Abstract
Background
Increased heart rate and a prolonged QT interval are important risk factors for cardiovascular morbidity and mortality, and can be influenced by the use of various medications, including tri/tetracyclic antidepressants (TCAs). We aim to identify genetic loci that modify the association between TCA use and RR and QT intervals.
Methods and Results
We conducted race/ethnic-specific genome-wide interaction analyses (with HapMap Phase II imputed reference panel imputation) of TCAs and resting RR and QT intervals in cohorts of European (n=45,706; n=1,417 TCA users), African (n=10,235; n=296 TCA users) and Hispanic/Latino (n=13,808; n=147 TCA users) ancestry, adjusted for clinical covariates. Among the populations of European ancestry, two genome-wide significant loci were identified for RR interval: rs6737205 in BRE (β = 56.3, Pinteraction = 3.9e−9) and rs9830388 in UBE2E2 (β = 25.2, Pinteraction = 1.7e−8). In Hispanic/Latino cohorts, rs2291477 in TGFBR3 significantly modified the association between TCAs and QT intervals (β = 9.3, Pinteraction = 2.55e−8). In the meta-analyses of the other ethnicities, these loci either were excluded from the meta-analyses (as part of quality control), or their effects did not reach the level of nominal statistical significance (Pinteraction > 0.05). No new variants were identified in these ethnicities. No additional loci were identified after inverse-variance-weighted meta-analysis of the three ancestries.
Conclusion
Among Europeans, TCA interactions with variants in BRE and UBE2E2, were identified in relation to RR intervals. Among Hispanic/Latinos, variants in TGFBR3 modified the relation between TCAs and QT intervals. Future studies are required to confirm our results.
Keywords: drug-gene interaction, Genome Wide Association Study, tri/tetracyclic antidepressants, RR interval, QT interval electrocardiography
Introduction
An increased resting heart rate and a prolonged QT interval are independent risk factors for cardiovascular morbidity and mortality[1–4]. To date, multiple medications have shown clinically significant effects on heart rate, the heart-rate corrected QT interval (QTc), or both[5–7]. For example, the tri/tetracyclic antidepressants (TCAs) have tachycardic and QT-prolonging effects originating from their anticholinergic properties (through antagonizing acetylcholine neurotransmitter signaling[5 7–11]). Despite drug safety warnings, particularly in at risk populations (e.g., the elderly), TCAs are still commonly prescribed in Western societies[12–14] for the treatment of depression, anxiety, insomnia, and neuropathic pain[12].
Both resting heart rate and QT interval duration are heritable, with hereditability estimates ranging from 55–77% for resting heart rate and 35–51% for QT intervals[15–16]. To date, multiple single nucleotide polymorphisms (SNPs) have been identified in genome-wide association studies of resting heart rate[17–19] and QT interval[20–21] among different ethnicities. However, the identified loci (21 for resting heart rate and 35 for QT interval duration[17–20]) explain only 0.8–0.9% and 8–10% of the total variance in these traits[17–20]. Inability to fully explain variance in heart rate and QT intervals may be related to the presence of gene-gene and gene-environment interactions[22]. To examine this possibility, a genome-wide, TCA-SNP interaction meta-analysis of QT was previously conducted in individuals of European ancestry within the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium[23]. However, no significant TCA-SNP interactions were identified, possibly due to the small number of TCA users in the study or its cross-sectional design[23]. Since then, new statistical methods have been developed to incorporate data from multiple visits[24], and additional cohorts of different ancestral origins have been included to increase statistical power.
The present effort collaboratively leverages these methods in a study designed to identify TCA-SNP interactions capable of explaining variation in heart rate (or RR interval) and QT, while also providing insights into the biology of tachycardic and QT-prolonging medications.
Methods
Study populations
The present study used data from 21 different cohorts of three ancestral populations (European [14 cohorts], African [5 cohorts], and Hispanic/Latino [2 cohorts, noting that “Hispanic/Latino” captures a diverse population][25]) that were assembled and analyzed by the Pharmacogenomics Working Group in the CHARGE consortium[26]. All cohorts conducted the analyses within their own study on the basis of a predefined protocol. Cohorts with genetic data were eligible to participate when data on medication use and on the study outcomes were both collected during the same visit. Genotype data had to be imputed with either the HapMap Phase 2[27] or 1000 Genomes reference panel[28]. One of the studies (Anglo-Scandinavian Cardiac Outcomes Trial [ASCOT]) did not record electrocardiograms (ECGs), and therefore participated in analyses on only RR, and not on QT. All studies were approved by local ethics committees, and all participants gave written informed consent. Cohort-specific descriptions of the study design can be found in the Supplementary Materials.
Inclusion and exclusion criteria
All participants with data on medication use and a high quality ECG (when available), and who were successfully genotyped, were eligible for inclusion in the analyses. Participants with atrial fibrillation, a pacemaker, and/or second or third degree atrioventricular block were excluded from the analyses, as were participants with heart failure or a QRS duration ≥120 milliseconds (ms).
Drug exposure assessment
Most cohorts collected information on medication use by inventory (Supplementary Table 1). However, the Rotterdam Study (RS) defined medication use on the basis of pharmacy dispensing data (from 1991 onwards). For these individuals, exposure was defined as a prescription filled for a medication of interest within 30 days preceding the ECG recording. Cohorts were asked to define exposures to the following medications (or medication classes): TCAs (ATC code “N06AA”), beta-blocking agents (ATC code “C07”), verapamil (ATC code “C08DA01”), diltiazem (ATC code “C08DB01”), and medications known to definitely prolong QT intervals or that are generally accepted to increase the risk of torsade de pointes. Categorization of medications as “definite” for QT prolongation was based on classification from the Arizona Center for Education and Research on Therapeutics (UAZ CERT) as of March 2008[29].
Assessment of QT and RR interval
In each cohort, research technicians recorded a standard 12-lead ECG or pulse rate (in the case of ASCOT) in the resting state for each participant (Supplementary Table 2). Almost all cohorts measured RR and QT intervals automatically, to decrease measurement error and inter-individual variation. Studies conducted all analyses longitudinally, allowing multiple visits per participant in the analyses when multiple ECGs were available (and when data on medication use were also collected).
Genotyping and imputation
Genome-wide SNP genotyping was performed within each cohort separately, using commercially available genotyping arrays from Affymetrix (Santa Clara, CA, USA) or Illumina (San Diego, CA, USA; Supplementary Table 3). Duplicates and samples with gender mismatches were excluded from all studies. First-degree relatives were excluded from all studies, except for the family-based Framingham Heart Study (FHS), Jackson Heart Study (JHS), and Hispanic Community Health Study/Study of Latinos (HCHS/SOL); HCHS/SOL investigators also used methods that accounted for admixture, population structure, and Hardy-Weinberg-departures, when estimating kinship coefficients[25]. Cohort-specific thresholds for genotyping call rates ranged from 95% to 99%. To increase homogeneity between cohorts with respect to the SNPs genotyped by the different platforms, as well as to increase coverage, summary results were based on SNPs from the HapMap Phase 2 (build 36) reference population[27], given the uniform availability of HapMap2-imputed SNPs and the computational burdens associated with performing analyses for reference panels with much larger numbers of SNPs.
Genome-wide TCA-SNP interaction analyses and meta-analysis
The statistical approaches used to estimate TCA-SNP interactions on RR or QT intervals depended on the study design (e.g., family-based) and the availability of ECG and medication data (e.g., cross-sectional or longitudinal). Cohorts with longitudinal ECG and medication data (e.g., Atherosclerosis Risk in Communities Study, Cardiovascular Health Study, RS, and Women’s Health Initiative [WHI]) used generalized estimating equations (GEE)[30] with independent working correlation structure. The family-based FHS and HCHS/SOL studies used linear mixed models that accounted for relatedness, sampling design (HCHS/SOL), and heterogeneity of outcome variance by drug use (HCHS/SOL). Cohorts with unrelated participants and with cross-sectional assessment of ECG and drug data used linear regression models with robust standard errors, as implemented in the ProbABEL software package[31] or in the “bosswithDF” package as implemented in the R statistical environment. Assuming that exposure to TCAs varies randomly across within-person visits for the analyses on RR, we had a power of 0.91 to observe interaction effects of at least 35 milliseconds for variants with a minor allele frequency (MAF) of at least 0.25 (Supplementary Table 4). For QT, we had a power 0.91 to observe interaction effects of at least 7 milliseconds for a MAF of at least 0.25.
TCA-SNP interaction analyses on both RR and QT were adjusted for age and sex. The analyses of RR were additionally adjusted for the use of beta-blocking agents, verapamil, and diltiazem. Similarly, analyses of QT were additionally adjusted for the use of medications that definitely prolong the QT interval and for the resting RR interval. Studies also adjusted for study-specific covariates, as necessary (e.g., study site and principal components).
The robust standard error estimates led to inflated type I errors when the number of participants exposed to the drug and the MAF were both small[24]. We addressed this potential for false-positive results by incorporating variability in the standard error estimates, through use of a t-reference distribution with degrees of freedom approximated via Satterthwaite’s methods[32–33]. However, at the lowest combinations of minor allele frequency and use of TCAs, the variability of the standard errors was poorly estimated, requiring exclusion of SNPs where 2*(number of exposed participants)*MAF*imputation quality < 10, as described previously [24]. An inverse-variance-weighted meta-analysis was then performed with genomic control using METAL, to combine the results from the different studies [34]. To avoid high type I errors from robust standard error estimates, standard errors were “corrected” using the t-distribution-based P-values. These “corrected” standard error estimates were used as inputs for the inverse-variance-weighted meta-analysis. Meta-analyses were performed for each ethnic group separately and for all ethnic groups together. To be considered in our study, SNPs had to be present, after quality control, in at least three cohorts (two cohorts in case of the Hispanic/Latino meta-analysis).
A two-sided P-value <5e−8 for TCA-SNP interactions was considered statistically significant in the genome-wide association analyses. Detailed summary results of the ethnic-specific analyses (including rs numbers, MAF values, effect sizes, and P-value) are available through dbGaP (https://www.ncbi.nlm.nih.gov/gap).
Evaluation of previously identified SNPs associated with resting heart rate and QT intervals
Within our European ancestry meta-analysis, we evaluated SNPs that were previously found to have main effects on heart rate or QT in the GWAS of European ancestry as done with HapMap Phase II imputed reference panel imputation [17–20]. From the European GWAS meta-analysis, we extracted all SNPs that had statistically significant effects on heart rate or QT interval (P-value <5e−8) and were present in at least three cohorts (after all quality control steps). We adjusted the P-value threshold for statistical significance using the Bonferroni correction: 2.38e−3 for RR intervals (21 independent loci) and 1.43e−3 for QT intervals (35 independent loci).
