Abstract
Background & Aims
Stearoyl-CoA desaturase (SCD) synthesizes monounsaturated fatty acids (MUFAs) and has been associated with development of metabolic syndrome, tumorigenesis, and stem cell characteristics. We investigated whether and how SCD promotes liver fibrosis and tumor development in mice.
Methods
Rodent primary hepatic stellate cells (HSCs), mouse liver tumor-initiating stem cell-like cells (TICs), and human hepatocellular carcinoma (HCC) cell lines were exposed to Wnt signaling inhibitors and changes in gene expression patterns were analyzed. We assessed the functions of SCD by pharmacologic and conditional genetic manipulation in mice with hepatotoxic or cholestatic induction of liver fibrosis, orthotopic transplants of TICs, or liver tumors induced by administration of diethyl nitrosamine. We performed bioinformatic analyses of SCD expression in HCC vs non-tumor liver samples collected from patients, and correlate levels with HCC stage and patient mortality. We performed nano-bead pull-down assays, liquid chromatography-mass spectrometry, computational modeling, and ribonucleoprotein immunoprecipitation analyses to identify MUFA-interacting proteins. We examined the effects of SCD inhibition on Wnt signaling, including expression and stability of low-density lipoprotein receptor-related proteins 5 and 6 (LRP5 and LRP6), by immunoblot and quantitative PCR analyses.
Results
SCD was overexpressed in activated HSC and HCC cells from patients; levels of SCD mRNA correlated with HCC stage and patient survival time. In rodent HSCs and TICs, the Wnt effector beta-catenin increased sterol regulatory element binding protein 1-dependent transcription of Scd, and beta-catenin was in return stabilized by MUFAs generated by SCD. This loop required MUFA inhibition of binding of RAS-related nuclear protein 1 to transportin 1 and reduced nuclear import of elav-like protein 1 (ELAVL1), increasing cytosolic levels of ELAVL1 and ELAVL1-mediated stabilization of mRNAs encoding LRP5 and LRP6. Genetic disruption of Scd and pharmacologic inhibitors of SCD reduced HSC activation and TIC self-renewal and attenuated liver fibrosis and tumorigenesis in mice. Conditional disruption of Scd2 in activated HSCs prevented growth of tumors from TICs and reduced formation of diethyl nitrosamine-induced liver tumors in mice.
Conclusions
In rodent HSCs and TICs, we found Scd expression to be regulated by Wnt–beta-catenin signaling and MUFAs produced by SCD to provide a positive-feedback loop that amplifies Wnt signaling via stabilization of Lrp5 and Lrp6 mRNAs, leading to liver fibrosis and tumor growth. SCD expressed by HSCs promoted liver tumor development. SCD expression was increased in HCCs from patients compared with non-tumor tissue, and correlated positively with tumor state and inversely with patient survival time.
Keywords: HCC, cancer stem cells, HuR, hepatocarcinogenesis
Introduction
Cirrhosis with excessive accumulation of extracellular matrix predisposes the liver to the development of hepatocellular carcinoma (HCC)1, suggesting the microenvironment of fibrotic liver promotes tumor development2. Although many studies have supported this notion, underlying mechanisms are still elusive. We have approached this question by focusing on the canonical Wnt pathway, a pathway common in both HSCs and tumor-initiating stem cell-like cells (TICs) or HCC cells for their cell fate regulation.
Activation of the canonical Wnt pathway is a hallmark of both liver fibrogenesis and carcinogenesis3. The Wnt gene family consists of structurally related genes which encode secreted proteins which regulate cell fate by binding to their Frizzled (Fzd)/low-density lipoprotein-receptor-related protein (LRP)5/6 receptor complex. This binding causes phosphorylation of dishevelled (Dvl) which in turn recruits Axin and glycogen synthase kinase 3 (GSK3) via LRP5/6, allowing stabilization and nuclear translocation of β-catenin, and stimulation of WNT target genes4. Induction of such classical β-catenin target genes enhances cell proliferation and survival, and promotes fibrogenesis5 and carcinogenesis3 which in the liver, are primarily mediated by activated HSCs (aHSCs)6 and hepatocyte-derived transformed cells7 or TICs7, 8, respectively. However, it is unknown whether and how Wnt-activation in aHSCs promotes liver tumorigenesis in which gain-of-function mutations of β-catenin is frequent.
To approach this question, we screened for Wnt-dependent genes in rat aHSCs by using three Wnt inhibitors and microarray analysis. This has identified stearoyl-CoA desaturases (Scd) as Wnt-target genes in aHSCs. SCD is a family of Δ9 fatty acid desaturases which generate monounsaturated fatty acids (MUFA), palmitoleate (POA,16:1) and oleate (OA,18:1) from palmitate (PA,16:0) and stearate (SA,18:0), respectively. The mouse has four Scd genes, Scd1-4 with Scd1 being predominant in adult liver and Scd2 in embryonic liver9. Humans express only SCD (also known as SCD1), as the major form in various tissues, with SCD5 being expressed in brain and pancreas10. SCD has pleiotropic effects on obesity, insulin resistance, inflammation, and cancer development, and is required for production of active palmitoleated Wnt proteins11. SCD is upregulated in fibrosis12 and facilitates cancer cell stemness13 by unknown mechanisms.
Here, we identify Scd1 and Scd2 as Wnt/β-catenin target genes which are transactivated by β-catenin interaction with SREBP-1c bound to the novel SRE site in the proximal promoters. Silencing of Scd2, the major isoenzyme in rodent HSCs, restores the HSC differentiation gene Pparγ and reverts aHSCs to quiescent cells. Mice bearing α1(I) collagen promoter-Cre (Col1a1-Cre) and Scd2flox/flox are resistant to HSC activation and liver fibrosis induced by CCl4 and bile duct ligation (BDL). In parallel, we demonstrate that Scd2 knockdown in TICs or pharmacologic SCD inhibition of human HCC Huh7 cells inhibits proliferation and self-renewal in vitro, and prevents TIC-initiated tumorigenesis in mice. Further, Scd2 expression by aHSCs is responsible for aHSC-mediated tumor promotion in both TIC-derived and DEN-induced tumorigenesis in mice. Moreover, SCD facilitates a novel positive feedback loop for β-catenin via MUFA-dependent LRP5/6 expression, which serves to activate both fibrogenic HSCs and oncogenic TICs.
Methods (More in Suppl. information)
Study approvals, primary HSCs, liver TICs and HCC cell lines
This study was approved by the IACUC (#13513) and IRB (#1992-048) of the Univ of Southern Calif and conducted according to the NIH guidelines. HSCs were isolated from normal male Wistar rats or rats subjected to 10-day BDL or 6 biweekly injections of CCl4 as described5. The purity of HSCs isolated exceeded 95% as determined by UV-excited autofluoresence. TICs were isolated from an alcohol/HCV-induced HCC mouse model as described8. Huh7 is a HCC cell line available from Japanese Collection of Research Bioresources Cell Bank. Hep3B and PLC/PRF/5 are HCC cell lines from ATCC. Liver TICs were transduced with lentiviral shRNA targeting Scd2 (Scd2-KD) (Sigma) or shRNA targeting GFP (GFP-KD) (Sigma) at MOI = 50. Five different shRNAs were tested. Stably transduced TICs were selected by puromycin, and then subjected to limiting dilution to obtain a clone which was subsequently expanded.
Wnt inhibitor treatment of HSCs or liver TICs
Fully activated primary rat HSCs were treated with: 1) adenovirus expressing DKK-1 (Ad.DKK-1) vs. Ad.GFP (control) at a multiplicity of infection (MOI) of 200 for 24 h in low glucose DMEM with 5% FBS and antibiotics, and then maintained for an additional 24 h in serum-free medium; 2) ICG-001 (10 μM) vs. vehicle for 24 h in serum-free medium or 92 h; or 3) FJ9 (200 μM) vs. vehicle for 24 h or 92 h in serum-free medium. Morphological effects were examined by phase contrast microscopy and lipid storage by oil red O staining.
