Abstract
The human relaxin peptide family consists of seven cystine‐rich peptides, four of which are known to signal through relaxin family peptide receptors, RXFP1–4. As these peptides play a vital role physiologically and in various diseases, they are of considerable importance for drug discovery and development. Detailed structure–activity relationship (SAR) studies towards understanding the role of important residues in each of these peptides have been reported over the years and utilized for the design of antagonists and minimized agonist variants. This review summarizes the current knowledge of the SAR of human relaxin 2 (H2 relaxin), human relaxin 3 (H3 relaxin), human insulin‐like peptide 3 (INSL3) and human insulin‐like peptide 5 (INSL5).
Linked Articles
This article is part of a themed section on Recent Progress in the Understanding of Relaxin Family Peptides and their Receptors. To view the other articles in this section visit http://onlinelibrary.wiley.com/doi/10.1111/bph.v174.10/issuetoc
Abbreviations
- H2 relaxin
human relaxin‐2
- H3 relaxin
human relaxin‐3
- IGF
insulin‐like growth factor
- INSL
insulin‐like peptide
- LRR
leucine‐rich repeat
- RXFP1–4
relaxin‐family peptide receptor 1–4
- SAR
structure–activity relationship
- TM
transmembrane
Tables of Links
| TARGETS | |
|---|---|
| GPCRs a | Enzymes b |
| RXFP1 receptor | ERK1 |
| RXFP2 receptor | ERK2 |
| RXFP3 receptor | |
| RXFP4 receptor |
These Tables list key protein targets and ligands in this article that are hyperlinked to corresponding entries in http://www.guidetopharmacology.org, the common portal for data from the IUPHAR/BPS Guide to PHARMACOLOGY (Southan et al., 2016), and are permanently archived in the Concise Guide to PHARMACOLOGY 2015/16 (a,bAlexander et al., 2015a,b).
Introduction
In the late 1970s, relaxin and insulin, two functionally diverse peptide hormones, were shown to be identical with respect to the distribution of its three disulfide bonds, which gave rise to the concept of the insulin superfamily (Schwabe and McDonald, 1977). Evolutionary studies revealed that insulin/relaxin‐like peptides are ubiquitously found in vertebrates and non‐vertebrates (Hsu, 2003; Wilkinson et al., 2005; Wilkinson and Bathgate, 2007) In humans, the insulin superfamily consists of insulin, insulin‐like growth factors (IGF‐1 and IGF‐2) and seven relaxin family peptides. The latter consists of human relaxin 1 (H1 relaxin), human relaxin 2 (H2 relaxin), human relaxin 3 (H3 relaxin), insulin‐like peptide 3 (INSL3 also known as relaxin‐like factor or Leydig insulin‐like peptide), insulin‐like peptide 4 (INSL4 or placentin), insulin‐like peptide 5 (INSL5) and insulin‐like peptide 6 (INSL6) (Hudson et al., 1983; Crawford et al., 1984; Adham et al., 1993; Chassin et al., 1995; Conklin et al., 1999; Chang et al., 2000; Bathgate et al., 2002; Alexander et al., 2015a) (Figure 1).
Figure 1.

Comparison of relaxin family peptides. (A) Primary sequences of relaxin family peptide in humans. To illustrate the conserved features, the amino acid types are colour coded according to their nature with basic residues in blue (Arg, Lys and His), acidic residues in red (Glu and Asp), hydrophobic residues in green (Ala, Val, Ile, Leu, Phe, Trp, Tyr, Pro and Met), hydrophilic in black (Ser, Thr, Asn and Gln) and glycine in light blue. The cysteine residues are highlighted in yellow, and the disulfide bonds shown by solid lines. (B) Superposition of the NMR structures of four relaxin family peptides. H2 relaxin is shown in cyan (PDB: 2MV1), H3 relaxin is shown in blue (PDB: 2FHW), INSL3 is shown in red (PDB: 2H8B) and INSL5 is shown in green (PDB: 2KBC). The A‐ and B‐chain N‐ and C‐termini are labelled A (N), A (C), B (N) and B (C) respectively.
While in higher primates (e.g. humans and great apes) there are two forms of relaxin (relaxin 1 and relaxin 2), which are thought to be a consequence of gene duplication during primate evolution, other mammals (e.g. mouse and rat) have only one relaxin gene that produces a relaxin hormone, equivalent to relaxin 2 in humans (H2 relaxin). Early studies confirmed that relaxin (equivalent to relaxin 2 in humans) is synthesized as a pre‐prohormone with four distinct regions: a signal peptide, a Β‐chain, a C‐chain and a COOH‐terminal A‐chain (Hudson et al., 1981; Haley et al., 1982). Like relaxin (equivalent to relaxin 2 in humans), relaxin 3 and INSL3 peptides are expressed as pre‐prohormones that after oxidative folding and proteolytic processing result in mature hormones with two chains (A and B) linked by two inter‐chain disulfide bonds and one intra‐A‐chain disulfide bond (Büllesbach and Schwabe, 2002; Liu et al., 2003).