The 21 SNPs associated with RR intervals and the 35 SNPs associated with QT intervals from the meta-analysis in Europeans were further used to calculate a combined multi-locus effect estimate on the TCA-SNP interaction. The resulting multi-locus effect can be interpreted as a Mendelian randomization analysis to assess whether a high resting heart rate and prolonged QT interval are causal effect modifiers of TCA-induced increases in heart rate or QT intervals[35]. This data-driven inverse-variance weighted approach[36] has been implemented in the “gtx” statistical package in the R statistical software environment[37].
Results
Study characteristics
The number of TCA users for each ethnic group were: Europeans, 1,417 (out of 45,706); African Americans, 295 (out of 10,235); and Hispanics/Latinos, 174 (out of 13,808) (Table 1). Cohorts had a mean age ranging from 40.2 (FHS) to 75.3 (Prospective Study of Pravastatin in the Elderly at Risk), and the percentage of included women ranged from 17.8% (ASCOT) to 100% (WHI). Mean RR intervals ranged from 875 (RS1) to 981 (FHS) ms, and QT intervals ranged from 397 (RS1) to 416 (HCHS/SOL) ms.
Table 1.
Study characteristics
| Cohorts | Nexposed | Ntotal | RR in ms, mean (SD) | QT in ms, mean (SD) | Age in years, mean (SD) | Females, % | QTdef, % | Beta blockers, % | Verapamil, % | Diltiazem, % |
|---|---|---|---|---|---|---|---|---|---|---|
| AGES | 67 | 1,976 | 938 (153) | 406 (33.6) | 74.6 (4.7) | 64.2 | 3.0 | 13.7 | 1.7 | 3.5 |
| ARIC | 343 | 8,132 | 929 (138) | 399 (28.8) | 54.0 (5.7) | 53.0 | 3.4 | 10.2 | 1.8 | 2.0 |
| ASCOT | 167 | 3,755 | 877 (153) | -- | 63.6 (8.1) | 17.8 | -- | 45.1 | 0.3 | 1.5 |
| CHS | 165 | 2,893 | 953 (151) | 414 (32.2) | 72.1 (5.2) | 62.8 | 3.3 | 10.8 | 3.4 | 3.3 |
| FHS | 56 | 3,168 | 981 (159) | 414 (29.9) | 40.2 (8.8) | 39.5 | 0.3 | 3.5 | NA | NA |
| Health ABC | 43 | 1,442 | 952 (153) | 414 (32.3) | 73.7 (2.8) | 49.4 | 3.5 | 13.8 | 3.5 | 5.7 |
| MESA | 55 | 2,384 | 977 (149) | 412 (29.3) | 62.3 (10.1) | 52.1 | 0.9 | 7.1 | 0.3 | 0.1 |
| NEO | 84 | 5,366 | 940 (150) | 406 (29.3) | 55.9 (5.9) | 47.0 | 1.4 | 12.6 | 0.3 | 0.2 |
| PROSPER | 151 | 4,555 | 932 (163) | 414 (36.0) | 75.3 (3.3) | 46.6 | 2.6 | 20.2 | 2.1 | 5.4 |
| RS1 | 85 | 4,805 | 875 (148) | 397 (28.8) | 68.8 (8.6) | 60.2 | 2.6 | 15.3 | 0.7 | 1.6 |
| RS2 | 31 | 1,889 | 886 (140) | 406 (28.6) | 65.0 (7.6) | 56.6 | 2.3 | 15.5 | 0.3 | 1.2 |
| RS3 | 23 | 1,950 | 881 (131) | 401 (26.0) | 56.0 (5.7) | 54.1 | 1.1 | 10.5 | 0.1 | 0.4 |
| WHI GARNET | 60 | 1,391 | 928 (139) | 401 (30.3) | 65.0 (6.8) | 100.0 | 1.4 | 11.4 | 2.4 | 2.1 |
| WHI MOPMAP | 87 | 2,000 | 929 (135) | 402 (30.1) | 63.0 (6.6) | 100.0 | 0.9 | 13.3 | 1.9 | 1.9 |
| Summary | 1417 | 45,706 | 875–981 | 397–414 | 40.2–75.3 | 17.8–100 | 0.3–3.5 | 3.5–45.1 | 0.3–3.5 | 0.1–5.7 |
| African Americans | ||||||||||
| ARIC | 114 | 2,191 | 927 (151) | 400 (32.8) | 53.0 (5.8) | 62.4 | 2.8 | 9.8 | 3.5 | 1.7 |
| CHS | 24 | 707 | 921 (166) | 409 (35.3) | 72.6 (5.6) | 64.6 | 2.9 | 11.2 | 5.8 | 6.2 |
| Health ABC | 24 | 1,014 | 932 (143) | 411 (34.8) | 73.4 (2.9) | 57.6 | 3.1 | 10.1 | 5.9 | 6.3 |
| JHS | 35 | 2,096 | 948 (150) | 411 (30.1) | 49.5 (11.8) | 60.5 | 1.3 | 8.6 | 2.7 | 3.1 |
| WHI CT SHARe | 98 | 4,227 | 921 (149) | 401 (33.8) | 61.0 (6.8) | 100.0 | 1.3 | 11.7 | 4.8 | 4.0 |
| Summary | 295 | 10,235 | 921–948 | 400–411 | 49.5–73.4 | 57.6–100 | 1.3–3.1 | 8.6–11.7 | 2.7–5.9 | 1.7–6.3 |
| Hispanic/Latinos | ||||||||||
| WHI CT SHARe | 41 | 1,784 | 928 (133) | 402 (29.7) | 60.0 (6.4) | 100.0 | 1.0 | 8.2 | 2.5 | 1.8 |
| HCHS/SOL | 133 | 12,024 | 975 (143) | 416 (28.4) | 45.7 (13.8) | 59.5 | 0.7 | 6.4 | 0.4 | 0.3 |
| Summary | 174 | 13,808 | 928–975 | 402–416 | 45.7–60.0 | 59.5–100 | 0.7–1.0 | 6.4–8.2 | 0.4–2.5 | 0.3–1.8 |
| Total summary | 1886 | 69,749 | 875–981 | 397–416 | 40.2–75.3 | 17.8–100 | 0.3–3.5 | 3.5–45.1 | 0.3–5.9 | 0.1–6.3 |
Abbreviations: AGES, Age, Gene/Environment Susceptibility – Reykjavik Study; ARIC, Atherosclerosis Risk in Communities Study; ASCOT, Anglo-Scandinavian Cardiac Outcomes Trial; CHS, Cardiovascular Health Study; FHS, Framingham Heart Study; GARNET, Genome-wide Association Research Network into Effects of Treatment; HCHS/SOL, Hispanic Community Health Study/Study of Latinos; Health ABC, Health, Aging, and Body Composition Study; JHS, Jackson Heart Study; MESA, Multi-Ethnic Study of Atherosclerosis; MOPMAP, Modification of PM-Mediated Arrhythmogenesis in Populations; NA, not available. NEO, Netherlands Epidemiology of Obesity; Nexposed, number of participants using tricyclic antidepressants; Ntotal, total number of participants contributing in the analysis; PROSPER, Prospective Study of Pravastatin in the Elderly at Risk; RS, Rotterdam Study; SD, standard deviation; SHARe, Single nucleotide polymorphism (SNP) Health Association Resource project; WHI CT, Women’s Health Initiative Clinical Trials.
Genome-wide interaction analysis between tricyclic antidepressants and RR intervals
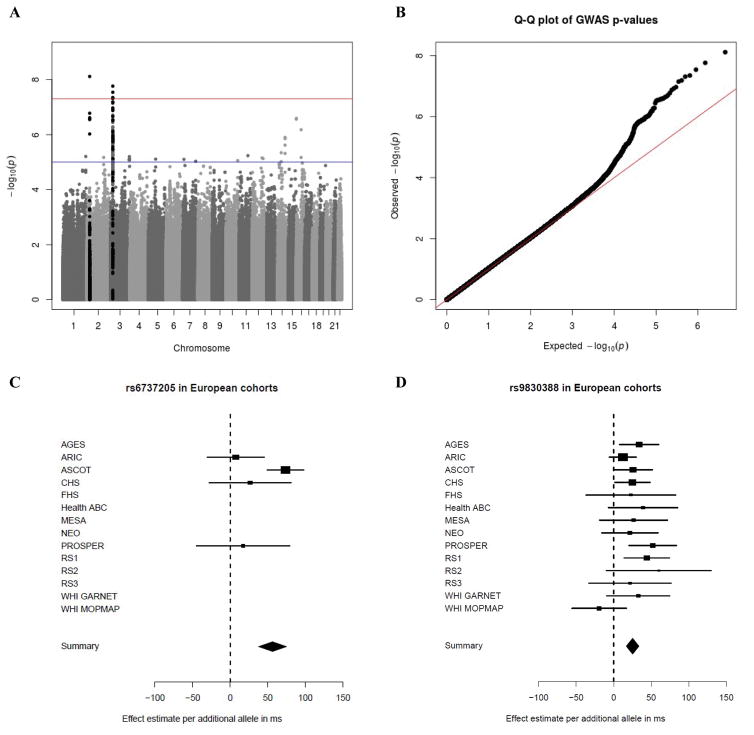
Within the cohorts of European ancestry, two independent loci reached statistical significance (Table 2; Figure 1A). A q-q plot of the meta-analysis in European cohorts is presented in Figure 1B. The top independent loci comprised the rs6737205 polymorphism on chromosome 2 (Figure 1C), and the rs9830388 polymorphism on chromosome 3 (Figure 1D). Variant rs6737205 (passed quality control in four European cohorts) mapped within the BRE gene and was associated with a 56.3 ms prolongation of the RR interval in TCA users, beyond the difference attributed to the allele among nonusers (Effect allele frequency [EAF]: 0.94; P-value = 7.66e−9). Variant rs9830388 (passed quality control in all European cohorts), mapped within the UBE2E2 gene, and was associated with a 25.2 ms longer RR interval in TCA users, beyond the difference attributed to the allele among nonusers (EAF: 0.51; P-value = 1.72e−8). Furthermore, rs11877129 (passed quality control in 11 studies) mapped within the ABCA3 gene and was suggestively (P-value < 1e−7) associated with a 36.7 ms longer RR interval in TCA users, beyond the difference attributed to the allele among nonusers (EAF: 0.09; P-value = 2.57e−7). There was no significant heterogeneity between studies in the observed estimates (P-values > 0.05). These three SNPs, however, did not reach nominal statistical significance in the meta-analyses of the African American cohorts and the Hispanic/Latino cohorts (Table 2, Supplementary Figure 1). Regional plots are presented in Supplementary Figure 2. A meta-analysis of the three ethnicities together did not yield any additional loci with statistically significant effects (Supplementary Figure 3).
Table 2.
Most significant independent loci in the genome-wide interaction analysis between tricyclic antidepressants and RR interval
| SNP | CHR | Mapped gene | Ethnicity | EAF | N studies | Effect allele | Betaint | SEint | Pint | Phet |
|---|---|---|---|---|---|---|---|---|---|---|
| rs6737205 | 2 | BRE | EA | 0.94 | 4 | A | 56.3 | 9.7 | 7.66e−9 | 0.16 |
| AA | Did not pass quality control | |||||||||
| HSP | Did not pass quality control | |||||||||
| rs9830388 | 3 | UBE2E2 | EA | 0.51 | 14 | A | 25.2 | 4.5 | 1.72e−8 | 0.44 |
| AA | 0.30 | 5 | A | 18.1 | 11.5 | 0.11 | 0.72 | |||
| HSP | 0.55 | 2 | A | −8.2 | 12.4 | 0.51 | 0.44 | |||
| rs11867129 | 16 | ABCA3 | EA | 0.09 | 11 | T | 35.7 | 6.9 | 2.57e−7 | 0.05 |
| AA | Did not pass quality control | |||||||||
| HSP | 0.41 | 2 | T | 4.5 | 13.5 | 0.74 | 0.12 | |||
Abbreviations: AA, African Americans; ABCA3, ATP binding cassette subfamily A member 3; BRE, brain and reproductive organ-expressed; CHR, chromosome; EA, Europeans ; EAF, effective allele frequency; HSP, Hispanic/Latinos; N, number of studies included in the meta-analysis after quality control. Phet, P-value for heterogeneity between studies; Pint, P-value of the interaction; SEint, standard error of the interaction; UBE2E2, ubiquitin conjugating enzyme E2E 2. Only independent genetic variants with a P-value for interaction below 5e−7 are displayed.
Figure 1. Genome-wide interaction analysis between tricyclic antidepressants and RR interval sin European cohorts.
Abbreviations: AGES, Age, Gene/Environment Susceptibility – Reykjavik Study; ARIC, Atherosclerosis Risk in Communities Study; ASCOT, Anglo-Scandinavian Cardiac Outcomes Trial; CHS, Cardiovascular Health Study; FHS, Framingham Heart Study; GARNET, Genome-wide Association Research Network into Effects of Treatment; HCHS/SOL, Hispanic Community Health Study/Study of Latinos; Health ABC, Health, Aging, and Body Composition Study; JHS, Jackson Heart Study; MESA, Multi-Ethnic Study of Atherosclerosis; MOPMAP, Modification of PM-Mediated Arrhythmogenesis in Populations; NEO, Netherlands Epidemiology of Obesity; Nexposed, number of independent participants using tricyclic antidepressants; PROSPER, Prospective Study of Pravastatin in the Elderly at Risk; RS, Rotterdam Study; SD, standard deviation; SHARe, WHI CT, Women’s Health Initiative Clinical Trials. A) –Log(p) plot of all SNPs present in at least 3 European cohorts and passing all quality control steps. In black are all SNPs within a 40 kb distance from the top result on chromosomes 2 and 3. B) Q-Q plot of the meta-analysis in European cohorts. λ = 1.031. C) Cohort-specific and meta-analysis estimate for rs6737205 on chromosome 2. Results are presented as the effect estimate of the interaction between rs6737205 and TCA-use status on RR intervals (with the 95% confidence interval). D) Cohort-specific and meta-analysis estimate for rs9830388 on chromosome 3. Results are presented as the effect estimate of the interaction between rs9830388 and TCA-use status on RR intervals (with the 95% confidence interval).
Genome-wide interaction analysis between tricyclic antidepressants and QT intervals
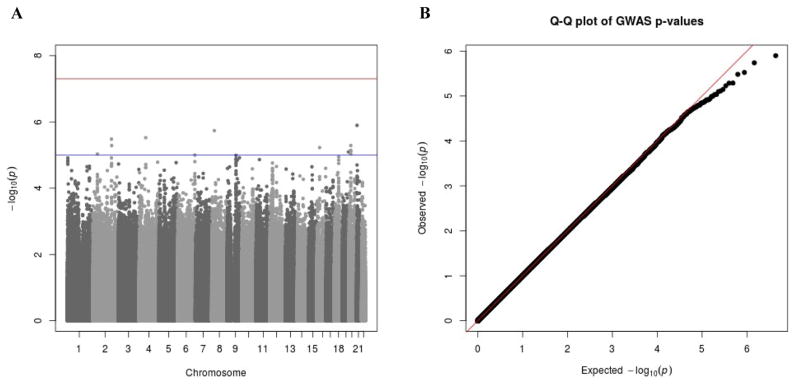
Results of the QT meta-analysis in the cohorts of European ancestry are presented in Figure 2. Within this analysis, no significant TCA-SNP interactions were observed. There was one locus in the Hispanic/Latino meta-analysis that reached genome-wide significance, represented by variant rs2291477 (which mapped to TGFBR3; β = 9.3; EAF: 0.88; P-value = 2.55e−8; Supplementary Figure 4/Supplementary Table 5). However, effects of this locus either did not reach nominal statistical significance or did not pass quality control in the European and African American meta-analyses (P-values > 0.05). Furthermore, no genome-wide significant TCA-SNP interactions were observed in the meta-analysis of the three ethnicities together (Supplementary Figure 5).
Figure 2. Genome-wide interaction analysis between tricyclic antidepressants and QT intervals in European cohorts.
A) –Log(p) plot of all SNPs present in at least 3 European cohorts and passing all quality control steps. B) Q-Q plot of the meta-analysis in European cohorts. λ = 1.022.
Previously identified loci for heart rate and QT intervals
The TCA-SNP interactions on RR and QT for SNPs that were previously associated with heart rate and QT are presented in Supplementary Tables 6 and 7. None of the loci previously associated with heart rate or QT showed TCA-SNP interactions on RR or QT respectively (after Bonferroni correction for the number of SNPs included). Furthermore, multi-locus effects of all loci for RR and QT were not statistically significant (Supplementary Figures 6 and 7; P-values = 0.35 and 0.74 for RR and QT, respectively).
Discussion
In a study population of 45,706 European individuals, among whom 1,417 individuals used a TCA at the moment of an ECG recording, we identified two independent loci (and one suggestive locus) that modified the association between TCAs and RR intervals. The significant loci were represented by variants rs6737205 (BRE) and rs9830388 (UBE2E2), and the suggestive locus by variant rs11867129 (ABCA3). As it is well-known that the anticholinergic activity of TCAs may increase heart rates and thus decrease the RR interval, these findings may have a biological basis. Although the variance explained by our findings was not calculated, it is likely modest in view of the low number of exposed individuals. The three loci (BRE, UBE2E2, and ABCA3) either were excluded from the meta-analyses of African and Hispanics/Latino ancestry participants (as part of quality control, e.g., DF <10) or their effects were not nominally significant in the meta-analyses (Pinteraction >0.05). There were no genome-wide significant loci that modified the association between TCA use and QT intervals in cohorts of European and African American ancestry, although one locus had an effect in Hispanic/Latino cohorts (TGFBR3, represented by rs2291477). None of the loci previously observed to be associated with RR and QT intervals modified TCA effects on their intervals. Furthermore, there was no multi-locus effect of variants previously associated with heart rate or QT intervals on TCA-SNP interactions with RR or QT intervals.
Genetic variation in BRE has not been related to any study outcome in prior GWAS reports. The association detected in our meta-analysis appeared to be driven by the estimate observed in ASCOT, although the P-value for heterogeneity among studies was not statistically significant. However, the directions of possible BRE effects were similar in the other three studies in which the BRE variant passed quality control. ASCOT was the only cohort that examined RR without ECGs, but this difference should have increased inter-individual variation and would decrease statistical power. Therefore, future replication studies of this variant are warranted.
Genetic variation in UBE2E2 was previously identified in GWAS reports on type 2 diabetes mellitus[38] and motion sickness[39]. To the best of our knowledge, no studies have been published on genetic variation in UBE2E2 in relation to cardiac conduction (e.g., heart rate or QT intervals) or pharmacological responses to medications. However, variant rs9830388 was associated with mRNA expression levels of UBE2E2 in blood, based on eQTL data[40–41]. Although significant in Europeans, the results on UBE2E2 were not generalized in African and Hispanic/Latino ancestry cohorts, perhaps due to limited sample sizes or the smaller effect size. Also, the linkage structure in this part of the genome may differ among ethnicities, as seen in the regional plots.
Genetic variation in ABCA3, which produced a suggestive TCA-SNP interaction for RR intervals, has been previously described in relation to the pharmacological response to imatinib in chronic myeloid leukemia [42]. However, we did not find evidence in the literature that ABCA3 is related to the electrophysiology of the heart.
Despite increasing our sample size and adding repeated ECG assessments to our previous effort on this research topic[23], no significant TCA-SNP interactions were identified for the QT interval in the meta-analysis of European cohorts (for which we had the largest number of TCA users). The single locus with a significant effect in the Hispanic/Latino meta-analysis (represented by rs2291477 in TGFBR3) did not have a nominally significant effect among Europeans and African Americans. The observation may suggest that TGFBR3 influences the QT interval specifically in Hispanic/Latino populations. Alternatively, effects of the TCA-SNP interactions for QT intervals were too small (if any) to be detected with the available sample size. Previously, this variant was identified in GWAS in relation to optic disc morphology[43], but not cardiac conduction. Therefore, potential explanations for the results of our study include lack of TCA-SNP interactions for QT intervals or presence of only small effects that could not be detected, even with the large sample size amassed in this study.
Earlier it was shown that TCA effects on QTc are related to the anticholinergic effects of TCAs on heart rate [12]. When the QT interval was corrected for heart rate by methods other than Bazett’s formula, the association between TCA use and QT intervals diminished. Moreover, when using the statistical model adopted herein, the effect of TCAs on QT intervals was shown to be negligible [12]. Furthermore, prescribed TCA doses and duration of TCA use can be variable among individuals and cohorts. Such variability can increase heterogeneity, making it more difficult to observe significant TCA-SNP interactions on QT intervals. However, it remains possible that TCAs increase the QT interval duration in rare cases (e.g., in association with low frequency genetic variants).
In addition, we showed that neither any genetic variants previously associated with a higher resting heart rate or QT intervals, nor a multi-locus score of these variants significantly modified the association between TCAs and resting RR or QT intervals[17–20]. The results of the multi-locus score analysis can be largely interpreted as a Mendelian randomization analysis, estimating whether a high resting heart rate or prolonged QT interval modified the TCA-SNP interactions. The results may indicate that participants with higher resting heart rates or prolonged QT intervals are not at higher risk for further TCA-induced increases in resting heart rate or QT duration. However, the variance explained by SNPs associated with mainly resting heart rate is low (0.8–0.9%)[17], which could affect the validity of this assumption.
A limitation of the study was the relatively low number of TCA users (especially in non-European ancestry cohorts), although our study is the largest effort assessing TCA-SNP interactions on RR and QT intervals to date. With limited power TCA interactions with relatively low frequency SNPs or in small samples could have been missed. Attempts at replication in non-European ancestry cohorts could have been underpowered. Second, our study was unable to account for prescribed TCA dosages, duration of use, and treatment adherence, which likely vary among cohorts from different countries. Such differences may have resulted in heterogeneity among studies. Third, results on RR from the European ancestry meta-analysis were not replicated in independent cohorts of a different ancestry. Fourth, one cohort determined RR intervals from heart rates (ASCOT). This method would increase RR measurement error, but independent of TCA exposures and genotypes. Any misclassification would have led to findings in the direction of the zero hypothesis. Furthermore, previously no heterogeneity in results was observed when comparing RR intervals from ECGs and pulse recordings [17]. Fifth, we were not able to adjust for potential confounders of SNP-TCA interactions. However, confounders with strong effects on the drug-outcome association were shown to only modestly bias the results [44]. And last, TCA doses may have been titrated to provide optimal blood concentrations, according to participant CYP2D6 and CYP2C19 genotypes. Because we defined TCA exposures as present or absent, SNP-TCA interactions caused by pharmacokinetic genes might have been missed.
For the present study, a well-powered, independent replication was not feasible, due to the limited availability of populations with high quality ECGs (twelve-lead), reliable assessment of drug use, and genome-wide SNP data. Our discovery effort should therefore be viewed as hypothesis generating. Future studies should attempt to replicate and validate our findings, to understand the pharmacological mechanisms involved.
In summary, we identified 2 independent genetic loci (BRE and UBE2E2) that modify the association between TCAs and heart rate in populations of European ancestry, and 1 locus that modifies the association between TCAs and QT intervals in Hispanic/Latino ancestry populations. If replicated and validated, these results may provide new insights into biological mechanisms underlying the effect of TCAs on heart rate.
Supplementary Material
Acknowledgments
Sources of Funding
Age, Gene/Environment Susceptibility – Reykjavik Study (AGES): This study has been funded by NIH contracts N01-AG-1-2100 and 271201200022C, the NIA Intramural Research Program, Hjartavernd (the Icelandic Heart Association), and the Althingi (the Icelandic Parliament). The study is approved by the Icelandic National Bioethics Committee, VSN: 00-063. The researchers are indebted to the participants for their willingness to participate in the study.
Atherosclerosis Risk in Communities (ARIC): The Atherosclerosis Risk in Communities Study is carried out as a collaborative study supported by National Heart, Lung and Blood Institute Contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C and HHSN268201100012C), R01HL087641, R01HL59367 and R01HL086694; National Human Genome Research Institute Contract U01HG004402; and National Institutes of Health Contract HHSN268200625226C. We thank the staff and participants of the ARIC study for their important contributions. Infrastructure was partly supported by Grant No. UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research.
Anglo-Scandinavian Cardiac Outcomes Trial United Kingdom (ASCOT): This trial was funded by an investigator-initiated grant from Pfizer, USA. The study was investigator led and was conducted, analysed and reported independently of the company. Genotyping was funded by the National Institutes for Health Research (NIHR) as part of the portfolio of translational research of the NIHR Cardiovascular Biomedical Research Unit at Barts and the London, QMUL, the NIHR Biomedical Research Centre at Imperial College, the International Centre for Circulatory Health Charity and the Medical Research Council through G952010. On behalf of the ASCOT investigators, we thank all ASCOT trial participants, physicians, nurses, and practices in the participating countries for their important contribution to the study. Helen R Warren, Mark J Caulfield and Patricia B Munrow wish to acknowledge support from the NIHR Cardiovascular Biomedical Unit at Barts and the London, Queen Mary University of London, UK.
Cardiovascular Health Study (CHS): This CHS research was supported by NHLBI contracts HHSN268201200036C, HHSN268200800007C, N01HC55222, N01HC85079, N01HC85080, N01HC85081, N01HC85082, N01HC85083, N01HC85086; and NHLBI grants U01HL080295, R01HL087652, R01HL105756, R01HL103612, R01HL120393, and R01HL085251with additional contribution from the National Institute of Neurological Disorders and Stroke (NINDS). Additional support was provided through R01AG023629 from the National Institute on Aging (NIA). A full list of principal CHS investigators and institutions can be found at CHS-NHLBI.org. The provision of genotyping data was supported in part by the National Center for Advancing Translational Sciences, CTSI grant UL1TR000124, and the National Institute of Diabetes and Digestive and Kidney Disease Diabetes Research Center (DRC) grant DK063491 to the Southern California Diabetes Endocrinology Research Center.
Framingham Heart Study (FHS): FHS work was supported by the National Heart Lung and Blood Institute of the National Institutes of Health and Boston University School of Medicine (Contract No. N01-HC-25195 and Contract No. HHSN268201500001I), its contract with Affymetrix for genotyping services (Contract No. N02-HL-6-4278), based on analyses by FHS investigators participating in the SNP Health Association Resource (SHARe) project. A portion of this research was conducted using the Linux Cluster for Genetic Analysis (LinGA-II), funded by the Robert Dawson Evans Endowment of the Department of Medicine at Boston University School of Medicine and Boston Medical Center. Additional support for these analyses was provided by R01HL103612 (PI Psaty, subcontract PI, Vasan). Measurement of the Gen 3 ECGs was supported by grants from the Doris Duke Charitable Foundation and the Burroughs Wellcome Fund (Newton-Cheh) and the NIH (HL080025, Newton-Cheh).
Health, Aging, and Body Composition Study (Health ABC): This research was supported by NIA Contracts N01AG62101, N01AG62103 and N01AG62106. The genome-wide association study was funded by NIA Grant 1R01AG032098-01A1 to Wake Forest University Health Sciences and genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University, Contract No. HHSN268200782096C. This research was supported in part by the Intramural Research Program of the NIH, National Institute on Aging.
Hispanic Community Health Study/Study of Latinos (HCHS/SOL): We thank the participants and staff of the HCHS/SOL study for their contributions to this study. The baseline examination of HCHS/SOL was carried out as a collaborative study supported by contracts from the National Heart, Lung, and Blood Institute (NHLBI) to the University of North Carolina (N01-HC65233), University of Miami (N01-HC65234), Albert Einstein College of Medicine (N01-HC65235), Northwestern University (N01-HC65236), and San Diego State University (N01-HC65237). The following Institutes/Centers/Offices contributed to the first phase of HCHS/SOL through a transfer of funds to the NHLBI: National Institute on Minority Health and Health Disparities, National Institute on Deafness and Other Communication Disorders, National Institute of Dental and Craniofacial Research (NIDCR), National Institute of Diabetes and Digestive and Kidney Diseases, National Institute of Neurological Disorders and Stroke, NIH Institution-Office of Dietary Supplements. The Genetic Analysis Center at University of Washington was supported by NHLBI and NIDCR contracts (HHSN268201300005C AM03 and MOD03). Genotyping efforts were supported by NHLBI HSN 26220/20054C, NCATS CTSI grant UL1TR000124, and NIDDK Diabetes Research Center (DRC) grant DK063491.
Jackson Heart Study (JHS): We thank the JHS participants and staff for their contributions to this work. The JHS is supported by contracts HHSN268201300046C, HHSN268201300047C, HSN268201300048C, HHSN268201300049C, HHSN268201300050C from the National Heart, Lung, and Blood Institute and the National Institute on Minority Health and Health Disparities.
Multi-Ethnic Study of Atherosclerosis (MESA): This research was supported by contracts HHSN2682015000031, N01-HC-95159, N01-HC-95160, N01-HC-95161, N01-HC-95162, N01-HC-95163, N01-HC-95164, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169 and by grants UL1-TR-000040, UL1-TR-001079, and UL1-RR-025005 from NCRR. Funding for MESA Family was provided by grants R01-HL-071205, R01-HL-071051, R01-HL-071250, R01-HL-071251, R01-HL-071252, R01-HL-071258, and R01-HL-071259, and by UL1-RR-025005 and UL1RR033176 from NCRR. Funding for MESA SHARe genotyping was provided by NHLBI Contract N02-HL-6-4278. The provision of genotyping data was supported in part by the National Center for Advancing Translational Sciences, CTSI grant UL1TR000124, and the National Institute of Diabetes and Digestive and Kidney Disease Diabetes Research Center (DRC) grant DK063491 to the Southern California Diabetes Endocrinology Research Center. Further information can be found at http://www.mesa-nhlbi.org and http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000209.v13.p3.
Netherlands Epidemiology of Obesity (NEO): The authors of the NEO study thank all individuals who participated in the Netherlands Epidemiology in Obesity study, all participating general practitioners for inviting eligible participants and all research nurses for collection of the data. We thank the NEO study group, Petra Noordijk and Ingeborg de Jonge for the coordination, lab and data management of the NEO study. The genotyping in the NEO study was supported by the Centre National de Génotypage (Paris, France), headed by Jean-Francois Deleuze.
Prospective Study of Pravastatin in the Elderly at Risk (PROSPER): The PROSPER study was supported by an investigator initiated grant obtained from Bristol-Myers Squibb. Professor Dr J W Jukema is an Established Clinical Investigator of the Netherlands Heart Foundation (Grant No. 2001 D 032). Support for genotyping was provided by the seventh framework program of the European commission (Grant No. 223004) and by the Netherlands Genomics Initiative (Netherlands Consortium for Healthy Aging Grant 050-060-810).
Rotterdam Study (RS): The RS is supported by the Erasmus Medical Center and Erasmus University Rotterdam; The Netherlands Organization for Scientific Research; The Netherlands Organization for Health Research and Development (ZonMw); the Research Institute for Diseases in the Elderly; The Netherlands Heart Foundation; the Ministry of Education, Culture and Science; the Ministry of Health Welfare and Sports; the European Commission; and the Municipality of Rotterdam. Support for genotyping was provided by The Netherlands Organization for Scientific Research (NWO) (175.010.2005.011, 911.03.012) and Research Institute for Diseases in the Elderly (RIDE). This study was supported by The Netherlands Genomics Initiative (NGI)/Netherlands Organization for Scientific Research (NWO) Project No. 050-060-810. This collaborative effort was supported by an award from the National Heart, Lung and Blood Institute (R01-HL-103612, PI BMP). CLA was supported in part by Grant R00-HL-098458 from the National Heart, Lung, and Blood Institute.
Women’s Health Initiative (WHI): WHI program is funded by the National Heart, Lung, and Blood Institute, National Institutes of Health, U.S. Department of Health and Human Services through contracts HHSN268201600018C, HHSN268201600001C, HHSN268201600002C, HHSN268201600003C, and HHSN268201600004C. The authors thank the WHI investigators and staff for their dedication, and the study participants for making the program possible. A full listing of WHI investigators can be found at: http://www.whi.org/researchers/Documents%20%20Write%20a%20Paper/WHI%20Investigator%20Long%20List.pdf
Women’s Health Initiative – GWAS of Treatment Response in Randomized Clinical Trials (WHI GARNET): Within the Genomics and Randomized Trials Network, a GWAS of Hormone Treatment and CVD and Metabolic Outcomes in the WHI was funded by the National Human Genome Research Institute, National Institutes of Health, U.S. Department of Health and Human Services through cooperative agreement U01HG005152 (Reiner). All contributors to GARNET science are listed @https://www.garnetstudy.org/Home. Dr. Busch was supported in part by a grant from the National Cancer Institute (5T32CA009001).
Women’s Health Initiative – Modification of Particulate Matter-Mediated Arrythmogenesis in Populations (WHI MOPMAP): The Modification of PM-Mediated Arrhythmogenesis in Populations was funded by the National Institute of Environmental Health Sciences, National Institutes of Health, U.S. Department of Health and Human Services through grant R01ES017794 (Whitsel).
Women’s Health Initiative – The SNP Health Associations Resource (WHI SHARe): The SNP Health Association Resource project was funded by the National Heart, Lung and Blood Institute, National Institutes of Health, U.S. Department of Health and Human Services through contract N02HL64278 (Kooperberg).
Footnotes
Disclosures
None.
References
- 1.Saxena A, Minton D, Lee DC, Sui X, Fayad R, Lavie CJ, Blair SN. Protective role of resting heart rate on all-cause and cardiovascular disease mortality. Mayo Clinic proceedings. 2013;88(12):1420–6. doi: 10.1016/j.mayocp.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kannel WB, Kannel C, Paffenbarger RS, Jr, Cupples LA. Heart rate and cardiovascular mortality: the Framingham Study. Am Heart J. 1987;113(6):1489–94. doi: 10.1016/0002-8703(87)90666-1. [DOI] [PubMed] [Google Scholar]
- 3.Nanchen D, Leening MJ, Locatelli I, Cornuz J, Kors JA, Heeringa J, Deckers JW, Hofman A, Franco OH, Stricker BH, Witteman JC, Dehghan A. Resting heart rate and the risk of heart failure in healthy adults: the Rotterdam Study. Circ Heart Fail. 2013;6(3):403–10. doi: 10.1161/CIRCHEARTFAILURE.112.000171. [DOI] [PubMed] [Google Scholar]
- 4.Straus SM, Kors JA, De Bruin ML, van der Hooft CS, Hofman A, Heeringa J, Deckers JW, Kingma JH, Sturkenboom MC, Stricker BH, Witteman JC. Prolonged QTc interval and risk of sudden cardiac death in a population of older adults. Journal of the American College of Cardiology. 2006;47(2):362–7. doi: 10.1016/j.jacc.2005.08.067. [DOI] [PubMed] [Google Scholar]
- 5. [Assessed on August 22, 2014];Anticholinergic drugs. available at: http://www.uea.ac.uk/mac/comm/media/press/2011/june/anticholinergics+study+drug+list.
- 6.Roden DM. Drug-induced prolongation of the QT interval. N Engl J Med. 2004;350(10):1013–22. doi: 10.1056/NEJMra032426. [DOI] [PubMed] [Google Scholar]
- 7.Woosley RL. [Assessed on August 22, 2014];Drugs that prolong the QTc interval and/or induce torsade de pointes. Available at http://www.crediblemeds.org/everyone/composite-list-all-qtdrugs/
- 8.van Noord C, Straus SM, Sturkenboom MC, Hofman A, Aarnoudse AJ, Bagnardi V, Kors JA, Newton-Cheh C, Witteman JC, Stricker BH. Psychotropic drugs associated with corrected QT interval prolongation. J Clin Psychopharmacol. 2009;29(1):9–15. doi: 10.1097/JCP.0b013e318191c6a8. [DOI] [PubMed] [Google Scholar]
- 9.Castro VM, Clements CC, Murphy SN, Gainer VS, Fava M, Weilburg JB, Erb JL, Churchill SE, Kohane IS, Iosifescu DV, Smoller JW, Perlis RH. QT interval and antidepressant use: a cross sectional study of electronic health records. BMJ. 2013;346:f288. doi: 10.1136/bmj.f288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reilly JG, Ayis SA, Ferrier IN, Jones SJ, Thomas SH. QTc-interval abnormalities and psychotropic drug therapy in psychiatric patients. Lancet. 2000;355(9209):1048–52. doi: 10.1016/s0140-6736(00)02035-3. [DOI] [PubMed] [Google Scholar]
- 11.Vieweg WV, Wood MA. Tricyclic antidepressants, QT interval prolongation, and torsade de pointes. Psychosomatics. 2004;45(5):371–7. doi: 10.1176/appi.psy.45.5.371. [DOI] [PubMed] [Google Scholar]
- 12.Noordam R, van den Berg ME, Niemeijer MN, Aarts N, Leening MJ, Deckers JW, Hofman A, Rijnbeek PR, Eijgelsheim M, Kors JA, Stricker BH, Visser LE. Assessing Prolongation of the Heart Rate Corrected QT Interval in Users of Tricyclic Antidepressants: Advice to Use Fridericia Rather Than Bazett's Correction. Journal of clinical psychopharmacology. 2015;35(3):260–5. doi: 10.1097/JCP.0000000000000321. [DOI] [PubMed] [Google Scholar]
- 13.Aarts N, Noordam R, Hofman A, Tiemeier H, Stricker BH, Visser LE. Utilization patterns of antidepressants between 1991 and 2011 in a population-based cohort of middle-aged and elderly. European psychiatry : the journal of the Association of European Psychiatrists. 2014;29(6):365–70. doi: 10.1016/j.eurpsy.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 14.Hansen DG, Rosholm JU, Gichangi A, Vach W. Increased use of antidepressants at the end of life: population-based study among people aged 65 years and above. Age and ageing. 2007;36(4):449–54. doi: 10.1093/ageing/afm056. [DOI] [PubMed] [Google Scholar]
- 15.Russell MW, Law I, Sholinsky P, Fabsitz RR. Heritability of ECG measurements in adult male twins. Journal of electrocardiology. 1998;30(Suppl):64–8. doi: 10.1016/s0022-0736(98)80034-4. [DOI] [PubMed] [Google Scholar]
- 16.Dalageorgou C, Ge D, Jamshidi Y, Nolte IM, Riese H, Savelieva I, Carter ND, Spector TD, Snieder H. Heritability of QT interval: how much is explained by genes for resting heart rate? Journal of cardiovascular electrophysiology. 2008;19(4):386–91. doi: 10.1111/j.1540-8167.2007.01030.x. [DOI] [PubMed] [Google Scholar]
- 17.den Hoed M, Eijgelsheim M, Esko T, Brundel BJ, Peal DS, Evans DM, Nolte IM, Segre AV, Holm H, Handsaker RE, Westra HJ, Johnson T, Isaacs A, Yang J, Lundby A, Zhao JH, Kim YJ, Go MJ, Almgren P, Bochud M, Boucher G, Cornelis MC, Gudbjartsson D, Hadley D, van der Harst P, Hayward C, den Heijer M, Igl W, Jackson AU, Kutalik Z, Luan J, Kemp JP, Kristiansson K, Ladenvall C, Lorentzon M, Montasser ME, Njajou OT, O'Reilly PF, Padmanabhan S, St Pourcain B, Rankinen T, Salo P, Tanaka T, Timpson NJ, Vitart V, Waite L, Wheeler W, Zhang W, Draisma HH, Feitosa MF, Kerr KF, Lind PA, Mihailov E, Onland-Moret NC, Song C, Weedon MN, Xie W, Yengo L, Absher D, Albert CM, Alonso A, Arking DE, de Bakker PI, Balkau B, Barlassina C, Benaglio P, Bis JC, Bouatia-Naji N, Brage S, Chanock SJ, Chines PS, Chung M, Darbar D, Dina C, Dorr M, Elliott P, Felix SB, Fischer K, Fuchsberger C, de Geus EJ, Goyette P, Gudnason V, Harris TB, Hartikainen AL, Havulinna AS, Heckbert SR, Hicks AA, Hofman A, Holewijn S, Hoogstra-Berends F, Hottenga JJ, Jensen MK, Johansson A, Junttila J, Kaab S, Kanon B, Ketkar S, Khaw KT, Knowles JW, Kooner AS, Kors JA, Kumari M, Milani L, Laiho P, Lakatta EG, Langenberg C, Leusink M, Liu Y, Luben RN, Lunetta KL, Lynch SN, Markus MR, Marques-Vidal P, Mateo Leach I, McArdle WL, McCarroll SA, Medland SE, Miller KA, Montgomery GW, Morrison AC, Muller-Nurasyid M, Navarro P, Nelis M, O'Connell JR, O'Donnell CJ, Ong KK, Newman AB, Peters A, Polasek O, Pouta A, Pramstaller PP, Psaty BM, Rao DC, Ring SM, Rossin EJ, Rudan D, Sanna S, Scott RA, Sehmi JS, Sharp S, Shin JT, Singleton AB, Smith AV, Soranzo N, Spector TD, Stewart C, Stringham HM, Tarasov KV, Uitterlinden AG, Vandenput L, Hwang SJ, Whitfield JB, Wijmenga C, Wild SH, Willemsen G, Wilson JF, Witteman JC, Wong A, Wong Q, Jamshidi Y, Zitting P, Boer JM, Boomsma DI, Borecki IB, van Duijn CM, Ekelund U, Forouhi NG, Froguel P, Hingorani A, Ingelsson E, Kivimaki M, Kronmal RA, Kuh D, Lind L, Martin NG, Oostra BA, Pedersen NL, Quertermous T, Rotter JI, van der Schouw YT, Verschuren WM, Walker M, Albanes D, Arnar DO, Assimes TL, Bandinelli S, Boehnke M, de Boer RA, Bouchard C, Caulfield WL, Chambers JC, Curhan G, Cusi D, Eriksson J, Ferrucci L, van Gilst WH, Glorioso N, de Graaf J, Groop L, Gyllensten U, Hsueh WC, Hu FB, Huikuri HV, Hunter DJ, Iribarren C, Isomaa B, Jarvelin MR, Jula A, Kahonen M, Kiemeney LA, van der Klauw MM, Kooner JS, Kraft P, Iacoviello L, Lehtimaki T, Lokki ML, Mitchell BD, Navis G, Nieminen MS, Ohlsson C, Poulter NR, Qi L, Raitakari OT, Rimm EB, Rioux JD, Rizzi F, Rudan I, Salomaa V, Sever PS, Shields DC, Shuldiner AR, Sinisalo J, Stanton AV, Stolk RP, Strachan DP, Tardif JC, Thorsteinsdottir U, Tuomilehto J, van Veldhuisen DJ, Virtamo J, Viikari J, Vollenweider P, Waeber G, Widen E, Cho YS, Olsen JV, Visscher PM, Willer C, Franke L, Global BC, Erdmann J, Thompson JR, Pfeufer A, Sotoodehnia N, Newton-Cheh C, Ellinor PT, Stricker BH, Metspalu A, Perola M, Beckmann JS, Smith GD, Stefansson K, Wareham NJ, Munroe PB, Sibon OC, Milan DJ, Snieder H, Samani NJ, Loos RJ Consortium CA, Consortium PG, Consortium QG, Consortium C-A, Consortium Q-I. Identification of heart rate-associated loci and their effects on cardiac conduction and rhythm disorders. Nature genetics. 2013;45(6):621–31. doi: 10.1038/ng.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deo R, Nalls MA, Avery CL, Smith JG, Evans DS, Keller MF, Butler AM, Buxbaum SG, Li G, Miguel Quibrera P, Smith EN, Tanaka T, Akylbekova EL, Alonso A, Arking DE, Benjamin EJ, Berenson GS, Bis JC, Chen LY, Chen W, Cummings SR, Ellinor PT, Evans MK, Ferrucci L, Fox ER, Heckbert SR, Heiss G, Hsueh WC, Kerr KF, Limacher MC, Liu Y, Lubitz SA, Magnani JW, Mehra R, Marcus GM, Murray SS, Newman AB, Njajou O, North KE, Paltoo DN, Psaty BM, Redline SS, Reiner AP, Robinson JG, Rotter JI, Samdarshi TE, Schnabel RB, Schork NJ, Singleton AB, Siscovick D, Soliman EZ, Sotoodehnia N, Srinivasan SR, Taylor HA, Trevisan M, Zhang Z, Zonderman AB, Newton-Cheh C, Whitsel EA. Common genetic variation near the connexin-43 gene is associated with resting heart rate in African Americans: a genome-wide association study of 13,372 participants. Heart rhythm : the official journal of the Heart Rhythm Society. 2013;10(3):401–8. doi: 10.1016/j.hrthm.2012.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mezzavilla M, Iorio A, Bobbo M, D'Eustacchio A, Merlo M, Gasparini P, Ulivi S, Sinagra G. Insight into genetic determinants of resting heart rate. Gene. 2014;545(1):170–4. doi: 10.1016/j.gene.2014.03.045. [DOI] [PubMed] [Google Scholar]
- 20.Arking DE, Pulit SL, Crotti L, van der Harst P, Munroe PB, Koopmann TT, Sotoodehnia N, Rossin EJ, Morley M, Wang X, Johnson AD, Lundby A, Gudbjartsson DF, Noseworthy PA, Eijgelsheim M, Bradford Y, Tarasov KV, Dorr M, Muller-Nurasyid M, Lahtinen AM, Nolte IM, Smith AV, Bis JC, Isaacs A, Newhouse SJ, Evans DS, Post WS, Waggott D, Lyytikainen LP, Hicks AA, Eisele L, Ellinghaus D, Hayward C, Navarro P, Ulivi S, Tanaka T, Tester DJ, Chatel S, Gustafsson S, Kumari M, Morris RW, Naluai AT, Padmanabhan S, Kluttig A, Strohmer B, Panayiotou AG, Torres M, Knoflach M, Hubacek JA, Slowikowski K, Raychaudhuri S, Kumar RD, Harris TB, Launer LJ, Shuldiner AR, Alonso A, Bader JS, Ehret G, Huang H, Kao WH, Strait JB, Macfarlane PW, Brown M, Caulfield MJ, Samani NJ, Kronenberg F, Willeit J, Smith JG, Greiser KH, Meyer Zu Schwabedissen H, Werdan K, Carella M, Zelante L, Heckbert SR, Psaty BM, Rotter JI, Kolcic I, Polasek O, Wright AF, Griffin M, Daly MJ, Dcct/Edic, Arnar DO, Holm H, Thorsteinsdottir U, Denny JC, Roden DM, Zuvich RL, Emilsson V, Plump AS, Larson MG, O'Donnell CJ, Yin X, Bobbo M, D'Adamo AP, Iorio A, Sinagra G, Carracedo A, Cummings SR, Nalls MA, Jula A, Kontula KK, Marjamaa A, Oikarinen L, Perola M, Porthan K, Erbel R, Hoffmann P, Jockel KH, Kalsch H, Nothen MM, den Hoed M, Loos RJ, Thelle DS, Gieger C, Meitinger T, Perz S, Peters A, Prucha H, Sinner MF, Waldenberger M, de Boer RA, Franke L, van der Vleuten PA, Beckmann BM, Martens E, Bardai A, Hofman N, Wilde AA, Behr ER, Dalageorgou C, Giudicessi JR, Medeiros-Domingo A, Barc J, Kyndt F, Probst V, Ghidoni A, Insolia R, Hamilton RM, Scherer SW, Brandimarto J, Margulies K, Moravec CE, del Greco MF, Fuchsberger C, O'Connell JR, Lee WK, Watt GC, Campbell H, Wild SH, El Mokhtari NE, Frey N, Asselbergs FW, Mateo Leach I, Navis G, van den Berg MP, van Veldhuisen DJ, Kellis M, Krijthe BP, Franco OH, Hofman A, Kors JA, Uitterlinden AG, Witteman JC, Kedenko L, Lamina C, Oostra BA, Abecasis GR, Lakatta EG, Mulas A, Orru M, Schlessinger D, Uda M, Markus MR, Volker U, Snieder H, Spector TD, Arnlov J, Lind L, Sundstrom J, Syvanen AC, Kivimaki M, Kahonen M, Mononen N, Raitakari OT, Viikari JS, Adamkova V, Kiechl S, Brion M, Nicolaides AN, Paulweber B, Haerting J, Dominiczak AF, Nyberg F, Whincup PH, Hingorani AD, Schott JJ, Bezzina CR, Ingelsson E, Ferrucci L, Gasparini P, Wilson JF, Rudan I, Franke A, Muhleisen TW, Pramstaller PP, Lehtimaki TJ, Paterson AD, Parsa A, Liu Y, van Duijn CM, Siscovick DS, Gudnason V, Jamshidi Y, Salomaa V, Felix SB, Sanna S, Ritchie MD, Stricker BH, Stefansson K, Boyer LA, Cappola TP, Olsen JV, Lage K, Schwartz PJ, Kaab S, Chakravarti A, Ackerman MJ, Pfeufer A, de Bakker PI, Newton-Cheh C e MC, Consortium CA, Consortium C, Consortium H. Genetic association study of QT interval highlights role for calcium signaling pathways in myocardial repolarization. Nature genetics. 2014;46(8):826–36. doi: 10.1038/ng.3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith JG, Avery CL, Evans DS, Nalls MA, Meng YA, Smith EN, Palmer C, Tanaka T, Mehra R, Butler AM, Young T, Buxbaum SG, Kerr KF, Berenson GS, Schnabel RB, Li G, Ellinor PT, Magnani JW, Chen W, Bis JC, Curb JD, Hsueh WC, Rotter JI, Liu Y, Newman AB, Limacher MC, North KE, Reiner AP, Quibrera PM, Schork NJ, Singleton AB, Psaty BM, Soliman EZ, Solomon AJ, Srinivasan SR, Alonso A, Wallace R, Redline S, Zhang ZM, Post WS, Zonderman AB, Taylor HA, Murray SS, Ferrucci L, Arking DE, Evans MK, Fox ER, Sotoodehnia N, Heckbert SR, Whitsel EA, Newton-Cheh C Care, consortia C. Impact of ancestry and common genetic variants on QT interval in African Americans. Circulation Cardiovascular genetics. 2012;5(6):647–55. doi: 10.1161/CIRCGENETICS.112.962787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TF, McCarroll SA, Visscher PM. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–53. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Avery CL, Sitlani CM, Arking DE, Arnett DK, Bis JC, Boerwinkle E, Buckley BM, Ida Chen YD, de Craen AJ, Eijgelsheim M, Enquobahrie D, Evans DS, Ford I, Garcia ME, Gudnason V, Harris TB, Heckbert SR, Hochner H, Hofman A, Hsueh WC, Isaacs A, Jukema JW, Knekt P, Kors JA, Krijthe BP, Kristiansson K, Laaksonen M, Liu Y, Li X, Macfarlane PW, Newton-Cheh C, Nieminen MS, Oostra BA, Peloso GM, Porthan K, Rice K, Rivadeneira FF, Rotter JI, Salomaa V, Sattar N, Siscovick DS, Slagboom PE, Smith AV, Sotoodehnia N, Stott DJ, Stricker BH, Sturmer T, Trompet S, Uitterlinden AG, van Duijn C, Westendorp RG, Witteman JC, Whitsel EA, Psaty BM. Drug-gene interactions and the search for missing heritability: a cross-sectional pharmacogenomics study of the QT interval. The pharmacogenomics journal. 2014;14(1):6–13. doi: 10.1038/tpj.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sitlani CM, Rice KM, Lumley T, McKnight B, Cupples LA, Avery CL, Noordam R, Stricker BH, Whitsel EA, Psaty BM. Generalized estimating equations for genome-wide association studies using longitudinal phenotype data. Statistics in medicine. 2015;34(1):118–30. doi: 10.1002/sim.6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Conomos MP, Laurie CA, Stilp AM, Gogarten SM, McHugh CP, Nelson SC, Sofer T, Fernandez-Rhodes L, Justice AE, Graff M, Young KL, Seyerle AA, Avery CL, Taylor KD, Rotter JI, Talavera GA, Daviglus ML, Wassertheil-Smoller S, Schneiderman N, Heiss G, Kaplan RC, Franceschini N, Reiner AP, Shaffer JR, Barr RG, Kerr KF, Browning SR, Browning BL, Weir BS, Aviles-Santa ML, Papanicolaou GJ, Lumley T, Szpiro AA, North KE, Rice K, Thornton TA, Laurie CC. Genetic Diversity and Association Studies in US Hispanic/Latino Populations: Applications in the Hispanic Community Health Study/Study of Latinos. American journal of human genetics. 2016;98(1):165–84. doi: 10.1016/j.ajhg.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Psaty BM, O'Donnell CJ, Gudnason V, Lunetta KL, Folsom AR, Rotter JI, Uitterlinden AG, Harris TB, Witteman JC, Boerwinkle E Consortium C. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: Design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circulation Cardiovascular genetics. 2009;2(1):73–80. doi: 10.1161/CIRCGENETICS.108.829747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.International HapMap C. The International HapMap Project. Nature. 2003;426(6968):789–96. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 28.1000 Genomes Project Consortium. Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arizona Center for Education and Research on Therapeutics. Drugs that Prolong the QT Interval CredibleMeds-AZCERT: AZ. USA: 2011. [assessed on February 2016]. http://www.azcert.org/ [Google Scholar]
- 30.Zeger SL, Liang KY. Longitudinal data analysis for discrete and continuous outcomes. Biometrics. 1986;42(1):121–30. [PubMed] [Google Scholar]
- 31.Aulchenko YS, Struchalin MV, van Duijn CM. ProbABEL package for genome-wide association analysis of imputed data. BMC bioinformatics. 2010;11:134. doi: 10.1186/1471-2105-11-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Satterthwaite FE. An approximate distribution of estimates of variance components. Biometrics. 1946;2(6):110–4. [PubMed] [Google Scholar]
- 33.Pan W, Wall MM. Small-sample adjustments in using the sandwich variance estimator in generalized estimating equations. Stat Med. 2002;21(10):1429–41. doi: 10.1002/sim.1142. [DOI] [PubMed] [Google Scholar]
- 34.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey Smith G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Statistics in medicine. 2008;27(8):1133–63. doi: 10.1002/sim.3034. [DOI] [PubMed] [Google Scholar]
- 36.Dastani Z, Hivert MF, Timpson N, Perry JR, Yuan X, Scott RA, Henneman P, Heid IM, Kizer JR, Lyytikainen LP, Fuchsberger C, Tanaka T, Morris AP, Small K, Isaacs A, Beekman M, Coassin S, Lohman K, Qi L, Kanoni S, Pankow JS, Uh HW, Wu Y, Bidulescu A, Rasmussen-Torvik LJ, Greenwood CM, Ladouceur M, Grimsby J, Manning AK, Liu CT, Kooner J, Mooser VE, Vollenweider P, Kapur KA, Chambers J, Wareham NJ, Langenberg C, Frants R, Willems-Vandijk K, Oostra BA, Willems SM, Lamina C, Winkler TW, Psaty BM, Tracy RP, Brody J, Chen I, Viikari J, Kahonen M, Pramstaller PP, Evans DM, St Pourcain B, Sattar N, Wood AR, Bandinelli S, Carlson OD, Egan JM, Bohringer S, van Heemst D, Kedenko L, Kristiansson K, Nuotio ML, Loo BM, Harris T, Garcia M, Kanaya A, Haun M, Klopp N, Wichmann HE, Deloukas P, Katsareli E, Couper DJ, Duncan BB, Kloppenburg M, Adair LS, Borja JB, Mu TC, Wilson JG, Musani S, Guo X, Johnson T, Semple R, Teslovich TM, Allison MA, Redline S, Buxbaum SG, Mohlke KL, Meulenbelt I, Ballantyne CM, Dedoussis GV, Hu FB, Liu Y, Paulweber B, Spector TD, Slagboom PE, Ferrucci L, Jula A, Perola M, Raitakari O, Florez JC, Salomaa V, Eriksson JG, Frayling TM, Hicks AA, Lehtimaki T, Smith GD, Siscovick DS, Kronenberg F, van Duijn C, Loos RJ, Waterworth DM, Meigs JB, Dupuis J, Richards JB, Voight BF, Scott LJ, Steinthorsdottir V, Dina C, Welch RP, Zeggini E, Huth C, Aulchenko YS, Thorleifsson G, McCulloch LJ, Ferreira T, Grallert H, Amin N, Wu G, Willer CJ, Raychaudhuri S, McCarroll SA, Hofmann OM, Segre AV, van Hoek M, Navarro P, Ardlie K, Balkau B, Benediktsson R, Bennett AJ, Blagieva R, Boerwinkle E, Bonnycastle LL, Bostrom KB, Bravenboer B, Bumpstead S, Burtt NP, Charpentier G, Chines PS, Cornelis M, Crawford G, Doney AS, Elliott KS, Elliott AL, Erdos MR, Fox CS, Franklin CS, Ganser M, Gieger C, Grarup N, Green T, Griffin S, Groves CJ, Guiducci C, Hadjadj S, Hassanali N, Herder C, Isomaa B, Jackson AU, Johnson PR, Jorgensen T, Kao WH, Kong A, Kraft P, Kuusisto J, Lauritzen T, Li M, Lieverse A, Lindgren CM, Lyssenko V, Marre M, Meitinger T, Midthjell K, Morken MA, Narisu N, Nilsson P, Owen KR, Payne F, Petersen AK, Platou C, Proenca C, Prokopenko I, Rathmann W, Rayner NW, Robertson NR, Rocheleau G, Roden M, Sampson MJ, Saxena R, Shields BM, Shrader P, Sigurdsson G, Sparso T, Strassburger K, Stringham HM, Sun Q, Swift AJ, Thorand B, Tichet J, Tuomi T, van Dam RM, van Haeften TW, van Herpt T, van Vliet-Ostaptchouk JV, Walters GB, Weedon MN, Wijmenga C, Witteman J, Bergman RN, Cauchi S, Collins FS, Gloyn AL, Gyllensten U, Hansen T, Hide WA, Hitman GA, Hofman A, Hunter DJ, Hveem K, Laakso M, Morris AD, Palmer CN, Rudan I, Sijbrands E, Stein LD, Tuomilehto J, Uitterlinden A, Walker M, Watanabe RM, Abecasis GR, Boehm BO, Campbell H, Daly MJ, Hattersley AT, Pedersen O, Barroso I, Groop L, Sladek R, Thorsteinsdottir U, Wilson JF, Illig T, Froguel P, van Duijn CM, Stefansson K, Altshuler D, Boehnke M, McCarthy MI, Soranzo N, Wheeler E, Glazer NL, Bouatia-Naji N, Magi R, Randall J, Elliott P, Rybin D, Dehghan A, Hottenga JJ, Song K, Goel A, Lajunen T, Doney A, Cavalcanti-Proenca C, Kumari M, Timpson NJ, Zabena C, Ingelsson E, An P, O'Connell J, Luan J, Elliott A, McCarroll SA, Roccasecca RM, Pattou F, Sethupathy P, Ariyurek Y, Barter P, Beilby JP, Ben-Shlomo Y, Bergmann S, Bochud M, Bonnefond A, Borch-Johnsen K, Bottcher Y, Brunner E, Bumpstead SJ, Chen YD, Chines P, Clarke R, Coin LJ, Cooper MN, Crisponi L, Day IN, de Geus EJ, Delplanque J, Fedson AC, Fischer-Rosinsky A, Forouhi NG, Franzosi MG, Galan P, Goodarzi MO, Graessler J, Grundy S, Gwilliam R, Hallmans G, Hammond N, Han X, Hartikainen AL, Hayward C, Heath SC, Hercberg S, Hillman DR, Hingorani AD, Hui J, Hung J, Kaakinen M, Kaprio J, Kesaniemi YA, Kivimaki M, Knight B, Koskinen S, Kovacs P, Kyvik KO, Lathrop GM, Lawlor DA, Le Bacquer O, Lecoeur C, Li Y, Mahley R, Mangino M, Martinez-Larrad MT, McAteer JB, McPherson R, Meisinger C, Melzer D, Meyre D, Mitchell BD, Mukherjee S, Naitza S, Neville MJ, Orru M, Pakyz R, Paolisso G, Pattaro C, Pearson D, Peden JF, Pedersen NL, Pfeiffer AF, Pichler I, Polasek O, Posthuma D, Potter SC, Pouta A, Province MA, Rayner NW, Rice K, Ripatti S, Rivadeneira F, Rolandsson O, Sandbaek A, Sandhu M, Sanna S, Sayer AA, Scheet P, Seedorf U, Sharp SJ, Shields B, Sigurethsson G, Sijbrands EJ, Silveira A, Simpson L, Singleton A, Smith NL, Sovio U, Swift A, Syddall H, Syvanen AC, Tonjes A, Uitterlinden AG, van Dijk KW, Varma D, Visvikis-Siest S, Vitart V, Vogelzangs N, Waeber G, Wagner PJ, Walley A, Ward KL, Watkins H, Wild SH, Willemsen G, Witteman JC, Yarnell JW, Zelenika D, Zethelius B, Zhai G, Zhao JH, Zillikens MC, Borecki IB, Meneton P, Magnusson PK, Nathan DM, Williams GH, Silander K, Bornstein SR, Schwarz P, Spranger J, Karpe F, Shuldiner AR, Cooper C, Serrano-Rios M, Lind L, Palmer LJ, Hu FBs, Franks PW, Ebrahim S, Marmot M, Kao WH, Pramstaller PP, Wright AF, Stumvoll M, Hamsten A, Procardis C, Buchanan TA, Valle TT, Rotter JI, Penninx BW, Boomsma DI, Cao A, Scuteri A, Schlessinger D, Uda M, Ruokonen A, Jarvelin MR, Peltonen L, Mooser V, Sladek R, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Chasman DI, Johansen CT, Fouchier SW, Peloso GM, Barbalic M, Ricketts SL, Bis JC, Feitosa MF, Orho-Melander M, Melander O, Li X, Li M, Cho YS, Go MJ, Kim YJ, Lee JY, Park T, Kim K, Sim X, Ong RT, Croteau-Chonka DC, Lange LA, Smith JD, Ziegler A, Zhang W, Zee RY, Whitfield JB, Thompson JR, Surakka I, Spector TD, Smit JH, Sinisalo J, Scott J, Saharinen J, Sabatti C, Rose LM, Roberts R, Rieder M, Parker AN, Pare G, O'Donnell CJ, Nieminen MS, Nickerson DA, Montgomery GW, McArdle W, Masson D, Martin NG, Marroni F, Lucas G, Luben R, Lokki ML, Lettre G, Launer LJ, Lakatta EG, Laaksonen R, Kyvik KO, Konig IR, Khaw KT, Kaplan LM, Johansson A, Janssens AC, Igl W, Hovingh GK, Hengstenberg C, Havulinna AS, Hastie ND, Harris TB, Haritunians T, Hall AS, Groop LC, Gonzalez E, Freimer NB, Erdmann J, Ejebe KG, Doring A, Dominiczak AF, Demissie S, Deloukas P, de Faire U, Crawford G, Chen YD, Caulfield MJ, Boekholdt SM, Assimes TL, Quertermous T, Seielstad M, Wong TY, Tai ES, Feranil AB, Kuzawa CW, Taylor HA, Jr, Gabriel SB, Holm H, Gudnason V, Krauss RM, Ordovas JM, Munroe PB, Kooner JS, Tall AR, Hegele RA, Kastelein JJ, Schadt EE, Strachan DP, Reilly MP, Samani NJ, Schunkert H, Cupples LA, Sandhu MS, Ridker PM, Rader DJ, Kathiresan S investigators M, Consortium D, Consortium M, Investigators G, Consortium D, Consortium G, Global BPC, Consortium G. Novel loci for adiponectin levels and their influence on type 2 diabetes and metabolic traits: a multi-ethnic meta-analysis of 45,891 individuals. PLoS genetics. 2012;8(3):e1002607. doi: 10.1371/journal.pgen.1002607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson T. Technical report. The Comprehensive R Archive Network; 2013. [Last accessed 28 September 2015]. Efficient calculation for multi-SNP genetic risk scores. http://cran.r-project.org/web/packages/gtx/vignettes/ashg2012.pdf. 2015. [Google Scholar]
- 38.Mahajan A, Go MJ, Zhang W, Below JE, Gaulton KJ, Ferreira T, Horikoshi M, Johnson AD, Ng MC, Prokopenko I, Saleheen D, Wang X, Zeggini E, Abecasis GR, Adair LS, Almgren P, Atalay M, Aung T, Baldassarre D, Balkau B, Bao Y, Barnett AH, Barroso I, Basit A, Been LF, Beilby J, Bell GI, Benediktsson R, Bergman RN, Boehm BO, Boerwinkle E, Bonnycastle LL, Burtt N, Cai Q, Campbell H, Carey J, Cauchi S, Caulfield M, Chan JC, Chang LC, Chang TJ, Chang YC, Charpentier G, Chen CH, Chen H, Chen YT, Chia KS, Chidambaram M, Chines PS, Cho NH, Cho YM, Chuang LM, Collins FS, Cornelis MC, Couper DJ, Crenshaw AT, van Dam RM, Danesh J, Das D, de Faire U, Dedoussis G, Deloukas P, Dimas AS, Dina C, Doney AS, Donnelly PJ, Dorkhan M, van Duijn C, Dupuis J, Edkins S, Elliott P, Emilsson V, Erbel R, Eriksson JG, Escobedo J, Esko T, Eury E, Florez JC, Fontanillas P, Forouhi NG, Forsen T, Fox C, Fraser RM, Frayling TM, Froguel P, Frossard P, Gao Y, Gertow K, Gieger C, Gigante B, Grallert H, Grant GB, Grrop LC, Groves CJ, Grundberg E, Guiducci C, Hamsten A, Han BG, Hara K, Hassanali N, Hattersley AT, Hayward C, Hedman AK, Herder C, Hofman A, Holmen OL, Hovingh K, Hreidarsson AB, Hu C, Hu FB, Hui J, Humphries SE, Hunt SE, Hunter DJ, Hveem K, Hydrie ZI, Ikegami H, Illig T, Ingelsson E, Islam M, Isomaa B, Jackson AU, Jafar T, James A, Jia W, Jockel KH, Jonsson A, Jowett JB, Kadowaki T, Kang HM, Kanoni S, Kao WH, Kathiresan S, Kato N, Katulanda P, Keinanen-Kiukaanniemi KM, Kelly AM, Khan H, Khaw KT, Khor CC, Kim HL, Kim S, Kim YJ, Kinnunen L, Klopp N, Kong A, Korpi-Hyovalti E, Kowlessur S, Kraft P, Kravic J, Kristensen MM, Krithika S, Kumar A, Kumate J, Kuusisto J, Kwak SH, Laakso M, Lagou V, Lakka TA, Langenberg C, Langford C, Lawrence R, Leander K, Lee JM, Lee NR, Li M, Li X, Li Y, Liang J, Liju S, Lim WY, Lind L, Lindgren CM, Lindholm E, Liu CT, Liu JJ, Lobbens S, Long J, Loos RJ, Lu W, Luan J, Lyssenko V, Ma RC, Maeda S, Magi R, Mannisto S, Matthews DR, Meigs JB, Melander O, Metspalu A, Meyer J, Mirza G, Mihailov E, Moebus S, Mohan V, Mohlke KL, Morris AD, Muhleisen TW, Muller-Nurasyid M, Musk B, Nakamura J, Nakashima E, Navarro P, Ng PK, Nica AC, Nilsson PM, Njolstad I, Nothen MM, Ohnaka K, Ong TH, Owen KR, Palmer CN, Pankow JS, Park KS, Parkin M, Pechlivanis S, Pedersen NL, Peltonen L, Perry JR, Peters A, Pinidiyapathirage JM, Platou CG, Potter S, Price JF, Qi L, Radha V, Rallidis L, Rasheed A, Rathman W, Rauramaa R, Raychaudhuri S, Rayner NW, Rees SD, Rehnberg E, Ripatti S, Robertson N, Roden M, Rossin EJ, Rudan I, Rybin D, Saaristo TE, Salomaa V, Saltevo J, Samuel M, Sanghera DK, Saramies J, Scott J, Scott LJ, Scott RA, Segre AV, Sehmi J, Sennblad B, Shah N, Shah S, Shera AS, Shu XO, Shuldiner AR, Sigurdsson G, Sijbrands E, Silveira A, Sim X, Sivapalaratnam S, Small KS, So WY, Stancakova A, Stefansson K, Steinbach G, Steinthorsdottir V, Stirrups K, Strawbridge RJ, Stringham HM, Sun Q, Suo C, Syvanen AC, Takayanagi R, Takeuchi F, Tay WT, Teslovich TM, Thorand B, Thorleifsson G, Thorsteinsdottir U, Tikkanen E, Trakalo J, Tremoli E, Trip MD, Tsai FJ, Tuomi T, Tuomilehto J, Uitterlinden AG, Valladares-Salgado A, Vedantam S, Veglia F, Voight BF, Wang C, Wareham NJ, Wennauer R, Wickremasinghe AR, Wilsgaard T, Wilson JF, Wiltshire S, Winckler W, Wong TY, Wood AR, Wu JY, Wu Y, Yamamoto K, Yamauchi T, Yang M, Yengo L, Yokota M, Young R, Zabaneh D, Zhang F, Zhang R, Zheng W, Zimmet PZ, Altshuler D, Bowden DW, Cho YS, Cox NJ, Cruz M, Hanis CL, Kooner J, Lee JY, Seielstad M, Teo YY, Boehnke M, Parra EJ, Chambers JC, Tai ES, McCarthy MI, Morris AP. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nature genetics. 2014;46(3):234–44. doi: 10.1038/ng.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hromatka BS, Tung JY, Kiefer AK, Do CB, Hinds DA, Eriksson N. Genetic variants associated with motion sickness point to roles for inner ear development, neurological processes and glucose homeostasis. Human molecular genetics. 2015;24(9):2700–8. doi: 10.1093/hmg/ddv028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Westra HJ, Peters MJ, Esko T, Yaghootkar H, Schurmann C, Kettunen J, Christiansen MW, Fairfax BP, Schramm K, Powell JE, Zhernakova A, Zhernakova DV, Veldink JH, Van den Berg LH, Karjalainen J, Withoff S, Uitterlinden AG, Hofman A, Rivadeneira F, t Hoen PA, Reinmaa E, Fischer K, Nelis M, Milani L, Melzer D, Ferrucci L, Singleton AB, Hernandez DG, Nalls MA, Homuth G, Nauck M, Radke D, Volker U, Perola M, Salomaa V, Brody J, Suchy-Dicey A, Gharib SA, Enquobahrie DA, Lumley T, Montgomery GW, Makino S, Prokisch H, Herder C, Roden M, Grallert H, Meitinger T, Strauch K, Li Y, Jansen RC, Visscher PM, Knight JC, Psaty BM, Ripatti S, Teumer A, Frayling TM, Metspalu A, van Meurs JB, Franke L. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nature genetics. 2013;45(10):1238–43. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. [Assessed December 1, 2015];Blood eQTL browser. available at: http://genenetworknl/bloodeqtlbrowser/
- 42.de Lima LT, Bueno CT, Vivona D, Hirata RD, Hirata MH, Hungria VT, Chiattone CS, Zanichelli MA, de Chauffaille ML, Guerra-Shinohara EM. Relationship between SLCO1B3 and ABCA3 polymorphisms and imatinib response in chronic myeloid leukemia patients. Hematology. 2015;20(3):137–42. doi: 10.1179/1607845414Y.0000000181. [DOI] [PubMed] [Google Scholar]
- 43.Springelkamp H, Mishra A, Hysi PG, Gharahkhani P, Hohn R, Khor CC, Cooke Bailey JN, Luo X, Ramdas WD, Vithana E, Koh V, Yazar S, Xu L, Forward H, Kearns LS, Amin N, Iglesias AI, Sim KS, van Leeuwen EM, Demirkan A, van der Lee S, Loon SC, Rivadeneira F, Nag A, Sanfilippo PG, Schillert A, de Jong PT, Oostra BA, Uitterlinden AG, Hofman A, Zhou T, Burdon KP, Spector TD, Lackner KJ, Saw SM, Vingerling JR, Teo YY, Pasquale LR, Wolfs RC, Lemij HG, Tai ES, Jonas JB, Cheng CY, Aung T, Jansonius NM, Klaver CC, Craig JE, Young TL, Haines JL, MacGregor S, Mackey DA, Pfeiffer N, Wong TY, Wiggs JL, Hewitt AW, van Duijn CM, Hammond CJ Consortium N. Meta-analysis of Genome-Wide Association Studies Identifies Novel Loci Associated With Optic Disc Morphology. Genetic epidemiology. 2015;39(3):207–16. doi: 10.1002/gepi.21886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Avery CL, Der JS, Whitsel EA, Sturmer T. Comparison of study designs used to detect and characterize pharmacogenomic interactions in nonexperimental studies: a simulation study. Pharmacogenetics and genomics. 2014;24(3):146–55. doi: 10.1097/FPC.0000000000000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.