Microarray analysis
Total RNA was extracted from HSCs. Equal amounts of extracted HSC RNA from 3 to 4 different rats for each of the 6 treatment categories (Ad. DKK-1 vs. Ad.GFP; ICG-001 vs. vehicle; and FJ9 vs. vehicle) were respectively pooled together. After confirmation of RNA quantity, integrity, and purity, the resultant 6 samples were analyzed at USC/Children’s Hospital Los Angeles Genome Core Laboratory using Affymetrix (Santa Clara, CA) Rat Gene 1.0 ST Array kits according to the manufacturer’s fragmentation, labeling, hybridization, and signal capture protocols. Partek Genomics Suite software was used to analyze microarray data and construct a heatmap displaying relative fold changes in global gene expression between HSCs treated with Wnt inhibitors and respective controls.
Model of TIC-induced HCC in mice diet-fed alcohol
Immune-competent B6 mice were fed increasing amounts of ethanol (up to 6%) for 5 weeks, and then mice were orthotopically transplanted into the left lobe with 2 × 105 TICs stably transduced with lentiviral shRNA targeting Scd2 (Scd2-KD) versus a control clone transduced with lentiviral shRNA targeting GFP (GFP-KD). Mice were then continued on ethanol diet for additional 3 weeks before being euthanized, at which time livers were harvested and inspected for tumor growth. Tumor number and size were quantitated.
Genetic and pharmacologic SCD manipulations
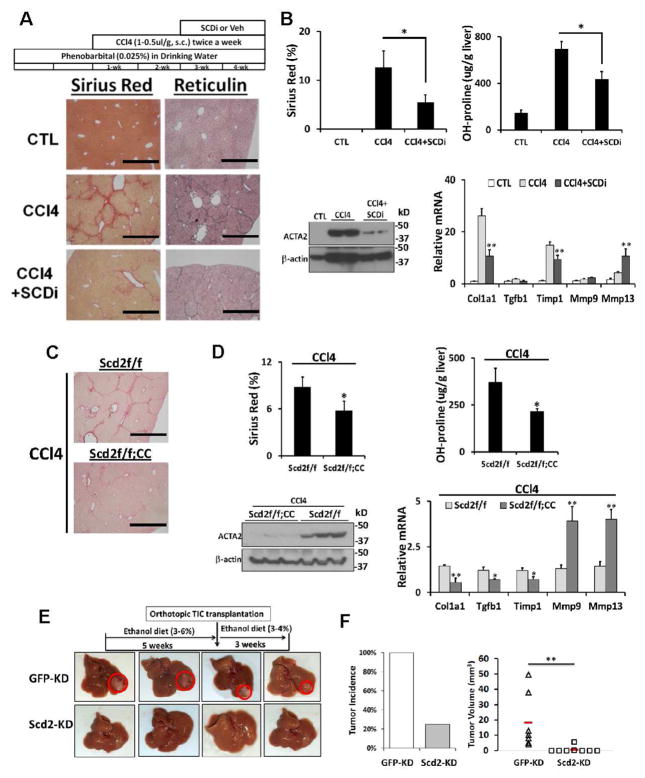
HSCs were also isolated from mice having the third exon of the Scd2 gene flanked by loxP sites (Scd2f/f). Scd2 ablation was achieved in culture by infecting the cells with adenovirus expressing Cre recombinase. Scd2f/f mice were crossed with α1(I)collagen promoter-Cre (CC) mice to generate the conditional KO mice (Scd2f/f;CC). Scd2f/f and Scd2f/f;CC mice were given phenobarbital (0.025%) in drinking water and injected with CCl4 (s.c., 1–2 μl/g) in mineral oil (1:1 dilution) or mineral oil only twice weekly for 4 wk. The mice were also subjected to aseptic BDL and liver fibrosis assessed on day 13. Pharmacologic inhibition of SCD with A939572 (BioVision, Inc.) was tested in CCl4 and BDL models. Livers were collected for morphometric analysis of Sirius red, reticulin staining, and immunoblot analysis of ACTA2. Blood was collected to measure plasma ALT activity as per assay manufacturer’s (Sigma) protocol. Hydroxyproline content of livers was determined by hydrolysis of liver proteins with HCl and ELISA assay (Cell Biolabs, Inc.). HSCs were isolated from 3 pairs of Scd2f/f;CC vs. Scd2f/f mice subjected to BDL for Scd2 and Scd1 mRNA qPCR, revealing selective 89% reduction in Scd2 in Scd2f/f;CC.
Statistical analysis
Data are presented as means ± SD or otherwise stated in figure legends. Statistical significance of difference was assessed by the Student’s t-test or Wilcoxon-Mann-Whitney U test with p value less than 0.05 considered statistically significant.
Results
Scd1/2 are Wnt target genes induced in activated HSCs
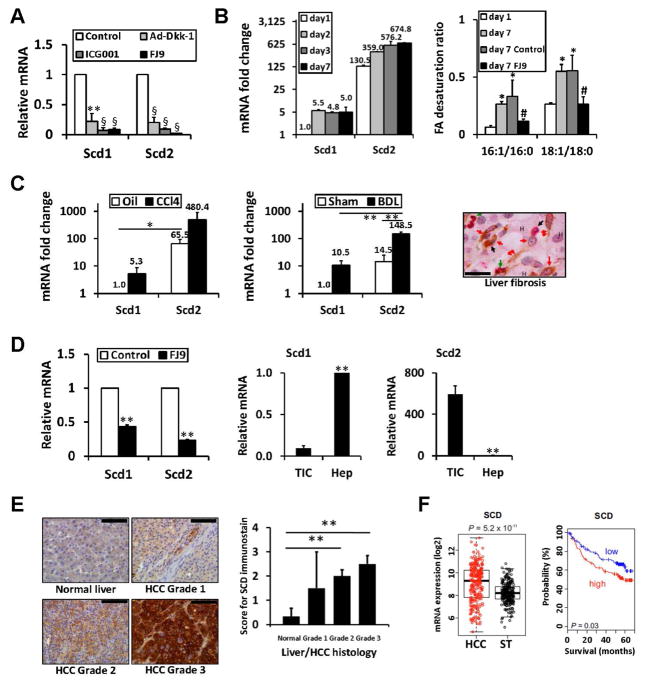
We discovered that Scd1 and Scd2 are putative Wnt target genes by microarray analysis of cultured rat HSCs treated with canonical Wnt inhibitors, DKK-1, ICG-001, or FJ9 (Fig. S1, Table S1) as validated by qPCR (Fig. 1A). We then examined the kinetics of Scd1 and Scd2 upregulation in rat HSCs undergoing activation in culture by TaqMan qPCR. Scd2 expression is >100 fold higher than Scd1 in quiescent HSCs cultured for 1 day and both genes are induced at day 2 when spontaneous HSC activation begins (Fig. 1B, left). To assess a biological readout of SCD in quiescent day 1 vs. fully-activated day 7 HSCs, we determined the cellular content of MUFA (16:1 and 18:1) produced by SCD and their precursor saturated fatty acids (16:0 and 18:0). The desaturation ratios (16:1/16:0 and 18:1/18:0), the readouts of SCD activity, are increased in day 7 aHSCs vs. day 1 cells and these increases are prevented by the Wnt inhibitor FJ9 (Fig. 1B right, Table S2). Next, we extended our analysis to aHSCs isolated from experimental rat liver fibrosis induced by CCl4 injections or BDL. Scd2 is upregulated 7–10 fold in HSCs isolated from these fibrosis models vs. respective controls (Fig. 1C left and middle).
Fig. 1. Scd2/SCD upregulated in liver fibrosis and cancers.
A. Scd 1 and Scd2, are repressed by three canonical Wnt inhibitors, Dkk1, FJ9, and ICG-001 in cultured rat HSCs, n=4. **p<0.01, §p<0.001. B. Left, Scd1 and Scd2 mRNA in HSCs during culture activation by TaqMan qPCR and expressed as fold change vs. Scd1 expression at day 1. Right, Increases in fatty acid desaturation ratios in day 7 cultured HSCs are abrogated by FJ9. *p<0.05 vs. day 1, #p<0.05 vs. vehicle control at day 7. C. Left and middle, Scd1 and Scd2 mRNA in HSCs from rat CCl4 (n=4) or BDL (n=3) liver fibrosis and respective controls (Oil and Sham) by TaqMan qPCR and expressed as fold change vs. controls. *p<0.05, **p<0.01. Right, HSCs (double red arrow) and myofibroblasts (red arrow) are positive for SCD (brown) and ACTA2 (red) staining in a liver section of alcoholic liver fibrosis patient. Macrophages (black arrow) and PMNs (green arrow) are also present. Hepatocytes are denoted by “H”. Scale bar= 40um. D. FJ9 suppresses Scd1 and Scd2 in mouse liver TICs (left). Scd2 expressed in TICs vs. Scd1 in normal hepatocytes (middle and right) (n=4). **p<0.01. E. SCD induction with the worsening grade of HCC as demonstrated by immunostaining (left) and its semi-quantitation (right) of 17 patients. **p<0.01. Scale bar=100um. F. SCD mRNA expression induced in HCC vs. non-tumor tissue in a patient cohort (n=242) (left) and correlates with mortality when patients are dichotomized by median expression value (right).
Wnt target genes Scd2 and SCD are induced in TICs and human HCC cell lines
SCD induction is also reported in malignancies14, 15. We examined the expression of Scd isoforms in mouse liver TICs isolated from liver tumors of alcohol-fed HCV NS5A transgenic mice, which exhibit self-renewal and tumor-initiating activities8. FJ9 significantly represses both Scd1 and Scd2 in TICs, supporting these genes are Wnt targets (Fig. 1D left). We reveal that Scd2 is the major form expressed in TICs while Scd1 is predominant in adult hepatocytes (Fig. 1D middle and right). SCD (a major isoform in man) is induced in 3 human HCC cell lines, Huh7 (~7 fold), PLC/PRF/5 (~8 fold), and Hep3B (~14 fold) vs. normal human hepatocytes obtained from 5 donors (Fig. S2A). FJ9 suppresses SCD in Huh7 (data not shown).
Translational relevance of SCD upregulation
We detected upregulated SCD in aHSCs and myofibroblasts positively stained for ACTA2 in liver section obtained from a patient with alcoholic liver disease (Fig. 1C right). SCD upregulation is also evident by immunostaining in accordance to advancing grade of HCC in 17 patients (Fig. 1E: left; right, semi-quantitative data for staining). Bioinformatic analysis of gene expression data show SCD expression correlates with clinical outcome in HCC at the population level. SCD is induced in tumor vs. non-tumor tissue in an HCC cohort (Fig. 1F left) and associated with increased mortality in this cohort (Fig. 1F right).
β-catenin interacts with SREBP-1c at novel SRE site to promote Scd transcription
We next investigated how canonical Wnt pathway induces Scd1 and Scd2. Both genes have no LEF/TCF sites within their promoters and are similarly regulated by a “novel” sterol-regulatory element (SRE) and two NF-Y sites which lie downstream of the novel SRE16. We used Scd1 promoter-reporter constructs without or with SRE or NF-Y deletion and mutations to map out a region responsive to FJ9 in the HSC line BSC. Results from these analyses (Fig. S3A) indeed point to the novel SRE and proximal NF-Y sites as potential targets of FJ9. SREBP-1c stimulates the promoter activity 12-fold, and FJ9 reduces this effect by 45% (Fig. S3B). SREBP-1c-induced Scd1 promoter activity is enhanced ~9-fold by co-expression of β-catenin (Fig. S3C). ChIP-qPCR analysis for the novel SRE and NF-Y sites demonstrates increased enrichments of SREBP-1c and β-catenin in day 7 aHSCs vs. day 1 cells (Fig. S3D). Finally, a re-ChIP analysis demonstrates increased β-catenin binding with SREBP-1c which is bound to the SRE/NF-Y site in day 7 aHSCs (Fig. S3E). We have obtained similar results with the Scd2 promoter (which also contains the novel SRE and both NF-Y sites) by β-catenin ChIP analysis (Fig. S3F), suggesting both genes are positively regulated by β-catenin via its interaction with SREBP-1c at the conserved novel SRE sites.
SCD is required for activation of HSCs and self-renewal of TICs, and stabilizes β-catenin in a manner dependent on MUFA
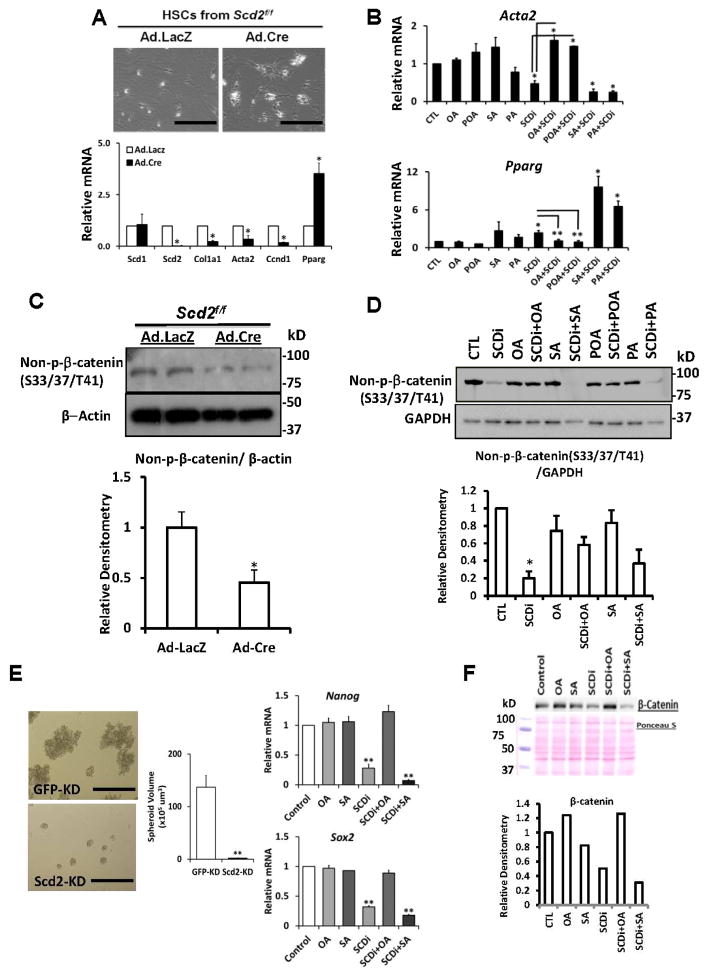
Next, we isolated HSCs from Scd2flox/flox mice and infected them in culture with adenovirus (Ad) that expresses Cre recombinase (Cre) or LacZ (control). Ad.LacZ-infected HSCs from Scd2flox/flox become fully-activated in culture (Fig. 2A top left). But Ad.Cre-infected, Scd2-ablated HSCs have retracted cell shape reminiscent of quiescent HSCs (Fig. 2A top right). qPCR confirms depletion of Scd2 mRNA with no effect on Scd1 and reveals suppression of HSC activation genes Col1a1 and Acta2, and up regulation of the HSC quiescence gene Pparγ (Fig. 2A bottom). We next pharmacologically inhibited SCD (SCDi) with a small piperidine-aryl urea-based inhibitor, A93957217. This manipulation also reverts aHSCs to quiescent cells (not shown). Moreover, this effect is prevented by addition of OA or POA, but not with SA or PA, suggesting that SCD activity and generation of MUFA are required for HSC activation. qPCR analysis confirms this notion by demonstration of Acta2 suppression and Pparγ upregulation by SCDi and abrogation of these effects by MUFA (Fig. 2B). Scd2 silencing also represses Wnt-dependent gene Ccnd1 (cyclin D1) (Fig. 2A bottom), suggesting SCD which is upregulated by β-catenin, may also support the Wnt pathway. Indeed, the non-phosphorylated, active form of β-catenin (non-p-β-catenin) is reduced in Scd2-silenced HSCs (Fig. 2C). SCDi treatment phenocopies this effect in a manner rescued by MUFA but not SA or PA (Fig. 2D). To perform similar functional analysis in the context of liver cancer, we isolated a TIC clone stably transduced with lentiviral shRNA targeting Scd2 (Scd2-KD) vs. a control clone transduced with a vector targeting GFP (GFP-KD). Scd2, not Scd1, is selectively knocked down (~80%) in Scd2-KD (Fig. S2B). Scd2 KD inhibits TIC self-renewal, as shown by a reduced volume of spheroids formed by Scd2-KD (Fig. 2E left and middle). Similarly, SCDi suppresses TIC self-renewal (Fig. S2C–E) and “stemness” genes such as Nanog and Sox2 and this repression is rescued by OA but not SA (Fig. 2E right). As shown for HSCs, β-catenin is reduced by SCDi and rescued by OA but not SA in mouse TICs (Fig. 2F). Collectively, these results demonstrate that MUFA, generated by Wnt-dependent SCD, provide a positive feedback loop to stabilize β-catenin in activation of both HSCs and TICs.
Fig. 2. SCD renders a positive-feedback loop for Wnt/β-catenin.
A. Expression of Cre via adenovirus (Ad.Cre, MOI=100) but not LacZ (Ad.LacZ) in HSCs from Scd2ff mice, reverts the cells to quiescent state (top, Scale bar=400um), suppresses Col1a1 and Acta2, and upregulates Pparγ (bottom). Ccnd1 is suppressed by Scd2 ablation (n=4). *p<0.05 vs. Ad.LacZ. B. Acta2 and Pparγ mRNA by qPCR, confirm SCDi effects and rescue with OA and POA but not with SA and PA. *p<0.05. C. Reduced stabilized β-catenin (non-phosphorylated at S33/S37/T41) in Scd2-silenced HSCs (top). A densitometric analysis of six experiments (bottom). *p<0.05. D. SCDi suppression of stabilized β-catenin in HSCs and its rescue with OA and POA but not with SA and PA. A densitometric analysis of three experiments. *p<0.05. E. Reduced spheroid size by Scd2-KD vs. GFP-KD TICs (left and middle) (n=3). **p<0.01. Scale bar=400um. SCDi (400 nM) represses Nanog and Sox2, and this repression is rescued by OA but not SA (right) (n=3). **p<0.01. F. SCDi suppresses β-catenin protein in TICs, the effect rescued by OA.
SCDi acts upstream of integrin-linked kinase
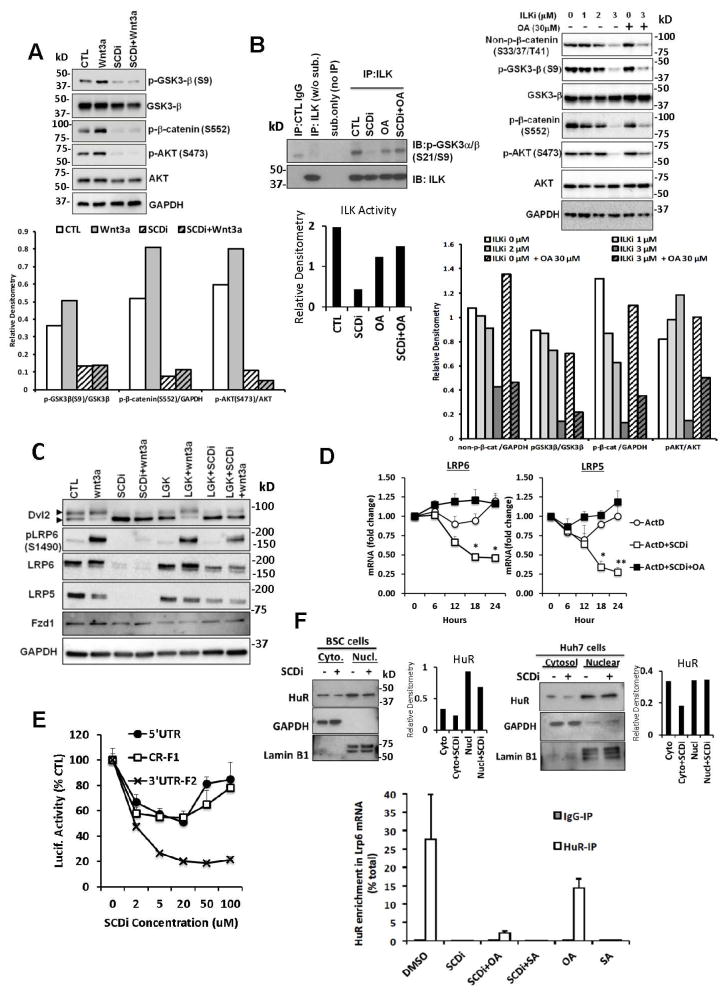
We next searched for a mechanism underlying the SCD-mediated positive loop for β-catenin. Integrin-linked kinase (ILK) is essential for HSC activation18 and implicated in tumorigenesis19. ILK participates in Wnt pathway20–22 by inhibiting GSK-3β23, 24, stabilizing β-catenin25, and modulating activation of AKT24. And these parameters are uniformly suppressed by SCDi (Fig. 2C and D, Fig. 3A), suggesting ILK may be a target of SCDi. Indeed, SCDi abrogates ILK activity in BSC as assessed by phosphorylation of GSK3α/β-GST fusion protein at S21 and S9 and this effect is rescued by OA (Fig. 3B left). An ILK inhibitor (ILKi) also reduces non-p β-catenin, p(S9)-GSK3β, and p(S552)-β-catenin, and p(S473)-AKT (Fig. 3B right). However, these effects are not rescued by OA, nor is cellular inactivation by ILKi (Fig. S4C–D), indicating that SCD acts upstream of ILK to promote Wnt pathway. In support, overexpression of ILK in SCDi-treated BSC does not fully rescue the effect (Fig. S4A–B). Similarly in TICs, ILKi suppresses spheroid formation and Sox2 and Nanog expression but these effects are not rescued by OA (Fig. S4E–G).
Fig. 3. SCD promotes LRP5/6 mRNA stability via ARE binding protein HuR.
A. SCDi reduces p(S9)-GSK3β, p(S552)-β-catenin, and p(S473)-AKT in BSCs without or with Wnt3a treatment. B. SCDi inhibits ILK activity in BSCs and this inhibition is rescued by OA (30 μM, left). ILK inhibitor (ILKi) reduces non-p β-catenin, p(S9)-GSK3β, and p(S552)-β-catenin, and p(S473)-AKT in BSC but these effects are not rescued by OA (right). C. Dvl2 phosphorylation induced by exogenous Wnt3a is inhibited by SCDi in BSC. Wnt3a-induced p-LRP6 is abrogated by SCDi due to a loss of LPR6 protein. D. With actinomycin D (ActD, 1 nM), LRP5/6 mRNA is degraded faster in BSC with SCDi vs. control. This effect of SCDi is rescued by OA. *p<0.05, **p<0.01. E. Luc-3′UTR-F2 is dose-dependently suppressed by SCDi to 20% of control in Huh7 cells. F. SCDi reduces HuR protein in cytosol of BSC (left top) and Huh7 cells (right top). SCDi or SA but not OA abrogates HuR binding to Lrp6 3′UTR ARE as determined by RIP analysis (bottom). Error bars represent SEM. *p<0.05
SCD promotes LRP5/6 mRNA stability and expression
We examined next phosphorylation of Dvl2, the most immediate downstream event of Wnt-FZD-LRP5/6 interactions, by assessing a mobility shift of Dvl2. In BSCs, immunoblot detects Dvl2 and a shifted band of p-Dvl2, suggesting activated Wnt pathway as expected of these cells (1st lane of Fig. 3C). SCDi reduces p-Dvl2 and increases unphosphorylated Dvl2 (3rd lane). The inhibitor of Porcupine, the acyltransferase responsible for lipid modification of Wnts, achieves a similar effect of reducing p-Dvl2 (LGK974, 5th lane), supporting the requirement of SCD in lipidation of endogenous Wnt ligands. Addition of exogenous Wnt3a increases p-Dvl2 and reduces non-p-Dvl2 (2nd lane) and this shift is still inhibited by SCDi (4th lane), supporting that SCDi’s effect is not just lipidation of endogenous Wnt ligands. We then analyzed p(S1490)-LRP6 as one of the multiple phosphorylation events for this receptor in the formation of the Wnt signalosome. Wnt3a-stimulated phosphorylation of LRP6 is blocked by SCDi (3rd lane), and this is due to loss of LRP6 protein (Fig. 3C). LRP5 protein is similarly lost by SCDi, suggesting that SCD is essential for expression of LRP5/6. Note LGK974 reduces p-LRP6 but not LRP protein. Similar LRP5/6 repression is also evident in SCDi-treated Huh7 cells (Fig. S5A) and TICs (data not shown). Both mRNAs and proteins declined after 12 h of SCDi and reached 10–40% of the control by 24 h in BSC (Fig. S5B and S5C left). Protein stability is not reduced by SCDi (Fig. S5C right). Lrp5/6 mRNA levels are increased in culture-activated HSCs and HSCs from the BDL model (Fig. S5D), supporting the in vivo relevance of LRP5/6 upregulation in aHSCs. Finally, Lrp5/6 mRNA stability analysis with actinomycin D (ActD) reveals both Lrp5/6 transcripts are destabilized by SCDi as compared to the cells treated with ActD alone, and these effects are rescued by OA treatment (Fig. 3D).
SCD supports LRP5/6 expression via the ARE binding protein HuR
In-silico algorithms (http://mirdb.org/miRDB/) for miRNA binding to Lrp6 mRNA 3′ UTR predict potential regulation by several microRNAs. But none of these miRNAs are regulated by SCDi as assessed by TaqMan-qPCR (Fig. S5E). The AU-rich element in 3′UTR mediates binding of proteins to regulate mRNA stability and translation. To assess this regulation in SCDi-treated cells, we used the Lrp6-luciferase reporter constructs with a 145nt 5′UTR (5′UTR), a coding region fragment (CR-F1), or a 3′UTR fragment containing an AU-rich element (ARE) (3′UTR-F2) of Lrp626. SCDi reduces the activity of both Luc-5′UTR and Luc-CR-F1 by 20–40% (Fig. 3E). However, Luc-3′UTR-F2 is dose-dependently and most conspicuously suppressed by SCDi by 80%, suggesting the primary role of the ARE in the 3′UTR-F2 in mediating SCDi’s suppressive effect. Among proteins known to bind the ARE, we focused our study on HuR which is overexpressed in HSC activation27 and tumors28. Further, HuR is recently shown to stabilize Lrp6 mRNA and stimulate its translation26. Indeed, SCDi reduces HuR protein in cytosol in BSC (Fig. 3F left top) and Huh7 cells (Fig. 3F right top). Further SCDi abrogates HuR binding to Lrp6 3′UTR ARE in Huh7 as shown by RIP analysis (Fig. 3F bottom), and this effect is slightly rescued with OA but not with SA. Importantly, the treatment with SA but not OA reproduces the effect of SCDi. These results support the notion that SCD inhibition or increased SA/OA resulting from SCD inhibition destabilizes Lrp6 mRNA due to reduced HuR binding to the ARE. We examined whether SCD affects the expression of Wnt3a and Wnt10b shown to be expressed by HSCs. Our qPCR analysis shows induction of these ligands by Scd2 ablation in HSCs, SCDi or SA treatment in TICs (Fig. S6).
TNPO1 and other importins as OA-interacting proteins
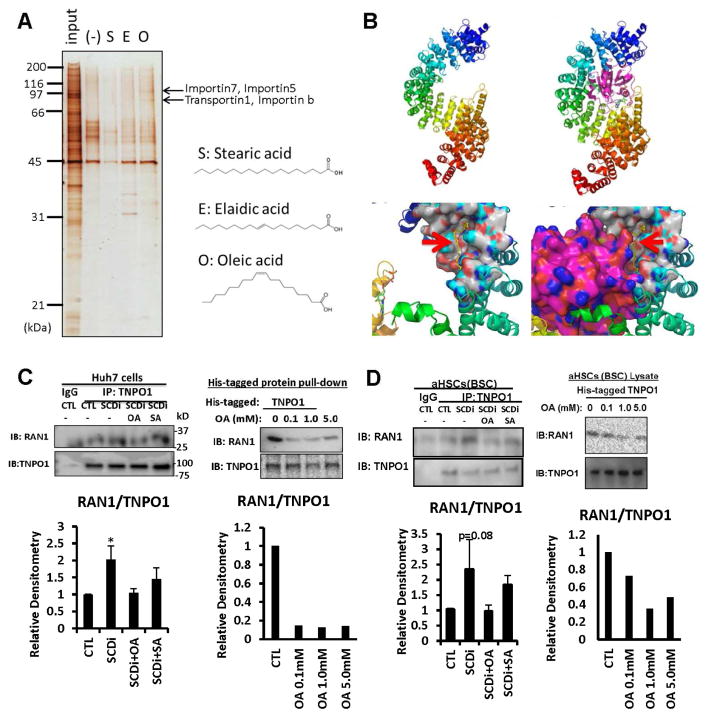
To gain molecular insights into how OA mediates the observed effects on HuR, we globally searched for cellular proteins which interacts with OA but not SA or elaidic acid (EA: the trans isomer of OA) by incubating TIC lysates with affinity nano-beads 29 conjugated with OA, SA, or EA. This analysis revealed proteins with apparent molecular weight of 95 and 120 kDa specifically bound to OA-conjugated beads (Fig. 4A). These bands were identified as importin family proteins TNPO1, importin 5, 7, or β by analysis by ESI-MS.
Fig. 4. OA interferes TNPO1-Ran1 GTPase binding.
A. Fatty acid interacting proteins were searched by incubating TIC lysates with nano-beads conjugated with O (oleic acid), S (stearic acid), or E (elaidic acid), differential gel display, and mass spectrometry analysis, revealing TNPO1, importin 5, 7, or β as putative OA-interacting proteins. B. Top, Three-dimensional structure of TNPO1 shown in ribbon representation. The molecule is colored in rainbow color with N-terminus in blue and the C-terminus in red. TNPO1 is shown in complex with Ran1 (in purple, right). Bottom, the Ran1 binding region at the N-terminus is shown in surface model. OA (red arrows) is shown in yellow stick model, and colors on binding surface are according to atoms (N-blue; O-red, C-white, H-cyan). C. Left, Co-IP shows SCDi increases TNPO1-Ran1 binding which is prevented by OA but not by SA in Huh7 cells. Right, OA suppresses the interaction of Ran1 with His-tagged TNPO1 in Huh7 cell lysate. D. Left, SCDi increases TNPO1-Ran1 binding which is prevented by OA but not by SA in BSC. Right, OA suppresses Ran1 interaction with His-tagged TNPO1 in BSC lysate.
Computational analysis predicts OA binds to TNPO1 and interferes with TNPO1-Ran1 GTPase interaction
We next performed computational analysis to understand the structural basis for OA binding to TNPO1 and importins. Importins, such as TNPO1, bind to the nuclear localization signal (NLS) of their respective cargoes such as HuR in the cytoplasm30. Given that HuR nuclear import is mediated by TNPO1 in a Ran-GTP dependent manner30, and this regulation is critical for controlling the HuR cytoplasmic pool and its actions on target mRNAs, we hypothesized that Ran1-GTP binding to TNPO1 may be interfered by OA, leading to impaired HuR nuclear import and increased cytosolic level of this protein. TNPO1 consists of a super helix of 20 HEAT repeat30. The X-ray crystallographic structure of TNPO1 with and without Ran1 has been determined (pdb code: 2bku31); Ran1 binds between N and C-termini of TNPO1, with maximum contacts at the N-terminus (Fig. 4B top). Sitemap (Schrodinger, Inc., San Diego) was used to identify OA binding at two sites at the TNPO1 N-terminus (Site 1: Site score: 0.970; DScore: 0.997; Volume: 267.8Å3; Site 2: Site score: 0.956; DScore: 1.047; Volume: 104.6Å3). The small molecule OA, was prepared for docking using ligprep and docked to the two putative sites using Glide in extra-precision (XP) mode. The final glide score was −4.47 and −2.60 Kcal/mol for sites 1 and 2, respectively. Site 1 was located at the interface of Ran1 and TNPO1 (red arrows, Fig. 4B bottom). Site 2 was also located at the N-terminus, but on the opposite side of Ran1-binding (not shown). The affinity at the binding Site 1 was estimated by short molecular simulation studies using Desmond, built in Schrodinger and ligand docking with Autodock32. Binding of OA to Site1 ranged from 77μM to 1mM.
OA inhibits TNPO1 and Ran1 binding
Based on the computational docking analysis results, we performed co-IP analysis for TNPO1 and Ran1 in Huh7 cells treated with SCDi with or without OA or SA. Indeed, SCDi treatment increases TNPO1-Ran1 binding which is prevented by OA but not SA at 30 μM (Fig. 4C left). Suppression of Ran1-TNPO1 complex formation by OA is achieved at a concentration of 0.1–1mM, approximating the OA affinity concentration estimated by computational modeling (Fig. 4C right). SCDi also increases TNPO1-Ran1 binding in BSC which is prevented by OA but not by SA (Fig. 4D left), and suppression of Ran1-TNPO1 complex formation by OA is achieved at ~1mM using a cell-free BSC lysate system (Fig. 4D right).
Pharmacologic SCD inhibition or genetic Scd2 ablation ameliorates liver fibrosis
We next tested whether SCDi inhibits liver fibrosis induced by CCl4 as schematically shown in Fig. 5A. After two weeks of CCl4 treatment, SCDi (A939572: 30 mg/kg, daily gavage, 5 days per week) or vehicle was administered for 2 weeks. SCDi reduced liver fibrosis by 40–50% as assessed by Sirius red morphometry and liver hydroxyproline content (Fig. 5A, B top), and decreased hepatic levels of α-smooth muscle actin (ACTA2) (Fig. 5B bottom left). SCDi also repressed Col1a1, Tgfb1, Timp1 but induced Mmp9 and Mmp13 (Fig. 5B bottom right), supporting its anti-fibrotic effect. To genetically target Scd2 ablation in aHSCs in this model, we crossed Scd2 floxed mice (Scd2ff) with mice carrying the Cre gene under the promoter of Col1a1 (CC). Using this Cre mouse, aHSCs in CCl4 liver fibrosis have previously been effectively targeted33. Both Scd2ff and Scd2ff;CC male mice were subjected to the same 4-week CCl4 regimen. The Sirius red staining morphometry and hydroxyproline content reveal that liver fibrosis was reduced by ~40% in Scd2ff;CC mice vs. Scd2ff mice (Fig. 5C and 5D top), and expression of ACTA2 was also abrogated in the former group (Fig. 5D bottom left). Conditional Scd2 ablation also repressed Col1a1, Tgfb1, Timp1 and induced Mmp9 and Mmp13 (Fig. 5D bottom right). Changes in major immune cell subsets of T-, B-, and myeloid cells were assessed by qPCR for CD3, CD20, CD68, respectively, and showed no differences between the groups (Fig. S7C). Similar anti-fibrotic effects of conditional Scd2 ablation were also observed in BDL-induced fibrosis (Fig. S7A–B), suggesting that targeting Scd2 in aHSCs is therapeutic for both hepatotoxic and cholestatic liver fibrosis in mice.
Fig. 5. Importance of Scd2 in liver fibrosis.
A. SCDi treatment reduces CCl4 liver fibrosis assessed by Sirius red and reticulin staining vs. vehicle-treated mice (n=6 each). Scale bar=1mm. B. SCDi reduces Sirius red morphometry and liver hydroxyproline content (top), hepatic ACTA2 (bottom left), and Col1a1, Tgfb1, Timp1 mRNA while inducing Mmp9 and Mmp13 (bottom right). C. CCl4-induced liver fibrosis is attenuated in mice with conditional Scd2 knockout (Scd2ff;CC) vs. Scd2ff control mice. D. Conditional Scd2 deficiency inhibits Sirius red staining morphometry and hydroxyproline content (top), hepatic ACTA2 (bottom left), and Col1a1, Tgfb1, Timp1 mRNA while inducing Mmp9 and Mmp13 (bottom right). E and F. Scd2 KD inhibits TIC-induced tumorigenesis in vivo. TICs with Scd2 KD or GFP-KD (control) were orthotopically transplanted in ethanol-fed B6 mice. Red horizontal bars denote group means. **p<0.01. Error bars represent SEM.
Knockdown of Scd2 ameliorates TIC-induced tumorigenesis in mice
We tested the in vivo effect of targeted Scd2 KD on TIC-initiated liver tumorigenesis. Immune-competent B6 mice were fed an ethanol-containing diet for 5 weeks, and then mice were orthotopically transplanted into the left lobe with 2 × 105 Scd2-KD or GFP-KD TICs and continued on ethanol diet for additional 3 weeks before euthanized (Fig. 5E). All mice with GFP-KD TIC transplantation developed liver tumors, but only 2 out of 8 mice transplanted with Scd2-KD TICs had visible tumors (Fig. 5F left). Further, the Scd2-KD-derived tumors were significantly smaller than GFP-KD-derived tumors (Fig. 5F right), demonstrating the importance of Scd2 in TIC-initiated liver tumorigenesis.
SCD2 is responsible for HSC promotion of TIC- or DEN-induced liver tumor development
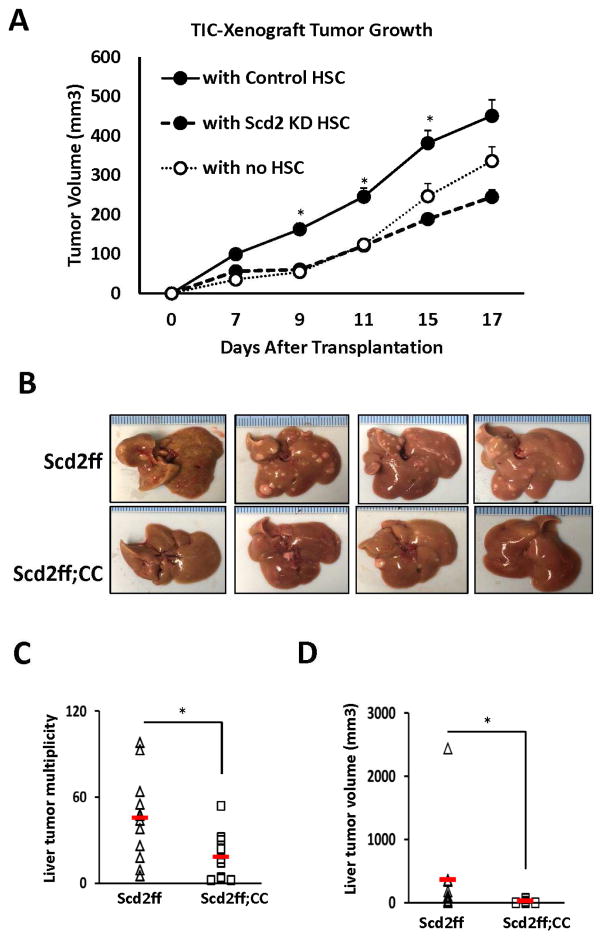
Having shown the functional importance of SCD2 in both HSCs and TICs, we next tested whether SCD2 in HSCs supports TIC-initiated tumorigenesis by co-transplantation of TICs in nude mice with HSCs isolated from Scd2ff mice and infected with Ad.Cre to silence Scd2 or Ad. LacZ to serve as a control. Compared to tumor growth initiated by TICs alone, co-transplantation of the control HSCs, significantly promotes tumor growth. However, HSCs with Scd2 KD fails to render this promotion (Fig. 6A), indicating the pivotal role of SCD2 in HSCs in promoting TIC-initiated tumor growth. Finally, we have extended testing this notion to the whole animal model with DEN-induced liver carcinogenesis. For this, we used Scd2ff vs. Scd2ff;CC mice as studied for liver fibrosis but subjected to DEN injection followed by feeding for 4 months, the Western alcohol diet which we believe to render a more clinically relevant mode of promotion than providing phenobarbital. Using this model, Scd2ff mice develop visible tumors with the multiplicity of 45±8.4 and total tumor volume of 360.9±193mm3. In contrast, the tumor multiplicity and volume in Scd2ff;CC are significantly reduced to 18±5.4 and 21.5±7.9mm3 (Fig. 6B–D), respectively, validating in vivo the importance of HSC SCD2 in tumor promotion.
Fig. 6. SCD2 confers HSC-mediated promotion of TIC-dependent tumorigenesis and DEN-induced liver tumor development in mice.
A. TIC-initiated tumor growth in nude mice is promoted when co-xenografted (1:1) with control HSCs (n=6) but not with Scd2 KD HSCs (n=6) vs. TICs transplanted with no HSCs (n=6). *p<0.05 vs. TICs with Scd2 KD HSCs. B. Representative photos of DEN-induced and Western alcohol diet-promoted liver tumor in Scd2ff mice which are reduced in conditional Scd2 knockout mice (Scd2ff;CC). C and D. SCD2 deficiency in aHSCs (Scd2ff;CC, n=10 vs. Scd2ff, n=12) significantly reduces multiplicity and tumor volume. Red horizontal bars and error bars denote group means and SEM. *p<0.05.
Discussion
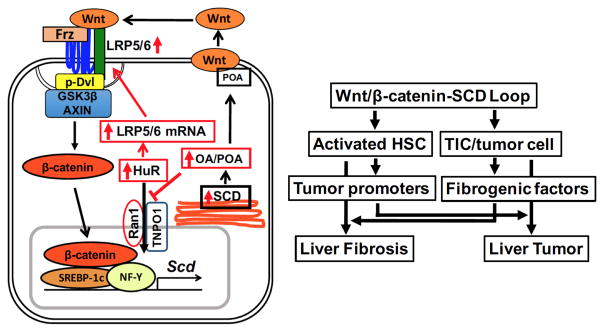
Several underlying mechanisms for the positive feedback loop for canonical Wnt pathway have been identified as means of amplifying growth stimulatory signals in malignant or stem cells. These include auto-induction of Wnt pathway components including Wnt ligand itself34; Wnt/β-catenin-mediated induction of phospholipase D which in turn enhances β-catenin/TCF4 complex formation35; β-catenin/TCF-mediated upregulation of miR-29a which targets negative regulators such as DKK1 and secreted frizzled related protein 236; and Wnt-induced c-Myc upregulating polycomb group protein BMI1 which transcriptionally auto-activates itself and represses DKK137. Our study now adds a newly discovered positive feed-forward loop involving Wnt-dependent expression of SCD which stabilizes Lrp5/6 mRNA via HuR (Fig. 7, left). This novel mechanism is essential for signal transduction of Wnt ligands originating from other or own cells because without it de novo expression of LRP5/6 and activation of canonical Wnt pathway will be lost. It is also elegantly coupled to the previously described SCD-Wnt release pathway to maximize the positive loop by promoting both release and signal transduction of Wnt. Central to SCD-dependent regulation of Lrp5/6 mRNA stability is HuR whose cytoplasmic level appears to be maintained by MUFA’s ability to block TNPO1-Ran1 binding and impair HuR nuclear import by TNPO1. This finding has far reaching implications beyond the regulation of LRP5/6 expression, as HuR controls transcripts of many fibrogenic and tumor-promoter genes28. Thus, the SCD-mediated increase in the cytoplasmic HuR pool, may upregulate these genes posttranscriptionally to promote liver fibrosis and cancer while amplifying canonical Wnt pathway via LRP5/6 expression. Indeed, many of these HuR-regulated genes are also Wnt-target genes, and this network of regulation may serve to maximize the stability and translation of Wnt-target gene transcripts.
Fig. 7. Schematic diagrams of a novel Wnt/β-catenin-SCD-LRP positive loop and cellular crosstak.
Left, Wnt/β-catenin-dependent SCD expression leads to HuR-mediated LRP5/6 mRNA stabilization via MUFA (OA, POA) inhibition of HuR nuclear transport by TNPO1 and Ran1. Right, the positive loop individually activates HSCs and TICs and also promotes crosstalk between the two cell types, underlying the link between liver fibrosis and tumor.
We have shown that the Wnt-SCD-LRP loop individually activates HSCs and TICs/HCC cells and induces liver fibrosis and cancer. More importantly, it is also shown to render a positive crosstalk between the two cell types (Fig. 7, right). As such conditional Scd2 knockout in aHSCs abrogates TIC-dependent tumorigenesis and DEN-induced liver tumor promotion, suggesting the positive loop in aHSCs critically dictates expression of tumor promoters and microenvironment conducive to tumor progression. Conversely, TICs and tumor cells activated by the amplified Wnt pathway, likely release fibrogenic mediators to promote liver fibrosis as stromal reactions to cancer are well recognized. This mutual crosstalk facilitated by cross-amplification of the Wnt/β-catenin pathway, may serve as a mechanism underlying the well-known clinical link between cirrhosis and HCC. Aberrant activation of the Wnt pathway is common in HCC, and SCD upregulation is not limited to HCC15, 38 but also evident in tumorigenesis of multiple types14 via mechanisms possibly involving β-catenin signaling39. Moreover, the demonstrated Wnt-SCD-LRP loop may explain the ability of SCD to induce stemness in human lung TICs13 and pluripotent stem cells40. To this end, this positive loop may be considered as a new therapeutic target for TICs or cancer cells with stemness which confer chemoresistance. As SCD is also implicated in obesity and fatty liver, SCD inhibition may particularly be an attractive therapeutic option for liver fibrosis and cancer arising from this etiological background. A most recent anti-HCV clinical trial for SCD1 inhibitor has shown no significant side effects41. PUFA supplementation may be considered to prevent PUFA deficiency associated with SCD inhibition.
Supplementary Material
Acknowledgments
Grant Support: NIH P50AA011999, U01AA018663, R24AA12885, and Medical Research Service of Department of Veterans Affairs (5I01BX001991) to HT, P50AA011999 and ACS IRG-58-007-54 Pilot Projects to KL, and NIH R01DK062388 and USDA Hatch W2005 to JMN.
Footnotes
Disclosures: The authors have declared that no conflict of interest exists.
Transcript Profiling: Gene Expression Omnibus (GEO) Accession #: GSE76330
Author contributions: KL, SMK, FC, and EH made equal contributions to execution of experiments and generation of a majority of data. YK, RH, LQ, RY, RW, SF, JX, RM, LM, JSL, and HT executed experiments and generated data. NF, JYW, and JMN provided material support. HT developed a study and experimental designs and interpreted data with KL and the collaborators. KL and HT drafted and finalized the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fattovich G, Stroffolini T, Zagni I, et al. Hepatocellular carcinoma in cirrhosis: incidence and risk factors. Gastroenterology. 2004;127:S35–50. doi: 10.1053/j.gastro.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 2.Coulouarn C, Clément B. Stellate cells and the development of liver cancer: therapeutic potential of targeting the stroma. J Hepatol. 2014;60:1306–9. doi: 10.1016/j.jhep.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 3.Monga SP. β-Catenin Signaling and Roles in Liver Homeostasis, Injury, and Tumorigenesis. Gastroenterology. 2015;148:1294–310. doi: 10.1053/j.gastro.2015.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell. 2012;149:1192–205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 5.Cheng JH, She H, Han YP, et al. Wnt antagonism inhibits hepatic stellate cell activation and liver fibrosis. Am J Physiol Gastrointest Liver Physiol. 2008;294:G39–49. doi: 10.1152/ajpgi.00263.2007. [DOI] [PubMed] [Google Scholar]
- 6.Friedman SL. Hepatic stellate cells: protean, multifunctional, and enigmatic cells of the liver. Physiol Rev. 2008;88:125–72. doi: 10.1152/physrev.00013.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He G, Dhar D, Nakagawa H, et al. Identification of liver cancer progenitors whose malignant progression depends on autocrine IL-6 signaling. Cell. 2013;155:384–96. doi: 10.1016/j.cell.2013.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen CL, Tsukamoto H, Liu JC, et al. Reciprocal regulation by TLR4 and TGF-β in tumor-initiating stem-like cells. J Clin Invest. 2013;123:2832–49. doi: 10.1172/JCI65859. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Miyazaki M, Dobrzyn A, Elias PM, et al. Stearoyl-CoA desaturase-2 gene expression is required for lipid synthesis during early skin and liver development. Proc Natl Acad Sci U S A. 2005;102:12501–6. doi: 10.1073/pnas.0503132102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J, Yu L, Schmidt RE, et al. Characterization of HSCD5, a novel human stearoyl-CoA desaturase unique to primates. Biochem Biophys Res Commun. 2005;332:735–42. doi: 10.1016/j.bbrc.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 11.Rios-Esteves J, Resh MD. Stearoyl CoA desaturase is required to produce active, lipid-modified Wnt proteins. Cell Rep. 2013;4:1072–81. doi: 10.1016/j.celrep.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Menghini R, Menini S, Amoruso R, et al. Tissue inhibitor of metalloproteinase 3 deficiency causes hepatic steatosis and adipose tissue inflammation in mice. Gastroenterology. 2009;136:663–72. e4. doi: 10.1053/j.gastro.2008.10.079. [DOI] [PubMed] [Google Scholar]
- 13.Noto A, Raffa S, De Vitis C, et al. Stearoyl-CoA desaturase-1 is a key factor for lung cancer-initiating cells. Cell Death Dis. 2013;4:e947. doi: 10.1038/cddis.2013.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roongta UV, Pabalan JG, Wang X, et al. Cancer cell dependence on unsaturated fatty acids implicates stearoyl-CoA desaturase as a target for cancer therapy. Mol Cancer Res. 2011;9:1551–61. doi: 10.1158/1541-7786.MCR-11-0126. [DOI] [PubMed] [Google Scholar]
- 15.Muir K, Hazim A, He Y, et al. Proteomic and lipidomic signatures of lipid metabolism in NASH-associated hepatocellular carcinoma. Cancer Res. 2013;73:4722–31. doi: 10.1158/0008-5472.CAN-12-3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tabor DE, Kim JB, Spiegelman BM, et al. Identification of conserved cis-elements and transcription factors required for sterol-regulated transcription of stearoyl-CoA desaturase 1 and 2. J Biol Chem. 1999;274:20603–10. doi: 10.1074/jbc.274.29.20603. [DOI] [PubMed] [Google Scholar]
- 17.Xin Z, Zhao H, Serby MD, et al. Discovery of piperidine-aryl urea-based stearoyl-CoA desaturase 1 inhibitors. Bioorg Med Chem Lett. 2008;18:4298–302. doi: 10.1016/j.bmcl.2008.06.088. [DOI] [PubMed] [Google Scholar]
- 18.Shafiei MS, Rockey DC. The function of integrin-linked kinase in normal and activated stellate cells: implications for fibrogenesis in wound healing. Lab Invest. 2012;92:305–16. doi: 10.1038/labinvest.2011.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Serrano I, McDonald PC, Lock F, et al. Inactivation of the Hippo tumour suppressor pathway by integrin-linked kinase. Nat Commun. 2013;4:2976. doi: 10.1038/ncomms3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oloumi A, Syam S, Dedhar S. Modulation of Wnt3a-mediated nuclear beta-catenin accumulation and activation by integrin-linked kinase in mammalian cells. Oncogene. 2006;25:7747–57. doi: 10.1038/sj.onc.1209752. [DOI] [PubMed] [Google Scholar]
- 21.Naves MA, Requiao-Moura LR, Soares MF, et al. Podocyte Wnt/β-catenin pathway is activated by integrin-linked kinase in clinical and experimental focal segmental glomerulosclerosis. J Nephrology. 2012;25:401–409. doi: 10.5301/jn.5000017. [DOI] [PubMed] [Google Scholar]
- 22.Wu X, Wang J, Jiang H, et al. Wnt3a activates β1-integrin and regulates migration and adhesion of vascular smooth muscle cells. Mol Med Rep. 2014;9:1159–1164. doi: 10.3892/mmr.2014.1937. [DOI] [PubMed] [Google Scholar]
- 23.D’Amico M, Hulit J, Amanatullah DF, et al. The integrin-linked kinase regulates the cyclin D1 gene through glycogen synthase kinase 3beta and cAMP-responsive element-binding protein-dependent pathways. J Biol Chem. 2000;275:32649–32657. doi: 10.1074/jbc.M000643200. [DOI] [PubMed] [Google Scholar]
- 24.Delcommenne M, Tan C, Gray V, et al. Phosphoinositide-3-OH kinase-dependent regulation of glycogen synthase kinase 3 and protein kinase B/AKT by the integrin-linked kinase. Proc Natl Acad Sci U S A. 1998;95:11211–6. doi: 10.1073/pnas.95.19.11211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Persad S, Troussard AA, McPhee TR, et al. Tumor suppressor PTEN inhibits nuclear accumulation of beta-catenin and T cell/lymphoid enhancer factor 1-mediated transcriptional activation. J Cell Biol. 2001;153:1161–74. doi: 10.1083/jcb.153.6.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu L, Christodoulou-Vafeiadou E, Rao JN, et al. RNA-binding protein HuR promotes growth of small intestinal mucosa by activating the Wnt signaling pathway. Mol Biol Cell. 2014;25:3308–18. doi: 10.1091/mbc.E14-03-0853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woodhoo A, Iruarrizaga-Lejarreta M, Beraza N, et al. Human antigen R contributes to hepatic stellate cell activation and liver fibrosis. Hepatology. 2012;56:1870–82. doi: 10.1002/hep.25828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J, Guo Y, Chu H, et al. Multiple functions of the RNA-binding protein HuR in cancer progression, treatment responses and prognosis. Int J Mol Sci. 2013;14:10015–41. doi: 10.3390/ijms140510015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kabe Y, Ohmori M, Shinouchi K, et al. Porphyrin accumulation in mitochondria is mediated by 2-oxoglutarate carrier. J Biol Chem. 2006;281:31729–35. doi: 10.1074/jbc.M604729200. [DOI] [PubMed] [Google Scholar]
- 30.Twyffels L, Gueydan C, Kruys V. Transportin-1 and Transportin-2: protein nuclear import and beyond. FEBS Lett. 2014;588:1857–68. doi: 10.1016/j.febslet.2014.04.023. [DOI] [PubMed] [Google Scholar]
- 31.Lee SJ, Matsuura Y, Liu SM, et al. Structural basis for nuclear import complex dissociation by RanGTP. Nature. 2005;435:693–6. doi: 10.1038/nature03578. [DOI] [PubMed] [Google Scholar]
- 32.Morris GM, Huey R, Lindstrom W, et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–91. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kisseleva T, Cong M, Paik Y, et al. Myofibroblasts revert to an inactive phenotype during regression of liver fibrosis. Proc Natl Acad Sci U S A. 2012;109:9448–53. doi: 10.1073/pnas.1201840109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deb A, Davis BH, Guo J, et al. SFRP2 regulates cardiomyogenic differentiation by inhibiting a positive transcriptional autofeedback loop of Wnt3a. Stem Cells. 2008;26:35–44. doi: 10.1634/stemcells.2007-0475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kang DW, Lee SH, Yoon JW, et al. Phospholipase D1 drives a positive feedback loop to reinforce the Wnt/beta-catenin/TCF signaling axis. Cancer Res. 2010;70:4233–42. doi: 10.1158/0008-5472.CAN-09-3470. [DOI] [PubMed] [Google Scholar]
- 36.Kapinas K, Kessler C, Ricks T, et al. miR-29 modulates Wnt signaling in human osteoblasts through a positive feedback loop. J Biol Chem. 2010;285:25221–31. doi: 10.1074/jbc.M110.116137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cho JH, Dimri M, Dimri GP. A positive feedback loop regulates the expression of polycomb group protein BMI1 via WNT signaling pathway. J Biol Chem. 2013;288:3406–18. doi: 10.1074/jbc.M112.422931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bansal S, Berk M, Alkhouri N, et al. Stearoyl-CoA desaturase plays an important role in proliferation and chemoresistance in human hepatocellular carcinoma. J Surg Res. 2014;186:29–38. doi: 10.1016/j.jss.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mauvoisin D, Charfi C, Lounis AM, et al. Decreasing stearoyl-CoA desaturase-1 expression inhibits β-catenin signaling in breast cancer cells. Cancer Sci. 2013;104:36–42. doi: 10.1111/cas.12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ben-David U, Gan QF, Golan-Lev T, et al. Selective elimination of human pluripotent stem cells by an oleate synthesis inhibitor discovered in a high-throughput screen. Cell Stem Cell. 2013;12:167–79. doi: 10.1016/j.stem.2012.11.015. [DOI] [PubMed] [Google Scholar]
- 41.Nio Y, Hasegawa H, Okumura H, et al. Liver-specific nono-unsaturated fatty acid synthase-1 inhibitor for anti-hepatitis C treatment. Antiviral Res. 2016;132:262–7. doi: 10.1016/j.antiviral.2016.07.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.