The relaxin peptides act upon GPCRs that are now known as relaxin family peptide (RXFP) receptors (Bathgate et al., 2006a; Alexander et al., 2015a). H2 relaxin, INSL3, H3 relaxin and INSL5 signal through RXFP1 (Hsu et al., 2002), RXFP2 (Kumagai et al., 2002), RXFP3 (Liu et al., 2003) and RXFP4 (Liu et al., 2005b) receptors respectively (Figure 2). The native receptors for INSL4 and INSL6 are yet to be identified (Halls et al., 2007). H1 relaxin is the most recently evolved human peptide and is a pseudogene in other primate species. Notably, H2 relaxin is the ortholog of the relaxin‐1 gene found with relaxin‐3 in most other mammalian species (Wilkinson et al., 2005), and it is presumed that H1 relaxin is also a ligand of the RXFP1 receptor. All the family members of relaxin peptides and their target receptors (RXFPs) have broad physiological roles and are important clinically [reviewed in: (Bathgate et al., 2013b)]. Considering the vital roles of relaxin peptides (Table 1) in various diseases, they are of considerable importance for drug discovery and development.
Figure 2.
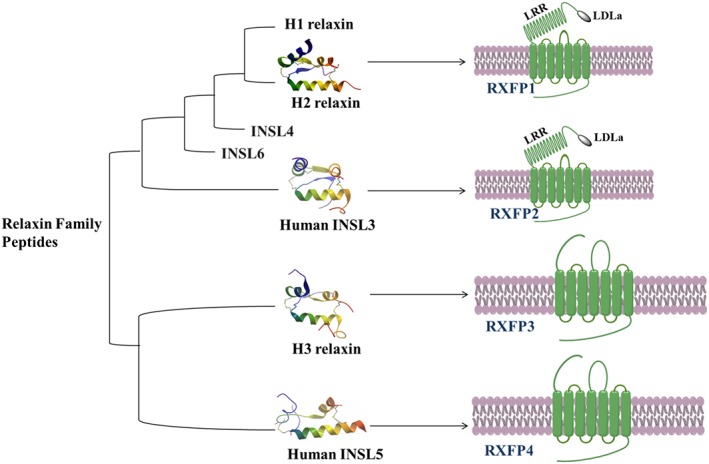
Peptide‐receptor pairing within the relaxin family peptides. H2 relaxin (PDB: 6RLX), H3 relaxin (PDB: 2FHW), INSL3 (PDB: 2H8B) and INSL5 (PDB: 2KBC). The receptors are models (generated with ChemDraw v15.0).
Table 1.
Physiological roles of relaxin family peptides in humans and other mammals and potential clinical significance
| Peptide–Receptor pair | Physiological role | Clinical significance |
|---|---|---|
| Relaxin 1 | Unknown | Unknown |
| Relaxin 2‐RXFP1 | Pregnancy roles | Heart disease |
| 1. Remodelling of the uterus throughout pregnancy | ||
| 2. Development of mammary nipple | Fibrosis | |
| Cardiovascular adaptations | ||
| INSL3‐RXFP2 | 1. Growth and development of the gubernaculum | Infertility |
| 2. Male and female germ cell survival | Birth control | |
| 3. Ovarian follicle function | ||
| Relaxin 3‐RXFP3 | Modulatory role in | Anxiety and depression |
| 1. Arousal | Drug addiction | |
| 2. Feeding | ||
| 3. Stress responses | ||
| 4. Cognition and drug addiction | ||
| INSL4 | Possible role in bone development | Unknown |
| INSL5‐RXFP4 | 1. Regulating appetite | Obesity |
| 2. Glucose metabolism | Anorexia | |
| Diabetes | ||
| INSL6 | Possible role in male fertility | Unknown |
All the relaxin family peptides share little sequence homology apart from the six cysteines that form three disulfide bonds (Figure 1A), and the overall fold of these peptides is similar (Figure 1B). However, the differences in their amino acid sequences have resulted in subtle changes in the length of the conserved helices and their functional selectivity towards their receptors. Detailed structure–activity relationship (SAR) studies towards understanding the role of residues in each of these chains have been reported over the years. This review summarizes the current knowledge of the SAR of H2 relaxin, H3 relaxin, INSL3 and INSL5 and how this information has been used to develop smaller peptide agonists and antagonists of RXFP receptors.
H2 relaxin
H2 relaxin is a pleiotropic hormone with key roles in reproductive and non‐reproductive processes including anti‐fibrotic (Samuel et al., 2011) and vasodilator effects (Du et al., 2010) (Table 1). Based on its cardio‐protective effects, the recombinant form of H2 relaxin is currently in Phase III clinical trials for the treatment of acute heart failure (Teerlink et al., 2009; 2013). These effects of H2 relaxin are mediated through the GPCR, RXFP1 (Hsu et al., 2002). H2 relaxin also cross‐reacts with the RXFP2 receptor (Kumagai et al., 2002), which is the cognate receptor for its related peptide, INSL3. However, most of the biological actions of H2 relaxin are thought to be mediated through RXFP1 receptors (Bathgate et al., 2013b), and there is no evidence in humans that H2 relaxin mediates any of its actions through RXFP2 receptors or that H2 relaxin is associated with the biological actions of INSL3. Rodent models are of no help in this regard as rodent relaxin does not activate the RXFP2 receptor (Bathgate et al., 2006b).
