Abstract
Parasites such as Plasmodium and Toxoplasma possess a vestigial plastid homologous to the chloroplasts of algae and plants. The plastid (known as the apicoplast; for apicomplexan plastid) is non-photosynthetic and very much reduced, but has clear endosymbiotic ancestry including a circular genome that encodes RNAs and proteins and a suite of bacterial biosynthetic pathways. Here we review the initial discovery of the apicoplast, and recount the major new insights into apicoplast origin, biogenesis and function. We conclude by examining how the apicoplast can be removed from malaria parasites in vitro, ultimately completing its reduction by chemical supplementation.
Keywords: Apicoplast, Endosymbiosis, Malaria, Plasmodium, Plastid, Toxoplasma
1. Introduction
The discovery of the vestigial plastid (apicoplast) in apicomplexan parasites such as malaria and Toxoplasma gondii gave us new insight into the origin of Phylum Apicomplexa, a large group of parasites. We now know that the group originated from photosynthetic ancestors, probably similar to modern dinoflagellate zooxanthellae. We have also learned that the apicoplast contains an ensemble of bacteria-like pathways to replicate and express its genome plus an anabolic capacity generating fatty acids, haem and isoprenoid precursors. Apicoplasts are essential, and perturbing them usually results in parasite death, thus making apicoplast metabolism an attractive target for drugs. Here, we focus on the discovery of the apicoplast, its origin and integration, the insights that tell what it does, and we examine how that knowledge has helped complete the reduction of the apicoplast that evolution didn’t quite manage.
2. Discovery of the apicoplast
The first inklings that the apicoplast even existed were images of circular, extrachromosomal DNA molecules in Plasmodium lophurae, a malarial parasite of ducks, published by Kilejian (1975). Similar circles were described from T. gondii by Borst et al. (1984). The cross-shaped configuration of the circles was due to the presence of inverted sequence repeats, which is a hallmark of plastid DNA (Williamson et al., 1985), but at the time, no one suspected that this genome could be from a plastid. Because malaria parasites were regarded as protozoa, the obvious presumption was that this small circular genome was the parasite’s mitochondrial genome (Kilejian, 1975; Williamson et al., 1985; Gardner et al., 1988). But all that changed when a linear molecule of 6 kb that encoded classical mitochondrial genes was subsequently found (Suplick et al., 1988; Aldritt et al., 1989; Vaidya et al., 1989; Feagin, 1992). Initial speculation held that both the linear and the circular genomes were mitochondrial, but sequence data from the circular genome challenged this concept in a strange and unanticipated way. Sequence of the 35 kb circle from Plasmodium falciparum proved that the genome did indeed have prokaryotic ancestry, but instead of the anticipated α-proteobacterial ancestry of the mitochondrial endosymbiont ancestor, the genes had closer ancestry to plastids of plants and algae (Gardner et al., 1991a,b, 1993, 1994).
Essentially the BLAST matches for the 35 kb sequences returning best matches to plastid genes were telling the mitochondrial DNA hunters that these aren’t the genes you’re looking for. Gradually they began to entertain the hypothesis that the circular genome was perhaps some cryptic relict from a plastid (Suplick et al., 1990; Gardner et al., 1991a,b; Wilson et al., 1991; Howe, 1992; Wilson, 1993; Williamson et al., 1994). Eventually, the complete sequence of the P. falciparum 35 kb circular genome revealed that it was plastid-like in every detail, except that it lacked any genes involved in photosynthesis (Wilson et al., 1996). At about the same time, electron microscope in situ hybridisation localised the 35 kb circular genome to a four membrane bound compartment in T. gondii that was clearly not the mitochondrion (McFadden et al., 1996; Köhler et al., 1997). The inescapable conclusion was that this compartment was a vestigial plastid – likely once photosynthetic – that persists in intracellular parasites of animals. How could this be?
3. Origin of the apicoplast
The evolutionary origin of the apicoplast was contentious for many years but is now firmly resolved. Initially there was disagreement about the number of bounding membranes, but it is now widely accepted that there are four (Fig. 1). The number of membranes is important to understand the evolutionary origin of the apicoplast, since more than two membranes is a hallmark of secondary endosymbiosis, in which the plastid is derived by eukaryote/ eukaryote endosymbiosis (Gould et al., 2008). In addition to the controversy about the number of membranes, a vigorous debate about the evolutionary affinity of the endosymbiont that became the apicoplast ensued (Wilson, 1993; Williamson et al., 1994; Köhler et al., 1997; McFadden et al., 1997; Funes et al., 2002, 2004; Cai et al., 2003; Waller et al., 2003; Waller and Keeling, 2006). Two alternatives were presented: the engulfed alga was either a member of the green algae (a relative of Chlamydomonas or Chlorella for instance), or was a red alga from a large group of seaweeds and unicellular algae with red pigmentation, perhaps the best known of which is Porphyra, the seaweed used to wrap savoury rice in the Japanese dish known as nori. The issue, which was important for an overall understanding of the origins and evolutionary history of this large parasitic group, remained moot until a genuine missing link came along in the form of a photosynthetic ancestor of the apicomplexan parasite lineage, namely Chromera velia (Moore et al., 2008).
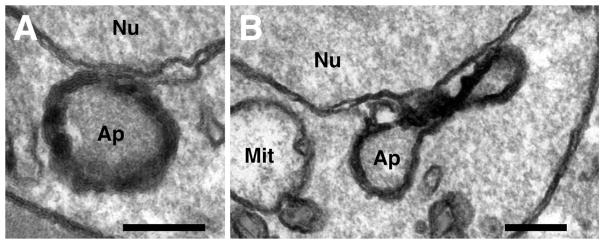
Fig. 1.
Transmission electron micrographs of the apicoplast in red blood cell stages of the human malaria parasite Plasmodium falciparum. (A) Cross section of an apicoplast (Ap) with four bounding membranes visible sitting close to the nucleus (Nu), which is bound by two membranes. (B) Longitudinal section of a dumbell shaped apicoplast (Ap) in close proximity to the mitochondrion (Mit) and the nucleus (Nu). Scale bars: 200 nm.
Chromera velia is one of the most important protist discoveries in decades. In addition to being a totally new group of algae (probably a new Phylum) with unique pigments, structure and ecology, it also served to clarify the origin of apicomplexan parasites and confirm the hypothesis that they arose from symbiotic algae that converted to parasitism (McFadden and Waller, 1997; Okamoto and McFadden, 2008). When the apicoplast was first identified, we proposed that an ancestor of the obligate intracellular parasites belonging to Phylum Apicomplexa was photosynthetic and lived as a mutualistic symbiont within invertebrate animals, similar to the way zooxanthellae dinoflagellates are symbionts in corals and other invertebrates (McFadden and Waller, 1997). Chromera velia is apparently the living descendant of such an ancestor (Okamoto and McFadden, 2008; Woo et al., 2015). Chromera velia is thought to live in corals, is sister to the Apicomplexa, and has a photosynthetic plastid bounded by four membranes (Moore et al., 2008; Janouskovec et al., 2010; Obornik et al., 2011). Importantly for the apicoplast origin debate, Chromera plastids share the same ancestry as apicoplasts and dinoflagellate plastids; Chromera (and related algae) undoubtedly harbour a red algal endosymbiont (Janouskovec et al., 2010). The apicoplast thus derives from an engulfed red alga and was acquired before dinoflagellates and the Apicomplexa diverged (Moore et al., 2008; Janouskovec et al., 2010). Since dinoflagellates diverged from the Apicomplexa at about the same time as animals arose, we can assume that the earliest invertebrates had symbionts, probably ancestors of modern symbiotic dinoflagellates and apicomplexan parasites, and that these symbionts were originally photosynthetic. At some stage a parasitic branch (the Apicomplexa) lost photosynthesis but (largely) retained the plastid, which we now know as the apicoplast. Intriguingly, symbiotic dinoflagellates reside within a membranous vacuole (the symbiosome) in animal host cells, and apicomplexan parasites reside within a membranous vacuole (the parasitophorous vacuole) in animal host cells. It will be interesting to learn whether there are homologies between how these vacuoles are established and maintained for their two different types of residents.
4. Integration of the apicoplast into the parasite
When the common ancestor of the Apicomplexa and dinoflagellates initially acquired its red algal endosymbiont, the alga would have been a fully autonomous, free-living eukaryotic organism. Today the endosymbiont is much reduced, having lost its nucleus, cytoplasm, wall and cytoskeleton. We understand a good deal about this reduction process thanks to the existence of two transitional forms, namely cryptomonads and chlorarachniophytes, in which the reduction process after secondary endosymbiosis is only partially complete (Douglas et al., 2001; Gilson et al., 2006; Curtis et al., 2012). Unlike these transition forms though, all that remains of the red algal endosymbiont that became the apicoplast is the genome and stroma of its plastid plus four bounding membranes. Counting from the inside, the four membranes are believed to represent the two membranes bounding the original red algal plastid, the plasma membrane of the endosymbiont (referred to as the periplastid membrane), and the outermost membrane is believed to derive from the host’s endomembrane, perhaps even representing the phagocytotic vacuole into which the original red alga symbiont was initially taken up. For an extended discussion of the multiple membranes readers are directed to Gould et al. (2015). Apicoplasts completely lack the pigment-loaded thylakoid membranes, which makes sense as they are non-photosynthetic. Clearly, there has been substantial reduction of the endosymbiont, and the theory of endosymbiosis holds that many of these reductive steps follow a pattern.
