartiles are represented by the top and the bottom of each box. Whisker lines indicate the maximum and minimum data point that are closer than 1.5 interquartile range of its nearest quartile. Circles indicate outliers. We used Welch's t-test for comparing the PIs between groups (n = 7 per age and genotype) and to zero. Significance levels represent p > 0.05 (not significant, n.s.), p ≤ 0.05 (*), p ≤ 0.01 (**), and p ≤ 0.001 (***). In a 2-choice assay, groups of wild-type flies of every age showed positive phototaxis, which decreases with age (indicated by positive PI values, which were significantly different from zero). glass mutants were photoneutral at all ages (their PIs were not significantly different from zero). Newly eclosed hazy mutants showed positive phototaxis, not different from that of wild-type flies (p = 0.67, median wild-type PI = 0.83). Five day old hazy mutants and wild-type flies show a decreased positive phototaxis, but their PIs are not different from each other (p = 0.30, median wild-type PI = 0.42). Ten day old hazy mutants were photoneutral, with their PIs comparable to zero (p = 0.08) or to glass mutants (p = 0.56), and significantly different from wild-type (median wild-type PI = 0.22).
Conclusions
Glass and Hazy play important roles in PR specification and maintenance.9,11,13,34 Here we have shown that Glass activates the expression of hazy in all PRs in the 3 visual organs of Drosophila—the compound eye, the ocelli and the Bolwig's organ—by binding to the same sites in the hazy promoter. Also, in agreement with a previous report, Glass is able to auto-activate its own expression,14 but we found no evidence that Hazy either activates glass or auto-activates its own expression. Together, our data favor a feed-forward mechanism in which Hazy acts downstream of Glass.9,35
The notion that Glass and Hazy function through a coherent feed-forward loop to activate PR genes is further supported by our previous findings that inducing the expression of Hazy partly rescues the glass mutant phenotype, and also that co-expressing Glass and Hazy together ectopically induces more phototransduction proteins than either Glass or Hazy alone.9 However, these findings were based on ectopic expression of both transcription factors in CNS cells in which they are normally not expressed and where we were not able to control their expression levels. Our analysis of the hazy mutant phenotype rather suggests that, by the end of terminal PR differentiation, Glass can induce the expression of most phototransduction components even in the absence of Hazy. Hazy itself is one of the targets of Glass and is required to maintain the expression of most phototransduction components throughout adult life. Thus, for the initial induction, Glass either directly activates PR-specific genes, or it interacts with other transcription factors to induce them. Hazy might be required for the initial induction of only a few genes, such as Rh6 or Trpl, but it is essential for the maintenance of most phototransduction components. Therefore, we anticipate that further research on the direct targets of Glass will reveal novel mechanisms for activating the expression of PR genes. Also, it will be interesting to explore further how Hazy functions in PR fate maintenance. Particularly, it remains unresolved whether Hazy is only required for maintaining PR gene expression or if it also functions to suppress PR dedifferentiation or degeneration.
Materials and methods
Fly stocks
In the present work w1118 (courtesy of R. Stocker) was used as wild-type control, and w1118; hazyhazy was used to study the hazy mutant phenotype. The hazyhazy mutant allele was provided by C. Desplan,13 and was isogenized by crossing it to Canton-S flies for 6 generations. Other stocks that we used are: hazy(wt)-GFP,9 hazy(gl1,2mut)-GFP,9 gl60j (Bloomington Drosophila Stock Center, No. 509),11 UAS-glass9 and UAS-hazy (courtesy of J. Bischof).37 As a reporter for glass we used glass-DsRed flies (courtesy of S. Kim), which are also called glass5.2-RHS.21 Flip-out misexpression experiments were performed as described previously9 by using hsFLP; tub(FRTcassette)Gal4, UAS-lacZ.nls (courtesy of E. Piddini).
Flies were raised at 25°C in a 12:12 hours light-dark cycle on cornmeal medium supplemented with molasses, fructose and yeast.
Antibody stainings
Immunohistochemistry was performed as previously described.9,17,38 Antibodies against proteins in the phototransduction cascade were kindly provided by N. Colley and S. Britt. To compare the expression of phototransduction proteins in control and hazy mutants, head sections from both genotypes were taken simultaneously, stained together on the same slide, and imaged with identical settings on a Leica SP5 confocal microscope.
Phototaxis assay
Our phototaxis analysis was based on a previous protocol.32 Briefly, newly eclosed flies were transferred each day to vials containing fresh food, which we used to stage them. For each experiment, we tested an average of 25.2 flies (ranging from 20 to 33). These flies were kept for 10 min in darkness, and then placed without anesthesia into a T-maze under red light conditions. Our set-up consisted of 2 tubes connected to each other, where a single LED illuminated from the end of one of the tubes with white light (SOLAROX, Germany, No. 50008300001). Light intensity was moderate: we measured 418.0739 µW/cm2 with a spectrometer (the emitted light spectrum possessed 2 intensity peaks: the first peak was at 444 nm with an intensity of 1.494 µW/cm2/nm and half-widths of 16 nm, and the second peak was at 585 nm with an intensity of 2.768 µW/cm2/nm and half-widths of 61.5 nm). We allowed the flies to move freely between both tubes for 2 minutes. Then, we counted the flies in the illuminated tube (L), in the dark tube (D), and those in the intersection between the 2 tubes (M). The preference index (PI) was calculated from the formula PI = (L − D)/(D + L + M). Data were analyzed in R with Welch's t-test.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank B. Bello, J. Bischof, S. Kim, R. Stocker, C. Desplan and the Bloomington Drosophila Stock Center for fly stocks. Also, we thank J. Jaeger, S. Britt, N. Colley and the Developmental Studies Hybridoma Bank at the University of Iowa for antibodies.
Funding
This work was funded by the Swiss National Science Foundation (31003A_149499 to S.G.S.) and the European Research Council (ERC-2012-StG 309832-PhotoNaviNet to S.G.S.).
References
- [1].Tsachaki M, Sprecher SG. Genetic and developmental mechanisms underlying the formation of the Drosophila compound eye. Dev Dyn 2012; 241:40-56; PMID:21932322; http://dx.doi.org/ 10.1002/dvdy.22738 [DOI] [PubMed] [Google Scholar]
- [2].Treisman JE. Retinal differentiation in Drosophila. Wiley Interdiscip Rev Dev Biol 2013; 2:545-57; PMID:24014422; http://dx.doi.org/ 10.1002/wdev.100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Potier D, Davie K, Hulselmans G, Naval Sanchez M, Haagen L, Huynh-Thu VA, Koldere D, Celik A, Geurts P, Christiaens V, et al.. Mapping gene regulatory networks in Drosophila eye development by large-scale transcriptome perturbations and motif inference. Cell Rep 2014; 9:2290-303; PMID:25533349; http://dx.doi.org/ 10.1016/j.celrep.2014.11.038 [DOI] [PubMed] [Google Scholar]
- [4].Halder G, Callaerts P, Gehring WJ. Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science 1995; 267:1788-92; PMID:7892602; http://dx.doi.org/ 10.1126/science.7892602 [DOI] [PubMed] [Google Scholar]
- [5].Pignoni F, Hu B, Zavitz KH, Xiao J, Garrity PA, Zipursky SL. The eye-specification proteins So and Eya form a complex and regulate multiple steps in Drosophila eye development. Cell 1997; 91:881-91; PMID:9428512; http://dx.doi.org/ 10.1016/S0092-8674(00)80480-8 [DOI] [PubMed] [Google Scholar]
- [6].Hoge MA. Another gene in the fourth chromosome of Drosophila. Am Nat 1915; 49:47-9; http://dx.doi.org/ 10.1086/279455 [DOI] [Google Scholar]
- [7].Silver SJ, Rebay I. Signaling circuitries in development: insights from the retinal determination gene network. Development 2005; 132:3-13; PMID:15590745; http://dx.doi.org/ 10.1242/dev.01539 [DOI] [PubMed] [Google Scholar]
- [8].Cheyette BN, Green PJ, Martin K, Garren H, Hartenstein V, Zipursky SL. The Drosophila sine oculis locus encodes a homeodomain-containing protein required for the development of the entire visual system. Neuron 1994; 12:977-96; PMID:7910468; http://dx.doi.org/ 10.1016/0896-6273(94)90308-5 [DOI] [PubMed] [Google Scholar]
- [9].Bernardo-Garcia FJ, Fritsch C, Sprecher SG. The transcription factor Glass links eye field specification with photoreceptor differentiation in Drosophila. Development 2016; 143:1413-23; PMID:26952983; http://dx.doi.org/ 10.1242/dev.128801 [DOI] [PubMed] [Google Scholar]
- [10].Naval-Sanchez M, Potier D, Haagen L, Sanchez M, Munck S, Van de Sande B, Casares F, Christiaens V, Aerts S. Comparative motif discovery combined with comparative transcriptomics yields accurate targetome and enhancer predictions. Genome Res 2013; 23:74-88; PMID:23070853; http://dx.doi.org/ 10.1101/gr.140426.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Moses K, Ellis MC, Rubin GM. The glass gene encodes a zinc-finger protein required by Drosophila photoreceptor cells. Nature 1989; 340:531-6; PMID:2770860; http://dx.doi.org/ 10.1038/340531a0 [DOI] [PubMed] [Google Scholar]
- [12].Liang X, Mahato S, Hemmerich C, Zelhof AC. Two temporal functions of Glass: Ommatidium patterning and photoreceptor differentiation. Dev Biol 2016; 414:4-20; PMID:27105580; http://dx.doi.org/ 10.1016/j.ydbio.2016.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zelhof AC, Koundakjian E, Scully AL, Hardy RW, Pounds L. Mutation of the photoreceptor specific homeodomain gene Pph13 results in defects in phototransduction and rhabdomere morphogenesis. Development 2003; 130:4383-92; PMID:12900454; http://dx.doi.org/ 10.1242/dev.00651 [DOI] [PubMed] [Google Scholar]
- [14].Moses K, Rubin GM. Glass encodes a site-specific DNA-binding protein that is regulated in response to positional signals in the developing Drosophila eye. Gen Dev 1991; 5:583-93; PMID:2010085; http://dx.doi.org/ 10.1101/gad.5.4.583 [DOI] [PubMed] [Google Scholar]
- [15].Ellis MC, O'Neill EM, Rubin GM. Expression of Drosophila glass protein and evidence for negative regulation of its activity in non-neuronal cells by another DNA-binding protein. Development 1993; 119:855-65; PMID:8187644 [DOI] [PubMed] [Google Scholar]
- [16].Mishra M, Oke A, Lebel C, McDonald EC, Plummer Z, Cook TA, Zelhof AC. Pph13 and orthodenticle define a dual regulatory pathway for photoreceptor cell morphogenesis and function. Development 2010; 137:2895-904; PMID:20667913; http://dx.doi.org/ 10.1242/dev.051722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Mishra AK, Bargmann BO, Tsachaki M, Fritsch C, Sprecher SG. Functional genomics identifies regulators of the phototransduction machinery in the Drosophila larval eye and adult ocelli. Dev Biol 2016; 410:164-77; PMID:26769100; http://dx.doi.org/ 10.1016/j.ydbio.2015.12.026 [DOI] [PubMed] [Google Scholar]
- [18].Mishra AK, Tsachaki M, Rister J, Ng J, Celik A, Sprecher SG. Binary cell fate decisions and fate transformation in the Drosophila larval eye. PLoS genetics 2013; 9:e1004027; PMID:24385925; http://dx.doi.org/ 10.1371/journal.pgen.1004027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Stark WS, Sapp R, Carlson SD. Ultrastructure of the ocellar visual system in normal and mutant Drosophila melanogaster. J Neurogenet 1989; 5:127-53; PMID:2500507; http://dx.doi.org/ 10.3109/01677068909066203 [DOI] [PubMed] [Google Scholar]
- [20].Schmucker D, Taubert H, Jackle H. Formation of the Drosophila larval photoreceptor organ and its neuronal differentiation require continuous Kruppel gene activity. Neuron 1992; 9:1025-39; PMID:1463605; http://dx.doi.org/ 10.1016/0896-6273(92)90063-J [DOI] [PubMed] [Google Scholar]
- [21].Park S, Bustamante EL, Antonova J, McLean GW, Kim SK. Specification of Drosophila corpora cardiaca neuroendocrine cells from mesoderm is regulated by Notch signaling. PLoS Genet 2011; 7:e1002241; PMID:21901108; http://dx.doi.org/ 10.1371/journal.pgen.1002241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Chou WH, Huber A, Bentrop J, Schulz S, Schwab K, Chadwell LV, Paulsen R, Britt SG. Patterning of the R7 and R8 photoreceptor cells of Drosophila: evidence for induced and default cell-fate specification. Development 1999; 126:607-16; PMID:9895309 [DOI] [PubMed] [Google Scholar]