SAR studies
Mutation and truncation studies
A comparison of amino acid sequences of relaxin from different species reveals that there are three similar residues, RB13, RB17 and I/VB20, in the mid‐region of the B‐chain (Büllesbach and Schwabe, 1988). A detailed SAR study revealed the importance of RB13 and RB17 (Figure 3) for the biological effects of H2 relaxin mediated through activation of the RXFP1 receptor (Büllesbach and Schwabe, 1991; Büllesbach et al., 1992). In addition, a third amino acid, IB20, located one further helical turn away was identified, suggesting a three‐point interaction site named a ‘relaxin binding cassette’ whereby B20 can be either I (Ile) or V (Val) (RB13XXX RB17XXI/VB20) (Büllesbach and Schwabe, 2000; 2005b).
Figure 3.
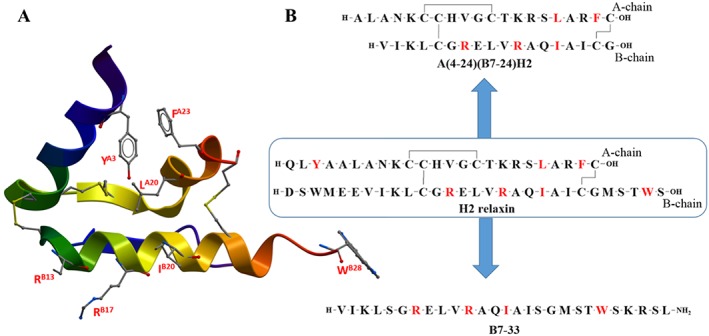
Summary of SAR of H2 relaxin. (A) X‐ray crystal structure of H2 relaxin (PDB: 6RLX) with important residue side chains (residue number labelled red). (B) Primary sequence of H2 relaxin, A(4‐24)(B7‐24)H2 and B7‐33 (important residues are coloured red).
The importance of the key residues was further highlighted by the finding that an H2 analogue, B‐R13/17K H2 relaxin (Table of Links) in which two arginine residues are substituted by lysine, showed very low affinity for RXFP1 receptors (Silvertown et al., 2007; Hossain et al., 2010). Interestingly, this peptide showed antagonistic properties in cells endogenously expressing RXFP1 receptors and in vivo (Silvertown et al., 2007; Hossain et al., 2010). This receptor‐binding cassette (RB13XXX RB17XXI/VB20) present within the mid‐region of the B‐chain is considered to be responsible for the primary binding interaction between H2 relaxin and the leucine‐rich repeat (LRR) region of the large extracellular domain of the RXFP1 receptor (Büllesbach and Schwabe, 2005b; Scott et al., 2009).
A secondary interaction involving the A‐chain of H2 relaxin and the transmembrane (TM) exoloops of the RXFP1 receptor has been suggested (Sudo et al., 2003; Hossain et al., 2008b; 2012; Diepenhorst et al., 2014; Sethi et al., 2016). Recent mutation and structural studies on RXFP1 receptors demonstrate that H2 relaxin binding to the linker between the LDLa module and the LRR domain stabilizes the α‐helical conformation of this linker and positions residues of the linker and the LDLa module to bind the TM domain to activate the RXFP1 receptor (Sethi et al., 2016). The potential A‐chain residues involved in this interaction and the potential interaction with the TM domain are unknown. Recent mutation studies of the A‐chain of the ligand indicated that binding is not driven by a single amino acid, although residues YA3, LA20 and FA23 (Figure 3) appeared to contribute (Chan et al., 2012). Interestingly, these residues are also important drivers of the affinity and activity of H2 relaxin for RXFP2 receptors with additional minor contributions from KA9, HA12, KA17, RA18 and RA22 (Chan et al., 2012) (Figure 3). A recent study also showed that, in addition to RB13XXX RB17XXIB20, the WB28 residue in the B‐chain contributes to the H2–RXFP2 but not the H2‐RXFP1 interaction (Chan et al., 2013).
Role of free C‐terminus
The effect of modifying the C‐terminus of both the A‐ and B‐chain of H2 relaxin on receptor binding and activation has been investigated (Haugaard‐Kedström et al., 2015). An amidated H2 relaxin (with both the A‐ and B‐chain C‐termini amidated) was chemically synthesized and compared with native H2 relaxin (with both the C‐termini as free acids). These peptides were found to be structurally very similar and equipotent at the RXFP1 receptor (Haugaard‐Kedström et al., 2015). Interestingly, the amide analogue did not self‐associate into the characteristic dimers, as seen in the crystal structure of native H2 relaxin (Eigenbrot et al., 1991). This indicates that the monomeric form of the hormone is pharmacologically active and that, unlike several other members of the family (Shabanpoor et al., 2013; Patil et al., 2016a), a free C‐terminus is not required for its activity on RXFP1 receptors (Haugaard‐Kedström et al., 2015).
Minimum active structure
Extensive truncation studies on H2 relaxin were undertaken to elucidate the minimum length required for its activity on the RXFP1 receptor. N‐terminal truncation of the A‐chain of up to four residues did not have a prominent effect on binding and activity. Further truncation caused reduced activity, but this was significantly restored by non‐native residue substitution (Hossain et al., 2008b). This indicated the importance of the α‐helix at the N‐termini of the A‐chain for structural integrity (Büllesbach and Schwabe, 1986; Büllesbach and Schwabe, 1987; Hossain et al., 2008b). Truncation of the B‐chain was tolerated by six residues from the N‐terminus and five residues from the C‐terminus (Hossain et al., 2011). These truncation studies resulted in the identification of A(4‐24)(B7‐24)H2 (also known as mini‐H2 relaxin) which is one‐third smaller in size (Hossain et al., 2011) compared with native H2 relaxin (Figure 3B). While this minimized peptide is a full agonist at RXFP1 receptors its potency is ~100‐fold lower than H2 relaxin (Hossain et al., 2011).
As most of the residues associated with high affinity binding are within the B‐chain, it was predicted that B‐chain‐only analogues could be developed. Preliminary single‐chain peptides were found to be notoriously insoluble and inactive (Del Borgo et al., 2005). However, by producing a B‐chain with an N‐terminal truncation and C‐terminal elongation, a soluble peptide B7‐33 was recently developed (Hossain et al., 2016) (Figure 3B). Initial testing in HEK‐293T cells overexpressing RXFP1 receptors and THP1 cells natively expressing RXFP1 receptors suggested that the peptide was a weak agonist. However, when the peptide was tested in rat renal myofibroblasts (from injured kidneys) and human cardiac fibroblasts (native RXFP1‐expressing cells) for its ability to promote matrix metalloproteinase 2 (MMP2), a collagen‐degrading enzyme, it was found to exhibit similar potency to H2 relaxin (Hossain et al., 2016). Importantly, like H2 relaxin, B7‐33 reversed lung and heart fibrosis in vivo in rat and mouse models (Hossain et al., 2016) and was a potent agonist in vivo in rodents. B7‐33 also potently activated ERK phosphorylation in both rat and human fibroblasts while only showing weak activity in HEK‐RXFP1 cells. Thus, B7‐33 seems to demonstrate fibroblast‐specific actions and may be a lead molecule for further development as a therapeutic for fibrosis and related disorders. This peptide also demonstrates that there is still much to be learnt as to the mechanism of action of relaxin in specific cell types.
H3 relaxin
H3 relaxin is a neuropeptide with its highest level of expression in the CNS, particularly in a specific nucleus in the hindbrain, the nucleus incertus (Bathgate et al., 2002). Recent work suggested that H3 relaxin has a modulatory role in arousal, feeding, stress responses, cognition and drug addiction (Table 1) (Ryan et al., 2013a,b; Smith et al., 2014). Although H3 relaxin can activate RXFP1 (Hsu et al., 2002) and RXFP4 (Liu et al., 2005b) receptors, it is believed that the actions of H3 relaxin in the brain are mediated through its cognate GPCR, RXFP3 (Liu et al., 2003). However, pharmacological studies in rodents must take the receptor cross‐reactivity into account; hence, much effort has been put into developing RXFP3 receptor‐selective agonists. Evidence from studies using such specific agonists suggest that the H3 relaxin/RXFP3 signalling system is a potential therapeutic target for psychiatric disorders, such as anxiety and depression, and drug addiction (Ryan et al., 2013a,b; Smith et al., 2014).
SAR studies
Mutation and truncation studies
There are distinct differences in the residues and modes of binding and activation of the different receptors – RXFP1, RXFP3 and RXFP4. Recent studies demonstrate that deletion of the N‐terminal seven residues of the B‐chain does not significantly affect the ligand binding activity for RXFP1, RXFP3 or RXFP4 receptors, thus confirming that the N‐terminus of the peptide does not play a critical role in binding (Kuei et al., 2007; Liu et al., 2009).
H3 relaxin binds to RXFP1 receptors using similar residues to H2 relaxin; hence, H3 relaxin contains an RXXXRXXI motif (RB12, RB16 and IB19), and mutation studies (Kuei et al., 2007; Liu et al., 2009) confirm that this is the driver of its high affinity binding to RXFP1 receptors (Büllesbach and Schwabe, 2000). In contrast, the binding of H3 relaxin to RXFP3 receptors requires two distinct domains within the peptide with the core B‐chain residues (RB8, RB12, IB15, RB16 and FB20) involved in binding and the C‐terminal RB26 and WB27 driving the activation the RXFP3 receptor (Kuei et al., 2007; Liu et al., 2009). The same residues, except RB12, are important for its interaction with the RXFP4 receptor. Figure 4A shows the solution NMR structure of H3 relaxin (Rosengren et al., 2006a) where the side chains of important residues are highlighted in red. Molecular modelling and receptor mutation studies reveal most of the complementary residues in the RXFP3 receptor. Specifically, RB12, RB16, RB26 and WB27 residues of H3 relaxin interact with E244, D145, D141 and W138 residues in the RXFP3 receptor respectively (Bathgate et al., 2013a; Hu et al., 2016b).
Figure 4.
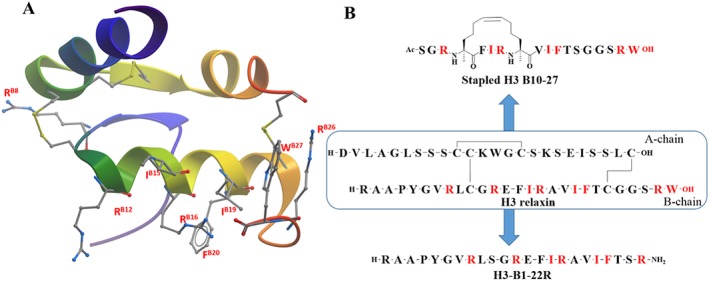
Summary of SAR of H3 relaxin. (A) NMR structure of H3 relaxin (PBD: 2FHW) with important residue side chains (residue number labelled red). (B) Primary sequences of H3 relaxin, stapled H3 B10‐27 and H3‐B1‐22R (important residues are coloured red).
In an attempt to develop a receptor‐specific agonist, the A‐chain of H3 relaxin was replaced with that from INSL5 and the resulting chimeric R3/I5 analogue exhibited selectivity for RXFP3 and RXFP4 receptors by substantially reducing the affinity for the RXFP1 receptor (Liu et al., 2005a) Additionally, by truncating the C‐terminus of the B‐chain of R3/I5 by five residues and adding a C‐terminal arginine, an RXFP3/4 antagonist, R3(BΔ23‐27)R/I5 chimeric peptide was achieved (Kuei et al., 2007).
Role of free C‐terminus
The effect of modifying the C‐terminus of H3 relaxin on receptor binding and activity has been investigated (Shabanpoor et al., 2013). Amidation of the C‐terminus of the B‐chain led to a significant drop in the binding and activity of the peptide at both RXFP3 and RXFP4 receptors. However, modification of the C‐terminus of the A‐chain did not have any effect on activity (Shabanpoor et al., 2013). It is clear that the key WB27 must be free acid and that these residues probably insert into the orthosteric binding pocket of RXFP3 and RXFP4 receptors in order to activate them. More specifically, molecular modelling and receptor mutations studies suggest that C‐terminal WB27 and RB26 residues of H3 relaxin interact with the W138 and E141 residues of the RXFP3 receptor respectively (Bathgate et al., 2013a; Hu et al., 2016b).
Minimum active structure
Extensive truncation studies on H3 relaxin were undertaken to understand the minimum length required for receptor activity. A minimized analogue of H3 relaxin, minimized relaxin‐3 analogue 2 (Table of Links) was achieved by removal of the intra‐molecular disulfide bond within the A‐chain and by truncation of the N‐terminus of the A‐chain (Shabanpoor et al., 2012). Like R3/I5, the resulting peptide also exhibited selectivity for RXFP3 and RXFP4 over RXFP1 receptors, and its activation of RXFP3 receptors in vivo led to increased feeding and significantly reduced anxiety‐like behaviour in adult rats (Shabanpoor et al., 2012).
Based on the current SAR knowledge, it is evident that the B‐chain of H3 relaxin contains all the residues important for both binding and activation of the RXFP3 receptor. This is further supported by the finding that the B‐chain alone of H3 relaxin is capable of binding to and activating the RXFP3 receptor (Liu et al., 2003), albeit with low affinity and potency. Thus, it is possible to engineer B‐chain‐only agonists and antagonists with high affinity and potency. Consequently, single‐B‐chain‐only analogues, stapled R3 B10‐27 (Hojo et al., 2016) and R3‐B1‐22R (Haugaard‐Kedström et al., 2011) have been designed and developed as a high affinity agonist and antagonist respectively. The stapled R3 B10‐27 agonist was demonstrated to be a full agonist for both cAMP inhibition and pERK activation (Hojo et al., 2016). Additionally, in vivo studies demonstrated that the stapled R3 B‐chain analogue significantly stimulated food intake to the same level as H3 relaxin (Hojo et al., 2016). In contrast, R3‐B1‐22R antagonized relaxin‐3/RXFP‐induced increase in feeding (Haugaard‐Kedström et al., 2011). Thus, stapled R3 B10‐27 and B1‐22R represent excellent tools for probing the pharmacology of H3 relaxin‐RXFP3 and are lead molecules for further development for potential clinical use.
Human INSL3
INSL3 is highly expressed in the Leydig cells of the testis (Adham, Burkhardt, Benahmed & Engel, 1993) and has a critical role in testis descent. INSL3 knockout mice are cryptorchid and as a result infertile (Nef and Parada, 1999). INSL3 plays an important role in the development of the gubernaculum and also seems to have a role in the maintenance of fertility in females (Spanel‐Borowski et al., 2001; Glister et al., 2013). The effects of INSL3 are mediated through the GPCR, RXFP2 (Kumagai et al., 2002).
SAR studies
Mutation and truncation studies
INSL3 like H2 relaxin binds to the LRR domain of the RXFP2 receptor using B‐chain residues, although these residues are different from those used by H2 relaxin to bind to RXFP1 receptors. More specifically, the residues HB12, RB16, VB19, RB20 and WB27 present are responsible for RXFP2 receptor binding (Büllesbach and Schwabe, 2006; Rosengren et al., 2006b). The corresponding residues in the LLR domain of the RXFP2 receptor that interact with the B‐chain of INSL3 were also determined and characterized by receptor mutation studies (Scott et al., 2007). INSL3 also requires the A‐chain for activity although unlike H2 relaxin the A‐chain contains a domain that is essential for activation but not binding. Hence, while no single amino acid side chain within the INSL3 A‐chain was found to be critical for RXFP2 receptor binding and activity, the potency of the INSL3 peptide was found to be driven predominantly by a cluster of four N‐terminal residues (residues A6–A9) in the A‐chain (Bathgate et al., 2012; Büllesbach and Schwabe, 2005a). Additionally, the amide bond between residues 8 and 9 (RA8 and YA9) at the N‐terminus of the A‐chain was reported to be important for activation of the RXFP2 receptor (Büllesbach and Schwabe, 2012). Truncations at the N‐terminus of INSL3 produced a high affinity RXFP2 antagonist (Büllesbach and Schwabe, 2005a). Figure 5 shows the solution of the NMR structure of INSL3 (Rosengren et al., 2006b).
Figure 5.
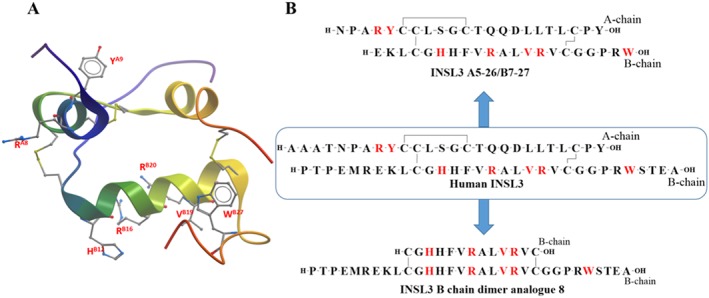
Summary of SAR of human INSL3. (A) NMR structure of human INSL3 (PDB: 2H8B) with important residue side chains (residue number labelled red). (B) Primary sequence of human INSL3, INSL3 A5‐26/B7‐27 and INSL3 B chain dimer analogue 8 (important residues are coloured red).
It is intriguing that H2 relaxin binds to and activates both RXFP1 and RXFP2 receptors whereas INSL3 is a selective ligand for RXFP2 receptors. Molecular modelling and receptor mutation studies suggest that H2 relaxin binds to the RXFP2 receptor utilizing a hybrid H2 relaxin/INSL3 binding site comprising some of the INSL3–RXFP2 interactions and also utilizing the partially conserved RXFP1–H2 relaxin binding site in the RXFP2 receptor (Scott et al., 2009).
Minimum active structure
The SAR data suggest that parts of both the A and B‐chain are necessary for agonistic activity on the RXFP2 receptor. Consequently, a minimized INSL3 was designed and synthesized, INSL3 A5‐26/B7‐27 (Figure 5), that retained near native agonistic activity (Bathgate et al., 2012). This peptide contains a truncated A‐ and B‐chain and is almost one‐quarter smaller than the native peptide and thus easier and cheaper to make and represents a potential lead peptide for further development as a fertility regulator.
As the B‐chain contains the key residues for binding the LRR domain, INSL3 B‐chain analogues have been designed and developed as functional antagonists. The lack of structural support from the A‐chain resulted in poor affinity of the B‐chain‐specific antagonist. Thus, a synthetic parallel dimer of the INSL3 B‐chain was developed that exhibited high affinity for the RXFP2 receptor (Shabanpoor et al., 2011) and antagonized INSL3‐mediated cAMP signalling through this receptor. Further refinement by truncation of 18 residues yielded a minimized analogue, INSL3:B10‐24/B1‐31, (Figure 5) that retained full binding affinity and INSL3 antagonism. This is an attractive lead for in vivo evaluation as an inhibitor of male and female fertility (Del Borgo et al., 2006).
Human INSL5
INSL5 was originally identified as a novel peptide hormone that is highly expressed in the gut (Conklin et al., 1999). Subsequent studies identified its cognate receptor as the GPCR, RXFP4 (Liu et al., 2005b). It is a hormonal product of colonic L‐cells, and recent data suggest that INSL5 is an orexigenic hormone that plays a physiological role in driving food intake (Table 1) (Grosse et al., 2014). In addition, another recent study suggests that INSL5 augments glucose‐stimulated insulin secretion from MIN6 pancreatic beta cells and GLP‐1 release from murine enteroendocrine GLUTag cells, indicating that INSL5–RXFP4 interaction may constitute a novel incretin axis involved in the regulation of metabolism and energy balance (Luo et al., 2015).
SAR study
Mutation and truncation studies
Human INSL5 is one of the most difficult peptides to produce chemically as both A and B chains were unusually resistant to standard synthesis protocols and required highly optimized conditions for their production (Hossain et al., 2008a). A preliminary SAR study, therefore, was carried out using a mouse homologue of INSL5, which was thought to be easier to assemble chemically for SAR studies. Interestingly, mouse INSL5 was found to be more potent than human INSL5 on human RXFP4 receptors (Belgi et al., 2011). On the basis of the H3 relaxin‐RXFP3 structure–function studies, two analogues of mouse INSL5 were produced (Belgi et al., 2013a,b). The analogue in which RB24 and WB25 were substituted with alanine completely abolished its interaction with the RXFP4 receptor (Belgi et al., 2013a,b). This observation is somewhat different to the H3 relaxin–RXFP3 interaction where mutation at equivalent residues of the ligand (or deletion of these residues) only partially reduced binding, but abolished activation (Kuei et al., 2007). A second analogue from this study in which three residues KB6, RB13 and YB18 were substituted with alanine showed a significant loss in activity, which correlated with binding, suggesting that KB6, RB13 and YB18 each play an important role in the binding of INSL5 to the RXFP4 receptor.
To elucidate the mode of interaction of INSL5 with the RXFP4 receptor, a charge‐exchange mutagenesis study was performed where the positively charged residues in the INSL5 B‐chain were substituted with negatively charged residues (Wang et al., 2014). This study confirmed that RB13 and RB23 play important roles in INSL5 binding to RXFP4 receptors and its activity. The solution for the NMR structure (Haugaard‐Jönsson et al., 2009) indicates that the B‐chain of INSL5 has an extended C‐terminal α‐helix compared with H3 relaxin (Figure 6A). In order to investigate the role of this extended α‐helix, an analogue where AB20 and SB21 were substituted by glycine was studied. The results indicated that a rigid α‐helical conformation at the C‐terminus of INSL5 B‐chain is important for its interaction with the RXFP4 receptor (Wang et al., 2014). While H3 relaxin cross‐reacts with other receptors including RXFP4 receptors, INSL5 is very selective (RXFP4) and does not interact with RXFP3 receptors. The mechanism by which INSL5 distinguishes RXFP4 and RXFP3 receptors has recently been identified (Hu et al., 2016a). The authors have identified four determinants (EB2, LB9, YB17 and a rigid B‐chain C‐terminus) on INSL5 that are responsible for its inactivity at RXFP3 receptors (Hu et al., 2016a).
Figure 6.
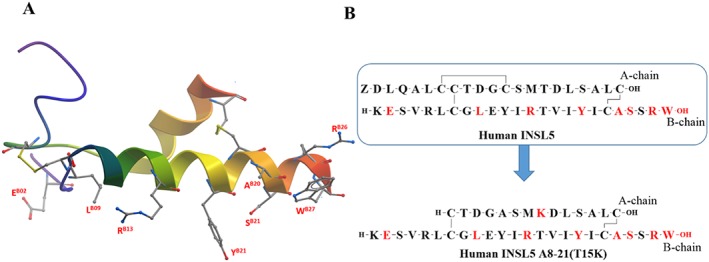
Summary of SAR of human INSL5. (A) NMR structure of human INSL5 (PDB: 2KBC) with important residue side chains (residue number labelled red). (B) Primary sequences of human INSL5 and hINSL5 A8‐21(T15K) (important residues are coloured red).
Role of the C‐terminus
The role of the C‐terminus of INSL5 in its binding to and activation of the RXFP4 receptor has been investigated (Belgi et al., 2011; Patil et al., 2016a). Modification of the C‐terminus of the A‐chain did not have any effect on activation. However, the free C‐terminus of the B‐chain is important for its activation of the RXFP4 receptor (Patil et al., 2016a), similar to what was seen for H3 relaxin at RXFP3 receptors. The loss of binding and activation due to C‐terminal WB25 mutation (Belgi et al., 2013a,b) or amidation (Patil et al., 2016a) suggest that the WB25 residue cannot be replaced or modified.
Minimum active structure
The recent SAR studies on mouse INSL5 led to the design and synthesis of a simplified mouse analogue (mouse INSL5: A8‐21) (Belgi et al., 2013a,b). This analogue showed similar affinity and potency to human INSL5. However, the human orthologue of this peptide (minimized INSL5 analogue 7; Table of Links)) was found to be significantly less potent (Belgi et al., 2013a,b). A specific residue, KA15, that contributed to the high potency of mouse INSL5 was identified, and a minimized human INSL5 orthologue human INSL5:A8‐21K15 was engineered (Figure 6B) (Patil et al., 2016b). This analogue exhibited near native hINSL5‐like affinity for the RXFP4 receptor and was a potent full agonist of both cAMP inhibition and pERK activation. The minimized analogue is much easier to assemble in large quantities and thus very useful as a readily available tool to analyse the physiological role of RXFP4 receptors.
SAR of H1 relaxin, INSL4 and INSL6
The physiological role of H1 relaxin is unclear. To date, a native human relaxin‐1 has not been isolated. However, a synthetic H1 relaxin was shown to have similar biological properties and potency to H2 relaxin at the RXFP1 receptor (Tan et al., 1998). There is little information available for INSL4–6, and also, their target receptors are yet to be identified. INSL4 is highly expressed in the placenta (Chassin et al., 1995) and was shown to play a role in bone development (Laurent et al., 1998). INSL6 is found predominantly in the testis (Lok et al., 2000), and INSL6 knockout mice showed a marked reduction in sperm numbers (Burnicka‐Turek et al., 2009) and motility suggesting a potential role of INSL6 in male fertility.
Conclusion
There is a considerable similarity in the tertiary structure between members of the relaxin family of peptides. However, the different peptides clearly utilize distinct residues and modes of interaction with their native receptors. Detailed SAR studies have identified key residues in the A‐ and B‐chain of the peptides involved in both binding and activation. The B‐chain of H3 relaxin is the sole determinant for its binding to and activation of RXFP3 receptors, and thus, it was possible to replicate the B‐chain and achieve a high affinity single‐chain agonist and antagonist. While H3 relaxin binds to and activates both RXFP3 and RXFP4 receptors in a similar manner, INSL5 interacts with the RXFP4 receptor in a different way. Although very little is known about the interaction of INSL5 with the RXFP4 receptor, current SAR data suggest that the B‐chain is responsible for both its binding to and activation of the RXFP4 receptor. Although a simplified RXFP4 agonist has been developed based on both the A‐ and B‐chain, a single‐chain RXFP4 ligand with high affinity might be not very far from reality. The B‐chain of INSL3 is important for its binding to the RXFP2 receptor, and the A‐chain is important for activation of this receptor. As a result, high affinity agonists with minimized A‐ and B‐chains and antagonists with B‐chain dimers have been developed. The interaction of H2 relaxin with the RXFP1 receptor has proved to be much more complex. Studies over many years have suggested that both the A‐ and B‐chain are essential for activity, which has made it difficult to produce smaller agonists and antagonists. However, the recent discovery that a B‐chain only analogue is a potent cell‐specific agonist of the RXFP1 receptor highlights that there is still a lot to learn about the interaction of H2 relaxin with the RXFP1 receptor. Further work in this area must take into account the clear cell type‐specific actions of relaxin analogues.
Author contributions
N.A.P, K.J.R., J.D.W., R.A.D.B. and M.A.H. wrote the manuscript. F.S. revised and edited the manuscript. R.A.D.B. and M.A.H. designed the lay out and sequences of the headlines of the manuscript. N.A.P and M.A.H. designed and made the figures.
Conflict of interest
The authors declare no conflicts of interest.
Acknowledgements
This research was partly funded by NHMRC (Australia) project grants (1023321, 1065481 and 1023078) and ARC linkage grant (LP120100654) to M.A.H., R.A.D.B., J.K.R. and J.D.W., a Florey Foundation Fellowship and Melbourne Research Grant Support Scheme (RGSS) awarded to M.A.H., a University of Melbourne Postgraduate Scholarship and Albert Shimmins postgraduate writing‐up award to N.A.P., Australian Future Fellowships to K.J.R. (FT130100890), an NHMRC Principal Research Fellowship to J.D.W. (GNT5018148) and NHMRC Senior Research Fellowship to R.A.D.B. (GNT1042650). We acknowledge financial support from Assoc. Prof. Anthony Hughes (Melbourne University) and Dr Johannes Grosse (Takeda Cambridge) through the Linkage grant project. Research at The Florey Institute of Neuroscience and Mental Health is supported by the Victorian Government Operational Infrastructure Support Program.
Patil, N. A. , Rosengren, K. J. , Separovic, F. , Wade, J. D. , Bathgate, R. A. D. , and Hossain, M. A. (2017) Relaxin family peptides: structure–activity relationship studies. British Journal of Pharmacology, 174: 950–961. doi: 10.1111/bph.13684.
Contributor Information
Ross A D Bathgate, Email: bathgate@florey.edu.au.
Mohammed Akhter Hossain, Email: akhter.hossain@florey.edu.au.
References
- Adham IM, Burkhardt E, Benahmed M, Engel W (1993). Cloning of a cDNA for a novel insulin‐like peptide of the testicular Leydig cells. J Biol Chem 268: 26668–26672. [PubMed] [Google Scholar]
- Alexander SPH, