ABSTRACT
The daily use by people of wireless communication devices has increased exponentially in the last decade, begetting concerns regarding its potential health hazards. Drosophila melanogaster four days-old adult female flies were exposed for 30 min to radiation emitted by a commercial mobile phone at a SAR of 0.15 W/kg and a SAE of 270 J/kg. ROS levels and apoptotic follicles were assayed in parallel with a genome-wide microarrays analysis. ROS cellular contents were found to increase by 1.6-fold (x), immediately after the end of exposure, in follicles of pre-choriogenic stages (germarium - stage 10), while sporadically generated apoptotic follicles (germarium 2b and stages 7–9) presented with an averaged 2x upregulation in their sub-population mass, 4 h after fly's irradiation with mobile device. Microarray analysis revealed 168 genes being differentially expressed, 2 h post-exposure, in response to radiofrequency (RF) electromagnetic field-radiation exposure (≥1.25x, P < 0.05) and associated with multiple and critical biological processes, such as basic metabolism and cellular subroutines related to stress response and apoptotic death. Exposure of adult flies to mobile-phone radiation for 30 min has an immediate impact on ROS production in animal's ovary, which seems to cause a global, systemic and non-targeted transcriptional reprogramming of gene expression, 2 h post-exposure, being finally followed by induction of apoptosis 4 h after the end of exposure. Conclusively, this unique type of pulsed radiation, mainly being derived from daily used mobile phones, seems capable of mobilizing critical cytopathic mechanisms, and altering fundamental genetic programs and networks in D. melanogaster.
KEYWORDS: apoptosis, cell death, drosophila, follicle, gene expression, microarrays, mobile-phone radiation, oogenesis, ovary, oxidative stress, ROS, transcription
Introduction
The rapidly increasing use of cellular phones, all around the world, over the past decades has aroused public concerns and fears regarding the potential health risks of non-ionizing electromagnetic radiation and fields. In response to these concerns, a recent review provides strong evidence regarding the oxidizing capacity of radiofrequencies (RFs), as 93% of the gathered experimental studies presented oxidative effects on a variety of biological systems (reviewed by Yakymenko et al.1). In mammalian cells, the major source of Reactive Oxygen Species (ROS) is the mitochondria. In vitro experiments have shown that, under normal conditions, approximately 1–3% of the oxygen used by the mitochondria is converted to hydrogen peroxide (H2O2).2 This percentage increases by various stimuli, such as low ATPase content or decreased mitochondrial ATP production.3 On the other hand, another cellular source of ROS are the NADH (Nicotinamide Adenine Dinucleotide) oxidases, which are membrane-associated enzymes that catalyze one-electron reduction of oxygen into superoxide radical using NADH as a donor of electron, thus producing powerful ROS species.4 Moreover, a positive feed-forward cycle between the aforementioned sources has been suggested.5 Both mitochondria and membrane NADH oxidases have been found to be critical sources of ROS generation under radiofrequency-radiation (RFR) exposure.6-8 Despite their origin, ROS, as they depend on effective concentration, may have a signal-transduction mediator role in cellular processes of survival or death.4 This dual ROS-dependent survival or death regulation in mammals, according to di Meo et al.,5 can be implemented either by transcription factors, or by activation of Mitogen Activated Protein Kinase (MAPK) cascades. Besides ROS content measurements,6 detection of apoptosis activation,9 individual gene expression modification10 and genome-wide transcriptional reprogramming have been previously reported in response to radiation emitted by mobile phones in various model systems in vitro and in vivo, albeit providing diverse conclusions.11-15
Hitherto, the diverse genetic backgrounds of model animal systems used, in combination with the various carrier frequencies, modulation schemes and field intensities, render the investigation of biological mechanisms that govern the molecular interactions of radiofrequencies (RFs) with living tissues and cells, a rather complicated and difficult task. Toward this direction, in the year 2001, it was proposed that proteomics and transcriptomics, together with other high-throughput screening platforms, could be successfully used to identify critical molecular targets of mobile-phone radiation and to address important biological questions that would be unfeasible to be answered by conventional technologies.16 Studies focusing on the potential impact of RF electromagnetic fields on gene expression profiling have been thoroughly evaluated, in the year 2011, by the International Agency for Research on Cancer (IARC), and they (RF fields) have been classified in the category of possible carcinogenic agents (group 2B).17,18
The biological model organism Drosophila melanogaster and, more specifically, the developmental process of oogenesis represent versatile, powerful and, in general, ideal experimental systems for studying in vivo a wide range of cellular processes, by molecular, genetic and morphological means, while they also appear to allow manifestations of unique behaviors, responses and sensitivities toward various environmental stress stimuli.19 Pioneering previous work on Drosophila melanogaster has demonstrated that short-term exposure to Extremely Low Frequency (ELF) Magnetic Fields (MFs) led to alterations in transcription activity of salivary gland cells.20-22 A more recent study, using transcriptomic analysis, revealed 439 upregulated genes and 874 downregulated genes following short-term exposure of male flies to ELFs, including genes involved in apoptosis, stress response and reproduction. In the same study, reproduction capacity was reduced by ELFs.23 Fecundity was also shown to be affected by RFs.24,25 As we have previously shown, exposure of adult flies to RF radiation apart from reduction of animal's fecundity also results in induction of apoptosis in the ovary and increase of ROS whole-body content.26-28 Thus, in an effort to unravel the complicated network that controls the interaction between non-ionizing radiation and ovarian cell sub-populations, we examined the critical cytopathic subroutines of oxidative stress and apoptotic death, in correlation with gene expression reprogramming, during early and middle oogenesis, after exposure of adult female flies to mobile-phone radiation.
Results
Experimental design
To study the potentially harmful effects of RF radiation on Drosophila ovarian tissue, 4 days-old adult female flies were irradiated with a GSM-1,800 MHz Talk mode mobile phone for 30 min. ROS-content upregulation and cell death induction were both examined in parallel with global-profiling patterns of gene transcription. ROS levels were measured immediately after the end of exposure, adapting a protocol of our earlier studies28 while apoptosis was detected 4 h later. Since previous time-course experiments have identified apoptosis to be activated 4 h post-exposure,29 it is this time-point of 4 h that was finally chosen for the present study. Based on our ROS- and apoptosis-related obtained data, a 2-h post-exposure, intermediate, time-point was chosen as the most suitable and informative one for the genome-wide gene-microarray analysis (Supplementary Fig. 1).
Mobile-phone radiation promotes ROS-content elevation and apoptosis induction, during early and middle Drosophila oogenesis
A 30-min mobile-phone exposure led to increased ROS levels in the nurse-cell cluster of developing follicles (Fig. 1A). A statistically significant upregulation of 1.6-fold (x) averaged value was detected in the pre-choriogenic stages of germarium (2b) up to stage 10 (Fig. 1B), whereas the choriogenic stages 11–14 presented with practically unalterable ROS levels (data not shown). Interestingly, RF radiation proved capable to induce a 2x increase in the number of follicles with fragmented DNA in the RF radiation-exposed group, as compared with the sham one, specifically at the two major developmental checkpoints of fly oogenesis, namely the germarium (2b) and stages 7–9 (Fig. 1C and D).
Figure 1.
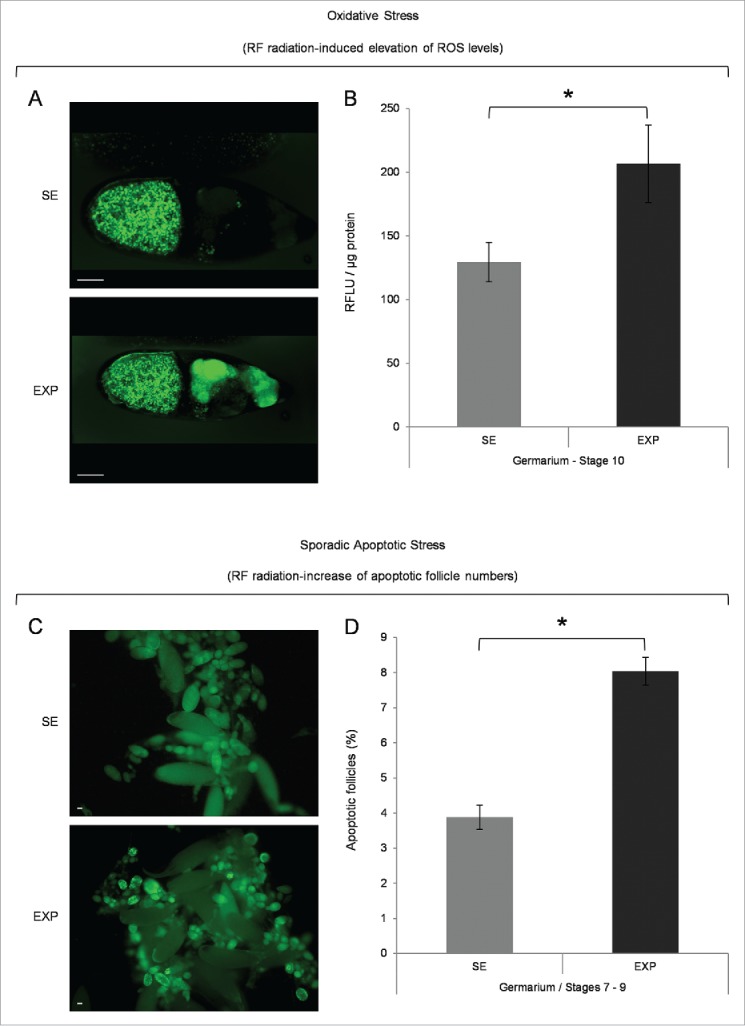
Mobile-phone radiation causes elevation of ROS cellular contents and activation of sporadic cell death, during early and middle oogenesis in Drosophila melanogaster. (A) Confocal laser scanning microscopy (CLSM) images illustrating representative ovarian follicles of stage 10, as they are stained with the general ROS-detector CM-H2DCFDA. Positively reagent-reacted nurse cells were detected only in ovaries of the exposed (EXP) flies (oocyte's green -spotty- signal emanates from auto-fluorescence of yolk granules). (B) Bar-graphs presenting ROS cellular levels ± SEM, as they are measured by suitably used fluorimetric assays, in control (SE: Sham Exposed) and truly exposed (EXP) follicles of all developmental stages from germarium (2b) up to stage 10. (C) Fluorescence microscopy images showing ovarian follicles from sham-exposed (SE) and truly exposed (EXP) flies, as they (follicles) are stained with the acridine orange apoptotic reagent. (D) The ratio of sporadically generated apoptotic follicles was estimated using the number of positively stained, with acridine orange, follicles, of early (germarium 2b) and middle (stages 7–9) oogenesis, against the total number of ovarian follicles of the same developmental stages ± SEM. Scale bars: 50 μm. N = 3, n = 3. SEM: Standard Error Mean. N: number of biological samples; n: number of biological experiments. *: P < 0.05.