The pattern of endosymbiont reduction is believed to occur in at least three steps: (i) the host must tap the endosymbiont’s resources, (ii) division of the endosymbionts must be regulated to optimise numbers, and (iii) the partners must somehow be locked together as a single unit of selection. Once these three steps are achieved, the endosymbiont can be regarded as a stable, integrated part of the host cell – a genuine organelle. The first step is believed to have occured through insertion of sugar transporters into the membrane(s) bounding the endosymbiont, allowing the host to tap photosynthate from its guest (Weber et al., 2006; Gross and Bhattacharya, 2009). Intriguingly, when the ancestor of the Apicomplexa converted to parasitism and stopped photosynthesising, this “tap” had to be switched to work in the opposite direction. The cytosol of the parasite now supplies the plastid rather than vice versa, and a useful analogy to this has been plant cells in darkness, which also have to “feed” their plastids when they can’t photosynthesise (Mullin et al., 2006).
Regulating endosymbiont division, thought to be the second step in taming the endosymbiont, is no less crucial. If the endosymbiont multiplies too rapidly within the host cell, it could overrun it. Conversely, if the host cells grow faster than their endosymbiont, not all hosts would inherit an endosymbiont and new endosymbionts would need to be acquired each generation. Again, a transition form in which partially integrated symbionts are only unilaterally inherited helps us understand this process (Okamoto and Inouye, 2005). We understand the division machinery, and dissemination of plant plastids during cytokinesis, quite well, and control is largely managed by ancient bacterial division genes now relocated by endosymbiotic gene transfer to the host nucleus (Okazaki et al., 2010). Apicoplasts however don’t use this same machinery, which is curious because much of it (FtsZ, ARC and Min systems) derives from bacterial binary fission and is evolutionarily conserved in derivative plastid of plants and algae (Vaishnava and Striepen, 2006). Apicoplasts do resemble plant plastids in using a dynamin like pinchase during division (Striepen et al., 2000; van Dooren et al., 2009), but how they divide and are distributed is not well understood. Apicoplasts have a physical connection between their genome and the centrioles in the cytoplasm, which neatly ensures crucial partitioning of the organelle genome into daughter cells, and this machinery likely works in similar ways in unicellular algae (Francia et al., 2012).
5. Protein translocation
The last step in the domestication of an endosymbiont is to lock it in; to prevent it from reverting to a free-living existence. We understand both the drivers and mechanisms of lock-in quite well. Selection favours lock-in whenever the partnership between endosymbiont and host is evolutionarily more fit than the independence. A simple lock-in scenario is for the endosymbiont to lose autonomy and become dependent on the host. Plastids and mitochondria are all only partially autonomous; none can survive solo. The obvious lock-in mechanism is loss of genetic autonomy of the endosymbiont by gene transfer, and plastids (including apicoplasts) have clearly lost DNA to the host cell nucleus (Martin and Herrmann, 1998; Howe et al., 2000; Huang et al., 2003; Martin, 2003). This loss is not detrimental so long as the plastid (even an incipient one) remains within its host, and the host supplies the plastid with the required gene products from the DNA the plastid has lost. In the case of the apicoplast about 480 genes in the P. falciparum nucleus derive from the endosymbiont, and their products are believed to be targeted into the apicoplast (Gardner et al., 2002; Foth and McFadden, 2003; Ralph et al., 2004).
The mechanism to restore essential factors to the plastid that are encoded by nuclear genes involves elaborate targeting mechanisms, nowhere more so than in the case of the four membrane-bound apicoplasts. Targeting is canonically achieved by address tags encoded within the cargo protein, and these tags engage with machinery and translocons that escort the gene product to its appropriate destination in the cell. For apicoplasts this tag is typically a two part N-terminal extension that is fortunately very distinctive in P. falciparum and Toxoplasma, allowing bioinformatic prediction of targeting (Foth et al., 2003). The trafficking and processing machinery from the parasite cytosol to the apicoplast is also quite well characterised with much of it deriving from the plastid of the red alga (van Dooren et al., 2002, 2008; McFadden and van Dooren, 2004; Kalanon and McFadden, 2008; Glaser et al., 2012; Sheiner et al., 2015) and some innovative retooling of endomembrane translocons to deal with the residual plasma membrane of the red alga (Sommer et al., 2007; Agrawal et al., 2009, 2013; Hempel et al., 2009; Spork et al., 2009; Bullmann et al., 2010; van Dooren and Striepen, 2013). Several proteins are also resident in the outer apicoplast membrane, but little is understood about how they get there other than that the mechanism differs from the canonical N-terminal tag system (Mullin et al., 2006; Karnataki et al., 2007b, 2009; Lim et al., 2009, 2016).
6. Function of the apicoplast
What does the apicoplast do? The short answer is that it makes things the parasite cannot do without. But exactly what these things are, and why the parasite cannot survive without them, are not fully clarified. Scrutiny of the apicoplast genome provided no real clue as to the purpose of the organelle (Wilson et al., 1996; Köhler et al., 1997). Photosynthesis had clearly been lost, but no functional pathways, beyond self-perpetuation, could be identified in the handful of genes encoded by the organelle genome (Wilson et al., 1996; Köhler et al., 1997). Meanwhile, pharmacological and genetic perturbation of the apicoplast was clearly shown to lead to parasite death (Fichera and Roos, 1997; He et al., 2001), robustly demonstrating that the organelle is essential. Efforts then turned to finding this essential function. It was here that the genome projects underway for T. gondii and P. falciparum became an invaluable resource. Genes for an apicoplast fatty acid biosynthesis (FAS) system indicated that the organelle probably made lipids (Waller et al., 1998), refuting the widely held belief that malaria parasites were unable to synthesise fatty acids de novo. As it turns out, malaria parasites do not make appreciable amounts of fatty acids during their blood stage (the stage analysed biochemically) and the machinery becomes active only in later life cycle phases (see below). An apicoplast pathway to synthesise isopentenyl diphosphate (IPP) – a precursor of isoprenoids for prenylation of proteins, ubiquinone side chains, dolichols and modification of tRNAs (Ralph et al., 2004) – was also identified from the genome data (Jomaa et al., 1999; Guggisberg et al., 2014a; Imlay and Odom, 2014; Armstrong et al., 2015; Imlay et al., 2015). Subsequent data mining revealed that the apicoplast of malaria parasites makes iron sulphur complexes (Seeber, 2002, 2003; Ralph et al., 2004) and cooperates with the mitochondrion in the synthesis of haem (Ralph et al., 2004; Sato et al., 2004; van Dooren et al., 2006; Nagaraj et al., 2008, 2009a,b; Shanmugam et al., 2010). These four anabolic roles for the apicoplast hinted that synthesis of crucial precursors was the essential role of the apicoplast, but the genes did not tell us exactly what was made and how it was essential. The genome did, however, allow construction of hypothetical maps of these plastid-like pathways (Ralph et al., 2004), and much of the detail of these maps has been confirmed through biochemical reconstructions, molecular genetics and metabolomics (reviewed in Lim and McFadden, 2010; Sheiner et al., 2013).
These anabolic apicoplast pathways are fuelled with phosphorylated three carbon sugars imported from the parasite’s cytosolic glycolytic pathway. Plastid-like transporters (plastidic phosphate translocators, pPTs), located in the membranes of the apicoplast, import reduced carbon compounds in a manner analogous to plant plastids in the dark (Mullin et al., 2006; Karnataki et al., 2007a; Brooks et al., 2010; Lim et al., 2010). Knock down of the apicoplast translocator gene in Toxoplasma resulted in immediate death of the parasites (Brooks et al., 2010). Curiously, there are two pPT-type translocators in the Plasmodium apicoplast that are apparently located in different membranes (Mullin et al., 2006), and the translocator in the outer membrane appears to be essential, whereas the one putatively located in the inner membrane is dispensable in rodent malaria, Plasmodium berghei (Banerjee et al., 2012). In summary, we now know that the apicoplast harbours a collection of plastid-like metabolic pathways that are supplied with carbon, energy and reducing power in a manner identical to a non-photosynthetic plant or algal plastid. The next challenge was to tease out which of these pathways is essential, when in the life cycle a pathway is essential, and why.
7. The apicoplast as a drug target
When first discovered, the apicoplast engendered much excitement as a novel target for therapeutic drugs (Gardner et al., 1991a, b; McFadden et al., 1996; Wilson et al., 1996). The cyanobacterial pathways therein, and the novel ways it was supplied with carbon, energy, reductants and proteins, are all very distant from human host metabolism and cellular processes, leaving room to design or discover specific inhibitors that would perturb the apicoplast but have no side effects (Jomaa et al., 1999; McFadden and Roos, 1999; Ralph et al., 2001, 2004; Gornicki, 2003; Seeber, 2003; Goodman and McFadden, 2007; Goodman et al., 2007).
The apicoplast is essentially a reduced bacterium inside the parasite. This modified bacterium, now an endosymbiont, possesses a circular genome that has to be replicated and expressed (Wilson et al., 1996). Numerous antibacterials target these processes, and many are parasiticidal, and several (clindamycin, azirthromycin and doxycycline) are used as an antimalarial therapy or prophylaxis (Wiesner et al., 2008). Here we examine the more recent insights.
Apicoplast type II fatty acid biosynthesis initially attracted intense drug attention when it was discovered (Waller et al., 1998). A flurry of reports described various putative inhibitors of FASII as parasiticidal, but in hindsight we now recognise that all these particular inhibitors are off-target (Shears et al., 2015). The crunch for FASII as a therapeutic drug target came with gene knockout studies showing that malaria parasites can do without vital enzymes of the fatty acid biosynthesis pathway during the red blood cell portion of their life cycle, implying that parasites do not need to synthesise fatty acids at this stage (Yu et al., 2008; Vaughan et al., 2009; Lindner et al., 2014). Nevertheless, FASII is crucial for the liver stage in rodent malarias and for the proper development of mosquito stage oocysts in human P. falciparum (Yu et al., 2008; Vaughan et al., 2009; van Schaijk et al., 2013).
Ablation of apicoplast FASII in the blood stages was the first hint that not all pathways would be essential in all malaria life cycle stages. By contrast, apicoplast fatty acid biosynthesis is essential in T. gondii in mice since ablation of the acyl carrier protein in vivo clears the infection (Mazumdar et al., 2006). It remains uncertain why so many bacterial FASII inhibitors kill blood stage malaria parasites even though they have mostly been shown to be off-target (Shears et al., 2015). One suggestion is that these compounds could be targeting cytosolic fatty acid elongases used to modify scavenged fatty acids (Shears et al., 2015). Another open question is whether FASII can be targeted in liver stages, where it is essential in rodent malarias (and probably human malaria parasites) for prophylaxis.
Following the revelation that apicoplast FASII is dispensable in three species of blood stage malaria parasites, it was also revealed that apicoplast-localised steps of the multi compartment haem biosynthesis pathway (Ralph et al., 2004) were similarly dispensable during the blood stage (Nagaraj et al., 2013; Ke et al., 2014; Sigala and Goldberg, 2014; Smith et al., 2014). In rodent and human malaria parasites the final enzyme for haem biosynthesis is essential for oocysts (Nagaraj et al., 2013; Ke et al., 2014; Sigala and Goldberg, 2014). Haem biosynthesis and FASII were thus off the table for malaria blood stage therapy. Nevertheless, the apicoplast was clearly essential because perturbation of its housekeeping activities kills the parasite (Dahl et al., 2006; Goodman et al., 2007; Dahl and Rosenthal, 2008; Goodman and McFadden, 2012). What could be the essential function?
8. Making the apicoplast disappear
Gene knock out studies showed that FASII and haem biosynthesis seemed unessential at the blood stage, and the role of Fe:S cluster synthesis is widely thought to be one of self-maintenance to generate reducing equivalents (Seeber, 2002, 2003; Ralph et al., 2004). This left IPP synthesis as the prime candidate for indispensable role of the apicoplast, during the blood stage at least. Moreover, isoprenoid precursor biosynthesis carried out by the prokaryotic MEP pathway is the only biosynthetic pathway conserved among apicomplexan parasites that harbor apicoplasts. Both Babesia and Theileria spp. contain an apicoplast, but their genomes lack type II fatty acid and haem biosynthetic genes and have significantly reduced suf genes required for Fe:S cluster biosynthesis (Brayton et al., 2007; Sato, 2011). Notably, both Babesia and Theileria parasites bypass liver stage infection in the mammalian host and only infect blood cells (typically, red blood cells in the case of Babesia and white blood cells in the case of Theileria). The reduction of apicoplast metabolism in Babesia and Theileria spp. may have been the first clue that the blood-stage function of the Plasmodium apicoplast is limited, since the survival of these closely-related apicomplexan parasites in blood cells only requires an intact isoprenoid precursor biosynthetic pathway and an Fe:S cluster biosynthetic pathway, albeit somewhat reduced. The latter is presumably to provide enzymes in the MEP pathway with cofactor and therefore is only required as long as the MEP pathway is essential. Further evolutionary evidence that the parasite apicoplast is “on its way out” is the complete loss of the apicoplast in Cryptosporidium spp. parasites (Zhu et al., 2000; Riordan et al., 2003; Abrahamsen et al., 2004; Huang et al., 2004). In fact, Cryptosporidium was the first and, until recently (Gornik et al., 2015), only known example of plastid loss in any eukaryote.
In the end, biochemistry accomplished what evolution had yet to complete in Plasmodium – the elimination of the apicoplast. It had been well established that inhibition of apicoplast genome expression by antibiotics, such as doxycycline, disrupts apicoplast replication and inheritance during parasite cell division (Dahl et al., 2006). As a consequence, antibiotic-treated parasites form daughter cells that lack apicoplasts and are not viable. Remarkably, these ‘apicoplast-minus’ parasites survive and continue to replicate when IPP, the product of isoprenoid precursor biosynthesis, is added to the growth media (Yeh and DeRisi, 2011). Thus IPP functionally replaces the apicoplast. This simple chemical rescue demonstrated that the only essential metabolic function of the apicoplast in blood-stage Plasmodium is the biosynthesis of isoprenoid precursors. IPP supplementation can apparently rescue parasites from any apicoplast dysfunction, making it a nifty tool to study apicoplast biology including identification of antimalarials with unknown targets as anti-apicoplast agents (Wu et al., 2015), and even perturbations of essential gene products (e.g. expression of a dominant-negative SufC, a member of the iron sulphur complex generating system in the apicoplast) (Gisselberg et al., 2013). Thus far it has only been possible to rescue in vitro cultured P. falciparum parasites, and the level of IPP required is remarkably high (200 μM) (Yeh and DeRisi, 2011). It remains to be established if this high concentration requirement in media is indicative of poor uptake by the parasites.
Given the evolutionary feat of taming an endosymbiont, and the complex cellular pathways required to maintain it, subsequently losing the organelle seems almost unimaginable. But that appears to be the trajectory of the apicoplast. A single metabolic product is the barrier to apicoplast loss in blood-stage Plasmodium and likely all life stages of Babesia and Theileria. The degeneration of the apicoplast can be compared with remnant mitochondria called mitosomes that have been described in several eukaryotes (Tovar et al., 1999; Riordan et al., 2003). Although mitochondria are often recognised as the “powerhouse” of the cell, their function in oxidative phosphorylation of ATP is often the first to go, and the mitosomes of Giardia and Trichomonas appear only to preserve Fe–S cluster biosynthesis, while lacking even a mitochondrial genome (van der Giezen and Tovar, 2005). Thus far only one organism is known to have lost the mitochondrion completely (Karnkowska et al., 2016). Similarly, a recent reconstruction of the evolution of apicoplast function by deciphering the pattern of loss of photosynthesis, plastid genomes, and plastid organelles amongst diverse organisms in this secondary plastid lineage showed that plastid retention is strongly linked to dependency on plastid isoprenoid precursor biosynthesis (Janouskovec et al., 2015; Waller et al., 2016). Once Cryptosporidium overcame this dependence by scavenging isoprenoid intermediates from the host, it lost its apicoplast (Bessoff et al., 2013). It was recently demonstrated that Hematodinium spp. also found a non-plastid source for isoprenoids and is the only other documented case of plastid loss (Gornik et al., 2015). Thus, isoprenoid precursor biosynthesis appears to be the “last person standing” in the devolution of the apicoplast. It is important to note however that the other biosynthesis pathways in Plasmodium apicoplasts (haem and FASII), make the organelle indispensable for completion of the life cycle and hence ability to spread from host to host. Until Plasmodium parasites can find a way to scavenge IPP, haem and fatty acids in sufficient quantities across each stage of their life cycle, the apicoplast will remain indispensable and therefore a vulnerability susceptible to drug attack.
Apicoplast IPP synthesis is also essential for gametocytogenesis in P. falciparum and subsequent development of the oocyst phase in mosquitoes (Wiley et al., 2015). Whether IPP synthesis is essential in liver stage human malarias, and therefore of potential utility as a prophylaxis target, is moot with one group claiming an effect for fosmidomycin in P. berghei liver stages (Nair et al., 2011) and another group claiming no effect (Baumeister et al., 2011).
For now, Plasmodium and other apicomplexan human pathogens (T. gondii, Babesia spp.) still require plastid isoprenoid precursor biosynthesis and are therefore obliged to retain their apicoplasts (Nair et al., 2011; Yeh and DeRisi, 2011; Agrawal et al., 2013). This makes the IPP synthesis pathway a viable drug target for treatment of acute malaria and other apicomplexan parasitic infections, with the exception of Cryptosporidium. Fosmidomycin, an inhibitor of the MEP pathway, has been tested in several clinical trials for malaria treatment (Borrmann et al., 2004a,b, 2005, 2006). Resistance to fosmidomycin has been selected for in vitro (Guggisberg et al., 2014b). Unexpectedly, resistance did not involve mutations in deoxyxylulose 5-phosphate reductoisomerase, the target of fosmidomycin, but an alteration in processing of the upstream substrate in the parasite cytoplasm (Guggisberg et al., 2014b). By developing therapies that prevent the apicoplast from fulfilling this role, we can take advantage of the parasite’s dependence on an organelle of extremely limited and vanishing function.
9. Conclusion
Since it was first identified in 1996, the apicoplast has revealed how the parasitic Phylum Apicomplexa arose, why antibacterials such as doxycycline kill malaria, and how an endosymbiotic alga can go from a photosynthetic asset to a vestigial organelle. Despite its diminishing role over evolutionary time, the apicoplast remains vital for making a few essential molecules unable to be scavenged in sufficient quantity from at least one of the hosts in the digenetic life cycle. Despite the fact that the apicoplast has a single, simple role in the blood stage of Plasmodium parasites (making IPP), it continues to have tremendous potential for therapeutic intervention because anything that blocks this role kills the parasite.
Acknowledgments
GIM is grateful for Program Grant and Project Grant support from the National Health and Medical Research Council of Australia and Discovery Grant support from the Australian Research Council. The Australian Red Cross generously supplied human red blood cells for our research.
References
- Abrahamsen MS, Templeton TJ, Enomoto S, Abrahante JE, Zhu G, Lancto CA, Deng M, Liu C, Widmer G, Tzipori S, Buck GA, Xu P, Bankier AT, Dear PH, Konfortov BA, Spriggs HF, Iyer L, Anantharaman V, Aravind L, Kapur V. Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science. 2004;304:441–445. doi: 10.1126/science.1094786. [DOI] [PubMed] [Google Scholar]
- Agrawal S, van Dooren GG, Beatty WL, Striepen B. Genetic evidence that an endosymbiont-derived endoplasmic reticulum-associated protein degradation (ERAD) system functions in import of apicoplast proteins. J Biol Chem. 2009;284:33683–33691. doi: 10.1074/jbc.M109.044024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal S, Chung DW, Ponts N, van Dooren GG, Prudhomme J, Brooks CF, Rodrigues EM, Tan JC, Ferdig MT, Striepen B, Le Roch KG. An apicoplast localized ubiquitylation system is required for the import of nuclear-encoded plastid proteins. PLoS Pathog. 2013;9:e1003426. doi: 10.1371/journal.ppat.1003426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldritt SM, Joseph JT, Wirth DF. Sequence identification of cytochrome b in Plasmodium gallinaceum. Mol Cell Biol. 1989;9:3614–3620. doi: 10.1128/mcb.9.9.3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong CM, Meyers DJ, Imlay LS, Freel Meyers C, Odom AR. Resistance to the antimicrobial agent fosmidomycin and an FR900098 prodrug through mutations in the deoxyxylulose phosphate reductoisomerase gene (dxr) Antimicrob Agents Chemother. 2015;59:5511–5519. doi: 10.1128/AAC.00602-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee T, Jaijyan DK, Surolia N, Singh AP, Surolia A. Apicoplast triose phosphate transporter (TPT) gene knockout is lethal for Plasmodium. Mol Biochem Parasitol. 2012;186:44–50. doi: 10.1016/j.molbiopara.2012.09.008. [DOI] [PubMed] [Google Scholar]
- Baumeister S, Wiesner J, Reichenberg A, Hintz M, Bietz S, Harb OS, Roos DS, Kordes M, Friesen J, Matuschewski K, Lingelbach K, Jomaa H, Seeber F. Fosmidomycin uptake into Plasmodium and Babesia-infected erythrocytes is facilitated by parasite-induced new permeability pathways. PLoS One. 2011;6:e19334. doi: 10.1371/journal.pone.0019334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessoff K, Sateriale A, Lee KK, Huston CD. Drug repurposing screen reveals FDA-approved inhibitors of human HMG-CoA reductase and isoprenoid synthesis that block Cryptosporidium parvum growth. Antimicrob Agents Chemother. 2013;57:1804–1814. doi: 10.1128/AAC.02460-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrmann S, Adegnika AA, Matsiegui PB, Issifou S, Schindler A, Mawili-Mboumba DP, Baranek T, Wiesner J, Jomaa H, Kremsner PG. Fosmidomycin-clindamycin for Plasmodium falciparum Infections in African children. J Infect Dis. 2004a;189:901–908. doi: 10.1086/381785. [DOI] [PubMed] [Google Scholar]
- Borrmann S, Issifou S, Esser G, Adegnika AA, Ramharter M, Matsiegui PB, Oyakhirome S, Mawili-Mboumba DP, Missinou MA, Kun JF, Jomaa H, Kremsner PG. Fosmidomycin-clindamycin for the treatment of Plasmodium falciparum malaria. J Infect Dis. 2004b;190:1534–1540. doi: 10.1086/424603. [DOI] [PubMed] [Google Scholar]
- Borrmann S, Adegnika AA, Moussavou F, Oyakhirome S, Esser G, Matsiegui PB, Ramharter M, Lundgren I, Kombila M, Issifou S, Hutchinson D, Wiesner J, Jomaa H, Kremsner PG. Short-course regimens of artesunate-fosmidomycin in treatment of uncomplicated Plasmodium falciparum malaria. Antimicrob Agents Chemother. 2005;49:3749–3754. doi: 10.1128/AAC.49.9.3749-3754.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrmann S, Lundgren I, Oyakhirome S, Impouma B, Matsiegui PB, Adegnika AA, Issifou S, Kun JF, Hutchinson D, Wiesner J, Jomaa H, Kremsner PG. Fosmidomycin plus clindamycin for treatment of pediatric patients aged 1 to 14 years with Plasmodium falciparum malaria. Antimicrob Agents Chemother. 2006;50:2713–2718. doi: 10.1128/AAC.00392-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borst P, Overdlve JP, Weijers PJ, Fase-Fowler F, Berg MVD. DNA circles with cruciforms from Isospora (Toxoplasma) gondii. Biochim Biophys Acta. 1984;781:100–111. doi: 10.1016/0167-4781(84)90128-3. [DOI] [PubMed] [Google Scholar]
- Brayton KA, Lau AO, Herndon DR, Hannick L, Kappmeyer LS, Berens SJ, Bidwell SL, Brown WC, Crabtree J, Fadrosh D, Feldblum T, Forberger HA, Haas BJ, Howell JM, Khouri H, Koo H, Mann DJ, Norimine J, Paulsen IT, Radune D, Ren Q, Smith RK, Jr, Suarez CE, White O, Wortman JR, Knowles DP, Jr, McElwain TF, Nene VM. Genome sequence of Babesia bovis and comparative analysis of apicomplexan hemoprotozoa. PLoS Pathog. 2007;3:1401–1413. doi: 10.1371/journal.ppat.0030148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks CF, Johnsen H, van Dooren GG, Muthalagi M, Lin SS, Bohne W, Fischer K, Striepen B. The Toxoplasma apicoplast phosphate translocator links cytosolic and apicoplast metabolism and is essential for parasite survival. Cell Host Microbe. 2010;7:62–73. doi: 10.1016/j.chom.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullmann L, Haarmann R, Mirus O, Bredemeier R, Hempel F, Maier UG, Schleiff E. Filling the gap, evolutionarily conserved Omp85 in plastids of chromalveolates. J Biol Chem. 2010;285:6848–6856. doi: 10.1074/jbc.M109.074807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Fuller AL, McDougald LR, Zhu G. Apicoplast genome of the coccidian Eimeria tenella. Gene. 2003;321:39–46. doi: 10.1016/j.gene.2003.08.008. [DOI] [PubMed] [Google Scholar]
- Curtis BA, Tanifuji G, Burki F, Gruber A, Irimia M, Maruyama S, Arias MC, Ball SG, Gile GH, Hirakawa Y, Hopkins JF, Kuo A, Rensing SA, Schmutz J, Symeonidi A, Elias M, Eveleigh RJ, Herman EK, Klute MJ, Nakayama T, Obornik M, Reyes-Prieto A, Armbrust EV, Aves SJ, Beiko RG, Coutinho P, Dacks JB, Durnford DG, Fast NM, Green BR, Grisdale CJ, Hempel F, Henrissat B, Hoppner MP, Ishida K, Kim E, Koreny L, Kroth PG, Liu Y, Malik SB, Maier UG, McRose D, Mock T, Neilson JA, Onodera NT, Poole AM, Pritham EJ, Richards TA, Rocap G, Roy SW, Sarai C, Schaack S, Shirato S, Slamovits CH, Spencer DF, Suzuki S, Worden AZ, Zauner S, Barry K, Bell C, Bharti AK, Crow JA, Grimwood J, Kramer R, Lindquist E, Lucas S, Salamov A, McFadden GI, Lane CE, Keeling PJ, Gray MW, Grigoriev IV, Archibald JM. Algal genomes reveal evolutionary mosaicism and the fate of nucleomorphs. Nature. 2012;492:59–65. doi: 10.1038/nature11681. [DOI] [PubMed] [Google Scholar]
- Dahl EL, Rosenthal PJ. Apicoplast translation, transcription and genome replication: targets for antimalarial antibiotics. Trends Parasitol. 2008;24:279–284. doi: 10.1016/j.pt.2008.03.007. [DOI] [PubMed] [Google Scholar]
- Dahl EL, Shock JL, Shenai BR, Gut J, DeRisi JL, Rosenthal PJ. Tetracyclines specifically target the apicoplast of the malaria parasite Plasmodium falciparum. Antimicrob Agents Chemother. 2006;50:3124–3131. doi: 10.1128/AAC.00394-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas S, Zauner S, Fraunholz M, Beaton M, Penny S, Deng L-T, Wu X, Reith M, Cavalier-Smith T, Maier U-G. The highly reduced genome of an enslaved algal nucleus. Nature. 2001;410:1091–1096. doi: 10.1038/35074092. [DOI] [PubMed] [Google Scholar]
- Feagin JE. The 6 kb element of Plasmodium falciparum encodes mitochondrial cytochrome genes. Mol Biochem Parasitol. 1992;52:145–148. doi: 10.1016/0166-6851(92)90046-m. [DOI] [PubMed] [Google Scholar]
- Fichera ME, Roos DS. A plastid organelle as a drug target in apicomplexan parasites. Nature. 1997;390:407–409. doi: 10.1038/37132. [DOI] [PubMed] [Google Scholar]
- Foth BJ, McFadden GI. The apicoplast: a plastid in Plasmodium falciparum and other Apicomplexan parasites. Int Rev Cytol. 2003;224:57–110. doi: 10.1016/s0074-7696(05)24003-2. [DOI] [PubMed] [Google Scholar]
- Foth BJ, Ralph SA, Tonkin CJ, Struck NS, Fraunholz M, Roos DS, Cowman AF, McFadden GI. Dissecting apicoplast targeting in the malaria parasite Plasmodium falciparum. Science. 2003;299:705–708. doi: 10.1126/science.1078599. [DOI] [PubMed] [Google Scholar]
- Francia ME, Jordan CN, Patel JD, Sheiner L, Demerly JL, Fellows JD, De Leon JC, Morrissette NS, Dubremetz J-F, Striepen B. Cell division in apicomplexan parasites is organized by a homolog of the striated rootletfiber of algal flagella. PLoS Biol. 2012;10:e1001444. doi: 10.1371/journal.pbio.1001444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funes S, Davidson E, Reyes-Prieto A, Magallon S, Herion P, King MP, Gonzalez-Halphen D. A green algal apicoplast ancestor. Science. 2002;298:2155. doi: 10.1126/science.1076003. [DOI] [PubMed] [Google Scholar]
- Funes S, Reyes-Prieto A, Perez-Martinez X, Gonzalez-Halphen D. On the evolutionary origins of apicoplasts: revisiting the rhodophyte vs. chlorophyte controversy. Microbes Infect. 2004;6:305–311. doi: 10.1016/j.micinf.2003.11.011. [DOI] [PubMed] [Google Scholar]
- Gardner MJ, Bates PA, Ling IT, Moore DJ, McCready S, Gunasekera MBR, Wilson RJM, Williamson DH. Mitochondrial DNA of the human malarial parasite Plasmodium falciparum. Mol Biochem Parasitol. 1988;31:11–18. doi: 10.1016/0166-6851(88)90140-5. [DOI] [PubMed] [Google Scholar]
- Gardner MJ, Feagin JE, Moore DJ, Spencer DF, Gray MW, Williamson DH, Wilson RJ. Organisation and expression of small subunit ribosomal RNA genes encoded by a 35-kilobase circular DNA in Plasmodium falciparum. Mol Biochem Parasitol. 1991a;48:77–88. doi: 10.1016/0166-6851(91)90166-4. [DOI] [PubMed] [Google Scholar]
- Gardner MJ, Williamson DH, Wilson RJM. A circular DNA in malaria parasites encodes an RNA polymerase like that of prokaryotes and chloroplasts. Mol Biochem Parasitol. 1991b;44:115–123. doi: 10.1016/0166-6851(91)90227-w. [DOI] [PubMed] [Google Scholar]
- Gardner MJ, Feagin JE, Moore DJ, Rangachari K, Williamson DH, Wilson RJ. Sequence and organization of large subunit rRNA genes from the extrachromosomal 35 kb circular DNA of the malaria parasite Plasmodium falciparum. Nucleic Acids Res. 1993;21:1067–1071. doi: 10.1093/nar/21.5.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner MJ, Goldman N, Barnett P, Moore PW, Rangachari K, Strath M, Whyte A, Williamson DH, Wilson RJ. Phylogenetic analysis of the rpoB gene from the plastid-like DNA of Plasmodium falciparum. Mol Biochem Parasitol. 1994;66:221–231. doi: 10.1016/0166-6851(94)90149-x. [DOI] [PubMed] [Google Scholar]
- Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, Paulsen IT, James K, Eisen JA, Rutherford K, Salzberg SL, Craig A, Kyes S, Chan MS, Nene V, Shallom SJ, Suh B, Peterson J, Angiuoli S, Pertea M, Allen J, Selengut J, Haft D, Mather MW, Vaidya AB, Martin DM, Fairlamb AH, Fraunholz MJ, Roos DS, Ralph SA, McFadden GI, Cummings LM, Subramanian GM, Mungall C, Venter JC, Carucci DJ, Hoffman SL, Newbold C, Davis RW, Fraser CM, Barrell B. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson PR, Su V, Slamovits CH, Reith ME, Keeling PJ, McFadden GI. Complete nucleotide sequence of the chlorarachniophyte nucleomorph: nature’s smallest nucleus. Proc Natl Acad Sci USA. 2006;103:9566–9571. doi: 10.1073/pnas.0600707103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gisselberg JE, Dellibovi-Ragheb TA, Matthews KA, Bosch G, Prigge ST. The suf iron-sulfur cluster synthesis pathway is required for apicoplast maintenance in malaria parasites. PLoS Pathog. 2013;9:e1003655. doi: 10.1371/journal.ppat.1003655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser S, van Dooren GG, Agrawal S, Brooks CF, McFadden GI, Striepen B, Higgins MK. Tic22 is an essential chaperone required for protein import into the apicoplast. J Biol Chem. 2012;287:39505–39512. doi: 10.1074/jbc.M112.405100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman CD, McFadden GI. Fatty acid biosynthesis as a drug target in apicomplexan parasites. Curr Drug Targets. 2007;8:15–30. doi: 10.2174/138945007779315579. [DOI] [PubMed] [Google Scholar]
- Goodman CD, McFadden GI. Targeting apicoplasts in malaria parasites. Curr Opin Ther Targets. 2012;17:1–12. doi: 10.1517/14728222.2013.739158. [DOI] [PubMed] [Google Scholar]
- Goodman CD, Su V, McFadden GI. The effects of anti-bacterials on the malaria parasite Plasmodium falciparum. Mol Biochem Parasitol. 2007;152:181–191. doi: 10.1016/j.molbiopara.2007.01.005. [DOI] [PubMed] [Google Scholar]
- Gornicki P. Apicoplast fatty acid biosynthesis as a target for medical intervention in apicomplexan parasites. Int J Parasitol. 2003;33:885–896. doi: 10.1016/s0020-7519(03)00133-4. [DOI] [PubMed] [Google Scholar]
- Gornik SG, Marsa F, Cassin AM, MacRae J, McConville MJ, Bacic A, McFadden GI, Pain A, Waller RF. Endosymbiosis undone: stepwise elimination of the plastid in the dinoflagellate Hematodinium. Proc Natl Acad Sci USA. 2015;112:5767–5772. doi: 10.1073/pnas.1423400112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SB, Waller RF, McFadden GI. Plastid evolution. Annu Rev Plant Biol. 2008;59:491–517. doi: 10.1146/annurev.arplant.59.032607.092915. [DOI] [PubMed] [Google Scholar]
- Gould SB, Maier UG, Martin WF. Protein import and the origin of red complex plastids. Curr Biol. 2015;25:R515–R521. doi: 10.1016/j.cub.2015.04.033. [DOI] [PubMed] [Google Scholar]
- Gross J, Bhattacharya D. Mitochondrial and plastid evolution in eukaryotes: an outsiders’ perspective. Nat Rev Genet. 2009;10:495–505. doi: 10.1038/nrg2610. [DOI] [PubMed] [Google Scholar]
- Guggisberg AM, Amthor RE, Odom AR. Isoprenoid biosynthesis in Plasmodium falciparum. Eukaryot Cell. 2014a;13:1348–1359. doi: 10.1128/EC.00160-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guggisberg AM, Park J, Edwards RL, Kelly ML, Hodge DM, Tolia NH, Odom AR. A sugar phosphatase regulates the methylerythritol phosphate (MEP) pathway in malaria parasites. Nat Commun. 2014b;5:4467. doi: 10.1038/ncomms5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He CY, Shaw MK, Pletcher CH, Striepen B, Tilney LG, Roos DS. A plastid segregation defect in the protozoan parasite Toxoplasma gondii. EMBO J. 2001;20:330–339. doi: 10.1093/emboj/20.3.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hempel F, Bullmann L, Lau J, Zauner S, Maier UG. ERAD-derived preprotein transport across the second outermost plastid membrane of diatoms. Mol Biol Evol. 2009;26:1781–1790. doi: 10.1093/molbev/msp079. [DOI] [PubMed] [Google Scholar]
- Howe CJ. Plastid origin of an extrachromosomal DNA molecule from Plasmodium, the causative agent of malaria. J Theor Biol. 1992;158:199–205. doi: 10.1016/s0022-5193(05)80718-0. [DOI] [PubMed] [Google Scholar]
- Howe CJ, Barbrook AC, Lockhart PJ. Organelle genes–do they jump or are they pushed? Trends Genet. 2000;16:65–66. doi: 10.1016/s0168-9525(99)01919-8. [DOI] [PubMed] [Google Scholar]
- Huang CY, Ayliffe MA, Timmis JN. Direct measurement of the transfer rate of chloroplast DNA into the nucleus. Nature. 2003;422:72–76. doi: 10.1038/nature01435. [DOI] [PubMed] [Google Scholar]
- Huang J, Mullapudi N, Lancto CA, Scott M, Abrahamsen MS, Kissinger JC. Phylogenomic evidence supports past endosymbiosis, intracellular and horizontal gene transfer in Cryptosporidium parvum. Genome Biol. 2004;5:R88. doi: 10.1186/gb-2004-5-11-r88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlay L, Odom AR. Isoprenoid metabolism in apicomplexan parasites. Curr Clin Microbiol Rep. 2014;1:37–50. doi: 10.1007/s40588-014-0006-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlay LS, Armstrong CM, Masters MC, Li T, Price KE, Edwards RL, Mann KM, Li LX, Stallings CL, Berry NG, O’Neill PM, Odom AR. Plasmodium IspD (2-C-methyl-D-erythritol 4-phosphate Cytidyltransferase), an essential and druggable antimalarial target. ACS Infect Dis. 2015;1:157–167. doi: 10.1021/id500047s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janouskovec J, Horak A, Obornik M, Lukes J, Keeling PJ. A common red algal origin of the apicomplexan, dinoflagellate, and heterokont plastids. Proc Natl Acad Sci USA. 2010;107:10949–10954. doi: 10.1073/pnas.1003335107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janouskovec J, Tikhonenkov DV, Burki F, Howe AT, Kolisko M, Mylnikov AP, Keeling PJ. Factors mediating plastid dependency and the origins of parasitism in apicomplexans and their close relatives. Proc Natl Acad Sci US A. 2015;112:10200–10207. doi: 10.1073/pnas.1423790112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jomaa H, Wiesner J, Sanderbrand S, Altincicek B, Weidemeyer C, Hintz M, Turbachova I, Eberl M, Zeidler J, Lichtenthaler HK, Soldati D, Beck E. Inhibitors of the nonmevalonate pathway of isoprenoid biosynthesis as antimalarial drugs. Science. 1999;285:1573–1576. doi: 10.1126/science.285.5433.1573. [DOI] [PubMed] [Google Scholar]
- Kalanon M, McFadden GI. The chloroplast protein translocation complexes of Chlamydomonas reinhardtii: a bioinformatic comparison of Toc and Tic components in plants, green algae and red algae. Genetics. 2008;179:95–112. doi: 10.1534/genetics.107.085704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnataki A, Derocher A, Coppens I, Nash C, Feagin JE, Parsons M. Cell cycle-regulated vesicular trafficking of Toxoplasma APT1, a protein localized to multiple apicoplast membranes. Mol Microbiol. 2007a;63:1653–1668. doi: 10.1111/j.1365-2958.2007.05619.x. [DOI] [PubMed] [Google Scholar]
- Karnataki A, Derocher AE, Coppens I, Feagin JE, Parsons M. A membrane protease is targeted to the relict plastid of Toxoplasma via an internal signal sequence. Traffic. 2007b;8:1543–1553. doi: 10.1111/j.1600-0854.2007.00637.x. [DOI] [PubMed] [Google Scholar]
- Karnataki A, DeRocher AE, Feagin JE, Parsons M. Sequential processing of the Toxoplasma apicoplast membrane protein FtsH1 in topologically distinct domains during intracellular trafficking. Mol Biochem Parasitol. 2009;166:126–133. doi: 10.1016/j.molbiopara.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnkowska A, Vacek V, Zubacova Z, Treitli SC, Petrzelkova R, Eme L, Novak L, Zarsky V, Barlow LD, Herman EK, Soukal P, Hroudova M, Dolezal P, Stairs CW, Roger AJ, Elias M, Dacks JB, Vlcek C, Hampl V. A eukaryote without a mitochondrial organelle. Curr Biol. 2016;26:1274–1284. doi: 10.1016/j.cub.2016.03.053. [DOI] [PubMed] [Google Scholar]
- Ke H, Sigala PA, Miura K, Morrisey JM, Mather MW, Crowley JR, Henderson JP, Goldberg DE, Long CA, Vaidya AB. The heme biosynthesis pathway is essential for Plasmodium falciparum development in mosquito stage but not in blood stages. J Biol Chem. 2014;289:34827–34837. doi: 10.1074/jbc.M114.615831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilejian A. Circular mitochondrial DNA from the avian malarial parasite Plasmodium lophurae. Biochim Biophys Acta. 1975;390:276–284. doi: 10.1016/0005-2787(75)90348-2. [DOI] [PubMed] [Google Scholar]
- Köhler S, Delwiche CF, Denny PW, Tilney LG, Webster P, Wilson RJM, Palmer JD, Roos DS. A plastid of probable green algal origin in apicomplexan parasites. Science. 1997;275:1485–1488. doi: 10.1126/science.275.5305.1485. [DOI] [PubMed] [Google Scholar]
- Lim L, McFadden GI. The evolution, metabolism and functions of the apicoplast. Philos Trans R Soc, B. 2010;365:749–763. doi: 10.1098/rstb.2009.0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim L, Kalanon M, McFadden GI. New proteins in the apicoplast membranes: time to rethink apicoplast protein targeting. Trends Parasitol. 2009;25:197–200. doi: 10.1016/j.pt.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Lim L, Linka M, Mullin KA, Weber AP, McFadden GI. The carbon and energy sources of the non-photosynthetic plastid in the malaria parasite. FEBS Lett. 2010;584:549–554. doi: 10.1016/j.febslet.2009.11.097. [DOI] [PubMed] [Google Scholar]
- Lim L, Sayers CP, Goodman CD, McFadden GI. Targeting of a transporter to the outer apicoplast membrane in the human malaria parasite Plasmodium falciparum. PLoS One. 2016;11:e0159603. doi: 10.1371/journal.pone.0159603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner SE, Sartain MJ, Hayes K, Harupa A, Moritz RL, Kappe SH, Vaughan AM. Enzymes involved in plastid-targeted phosphatidic acid synthesis are essential for Plasmodium yoelii liver-stage development. Mol Microbiol. 2014;91:679–693. doi: 10.1111/mmi.12485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W. Gene transfer from organelles to the nucleus: frequent and in big chunks. Proc Natl Acad Sci USA. 2003;100:8612–8614. doi: 10.1073/pnas.1633606100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W, Herrmann RG. Gene transfer from organelles to the nucleus: how much, what happens, and why? Plant Physiol. 1998;118:9–17. doi: 10.1104/pp.118.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumdar J, Wilson EH, Masek K, Hunter CA, Striepen B. Apicoplast fatty acid synthesis is essential for organelle biogenesis and parasite survival in Toxoplasma gondii. Proc Natl Acad Sci USA. 2006;103:13192–13197. doi: 10.1073/pnas.0603391103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden GI, Roos DS. Apicomplexan plastids as drug targets. Trends Microbiol. 1999;6:328–333. doi: 10.1016/s0966-842x(99)01547-4. [DOI] [PubMed] [Google Scholar]
- McFadden GI, van Dooren GG. Evolution: red algal genome affirms a common origin of all plastids. Curr Biol. 2004;14:R514–R516. doi: 10.1016/j.cub.2004.06.041. [DOI] [PubMed] [Google Scholar]
- McFadden GI, Waller RF. Plastids in parasites of humans. BioEssays. 1997;19:1033–1040. doi: 10.1002/bies.950191114. [DOI] [PubMed] [Google Scholar]
- McFadden GI, Reith M, Munholland J, Lang-Unnasch N. Plastid in human parasites. Nature. 1996;381:482. doi: 10.1038/381482a0. [DOI] [PubMed] [Google Scholar]
- McFadden GI, Waller RF, Reith M, Munholland J, Lang-Unnasch N. Plastids in apicomplexan parasites. Plant Syst Evol (Suppl) 1997;11:261–287. [Google Scholar]
- Moore RB, Obornik M, Janouskovec J, Chrudimsky T, Vancova M, Green DH, Wright SW, Davies NW, Bolch CJ, Heimann K, Slapeta J, Hoegh-Guldberg O, Logsdon JM, Carter DA. A photosynthetic alveolate closely related to apicomplexan parasites. Nature. 2008;451:959–963. doi: 10.1038/nature06635. [DOI] [PubMed] [Google Scholar]
- Mullin KA, Lim L, Ralph SA, Spurck TP, Handman E, McFadden GI. Membrane transporters in the relict plastid of malaria parasites. Proc Natl Acad Sci USA. 2006;103:9572–9577. doi: 10.1073/pnas.0602293103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraj VA, Arumugam R, Gopalakrishnan B, Jyothsna YS, Rangarajan PN, Padmanaban G. Unique properties of Plasmodium falciparum porphobilinogen deaminase. J Biol Chem. 2008;283:437–444. doi: 10.1074/jbc.M706861200. [DOI] [PubMed] [Google Scholar]
- Nagaraj VA, Arumugam R, Chandra NR, Prasad D, Rangarajan PN, Padmanaban G. Localisation of Plasmodium falciparum uroporphyrinogen III decarboxylase of the heme-biosynthetic pathway in the apicoplast and characterisation of its catalytic properties. Int J Parasitol. 2009a;39:559–568. doi: 10.1016/j.ijpara.2008.10.011. [DOI] [PubMed] [Google Scholar]
- Nagaraj VA, Prasad D, Rangarajan PN, Padmanaban G. Mitochondrial localization of functional ferrochelatase from Plasmodium falciparum. Mol Biochem Parasitol. 2009b;168:109–112. doi: 10.1016/j.molbiopara.2009.05.008. [DOI] [PubMed] [Google Scholar]
- Nagaraj VA, Sundaram B, Varadarajan NM, Subramani PA, Kalappa DM, Ghosh SK, Padmanaban G. Malaria parasite-synthesized heme is essential in the mosquito and liver stages and complements host heme in the blood stages of infection. PLoS Pathog. 2013;9:e1003522. doi: 10.1371/journal.ppat.1003522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair SC, Brooks CF, Goodman CD, Strurm A, McFadden GI, Sundriyal S, Anglin JL, Song Y, Moreno SN, Striepen B. Apicoplast isoprenoid precursor synthesis and the molecular basis of fosmidomycin resistance in Toxoplasma gondii. J Exp Med. 2011;208:1547–1559. doi: 10.1084/jem.20110039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obornik M, Vancova M, Lai DH, Janouskovec J, Keeling PJ, Lukes J. Morphology and ultrastructure of multiple life cycle stages of the photosynthetic relative of Apicomplexa, Chromera velia. Protist. 2011;162:115–130. doi: 10.1016/j.protis.2010.02.004. [DOI] [PubMed] [Google Scholar]
- Okamoto N, Inouye I. A secondary symbiosis in progress? Science. 2005;310:287. doi: 10.1126/science.1116125. [DOI] [PubMed] [Google Scholar]
- Okamoto N, McFadden GI. The mother of all parasites. Fut Microbiol. 2008;3:391–395. [Google Scholar]
- Okazaki K, Kabeya Y, Miyagishima S-Y. The evolution of the regulatory mechanism of chloroplast division. Plant Signal Behav. 2010;5:164–167. doi: 10.4161/psb.5.2.10461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph SA, D’Ombrain MC, McFadden GI. The apicoplast as an antimalarial drug target. Drug Resist Updat. 2001;4:145–151. doi: 10.1054/drup.2001.0205. [DOI] [PubMed] [Google Scholar]
- Ralph SA, van Dooren GG, Waller RF, Crawford MJ, Fraunholz MJ, Foth BJ, Tonkin CJ, Roos DS, McFadden GI. Metabolic maps and functions of the Plasmodium falciparum apicoplast. Nat Rev Microbiol. 2004;2:203–216. doi: 10.1038/nrmicro843. [DOI] [PubMed] [Google Scholar]
- Riordan CE, Ault JG, Langreth SG, Keithly JS. Cryptosporidium parvum Cpn60 targets a relict organelle. Curr Genet. 2003;44:138–147. doi: 10.1007/s00294-003-0432-1. [DOI] [PubMed] [Google Scholar]
- Sato S. The apicomplexan plastid and its evolution. Cell Mol Life Sci. 2011;68:1285–1296. doi: 10.1007/s00018-011-0646-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, Clough B, Coates L, Wilson RJ. Enzymes for heme biosynthesis are found in both the mitochondrion and plastid of the malaria parasite Plasmodium falciparum. Protist. 2004;155:117–125. doi: 10.1078/1434461000169. [DOI] [PubMed] [Google Scholar]
- Seeber F. Biogenesis of iron–sulphur clusters in amitochondriate and apicomplexan protists. Int J Parasitol. 2002;32:1207–1217. doi: 10.1016/s0020-7519(02)00022-x. [DOI] [PubMed] [Google Scholar]
- Seeber F. Biosynthetic pathways of plastid-derived organelles as potential drug targets against parasitic apicomplexa. Curr Drug Targets Immune Endocr Metabol Disord. 2003;3:99–109. doi: 10.2174/1568008033340261. [DOI] [PubMed] [Google Scholar]
- Shanmugam D, Wu B, Ramirez U, Jaffe EK, Roos DS. Plastid-associated porphobilinogen synthase from Toxoplasma gondii: kinetic and structural properties validate therapeutic potential. J Biol Chem. 2010;285:22122–22131. doi: 10.1074/jbc.M110.107243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shears MJ, Botte CY, McFadden GI. Fatty acid metabolism in the Plasmodium apicoplast: drugs, doubts and knockouts. Mol Biochem Parasitol. 2015;199:34–50. doi: 10.1016/j.molbiopara.2015.03.004. [DOI] [PubMed] [Google Scholar]
- Sheiner L, Vaidya AB, McFadden GI. The metabolic roles of the endosymbiotic organelles of Toxoplasma and Plasmodium spp. Curr Opin Microbiol. 2013;16:452–458. doi: 10.1016/j.mib.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheiner L, Fellows JD, Ovciarikova J, Brooks CF, Agrawal S, Holmes ZC, Bietz I, Flinner N, Heiny S, Mirus O, Przyborski JM, Striepen B. Toxoplasma gondii Toc75 functions in import of stromal but not peripheral apicoplast proteins. Traffic. 2015;16:1254–1269. doi: 10.1111/tra.12333. [DOI] [PubMed] [Google Scholar]
- Sigala PA, Goldberg DE. The peculiarities and paradoxes of Plasmodium heme metabolism. Annu Rev Microbiol. 2014;68:259–278. doi: 10.1146/annurev-micro-091313-103537. [DOI] [PubMed] [Google Scholar]
- Smith CM, Jerkovic A, Puy H, Winship I, Deybach JC, Gouya L, van Dooren G, Goodman CD, Sturm A, Manceau H, McFadden GI, David P, Mercereau-Puijalon O, Burgio G, McMorran BJ, Foote SJ. Red cells from ferrochelatase-deficient erythropoietic protoporphyria patients are resistant to growth of malarial parasites. Blood. 2014;125:534–541. doi: 10.1182/blood-2014-04-567149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer MS, Gould SB, Lehmann P, Gruber A, Przyborski JM, Maier UG. Der1-mediated preprotein import into the periplastid compartment of chromalveolates? Mol. Biol Evol. 2007;24:918–928. doi: 10.1093/molbev/msm008. [DOI] [PubMed] [Google Scholar]
- Spork S, Hiss JA, Mandel K, Sommer M, Kooij TW, Chu T, Schneider G, Maier UG, Przyborski JM. An unusual ERAD-like complex is targeted to the apicoplast of Plasmodium falciparum. Eukaryot Cell. 2009;8:1134–1145. doi: 10.1128/EC.00083-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Striepen B, Crawford MJ, Shaw MK, Tilney LG, Seeber F, Roos DS. The plastid of Toxoplasma gondii is divided by association with the centrosomes. J Cell Biol. 2000;151:1423–1434. doi: 10.1083/jcb.151.7.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suplick K, Akella R, Saul A, Vaidya AB. Molecular cloning and partial sequence of a 5.8 kilobase pair repetitive DNA from Plasmodium falciparum. Mol Biochem Parasitol. 1988;30:289–290. doi: 10.1016/0166-6851(88)90098-9. [DOI] [PubMed] [Google Scholar]
- Suplick K, Morrisey J, Vaidya AB. Complex transcription from the extrachromosomal DNA encoding mitochondrial functions of Plasmodium yoelii. Mol Cell Biol. 1990;10:6381–6388. doi: 10.1128/mcb.10.12.6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovar J, Fischer A, Clark CG. The mitosome, a novel organelle related to mitochondria in the amitochondrial parasite Entamoeba histolytica. Mol Microbiol. 1999;32:1013–1021. doi: 10.1046/j.1365-2958.1999.01414.x. [DOI] [PubMed] [Google Scholar]
- Vaidya AB, Akella R, Suplick K. Sequences similar to genes for two mitochondrial proteins and portions of ribosomal RNA in tandemly arrayed 6- kilobase-pair DNA of a malarial parasite. Mol Biochem Parasitol. 1989;35:97–108. doi: 10.1016/0166-6851(89)90112-6. [DOI] [PubMed] [Google Scholar]
- Vaishnava S, Striepen B. The cell biology of secondary endosymbiosis-how parasites build, divide and segregate the apicoplast. Mol Microbiol. 2006;61:1380– 1387. doi: 10.1111/j.1365-2958.2006.05343.x. [DOI] [PubMed] [Google Scholar]
- van der Giezen M, Tovar J. Degenerate mitochondria. EMBO Rep. 2005;6:525– 530. doi: 10.1038/sj.embor.7400440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dooren GG, Striepen B. The algal past and parasite present of the apicoplast. Annu Rev Microbiol. 2013;67:271–289. doi: 10.1146/annurev-micro-092412-155741. [DOI] [PubMed] [Google Scholar]
- van Dooren GG, Su V, D’Ombrain MC, McFadden GI. Processing of an apicoplast leader sequence in Plasmodium falciparum and the identification of a putative leader cleavage enzyme. J Biol Chem. 2002;277:23612–23619. doi: 10.1074/jbc.M201748200. [DOI] [PubMed] [Google Scholar]
- van Dooren GG, Stimmler LM, McFadden GI. Metabolic maps and functions of the Plasmodium mitochondrion. FEMS Microbiol Rev. 2006;30:596–630. doi: 10.1111/j.1574-6976.2006.00027.x. [DOI] [PubMed] [Google Scholar]
- van Dooren GG, Tomova C, Agrawal S, Humbel BM, Striepen B. Toxoplasma gondii Tic20 is essential for apicoplast protein import. Proc Natl Acad Sci USA. 2008;105:13574–13579. doi: 10.1073/pnas.0803862105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dooren GG, Reiff SB, Tomova C, Meissner M, Humbel BM, Striepen B. A novel dynamin-related protein has been recruited for apicoplast fission in Toxoplasma gondii. Curr Biol. 2009;19:267–276. doi: 10.1016/j.cub.2008.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Schaijk BC, Kumar TR, Vos MW, Richman A, van Gemert GJ, Li T, Eappen AG, Williamson KC, Morahan BJ, Fishbaugher M, Kennedy M, Camargo N, Khan SM, Janse CJ, Sim KL, Hoffman SL, Kappe SH, Sauerwein RW, Fidock DA, Vaughan AM. Type II fatty acid biosynthesis is essential for Plasmodium falciparum sporozoite development in the midgut of Anopheles mosquitoes. Eukaryot Cell. 2013 doi: 10.1128/EC.00264-13. http://dx.doi.org/10.1128/EC.00264-13. [DOI] [PMC free article] [PubMed]
- Vaughan AM, O’Neill MT, Tarun AS, Camargo N, Phuong TM, Aly AS, Cowman AF, Kappe SH. Type II fatty acid synthesis is essential only for malaria parasite late liver stage development. Cell Microbiol. 2009;11:506–520. doi: 10.1111/j.1462-5822.2008.01270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waller RF, Keeling PJ. Alveolate and chlorophycean mitochondrial cox2 genes split twice independently. Gene. 2006;383:33–37. doi: 10.1016/j.gene.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Waller RF, Keeling PJ, Donald RGK, Striepen B, Handman E, Lang-Unnasch N, Cowman AF, Besra GS, Roos DS, McFadden GI. Nuclear-encoded proteins target to the plastid in Toxoplasma gondii and Plasmodium falciparum. Proc Natl Acad Sci USA. 1998;95:12352–12357. doi: 10.1073/pnas.95.21.12352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waller RF, Keeling PJ, van Dooren GG, McFadden GI. Comment on “A green algal apicoplast ancestor”. Science. 2003;301:49. doi: 10.1126/science.1084684. [DOI] [PubMed] [Google Scholar]
- Waller RF, Gornik SG, Koreny L, Pain A. Metabolic pathway redundancy within the apicomplexan-dinoflagellate radiation argues against an ancient chromalveolate plastid. Commun Integr Biol. 2016;9:e1116653. doi: 10.1080/19420889.2015.1116653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber AP, Linka M, Bhattacharya D. Single, ancient origin of a plastid metabolite translocator family in Plantae from an endomembrane-derived ancestor. Eukaryot Cell. 2006;5:609–612. doi: 10.1128/EC.5.3.609-612.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesner J, Reichenberg A, Heinrich S, Schlitzer M, Jomaa H. The plastid-like organelle of apicomplexan parasites as drug target. Curr Pharm Des. 2008;14:855–871. doi: 10.2174/138161208784041105. [DOI] [PubMed] [Google Scholar]
- Wiley JD, Merino EF, Krai PM, McLean KJ, Tripathi AK, Vega-Rodriguez J, Jacobs-Lorena M, Klemba M, Cassera MB. Isoprenoid precursor biosynthesis is the essential metabolic role of the apicoplast during gametocytogenesis in Plasmodium falciparum. Eukaryot Cell. 2015;14:128–139. doi: 10.1128/EC.00198-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson DH, Wilson RJM, Bates RA, McCready S, Perler R, Qiang B-U. Nuclear and mitochondrial DNA of the primate parasite Plasmodium knowlesi. Mol Biochem Parasitol. 1985;14:199–209. doi: 10.1016/0166-6851(85)90038-6. [DOI] [PubMed] [Google Scholar]
- Williamson DH, Gardner MJ, Preiser P, Moore DJ, Rangachari K, Wilson RJ. The evolutionary origin of the 35 kb circular DNA of Plasmodium falciparum: new evidence supports a possible rhodophyte ancestry. Mol Gen Genet. 1994;243:249–252. doi: 10.1007/BF00280323. [DOI] [PubMed] [Google Scholar]
- Wilson I. Plastids better red than dead [letter] Nature. 1993;366:6456. doi: 10.1038/366638a0. [DOI] [PubMed] [Google Scholar]
- Wilson RJM, Gardner MJ, Feagin JE, Williamson DH. Have malaria parasites three genomes? Parasitol. Today. 1991;7:134–136. doi: 10.1016/0169-4758(91)90276-t. [DOI] [PubMed] [Google Scholar]
- Wilson RJM, Denny PW, Preiser PR, Rangachari K, Roberts K, Roy A, Whyte A, Strath M, Moore DJ, Moore PW, Williamson DH. Complete gene map of the plastid-like DNA of the malaria parasite Plasmodium falciparum. J Mol Biol. 1996;261:155–172. doi: 10.1006/jmbi.1996.0449. [DOI] [PubMed] [Google Scholar]
- Woo YH, Ansari H, Otto TD, Klinger CM, Kolisko M, Michalek J, Saxena A, Shanmugam D, Tayyrov A, Veluchamy A, Ali S, Bernal A, del Campo J, Cihlar J, Flegontov P, Gornik SG, Hajduskova E, Horak A, Janouskovec J, Katris NJ, Mast FD, Miranda-Saavedra D, Mourier T, Naeem R, Nair M, Panigrahi AK, Rawlings ND, Padron-Regalado E, Ramaprasad A, Samad N, Tomcala A, Wilkes J, Neafsey DE, Doerig C, Bowler C, Keeling PJ, Roos DS, Dacks JB, Templeton TJ, Waller RF, Lukes J, Obornik M, Pain A. Chromerid genomes reveal the evolutionary path from photosynthetic algae to obligate intracellular parasites. Elife. 2015;4:e06974. doi: 10.7554/eLife.06974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Herrera Z, Ebert D, Baska K, Cho SH, DeRisi JL, Yeh E. A chemical rescue screen identifies a Plasmodium falciparum apicoplast inhibitor targeting MEP isoprenoid precursor biosynthesis. Antimicrob Agents Chemother. 2015;59:356–364. doi: 10.1128/AAC.03342-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E, DeRisi J. Chemical rescue of malaria parasites lacking an apicoplast defines organelle function in blood-stage Plasmodium falciparum. PLoS Biol. 2011;9:e1001138. doi: 10.1371/journal.pbio.1001138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Kumar TR, Nkrumah LJ, Coppi A, Retzlaff S, Li CD, Kelly BJ, Moura PA, Lakshmanan V, Freundlich JS, Valderramos JC, Vilcheze C, Siedner M, Tsai JH, Falkard B, Sidhu AB, Purcell LA, Gratraud P, Kremer L, Waters AP, Schiehser G, Jacobus DP, Janse CJ, Ager A, Jacobs WR, Jr, Sacchettini JC, Heussler V, Sinnis P, Fidock DA. The fatty acid biosynthesis enzyme FabI plays a key role in the development of liver-stage malarial parasites. Cell Host Microbe. 2008;4:567–578. doi: 10.1016/j.chom.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, Marchewka MJ, Keithly JS. Cryptosporidium parvum appears to lack a plastid genome. Microbiology. 2000;146(Pt 2):315–321. doi: 10.1099/00221287-146-2-315. [DOI] [PubMed] [Google Scholar]