- [23].de Couet HG, Tanimura T. Monoclonal antibodies provide evidence that rhodopsin in the outer rhabdomeres of Drosophila melanogaster is not glycosylated. Europ J Cell Biol 1987; 44:50-6 [Google Scholar]
- [24].Wong F, Schaefer EL, Roop BC, LaMendola JN, Johnson-Seaton D, Shao D. Proper function of the Drosophila trp gene product during pupal development is important for normal visual transduction in the adult. Neuron 1989; 3:81-94; PMID:2482778; http://dx.doi.org/ 10.1016/0896-6273(89)90117-7 [DOI] [PubMed] [Google Scholar]
- [25].Dolph PJ, Ranganathan R, Colley NJ, Hardy RW, Socolich M, Zuker CS. Arrestin function in inactivation of G protein-coupled receptor rhodopsin in vivo. Science 1993; 260:1910-6; PMID:8316831; http://dx.doi.org/ 10.1126/science.8316831 [DOI] [PubMed] [Google Scholar]
- [26].Zhu L, McKay RR, Shortridge RD. Tissue-specific expression of phospholipase C encoded by the norpA gene of Drosophila melanogaster. J Biol Chem 1993; 268:15994-6001; PMID:8340420 [PubMed] [Google Scholar]
- [27].Lee YJ, Shah S, Suzuki E, Zars T, O'Day PM, Hyde DR. The Drosophila dgq gene encodes a G alpha protein that mediates phototransduction. Neuron 1994; 13:1143-57; PMID:7946351; http://dx.doi.org/ 10.1016/0896-6273(94)90052-3 [DOI] [PubMed] [Google Scholar]
- [28].Niemeyer BA, Suzuki E, Scott K, Jalink K, Zuker CS. The Drosophila light-activated conductance is composed of the two channels TRP and TRPL. Cell 1996; 85:651-9; PMID:8646774; http://dx.doi.org/ 10.1016/S0092-8674(00)81232-5 [DOI] [PubMed] [Google Scholar]
- [29].Shieh BH, Niemeyer B. A novel protein encoded by the InaD gene regulates recovery of visual transduction in Drosophila. Neuron 1995; 14:201-10; PMID:7826638; http://dx.doi.org/ 10.1016/0896-6273(95)90255-4 [DOI] [PubMed] [Google Scholar]
- [30].Montell C. Drosophila visual transduction. Trends Neurosci 2012; 35:356-63; PMID:22498302; http://dx.doi.org/ 10.1016/j.tins.2012.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Enuameh MS, Asriyan Y, Richards A, Christensen RG, Hall VL, Kazemian M, Zhu C, Pham H, Cheng Q, Blatti C, et al.. Global analysis of Drosophila Cys(2)-His(2) zinc finger proteins reveals a multitude of novel recognition motifs and binding determinants. Genome Res 2013; 23:928-40; PMID:23471540; http://dx.doi.org/ 10.1101/gr.151472.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Le Bourg E, Badia J. Decline in photopositive tendencies with age inDrosophila melanogaster (Diptera: Drosophilidae). J Insect Behav 1995; 8:835-45; http://dx.doi.org/ 10.1007/BF02009510 [DOI] [Google Scholar]
- [33].Pak WL, Grossfield J, White NV. Nonphototactic mutants in a study of vision of Drosophila. Nature 1969; 222:351-4; PMID:5782110; http://dx.doi.org/ 10.1038/222351a0 [DOI] [PubMed] [Google Scholar]
- [34].Rister J, Razzaq A, Boodram P, Desai N, Tsanis C, Chen H, Jukam D, Desplan C. Single–base pair differences in a shared motif determine differential Rhodopsin expression. Science 2015; 350:1258-61; PMID:26785491; http://dx.doi.org/ 10.1126/science.aab3417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Alon U. Network motifs: theory and experimental approaches. Nat Rev Genet 2007; 8:450-61; PMID:17510665; http://dx.doi.org/ 10.1038/nrg2102 [DOI] [PubMed] [Google Scholar]
- [36].Campos-Ortega JA, Hartenstein V. The embryonic development of Drosophila melanogaster. Berlin, Germany: Springer-Verlag, 1985. [Google Scholar]
- [37].Bischof J, Bjorklund M, Furger E, Schertel C, Taipale J, Basler K. A versatile platform for creating a comprehensive UAS-ORFeome library in Drosophila. Development 2013; 140:2434-42; PMID:23637332; http://dx.doi.org/ 10.1242/dev.088757 [DOI] [PubMed] [Google Scholar]
- [38].Rothwell WF, Sullivan W. Fluorescent analysis of Drosophila embryos In: Sullivan W, Ashburner M, Hawley RS, eds. Drosophila protocols. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press, 2000:141-57. [Google Scholar]