Genome-wide transcriptional alterations in Drosophila ovary after exposure to mobile-phone radiation
Thorough statistical analysis between the exposed and sham-exposed samples revealed that 168 genes altered their transcriptional expression patterns at a notable and biologically important level (≥1.25x, P < 0.05) (Supplementary Table 1). More specifically, 153 genes were shown to be significantly activated in the exposed group, whereas 15 ones were found to be notably repressed under the same treatment conditions (Fig. 2). Among the upregulated genes, 35 ones presented with more than 1.5x increase value; whereas 4 genes reduced their expression levels less than 0.66x in the exposed group, compared with the sham one (Fig. 3A). It seems that the majority of ovarian genes identified herein were upregulated after RF-radiation exposure, while only a small number of them were downregulated, therefore suggesting that the primary genome-wide response of fly ovary to mobile-phone radiation is the global gene activation.
Figure 2.
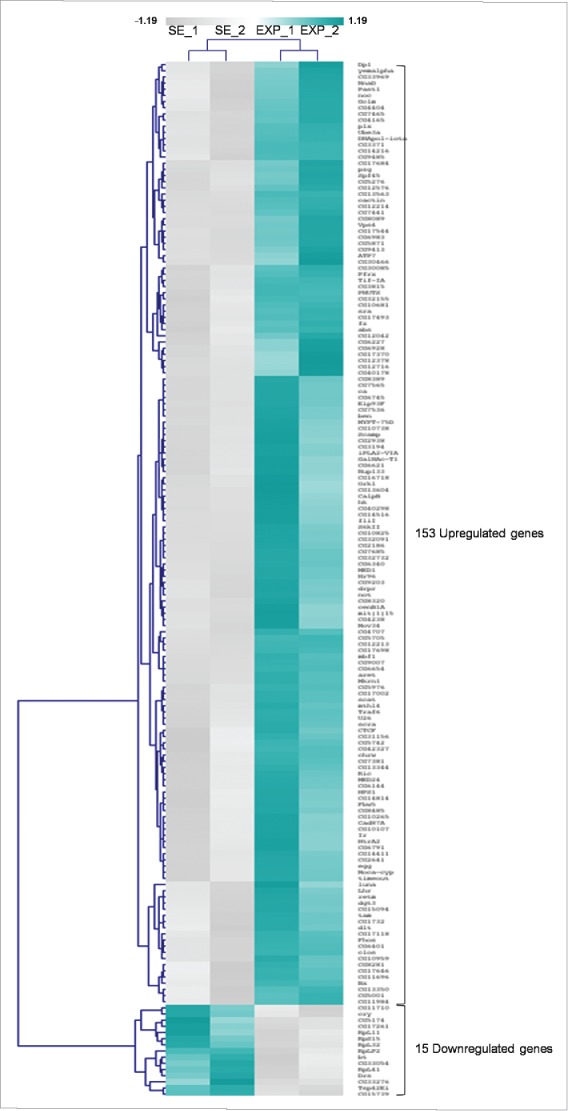
Mobile-phone radiation alters gene-expression profiling mainly through a transcriptional activation process: a genome-wide approach in fly ovary. Heat-map showing normalized expression levels of 168 identified DEGs (≥1.25 fold, P < 0.05), after application of a microarray-based technology, with their (DEGs) vast majority following an upregulated pattern of expression (compared with control conditions), in response to RF-radiation exposure. Gene-clustering was performed using Euclidean distance and average linkage-analysis software. Light-blue color indicates genes with upregulated levels of expression (153), while gray color specifies genes with downregulated levels of expression (15). SE: Sham Exposed. EXP: Exposed (truly). N = 2, n = 2. N: number of biological samples; n: number of biological experiments. *: P < 0.05.
Figure 3.
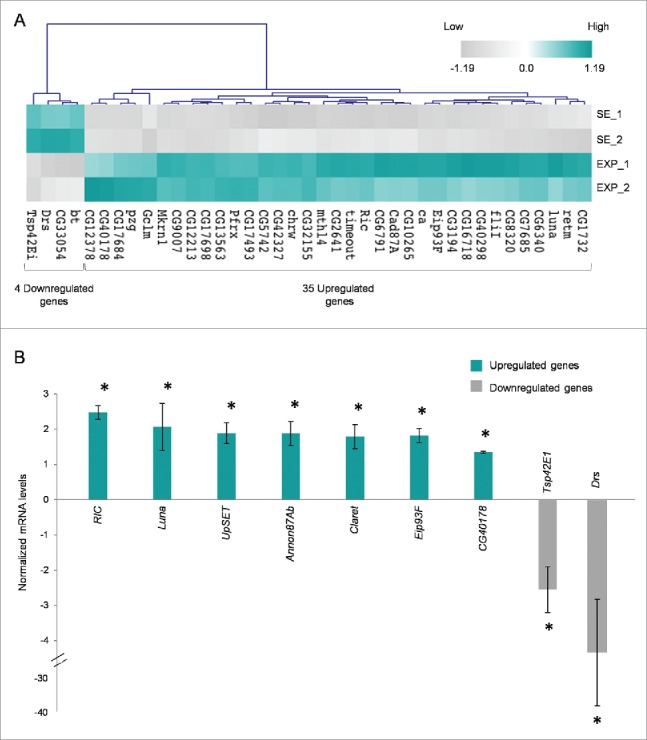
Mobile-phone radiation notably reprograms gene-expression activity in Drosophila melanogaster ovary. (A) Heat-map of DEGs being featured by prominent transcriptional perturbation (≥1.5x, P < 0.05), after RF radiation-induced stress in fly ovary. Gene-clustering was performed using 1.5x cut off, Euclidean distance and average linkage-analysis subroutine. Light-blue color indicates genes with upregulated levels of expression (35), while gray color specifies genes with downregulated levels of expression (4). SE: Sham Exposed. EXP: Exposed (truly). N = 2; n = 2. (B) Validation of gene-microarray data by suitably used RT-qPCR (real time-quantitative polymerase chain reaction) protocols. Bar graph presenting normalized mRNA levels ± SEM. N = 2; n = 3. SEM: Standard Error Mean. N: number of biological samples; n: number of biological experiments. *: P < 0.05.
Nine genes were selected for further verification of the microarray-platform analysis applied. Selection criteria were mainly based on gene's annotation bioinformatics data, to cover a variety of biological processes. In particular, FBgn0036398 (UpSET) and FBgn0040765 (Luna) were related to oogenesis, FBgn0013948 (Ecdysone-induced protein 93F – Eip93F), FBgn0000247 (Claret) and FBgn0260742 (Anon87A) to cell death, FBgn0017549 (Ras-related protein interacting with calmodulin – RIC) and FBgn0058178 (CG40178) to stress response, and FBgn0010381 (Drosomycin – Drs) to innate immune response, while the protein product of CG (Tetraspanin – Tsp42Ei) was previously identified as a plasma membrane component. RT-qPCR used protocols undoubtedly confirmed the microarray-derived expression profiling for all the 9 examined genes (Fig. 3B). RIC and Luna presented with more than 2x transcriptional induction in response to RF radiation, while UpSET, Annon87Ab, Claret and Eip93F followed a close to 2x upregulation of their gene-expression capacities under the same stress conditions. On the contrary, Tsp42E1 and Drs were both characterized by a significant RF radiation-directed reduction of their respective transcriptional activities, with Drs gene being strongly downregulated at an approximate level of 4x, therefore suggesting the plausible mobile-phone radiation-induced compromise of immune proficiency in fly ovary.
RF radiation-driven perturbation of transcriptional integrity was further analyzed through Gene Ontology (GO), DAVID and PANTHER bioinformatics resources, and the differentially expressed genes (DEGs), identified herein, were classified and reassembled into networks, according to the molecular function, cellular topology and biological process of their respective cognate products. By using the DAVID platform, its “Functional Annotation” subroutine and “Cellular Compartmentalization” category, revealed that only 3 proteins were recognized as “Extracellular Matrix” (Fold Enrichment 0.42) elements, whereas, regarding the “Intracellular Region” 7 proteins were identified in the “Cytosol” (Fold Enrichment 1.16) and 26 proteins were categorized as “Plasma Membrane” components (Fold Enrichment 0.74). Surprisingly, the majority of our DEGs (in response to RF radiation) encoded proteins that were tightly associated with organelles' structure and function (52 proteins, Fold Enrichment 1.12), while 31 of them were embraced under the description of “Nuclear proteins” (Fold Enrichment 1.27). Interestingly, there were also gene products recognized to be involved either in secretory and trafficking interrelated networks, such as the “Endosome,” the “Golgi Apparatus” and the “Endoplasmic Reticulum” or in organelles involved in translation and metabolism namely ribosome and mitochondria (Fig. 4A). All five DEGs encoding “Ribosomal Proteins” were downregulated in response to RF radiation (Supplementary Fig. 2A)
Figure 4.
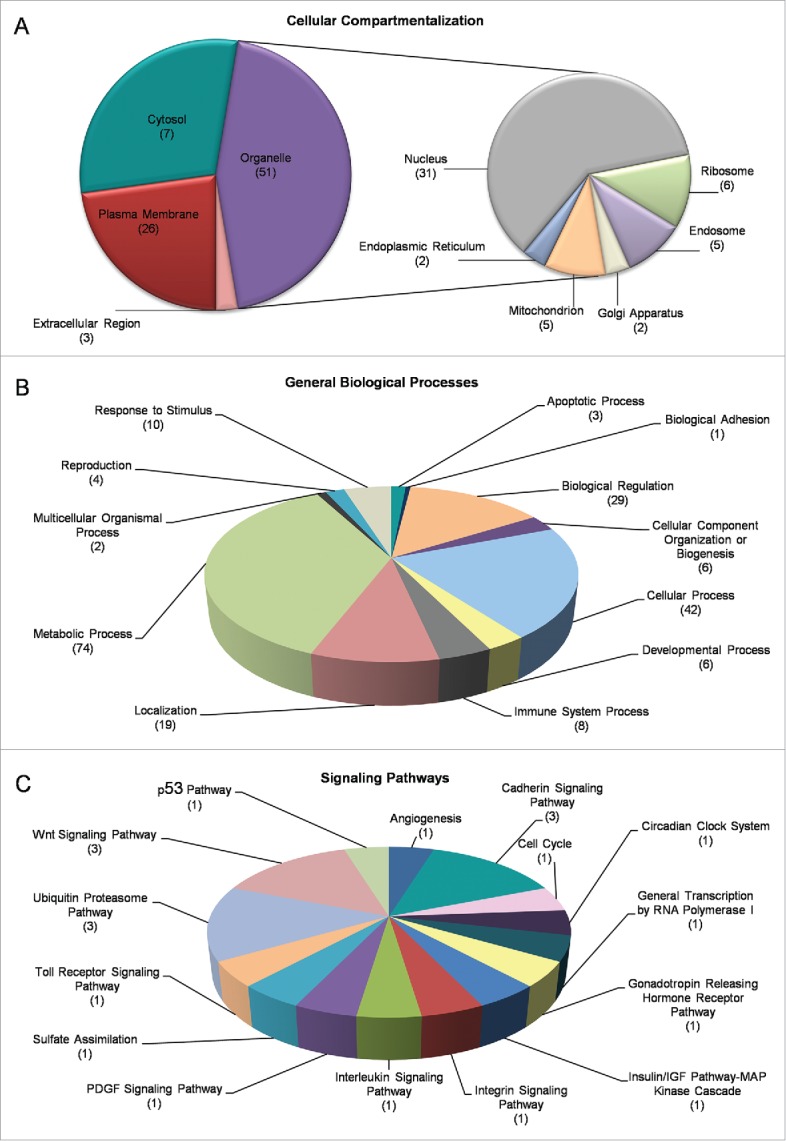
Gene Ontology (GO) and cluster classification analysis of DEGs. Drosophila-ovary genes that have proved to be differentially regulated and expressed by mobile-phone radiation were analyzed using the DAVID-GO bioinformatics subroutine, and they were sequentially categorized according to: (A) “Cellular Compartmentalization” and (B) “General Biological Processes.” (C) Using PANTHER bioinformatics platform, DEG transcripts were also grouped in multiple and diverse “Signaling Pathways.” The number of genes that could be classified in each different category is indicated in parenthesis.
RF radiation-specific DEGs also presented with certain features that clearly engage them (DEGs) in diverse biological processes, such as “Metabolism” (74 proteins), “Adhesion” (1 protein), “Apoptosis” (3 proteins), “Reproduction” (4 proteins), “Development” (6 proteins) and “Immunity” (8 proteins) (Fig. 4B), as well as in multiple signal transduction pathways, with “Ubiquitin/Proteasome” (3 proteins), “Wnt” (3 proteins) and “Cadherin” (3 proteins) being the most pronounced ones among all (Fig. 4C). According to GO annotations, the group “Cellular Process” (comprises proteins involved in “Vesicle-mediated Transport” (16), “Membrane Organization”/“Invagination” (14), “Endocytosis” (13), “Phagocytosis” (10) and “Engulfment” (9), while the category “Metabolic Process” mainly refers to catabolic pathways targeting protein degradation, such as “Proteolysis” (22), and “Modification-dependent Protein Catabolism” subroutines (12) (data not shown). KEGG pathway analysis revealed 5 out of 13 proteins having been identified by DAVID to critically contribute to “Endocytic Network” functions (Supplementary Fig. 2B).
Next, we classified our DEGs via the “Molecular Function” filter and two pivotal groups, namely the “Binding Interactions” and “Catalytic Activities,” could be reliably recognized. “Binding Interactions” included, among others, “Protein Binding” (81), “DNA Binding” (19), “RNA Binding” (15), “Zinc-ion Binding” (28), “Calcium-ion Binding” (8) and “ATP Binding” (15) protein members (Fig. 5A). “Catalytic Activities,” interestingly, embraced 38 gene products with “Hydrolase” (e.g. “Peptidase,” “Lipase,” “Phosphatase,” “Oxidoreductase” and “Lyase”) activity, 15 with “Transferase Activity” and 8 with “Transporter Activity.” Notably, 14 “Transcription Factors” and 7 “Receptors” were identified to be likely implicated in the cellular responses of fly ovary to mobile-phone radiation-induced stress (Fig. 5B).
Figure 5.
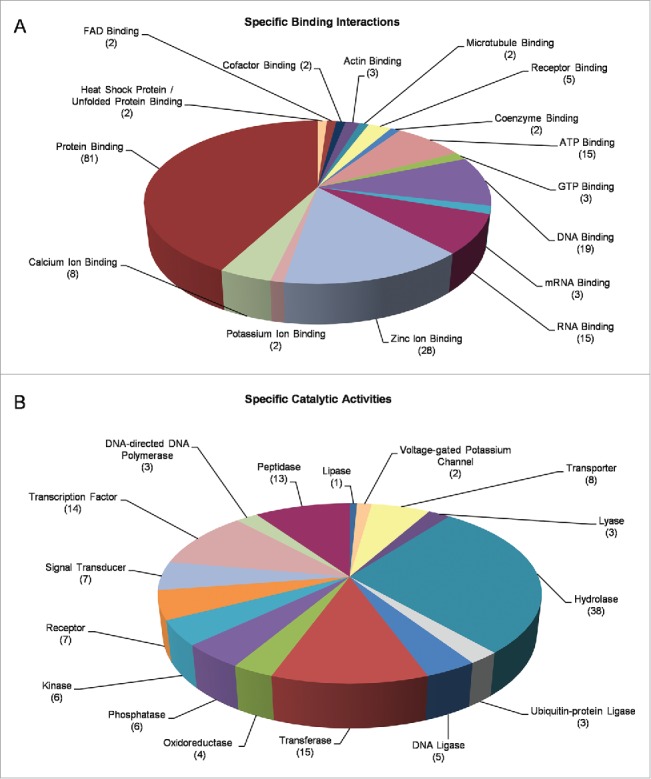
Molecular categorization of DEGs through specific functional annotation. Fly-ovary genes shown to be differentially controlled and expressed in response to mobile-phone radiation exposure were processed through engagement of DAVID-GO bioinformatics tool and they were further classified according to their: (A) “Specific Binding Interactions” and (B) “Specific Catalytic Activities.” The number of genes that could be embraced by each different category is indicated in parenthesis.
Taken together, our data strongly support the notion that RF-radiation exposure critically modulates the transcriptional capacity and expression of a diverse group of genes that orchestrate essential cellular functions and processes in Drosophila ovary. However, the majority of RF radiation-specific DEG products seem to be preferentially targeted and localized onto sub-cellular organelles. This likely indicates a novel, but still elusive, cardinal role of “organelome” (the assembly of all functional organelles in a cell) in ovarian cell-cluster transcriptome dynamics, and vice versa, during certain (e.g., mobile-phone radiation) stress conditions.
Functional categorization of RF radiation-specific DEGs into stress-related and apoptotic death-promoting groups
The operation of an RF radiation-driven cytotoxic network critically orchestrated by ROS and apoptotic determinants prompted us to further analyze our DEGs with regard to their contribution to stress response and cell death processes, by suitably using the powerful bioinformatics platforms of GO categories in Flybase, DAVID and FlyAtlas (http://flyatlas.org) interfaces. Fifteen genes were identified to be involved in cellular responses to stress, with special emphasis on genotoxic (DNA damage) and oxidative stress (Table 1). Surprisingly, Cryptochrome, whose protein product negatively regulates nucleic acid metabolism and DNA repair, and CG33276 gene, whose thiocarboxylated protein form is an oxidative-stress promoter, were both proved to be transcriptionally downregulated in response to RF radiation, thus suggesting the activation of counteractive mechanisms engaged in cellular detoxification from mobile-phone radiation-induced harms.
Table 1.
Stress-related DEGs upon mobile-phone radiation. Stress-related genes, transcriptionally modified by mobile-phone radiation in Drosophila melanogaster ovarian tissue (>1.25x, P < 0.05). Gene transcription upregulation, or downregulation, is shown (fold and arrows) for the exposed vs. Sham-exposed (control) fly groups.
| Gene Title | Official Gene Symbol | Flybase Clone ID | Molecular Function (GO, INTERPRO, Flybase, DAVID) | Fold (x) | Transcriptional Regulation Response | P value |
|---|---|---|---|---|---|---|
| Glutamate-cysteine ligase modifier subunit | Gclm | FBgn0046114 | Cellular response to DNA damage stimulus | 1.8 | 0.026 | |
| CG40178 | FBgn0058178 | Cell redox homeostasis | 1.78 | 0.038 | ||
| Ras-related protein interacting with calmodulin | Ric | FBgn0017549 | Response to oxidative stress | 1.74 | 0.030 | |
| Methuselah-like 4 | Mthl4 | FBgn0034219 | Response to stress | 1.57 | 0.015 | |
| Timeout | Timeout | FBgn0038118 | DNA damage checkpoint | 1.54 | 0.029 | |
| Subdued | CG16718 | FBgn0038721 | Detection of temperature stimulus involved in sensory perception of pain | 1.52 | 0.042 | |
| TNF-receptor-associated factor 6 | Traf6 | FBgn0026318 | Stress-activated protein kinase signaling cascade | 1.39 | 0.015 | |
| Hormone receptor-like in 96 | Hr96 | FBgn0015240 | Response to starvation | 1.38 | 0.013 | |
| Spf45 | CG17540 | FBgn0086683 | Response to DNA damage stimulus | 1.35 | 0.018 | |
| CG6227 | FBgn0030631 | DNA repair | 1.35 | 0.045 | ||
| Scamp | Scamp | FBgn0040285 | Response to stress, Response to heat | 1.34 | 0.034 | |
| DNApol-iota | DNApol-iota | FBgn0037554 | Cellular response to stress, Damaged DNA binding | 1.29 | 0.006 | |
| CG5001 | FBgn0031322 | Unfolded protein binding, Response to heat, Chaperonin | 1.27 | 0.038 | ||
| CG33276 | FBgn0053276 | Positive regulation of oxidative stress | 0.68 | 0.041 | ||
| Cryptochrome | Cry | FBgn0025680 | Negative regulation of DNA repair | 0.67 | 0.028 |
In parallel with the stress-related genes described in Table 1, there were also DEGs strongly associated with programmed cell death. Table 2 presents 10 genes being transcriptionally upregulated after mobile-phone radiation exposure, and also critically implicated in apoptotic and/or autophagic death. Interestingly, among them, besides the apoptotic ones, 4 genes were identified to promote autophagy, therefore suggesting the ability of apoptosis and autophagy to synergistically compel RF radiation-exposed fly-ovary cells to death. Altogether, these findings demonstrate that transcriptional expression of stress-, apoptosis- and autophagy-related genes can be notably increased upon exposure to mobile-phone radiation, and they can likely contribute to its damaging power in Drosophila ovary.
Table 2.
Cell-death-related DEGs upon mobile-phone radiation. Programmed cell death-related genes, transcriptionally activated by mobile-phone radiation in Drosophila melanogaster ovarian tissue (>1.25x, P < 0.05). Gene transcription upregulation is denoted, for the exposed vs. control fly groups, in the form of fold and arrows.
| Gene Title | Official Gene Symbol | Flybase Code | Molecular Function (GO, INTERPRO, Flybase, DAVID) | Fold (x) | Transcriptional Regulation Response | P value |
|---|---|---|---|---|---|---|
| Anon 87Ab | CG12213 | FBgn0260742 | Positive regulation of JAK/STAT | 1.75 | <0.001 | |
| Claret | Ca | FBgn0000247 | Regulation of autophagy | 1.64 | 0.015 | |
| Ecdysone-induced protein 93F | Eip93F | FBgn0013948 | Activation of caspase activity, Autophagic cell death | 1.57 | 0.015 | |
| TNF-receptor-associated factor 6 | Traf6 | FBgn0026318 | Cell death, Positive regulation of autophagy | 1.39 | 0.015 | |
| HTRA2-related serine protease | HtrA2 | FBgn0038233 | Regulation of programmed cell death | 1.37 | 0.046 | |
| Bendless | Ben | FBgn0000173 | Positive regulation of cell death, Positive regulation of JNK cascade | 1.34 | 0.020 | |
| Abstract | Abs | FBgn0015331 | Apoptosis, Programmed cell death | 1.32 | 0.031 | |
| Draper | Drpr | FBgn0027594 | Apoptotic cell clearance | 1.31 | 0.023 | |
| Mediator complex subunit 24 | MED24 | FBgn0035851 | Positive regulation of programmed cell death | 1.31 | 0.033 | |
| Vacuolar protein sorting 4 | Vps4 | FBgn0027605 | Autophagic vacuole fusion | 1.27 | 0.012 | |
| CG5001 | FBgn0031322 | Cell death, Apoptosis | 1.27 | 0.038 |
Identification and molecular wiring of DEG human orthologs into functional networks
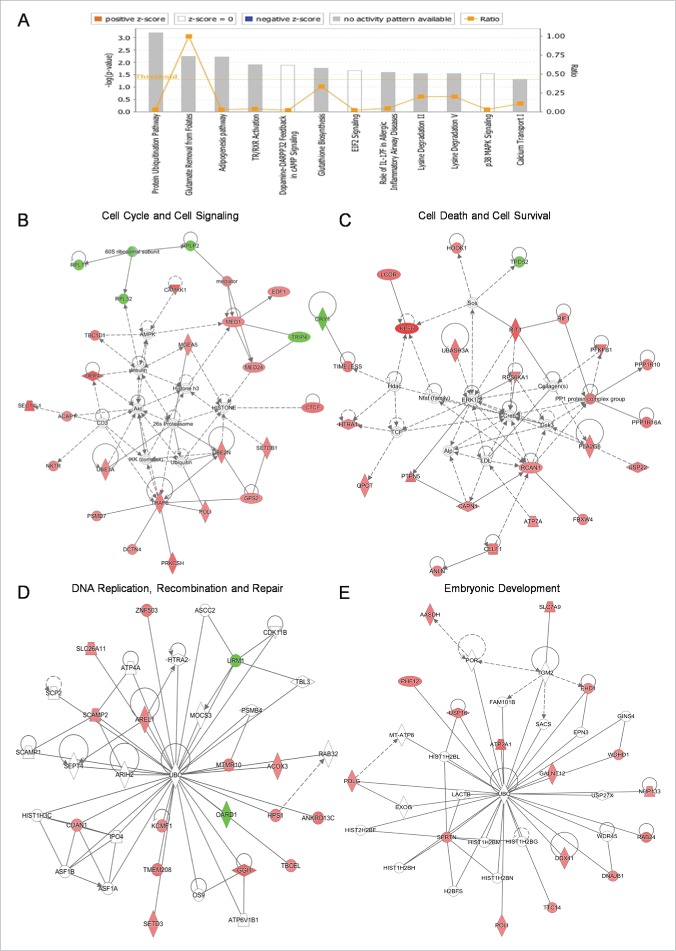
Through employment of PANTHER bioinformatics tool, we were able to identify 3 Drosophila DEGs whose human counterparts are strongly related to baneful diseases. In particular, the human orthologs of Frizzled and CG16718 Drosophila genes have been previously shown to critically control Alzheimer's disease,30,31 while Calpain B has been tightly linked to Huntington's disease.32,33 Hence, we performed a more detailed and thorough analysis based on human orthologs that, in general, are better characterized and more completely annotated. Human orthologs were identified via Mouse Genome Database (MGD; MGI 5th version; http://www.informatics.jax.org) platform34 From the list of RF radiation-modulated genes, 124 ones presented with a human ortholog each and, among them, 113 were transcriptionally upregulated, while only 11 genes followed a downregulation expression pattern in response to the applied stress. By using the Ingenuity Pathway Analysis (IPA) tool, it proved that these genes could be reliably categorized in diverse networks of cellular patho-physiology. p38-MAPK-dependent signaling (Supplementary Fig. 3A) and Glutathione biosynthesis (Supplementary Fig. 3B), known to essentially orchestrate stress responses, were among the principally altered pathways. Similar gene-product wiring protocols allowed the molecular reconstruction of 8 functional networks, mainly implicated in “EIF2 Signaling” and “Protein Ubiquitination” (Fig. 6A and B), in “Cell Cycle” and “Signal Transduction” (Fig. 6B and C), in “Cell Death” and “Cell Survival” (Fig. 6C), in “DNA Replication,” “DNA Recombination” and “DNA Repair” (Fig. 6D), and in “Embryonic Development” (Fig. 6E) processes, therefore unveiling the non-targeted, multifaceted and versatile detrimental power of mobile phone-emitting radiation over fly ovary and, in general, other critical (mitotically active) organs of living organisms of different evolutionary levels, including humans.
Figure 6.
Figure 6. Cataloguing and functional clustering of DEG human orthologs through engagement of Ingenuity Pathway Analysis (IPA) tool. (A) Bar-graphs denoting fly-ovary DEGs with statistically significant (P < 0.05) homologies to human counterparts and their cognate reconstructed protein pathways. Ratio is referred to the number of genes from our DEG list [comparison of their transcriptional activities between SE (sham exposed) and EXP (truly exposed) fly groups] divided by the total number of genes involved in each specific pathway that has been notably affected by flies' exposure to mobile-phone radiation. Threshold is determined at P value 0.05 [-log (0.05) = 1.30]. To the same direction, RF-radiation exposure proved able to differentially perturb the transcriptional-expression profiling of several genes (DEG human orthologs) being critically implicated in: (B) “Cell Cycle” - “Cell Signaling,” (C) “Cell Death” - “Cell Survival,” (D) “DNA Replication” - “DNA Recombination” - “DNA Repair” and (E) “Embryonic Development,” as clearly revealed through suitable application of IPA bioinformatics tool. Red color indicates the upregulated gene products, while green color specifies the downregulated ones. Figures were automatically produced via IPA-software protocols.
Discussion
The last few decades there is a constantly increasing concern for the adverse effects of non-ionizing radiation on human health. The aim of the present study was to systemically characterize cell-cytopathy and gene-expression responses following exposure of Drosophila melanogaster to short-term mobile-phone radiation. Nearly a 2x ROS-content elevation proved as an immediate cellular response of fly's ovary, and specifically its pre-choriogenic developing follicles, to 30-min, in vivo, exposure of adult flies to a commercial mobile phone emitting a GSM-Talk signal. Next, 4 h post-exposure, a notable increase in sporadic apoptotic cell death was detected during early (germarium 2b) and middle (stages 7–9) oogenesis. These findings not only reinforce our previous conclusions regarding the sensitivity of Drosophila ovarian tissue to RF radiation,27 but are also consistent with earlier studies which have reported ROS-level upregulation and apoptosis activation in the germ-line cells of male rats, after their in vivo exposure to an RF field of 900 MHz.35,36
Following a molecular-system and time-dependent specific rationale, we, next, investigated the transcriptional responses of ovarian cell sub-populations to RF radiation 2 h after the end of exposure, by a genome-wide gene-expression profiling analysis and suitable correlations of the obtained data with available databases. Microarray-platform results indicated that in vivo exposure of young-adult flies, for just 30 min, to radiation being emitted by a mobile phone of GSM 1,800 MHz, at a low, 6 min-average specific absorption rate (SAR) value of 0.15 W/kg, well below the permissible human-head exposure (1.8 W/kg), is able to induce critical alterations in the transcriptional dynamics and gene-expression activities of early- and middle-stage follicles during Drosophila oogenesis, under the specific exposure set up and conditions. One hundred and sixty eight genes (168) were differentially expressed after RF radiation-induced stress and they could be classified in diverse biological-process reassembled networks, thus suggesting that RF radiation operates as a non-targeted and multifaceted cytotoxic agent that can simultaneously deregulate various cellular functions, ultimately inducing ovary's systemic pathology not only in flies but likely in humans, as well. Similarly, Remondini et al., via employment of human cell lines and primary cells, have previously reported that RF radiation is capable to promote a variety of alterations in gene expression programs.37 Interestingly, our DEG products presented with a strong localization preference onto sub-cellular organelles, therefore suggesting that mobile-phone radiation may not target specific molecules but rather multi-protein systems, such as the trafficking machinery and its organelles, critically controlling ovary's nutritional, biosynthetic, metabolic, energetic and signaling requirements during Drosophila gametogenesis, embryogenesis, post-embryonic development and aging. Whether the organelle-specific DEGs orchestrate the RF radiation-induced cytopathic subroutines, or direct a counteractive -defense- network that detoxifies cells from molecular “radio-hazards” is an open and challenging issue that definitely deserves to be pursued and successfully resolved (paper in preparation).
Among our identified DEGs and their cognate protein products, ∼9% (15 genes) were characterized as stress-related and ∼6% (10 genes) as death-related determinants. Notably, certain components of stress-associated critical pathways, like the p38-MAPK and GSH (Glutathione) biosynthetic ones, proved to follow an RF radiation-dependent upregulation in their gene expression activities. These data, together with the induction of oxidative (ROS-level increase) and apoptotic cytotoxicity, observed in the pre-choriogenic ovarian follicles, strongly indicate that mobile-phone radiation represents a hazardous stress factor, with an intrinsic apoptotic capacity that can seriously harm Drosophila oogenesis. Indeed, bioinformatics analysis revealed annotations including “cellular response to stress,” “response to DNA damage stimulus,” “DNA repair,” “proteolysis,” “positive regulation of caspase activity” and “JNK cascade.” Similar observations were reported 2 h post-exposure of Drosophila larvae to 4,000R of X-rays ionizing radiation, although the recorded value was above 2x.38
The RIC fly homolog of mammalian Rit/Rin, with a conserved role as regulator of p38-MAPK response to oxidative stress39 and the CG40178 gene, likely implicated in cell redox homeostasis, as foreseen via bioinformatics-mediated annotation, were both upregulated, in a statistically significant manner, in the exposed female-fly, as compared with the control, group. Mobile-phone radiation could also stimulate the transcriptional activity of Bendless (Ben) gene, whose cognate protein functions as an E2 ubiquitin-conjugating enzyme for dTRAF2, while it (Ben) positively regulates JNK signaling-dependent tumor growth and invasion, cell death, oxidative-stress resistance, and longevity in flies.40 These findings are consistent with previous observations of Lee et al.,41 who reported ROS-content elevation and JNK activation in Drosophila upon exposure to mobile-phone radiation.
Remarkably, 4 of our RF radiation-induced gene products were classified in “Autophagy,” while ∼6% (10 genes) of all identified DEGs and their cognate proteins were recognized as “Phagocytosis”-related components. Upregulation of autophagic-related genes was also observed in brain regions of mice exposed to 835 MHz RF at a SAR of 4.0 W/kg for 5 h/day for 12 weeks.42 Both cellular subroutines of autophagy and phagocytosis have been previously reported to operate during programmed cell death in Drosophila ovary.43,44 Claret (Rab32) gene that encodes a protein essentially regulating lipid storage and autophagy45 and Eip93F, whose product also controls autophagic cell death in Drosophila,46 where both, herein, presented with notable upregulation in response to the applied stress. Moreover, the cognate gene of Draper (Drpr) protein that is required for follicle-cell engulfment and JNK-pathway activation47 was also shown with a transcriptional activation profiling in the exposed, vs. control, fly group. Taken together, our data indicate that the RF radiation-induced cell death in fly ovary, besides apoptosis, may also be critically mediated by inducible autophagy, as well, with both processes operating in a likely synergistic fashion.
In addition to cell death subroutines, bioinformatics data analysis revealed that RF radiation-specific DEGs could be categorized in multiple cellular processes regulating fly oogenesis, such as “Vesicle-mediated Transport,” “Membrane Organization”/“Invagination” and “Endocytosis,” which have proved to play major roles in follicle development, as they serve for yolk protein accumulation in the developing oocyte.48-50 Yolk proteins are the sole nutritional and energetic sources for embryogenesis in Drosophila, and any disruption of their trafficking integrity and metabolic homeostasis may result in severe impairment of embryonic and post-embryonic fly development. Among others, it may be this mobile-phone radiation-directed malfunction of yolk-protein metabolism that likely results in the observed reduction of Drosophila offspring-production efficiency, upon exposure to the particular stress, as it has been previously reported by our group.27 Similarly, perturbation of embryonic development and increased mortality were observed after radiation exposure of chicken embryos to a mobile phone, intermittently 3 h per day during the incubation period.51
Tight regulation of oxidative stress represents an essential process for survival and growth of the affected cells. Production or detoxification of ROS, and their downstream transducing signals, and in general redox balance and homeostasis, seem to implicate several transcription factors in humans, with AP-1, FOXOs (3/4), HIF-1α, NRF2, p53 and NF-κB (REL) being the most critical ones.52-56 Tohidi et al.57 showed RF-dependent upregulation of p53 and p21genes, underlying that RF radiation can control the expression levels of principle transcription factors. Through application of bioinformatics tool, several fly-ovary DEGs proved to carry putative binding sites of the aforementioned transcription factors in their cognate proximal [-1 to −500 bp from TSS (transcription start site)] promoters (Table 3), therefore indicating the decisive contribution of ROS engagement to specific (e.g., NRF2) transcription factor-dependent genome-wide reprogramming of ovarian-gene expression, upon Drosophila exposure to mobile-phone radiation. For example, among others, it may be the redox imbalance that triggers the AP-1- and NRF2-mediated upregulation of Mkrn1 gene expression (Figs. 2 and 3, and Table 3), after mobile-phone radiation-induced stress in fly ovary. Each DEG's transcriptional response to RF-radiation cytotoxic power likely results (without excluding alternative mechanisms) from a distinct combination of ROS-dependent (regulated at the post-translational level) transcription factors (e.g., AP-1/NRF2 or FOXOs/NF-κB), which all together may be capable to successfully orchestrate the ensuing sporadic apoptotic death of Drosophila-ovary cellular compartments observed upon mobile-phone radiation exposure (Fig. 1). In an effort to unravel the mechanism between ROS origin and ROS-dependent transcriptional regulation following mobile-phone radiation exposure, plants might be an alternative model system to examine. Given that mitochondria are not the primary source of ROS production outside the animal kingdom,58 three studies conducted in plants showed ROS increase by RF exposure,59-61 therefore opening new mechanistic windows for the inter-relations among ROS, RF and gene activity.
Table 3.
DEGs with putative binding sites of ROS-dependent transcription factors. Catalog of fly-ovary DEGs being characterized by the in silico presence (P < 10−4) of the, herein, denoted transcription-factor (AP-1, FOXO3, FOXO4, HIF-1α, NRF2, p53 and REL/NFκB) binding sites in their (DEGs) (semi-)proximal -respective- promoter regions [-1 to −500 bp, upstream (5′) from the transcription start site (TSS: +1)]. Each one of the, herein, examined transcription factors has been previously reported to critically control redox homeostasis under certain cellular settings and stress conditions [41–45]. Identification of binding sites was performed with FIMO algorithm from MEME suite (http://meme-suite.org/) [46] with default settings, using the motifs identified from JASPAR database (http://jaspar.genereg.net), a high-quality transcription factor-binding profile database. The number of times (x) a binding site is recognized in an examined-gene (DEG) promoter is described in its respective parenthesis (e.g., 2x or 3x), when required to be given.
| AP-1[41] | FOXO3[42] | FOXO4[42] | HIF-1α[43, 44] | NRF2[43, 44] | p53[42, 44] | REL (NFκB)[43–45] | |
|---|---|---|---|---|---|---|---|
| CG1732 | + (2x) | + | + | + | |||
| CG7726 (RpL11) | + | + | + (2x) | + | |||
| CG8389 | + (2x) | + | + (2x) | ||||
| CG3815 | + | + | + | ||||
| CG5170 (Dp1) | + (2x) | ||||||
| CG18468 (Lhr) | + (2x) | ||||||
| CG10107 | + | ||||||
| CG14513 (Yemα) | + (3x) | ||||||
| CG32019(Bt) | + | + | + | ||||
| CG2092 (Scra) | + | ||||||
| CG17697(Fz) | + | ||||||
| CG7184 (Mkrn1) | + | + | |||||
| CG6536 (Mthl4) | + | ||||||
| CG6401 | + | ||||||
| CG6190 (Ube3a) | + | ||||||
| CG30466 | + | ||||||
| CG17540 (Spf45) | + | ||||||
| CG14411 | + | ||||||
| CG1093(Plx) | + | ||||||
| CG10738 | + | ||||||
| CG5276 | + | + | + | ||||
| CG5871 | + | + | + (2x) | ||||
| CG8320 | + | ||||||
| CG6791 | + (2x) | ||||||
| CG32627 (NnaD) | + | ||||||
| CG11783 (Hr96) | + | ||||||
| CG2641 | + | + | + | ||||
| CG9485 | + | + | |||||
| CG7465 | + | + | |||||
| CG6977 (Cad87A) | + | + | |||||
| CG6958 (Nup133) | + (2x) | + (2x) | |||||
| CG6072 (Sra) | + | + | |||||
| CG5976 | + | + | |||||
| CG3870 (Chrw) | + | + | |||||
| CG32155 | + | + | |||||
| CG2186 | + | + | |||||
| CG17596 (S6kII) | + | + | |||||
| CG17370 | + (3x) | + (3x) | |||||
| CG14516 | + | + | |||||
| CG13350 | + | + | |||||
| CG10961 (Traf6) | + | + | |||||
| CG10959 | + | + | |||||
| CG10653 (Hk) | + | + | |||||
| CG12214 | + | + | + | ||||
| CG8332 (RpS15) | + | + | |||||
| CG7685 | + | + | |||||
| CG3194 | + | + | |||||
| CG1866 (Moca-cyp) | + | + | |||||
| CG17261 | + | + | |||||
| CG10265 | + | + | |||||
| CG5742 | + | ||||||
| CG9007 | + | ||||||
| CG6718 (IPLA2-VIA) | + | ||||||
| CG32315 (Dlt) | + | ||||||
| CG7162 (MED1) | + | ||||||
| CG6148 (Past1) | + |
Conclusively, our data strongly suggest that, in the ovarian developing follicles of D. melanogaster model system, pulsed, non-ionizing radiation, being emitted by commercially available mobile-phone devices, triggers genome-wide and non-targeted transcriptional perturbation of gene-expression profiling, likely through ROS-dependent and oxidative stress-mediated mechanisms, which, all together, can subsequently compel sensitive ovarian-cell sub-populations to sporadic apoptotic death. We expect that similar RF radiation-specific pathology could be likely observed in other critical fly organs, such as the brain, heart and intestine (paper in preparation), while complete or partial failure of sensitive organic systems (e.g., ones with strong mitotic and/or metabolic activities) in humans could be also associated or attributed (even to some extent) to mobile-phone wrong management, bad practice and/or overuse.
Materials and methods
Fly culture
The wild type Oregon R strain of Drosophila melanogaster was maintained at 24°C, as described previously.27 To start the experiment, newly emerged (2–4 h old after pupal eclosion) female flies (10 individuals per vial) were randomly collected and subsequently incubated in new vials with fresh medium at 24°C. Flies were maintained in opened, plastic vials, covered with cotton and fed on standard diet.
Exposure conditions
Four days-old, adult and synchronized female flies were exposed to a commercial cell phone GSM-Talk 1,800 MHz signal, for 30 min as described before.27 Flies were kept at 2 cm distance from the cell-phone. Sham-exposed (control) flies of the same age as the exposed ones were placed close to the same but switched off mobile phone for 30 min, which (phone) in this case was switched off. Cell phone was operating like a normal call with voice activation. Averaged E-field intensity value was measured at 10 V/m for 6 min, in accordance with guidelines and limits specified by the International Commission on Non-Ionizing Radiation Protection (ICNIRP).62 NARDA SRM3000 (Narda Safety Test Solutions, German) and Rohde & Swartz FSL/6 (Rohde & Schwarz GmbH & Co KG, German) spectrum analyzers were used via utilization of near-field probes. Power density was calculated at a level of 0.27 W/m2.
The conductivity value σ for fruit fly, according to Wang et al.,63 should be about 1.5 S/m at 1,800 MHz, which at 20°C results in a Specific Absorption Energy (SAE) of 270 J/kg at E = 10 V/m, density (ρ) = 1,000 kg/m3, and time (t) = 1,800 sec, according to the formula; SAE = (σ·E_RMSˆ2)/ρ t J/kg. SAR was calculated to be equal to 0.15 W/kg, according to the formula SAR = (σ·E_RMSˆ2)/ρ W/kg.
A recent publication by Zhang et al.64 concluded that thermal stress weakened the physiologic function and promoted the HSR (Heat Shock Response) and OS (Oxidative Stress) of flies. ELF-EMF aggravated damages and enhanced thermal stress-induced HSP and OS responses. Therefore, thermal stress and ELF-EMF elicited a synergistic effect. Thus, temperature was monitored during the whole time of exposure and no increase was observed during irradiation. Three replicates per experimental condition (30 female flies, in total) and three independent biological experiments were performed.
ROS detection
Female flies were anaesthetized and their ovaries were manually dissected in Phosphate-Buffered Saline (PBS) solution. Ovarian follicles were carefully collected and subsequently separated into two distinct groups. The first group contained follicles from all developmental stages up to stage 10, while the second one embraced follicles from stage 11 to 14. ROS cellular levels in the examined follicles were quantified via employment of a fluorimetric assay, using the CM-H2DCFDA (Life Technologies, USA) general ROS-species detector,65 as previously described.28 ROS imaging was performed after the incubation of Sham-exposed (control) and mobile phone-treated follicles with 1 μM of CM-H2DCFDA reagent, for 30 min, in the dark. CM-H2DCFDA ester was, next, removed and follicles were treated with PBS, for 20 min, in the dark, under shaking conditions. Finally, ovarian follicles were extensively washed three times, carefully placed on a vidal slide and thoroughly visualized under a Nikon Eclipse TE-2000U fluorescent microscope equipped with confocal laser compartments (Nikon Instruments, Japan).
Apoptotic assay: Acridine orange staining
Apoptotic ovarian follicles were detected as previously described,26 via employment of a staining protocol based on the acridine orange reagent (Life Technologies), which represents a specific dye widely used in Drosophila research as a reliable and valid indicator of apoptosis.66-68 Briefly, follicles were stained with 1.6 μM acridine orange dye (Invitrogen) in Ringer's solution for 5 min in the dark with constant shake. After recovery incubation for 5 min with Ringer’ s solution and three quick washes follicles were placed on a vidal slide and observed under a Nikon Eclipse TE-2000U fluorescent microscope. Percentage of sporadic dying follicles was calculated as the ratio of follicles, from germarium up to stage 10, presenting positive acridine orange staining signal (DNA fragmentation) to the total number of same-stage follicles.
RNA extraction, amplification and hybridization
Total RNA was isolated 2 h post-exposure. Sham-exposed (control) and mobile-phone radiation-exposed flies were subjected to light anesthesia (with ether) and ovaries were rapidly, but carefully, dissected in 1x Ringer's solution. Separated ovarian follicles of all developmental stages, up to stage 12, were collected and homogenized in Trizol reagent, and finally stored at −80°C, waiting for the gene-microarray protocol to be applied. To ensure repeatability and reduced background (transcriptional noise) levels, a large number of ovaries (180 per sample, for each experimental condition) were purified and used, while three independent experiments were performed, each time, to exclude random variations for all used protocols. Likewise, gene-microarray transcriptional assays were independently performed twice, to ensure the reliability and validity of the applied method itself. Follicles from three independent experiments per condition were pooled together, to form a single, unified and large sample containing approximately 11,500 follicles. Two sham-exposed and two authentically exposed samples were, next, processed for RNA extraction and subsequent gene-microarray analysis. RNA was isolated via chloroform treatment and was further purified following standard procedures (Nucleospin RNA isolation kit; Macherey-Nagel, Germany). RNA concentration and purity were determined on a NanoDrop 2,000 Spectrophotometer (NanoDrop Technologies, USA), while each preparation's integrity was assessed using Agilent 2,100 Bioanalyzer (Agilent Technologies, USA). Two hundred nanograms of total RNA were prepared for gene expression-profiling analysis with GeneChip® 3′ IVT Plus Reagent kit (Affymetrix, USA). Briefly, complementary RNA (cRNA) was generated by amplification and biotin-labeling of poly-[A+] RNA, according to Affymetrix recommended protocol. Twelve micrograms of purified cRNA were fragmented after incubation with 3′ Fragmentation Buffer (Affymetrix, USA), for 35 min, at 94°C and they were finally used to hybridize Affymetrix Drosophila Genome 2.0 Array Chip, according to manufacturer's instructions. The chip comprised 18,880 probe sets and was designed to analyze over 18,500 unique transcripts.
Microarray-data processing, normalization and statistical analysis
Microarray analysis was performed with the R statistical-environment version 2.13, using the Bioconductor package. MAS5.0 algorithm was applied for background correction, followed by array median normalization, and probes were log2 transformed. Presence/absence calls from MAS5.0 algorithm were used to classify probes/genes as expressed or non-expressed transcriptional entities. To discard probes representing the same RNA transcript, we selected ones with the highest average intensity value across all samples. Student's two-tailed t-test was used to identify differentially expressed genes with a cut-off value of P<0.05 and fold-change cut-off value of 1.25. Clustering of the differentially expressed genes was performed with Euclidean distance metric and average linkage in MeV (MultiExperiment Viewer) software. The microarray data (GSEXXXX) were submitted to GEO.
Functional annotation and pathway analysis
Gene ontology analysis was performed on all the differentially expressed genes using Drosophila Gene Ontology (GO) (http://www.geneontology.org) and Flybase (http://flybase.org). Additionally, DAVID (Database for Annotation, Visualization and Integrated Discovery) bioinformatics resources 6.7, NIAID/NIH (http://david.abcc.ncifcrf.gov),69,70 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway maps (http://www.genome.jp/kegg/pathway.html),71,72 Protein ANalysis THrough Evolutionary Relationships (PANTHER) classification system (http://www.pantherdb.org)73,74 and Ingenuity Pathway Analysis (IPA) (http://www.ingenuity.com) functional annotation online analysis tools were used for genes' classification with default settings. All bioinformatics analyses were performed with the list of 168 differentially expressed genes (DEGs), i.e. with both the upregulated and the downregulated ones, using the default settings of bioinformatics databases.
Real-time quantitative PCR
To confirm gene microarray-based transcriptional-profiling data, an aliquot of each RNA sample, previously having been analyzed by its respective microarray reaction, was used for further validation of the results, via a RT-qPCR (real time-quantitative polymerase chain reaction) approach. Next, DNA was digested, for 1 h, at 37°C, using HaeIII and DNase I (New England Biolabs, USA). Reverse transcription was performed on 8 μg of total RNA, using RevertAid H Minus Reverse Transcriptase (Fermentas, USA), following manufacturer's instructions. Real-time qPCR was performed with the use of gene-specific primers (Supplementary Table 2) and Maxima SYBR Green/ROX qPCR Master Mix (Thermo Fisher Scientific Inc., USA), on a Bio-Rad CFX 96 real-time system (Bio-Rad, USA). Relative mRNA expression levels were normalized to Actin5C gene of reference. Samples were duplicated (n = 2) and real-time qPCR was repeated 3 times for every gene (N = 3). Statistical analysis was performed by independent t-test, using the Welch approach.
Statistical analysis
Each experiment was performed three independent times (n: number of biological experiments) and each time samples were duplicated (N: number of biological samples), unless stated otherwise. Results are presented as Averaged Mean ± Standard Error Mean (SEM). Data were analyzed through IBM SPSS (Statistical Package for Social Sciences, v22) and presented a normal distribution as it was determined via Shapirov-Wilk test. Statistical significance between exposed and sham exposed groups was evaluated through Independent T-test, unless stated otherwise.
Supplementary Material
Disclosure of potential conflicts of interest
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Funding
This research has been co-financed by the European Union (European Social Fund – ESF) and Greek national funds through the Operational Program “Education and Lifelong Learning” of the National Strategic Reference Framework (NSRF) - Research Funding Program: THALIS –UOA- - MIS-375784: “Biological effects of non-ionizing electromagnetic radiation: a multidisciplinary approach” grant coordinated by L.H. Margaritis. A.K. Manta is supported by this grant for PhD fulfillment.
References
- [1].Yakymenko I, Tsybulin O, Sidorik E, Henshel D, Kyrylenko O, Kyrylenko S. Oxidative mechanisms of biological activity of low-intensity radiofrequency radiation. Electromagn Biol Med 2016; 35:186-202; PMID:26151230; http://dx.doi.org/ 10.3109/15368378.2015.1043557 [DOI] [PubMed] [Google Scholar]
- [2].Sastre J, Pallardo FV, Vina J. Mitochondrial oxidative stress plays a key role in aging and apoptosis. IUBMB Life 2000; 49:427-35; PMID:10902575; http://dx.doi.org/ 10.1080/152165400410281 [DOI] [PubMed] [Google Scholar]
- [3].Mracek T, Pecina P, Vojtiskova A, Kalous M, Sebesta O, Houstek J. Two components in pathogenic mechanism of mitochondrial ATPase deficiency: energy deprivation and ROS production. Exp Gerontol 2006; 41:683-7; PMID:16581217; http://dx.doi.org/ 10.1016/j.exger.2006.02.009 [DOI] [PubMed] [Google Scholar]
- [4].Droge W. Free radicals in the physiological control of cell function. Physiol Rev 2002; 82:47-95; PMID:11773609; http://dx.doi.org/ 10.1152/physrev.00018.2001 [DOI] [PubMed] [Google Scholar]
- [5].Di Meo S, Reed TT, Venditti P, Victor VM. Role of ROS and RNS Sources in Physiological and Pathological Conditions. Oxid Med Cell Longev 2016; 2016:1245049; PMID:27478531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].De Iuliis GN, Newey RJ, King BV, Aitken RJ. Mobile phone radiation induces reactive oxygen species production and DNA damage in human spermatozoa in vitro. PLoS One 2009; 4:e6446; PMID:19649291; http://dx.doi.org/ 10.1371/journal.pone.0006446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Burlaka A, Selyuk M, Gafurov M, Lukin S, Potaskalova V, Sidorik E. Changes in mitochondrial functioning with electromagnetic radiation of ultra high frequency as revealed by electron paramagnetic resonance methods. Int J Radiat Biol 2014; 90:357-62; PMID:24597749; http://dx.doi.org/ 10.3109/09553002.2014.899448 [DOI] [PubMed] [Google Scholar]
- [8].Friedman J, Kraus S, Hauptman Y, Schiff Y, Seger R. Mechanism of short-term ERK activation by electromagnetic fields at mobile phone frequencies. Biochem J 2007; 405:559-68; PMID:17456048; http://dx.doi.org/ 10.1042/BJ20061653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Zhao TY, Zou SP, Knapp PE. Exposure to cell phone radiation up-regulates apoptosis genes in primary cultures of neurons and astrocytes. Neurosci Lett 2007; 412:34-8; PMID:17187929; http://dx.doi.org/ 10.1016/j.neulet.2006.09.092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zalata A, El-Samanoudy AZ, Shaalan D, El-Baiomy Y, Mostafa T. In vitro effect of cell phone radiation on motility, DNA fragmentation and clusterin gene expression in human sperm. Int J Fertil Steril 2015; 9:129-36; PMID:25918601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Zeng Q, Chen G, Weng Y, Wang L, Chiang H, Lu D, Xu Z. Effects of global system for mobile communications 1800 MHz radiofrequency electromagnetic fields on gene and protein expression in MCF-7 cells. Proteomics 2006; 6:4732-8; PMID:16888767; http://dx.doi.org/ 10.1002/pmic.200600234 [DOI] [PubMed] [Google Scholar]
- [12].Whitehead TD, Moros EG, Brownstein BH, Roti Roti JL. The number of genes changing expression after chronic exposure to code division multiple access or frequency DMA radiofrequency radiation does not exceed the false-positive rate. Proteomics 2006; 6:4739-44; PMID:16933338; http://dx.doi.org/ 10.1002/pmic.200600051 [DOI] [PubMed] [Google Scholar]
- [13].Belyaev IY, Koch CB, Terenius O, Roxstrom-Lindquist K, Malmgren LO, W HS, Salford LG, Persson BR. Exposure of rat brain to 915 MHz GSM microwaves induces changes in gene expression but not double stranded DNA breaks or effects on chromatin conformation. Bioelectromagnetics 2006; 27:295-306; PMID:16511873; http://dx.doi.org/ 10.1002/bem.20216 [DOI] [PubMed] [Google Scholar]
- [14].McNamee JP, Bellier PV, Konkle AT, Thomas R, Wasoontarajaroen S, Lemay E, Gajda GB. Analysis of gene expression in mouse brain regions after exposure to 1.9 GHz radiofrequency fields. Int J Radiat Biol 2016; 92:338-50; PMID:27028625; http://dx.doi.org/ 10.3109/09553002.2016.1159353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Paparini A, Rossi P, Gianfranceschi G, Brugaletta V, Falsaperla R, De Luca P, Romano Spica V. No evidence of major transcriptional changes in the brain of mice exposed to 1800 MHz GSM signal. Bioelectromagnetics 2008; 29:312-23; PMID:18175331; http://dx.doi.org/ 10.1002/bem.20399 [DOI] [PubMed] [Google Scholar]
- [16].Leszczynski D. Mobile phones, precautionary principle, and future research. Lancet 2001; 358:1733; PMID:11728587; http://dx.doi.org/ 10.1016/S0140-6736(01)06757-5 [DOI] [PubMed] [Google Scholar]
- [17].Non-ionizing radiation, Part 2: Radiofrequency electromagnetic fields. IARC Monogr Eval Carcinog Risks Hum 2013; 102:1-460; PMID:24772662 [PMC free article] [PubMed] [Google Scholar]
- [18].Hardell L, Carlberg M. Using the Hill viewpoints from 1965 for evaluating strengths of evidence of the risk for brain tumors associated with use of mobile and cordless phones. Rev Environ Health 2013; 28:97-106; PMID:24192496; http://dx.doi.org/ 10.1515/reveh-2013-0006 [DOI] [PubMed] [Google Scholar]
- [19].Pritchett TL, Tanner EA, McCall K. Cracking open cell death in the Drosophila ovary. Apoptosis 2009; 14:969-79; PMID:19533361; http://dx.doi.org/ 10.1007/s10495-009-0369-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Goodman R, Weisbrot D, Uluc A, Henderson A. Transcription in Drosophila-melanogaster salivary-gland cells is altered following exposure to low-frequency electromagnetic-fields - Analysis of chromosome-3L and chromosome-X. Bioelectrochemistry and Bioenergetics 1992a; 343:311-318. [DOI] [PubMed] [Google Scholar]
- [21].Goodman R, Weisbrot D, Uluc A, Henderson A. Transcription in Drosophila melanogaster salivary gland cells is altered following exposure to low-frequency electromagnetic fields: analysis of chromosome 3R. Bioelectromagnetics 1992b; 13:111-8; PMID:1590811; http://dx.doi.org/ 10.1002/bem.2250130205 [DOI] [PubMed] [Google Scholar]
- [22].Weisbrot D, Uluc A, Henderson A, Goodman R. Transcription in Drosophila-melanogaster salivary-gland cells is altered following exposure to low-frequency electromagnetic-fields - analysis of chromosome-2R and chromosome-2L. Bioelectrochemistry and Bioenergetics 1993; 31:39-47; http://dx.doi.org/ 10.1016/0302-4598(93)86104-9 [DOI] [PubMed] [Google Scholar]
- [23].Li SS, Zhang ZY, Yang CJ, Lian HY, Cai P. Gene expression and reproductive abilities of male Drosophila melanogaster subjected to ELF-EMF exposure. Mutat Res 2013; 758:95-103; PMID:24157427; http://dx.doi.org/ 10.1016/j.mrgentox.2013.10.004 [DOI] [PubMed] [Google Scholar]
- [24].Panagopoulos DJ, Karabarbounis A, Margaritis LH. Effect of GSM 900-MHz mobile phone radiation on the reproductive capacity of Drosophila melanogaster. Electromagn Biol Med 2004; 23:29-43; http://dx.doi.org/ 10.1081/JBC-120039350 [DOI] [Google Scholar]
- [25].Weisbrot D, Lin H, Ye L, Blank M, Goodman R. Effects of mobile phone radiation on reproduction and development in Drosophila melanogaster. J Cell Biochem 2003; 89:48-55; PMID:12682907; http://dx.doi.org/ 10.1002/jcb.10480 [DOI] [PubMed] [Google Scholar]
- [26].Sagioglou NE MA, Giannarakis IK, Skouroliakou AS, Margaritis LH. Apoptotic cell death during Drosophila oogenesis is differentially increased by electromagnetic radiation depending on modulation, intensity and duration of exposure. Electromagn Biol Med 2016; 35:40-53; PMID:24236537; http://dx.doi.org/ 10.3109/15368378.2014.971959 [DOI] [PubMed] [Google Scholar]
- [27].Margaritis LH, Manta AK, Kokkaliaris KD, Schiza D, Alimisis K, Barkas G, Georgiou E, Giannakopoulou O, Kollia I, Kontogianni G, et al.. Drosophila oogenesis as a bio-marker responding to EMF sources. Electromagn Biol Med 2014; 33:165-89; PMID:23915130; http://dx.doi.org/ 10.3109/15368378.2013.800102 [DOI] [PubMed] [Google Scholar]
- [28].Manta AK, Stravopodis DJ, Papassideri IS, Margaritis LH. Reactive oxygen species elevation and recovery in Drosophila bodies and ovaries following short-term and long-term exposure to DECT base EMF. Electromagn Biol Med 2014; 33:118-31; PMID:23781995; http://dx.doi.org/ 10.3109/15368378.2013.791991 [DOI] [PubMed] [Google Scholar]
- [29].Chavdoula ED, Panagopoulos DJ, Margaritis LH. Comparison of biological effects between continuous and intermittent exposure to GSM-900-MHz mobile phone radiation: Detection of apoptotic cell-death features. Mutat Res 2010; 700:51-61; PMID:20472095; http://dx.doi.org/ 10.1016/j.mrgentox.2010.05.008 [DOI] [PubMed] [Google Scholar]
- [30].Chacon MA, Varela-Nallar L, Inestrosa NC. Frizzled-1 is involved in the neuroprotective effect of Wnt3a against Abeta oligomers. J Cell Physiol 2008; 217:215-27; PMID:18521822; http://dx.doi.org/ 10.1002/jcp.21497 [DOI] [PubMed] [Google Scholar]
- [31].Briones N, Dinu V. Data mining of high density genomic variant data for prediction of Alzheimer's disease risk. BMC Med Genet 2012; 13:7; PMID:22273362; http://dx.doi.org/ 10.1186/1471-2350-13-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kim YJ, Yi Y, Sapp E, Wang Y, Cuiffo B, Kegel KB, Qin ZH, Aronin N, DiFiglia M. Caspase 3-cleaved N-terminal fragments of wild-type and mutant huntingtin are present in normal and Huntington's disease brains, associate with membranes, and undergo calpain-dependent proteolysis. Proc Natl Acad Sci U S A 2001; 98:12784-9; PMID:11675509; http://dx.doi.org/ 10.1073/pnas.221451398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Gafni J, Ellerby LM. Calpain activation in Huntington's disease. J Neurosci 2002; 22:4842-9; PMID:12077181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Eppig JT, Blake JA, Bult CJ, Kadin JA, Richardson JE. The Mouse Genome Database (MGD): facilitating mouse as a model for human biology and disease. Nucleic Acids Res 2015; 43:D726-36; PMID:25348401; http://dx.doi.org/ 10.1093/nar/gku967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Kesari KK, Kumar S, Behari J. Effects of radiofrequency electromagnetic wave exposure from cellular phones on the reproductive pattern in male Wistar rats. Appl Biochem Biotechnol 2011; 164:546-59; PMID:21240569; http://dx.doi.org/ 10.1007/s12010-010-9156-0 [DOI] [PubMed] [Google Scholar]
- [36].Liu Q, Si T, Xu X, Liang F, Wang L, Pan S. Electromagnetic radiation at 900 MHz induces sperm apoptosis through bcl-2, bax and caspase-3 signaling pathways in rats. Reprod Health 2015; 12:65; PMID:26239320; http://dx.doi.org/ 10.1186/s12978-015-0062-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Remondini D, Nylund R, Reivinen J, Poulletier de Gannes F, Veyret B, Lagroye I, Haro E, Trillo MA, Capri M, Franceschi C, et al.. Gene expression changes in human cells after exposure to mobile phone microwaves. Proteomics 2006; 6:4745-54; PMID:16878293; http://dx.doi.org/ 10.1002/pmic.200500896 [DOI] [PubMed] [Google Scholar]
- [38].van Bergeijk P, Heimiller J, Uyetake L, Su TT. Genome-wide expression analysis identifies a modulator of ionizing radiation-induced p53-independent apoptosis in Drosophila melanogaster. PLoS One 2012; 7:e36539; PMID:22666323; http://dx.doi.org/ 10.1371/journal.pone.0036539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Cai W, Rudolph JL, Harrison SM, Jin L, Frantz AL, Harrison DA, Andres DA. An evolutionarily conserved Rit GTPase-p38 MAPK signaling pathway mediates oxidative stress resistance. Mol Biol Cell 2011; 22:3231-41; PMID:21737674; http://dx.doi.org/ 10.1091/mbc.E11-05-0400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Ma X, Li W, Yu H, Yang Y, Li M, Xue L, Xu T. Bendless modulates JNK-mediated cell death and migration in Drosophila. Cell Death Differ 2014; 21:407-15; PMID:24162658; http://dx.doi.org/ 10.1038/cdd.2013.154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Lee KS, Choi JS, Hong SY, Son TH, Yu K. Mobile phone electromagnetic radiation activates MAPK signaling and regulates viability in Drosophila. Bioelectromagnetics 2008; 29:371-9; PMID:18286519; http://dx.doi.org/ 10.1002/bem.20395 [DOI] [PubMed] [Google Scholar]
- [42].Kim JH, Huh YH, Kim HR. Induction of Autophagy in the Striatum and Hypothalamus of Mice after 835 MHz Radiofrequency Exposure. PLoS One 2016; 11:e0153308; PMID:27073885; http://dx.doi.org/ 10.1371/journal.pone.0153308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Jenkins VK, Timmons AK, McCall K. Diversity of cell death pathways: insight from the fly ovary. Trends Cell Biol 2013; 23:567-74; PMID:23968895; http://dx.doi.org/ 10.1016/j.tcb.2013.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Nezis IP, Lamark T, Velentzas AD, Rusten TE, Bjorkoy G, Johansen T, Papassideri IS, Stravopodis DJ, Margaritis LH, Stenmark H, et al.. Cell death during Drosophila melanogaster early oogenesis is mediated through autophagy. Autophagy 2009; 5:298-302; PMID:19066465; http://dx.doi.org/ 10.4161/auto.5.3.7454 [DOI] [PubMed] [Google Scholar]
- [45].Wang C, Liu Z, Huang X. Rab32 is important for autophagy and lipid storage in Drosophila. PLoS One 2012; 7:e32086; PMID:22348149; http://dx.doi.org/ 10.1371/journal.pone.0032086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Lee CY, Baehrecke EH. Steroid regulation of autophagic programmed cell death during development. Development 2001; 128:1443-55; PMID:11262243 [DOI] [PubMed] [Google Scholar]
- [47].Etchegaray JI, Timmons AK, Klein AP, Pritchett TL, Welch E, Meehan TL, Li C, McCall K. Draper acts through the JNK pathway to control synchronous engulfment of dying germline cells by follicular epithelial cells. Development 2012; 139:4029-39; PMID:22992958; http://dx.doi.org/ 10.1242/dev.082776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Postlethwait JH, Giorgi F. Vitellogenesis in insects. Dev Biol (N Y 1985) 1985; 1:85-126; PMID:3917207 [DOI] [PubMed] [Google Scholar]
- [49].DiMario PJ, Mahowald AP. Female sterile (1) yolkless: a recessive female sterile mutation in Drosophila melanogaster with depressed numbers of coated pits and coated vesicles within the developing oocytes. J Cell Biol 1987; 105:199-206; PMID:2886508; http://dx.doi.org/ 10.1083/jcb.105.1.199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Schonbaum CP, Perrino JJ, Mahowald AP. Regulation of the vitellogenin receptor during Drosophila melanogaster oogenesis. Mol Biol Cell 2000; 11:511-21; PMID:10679010; http://dx.doi.org/ 10.1091/mbc.11.2.511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Ye W, Wang F, Zhang W, Fang N, Zhao W, Wang J. Effect of Mobile Phone Radiation on Cardiovascular Development of Chick Embryo. Anat Histol Embryol 2016; 45:197-208; PMID:26171674; http://dx.doi.org/ 10.1111/ahe.12188 [DOI] [PubMed] [Google Scholar]
- [52].Thannickal VJ, Fanburg BL. Reactive oxygen species in cell signaling. Am J Physiol Lung Cell Mol Physiol 2000; 279:L1005-28; PMID:11076791 [DOI] [PubMed] [Google Scholar]
- [53].van der Horst A, Burgering BM. Stressing the role of FoxO proteins in lifespan and disease. Nat Rev Mol Cell Biol 2007; 8:440-50; PMID:17522590; http://dx.doi.org/ 10.1038/nrm2190 [DOI] [PubMed] [Google Scholar]
- [54].Gorlach A, Dimova EY, Petry A, Martinez-Ruiz A, Hernansanz-Agustin P, Rolo AP, Palmeira CM, Kietzmann T. Reactive oxygen species, nutrition, hypoxia and diseases: Problems solved? Redox Biol 2015; 6:372-85; PMID:26339717; http://dx.doi.org/ 10.1016/j.redox.2015.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Schieber M, Chandel NS. ROS function in redox signaling and oxidative stress. Curr Biol 2014; 24:R453-62; PMID:24845678; http://dx.doi.org/ 10.1016/j.cub.2014.03.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Morgan MJ, Liu ZG. Crosstalk of reactive oxygen species and NF-kappaB signaling. Cell Res 2011; 21:103-15; PMID:21187859; http://dx.doi.org/ 10.1038/cr.2010.178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Tohidi FZ, Toosi MHB, Azimian H, Khademi S, Fardid R, Sarab GA. The gene expression level of p53 and p21 in mouse brain exposed to radiofrequency field. International Journal of Radiation Research 2015; 13:337-343. [Google Scholar]
- [58].Tripathy BC, Oelmuller R. Reactive oxygen species generation and signaling in plants. Plant Signal Behav 2012; 7:1621-33; PMID:23072988; http://dx.doi.org/ 10.4161/psb.22455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Sharma VP, Singh HP, Kohli RK, Batish DR. Mobile phone radiation inhibits Vigna radiata (mung bean) root growth by inducing oxidative stress. Sci Total Environ 2009; 407:5543-7; PMID:19682728; http://dx.doi.org/ 10.1016/j.scitotenv.2009.07.006 [DOI] [PubMed] [Google Scholar]
- [60].Singh HP, Sharma VP, Batish DR, Kohli RK. Cell phone electromagnetic field radiations affect rhizogenesis through impairment of biochemical processes. Environ Monit Assess 2012; 184:1813-21; PMID:21562792; http://dx.doi.org/ 10.1007/s10661-011-2080-0 [DOI] [PubMed] [Google Scholar]
- [61].Tkalec M, Malaric K, Pevalek-Kozlina B. Exposure to radiofrequency radiation induces oxidative stress in duckweed Lemna minor L. Sci Total Environ 2007; 388:78-89; PMID:17825879; http://dx.doi.org/ 10.1016/j.scitotenv.2007.07.052 [DOI] [PubMed] [Google Scholar]
- [62].Radiofrequency electromagnetic fields (300 Hz-300 GHz) summary of an advisory report Health Council of The Netherlands: Radiofrequency Radiation Committee. Health Phys 1998; 75:51-5; PMID:9645665 [PubMed] [Google Scholar]
- [63].Wang Z, Seebauer EG. Temperature-dependent energy thresholds for ion-stimulated defect formation in solids. Phys Rev Lett 2005; 95:015501; PMID:16090628; http://dx.doi.org/ 10.1103/PhysRevLett.95.015501 [DOI] [PubMed] [Google Scholar]
- [64].Zhang ZY, Zhang J, Yang CJ, Lian HY, Yu H, Huang XM, Cai P. Coupling Mechanism of Electromagnetic Field and Thermal Stress on Drosophila melanogaster. PLoS One 2016; 11:e0162675; PMID:27611438; http://dx.doi.org/ 10.1371/journal.pone.0162675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Kristiansen KA, Jensen PE, Moller IM, Schulz A. Monitoring reactive oxygen species formation and localisation in living cells by use of the fluorescent probe CM-H(2)DCFDA and confocal laser microscopy. Physiol Plant 2009; 136:369-83; PMID:19493304; http://dx.doi.org/ 10.1111/j.1399-3054.2009.01243.x [DOI] [PubMed] [Google Scholar]
- [66].McCall K, Peterson JS. Detection of apoptosis in Drosophila. Methods Mol Biol 2004; 282:191-205; PMID:15105566 [DOI] [PubMed] [Google Scholar]
- [67].Arama E, Steller H. Detection of apoptosis by terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling and acridine orange in Drosophila embryos and adult male gonads. Nat Protoc 2006; 1:1725-31; PMID:17487155; http://dx.doi.org/ 10.1038/nprot.2006.235 [DOI] [PubMed] [Google Scholar]
- [68].Denton D, Mills K, Kumar S. Methods and protocols for studying cell death in Drosophila. Methods Enzymol 2008; 446:17-37; PMID:18603114; http://dx.doi.org/ 10.1016/S0076-6879(08)01602-9 [DOI] [PubMed] [Google Scholar]
- [69].Huang da W SB, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009; 37:1-13; PMID:19033363; http://dx.doi.org/ 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Huang da W SB, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4:44-57; PMID:19131956; http://dx.doi.org/ 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- [71].Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000; 28:27-30; PMID:10592173; http://dx.doi.org/ 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res 2014; 42:D199-205; PMID:24214961; http://dx.doi.org/ 10.1093/nar/gkt1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc 2013; 8:1551-66; PMID:23868073; http://dx.doi.org/ 10.1038/nprot.2013.092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Mi H, Muruganujan A, Thomas PD. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res 2013; 41:D377-86; PMID:23193289; http://dx.doi.org/ 10.1093/nar/gks1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.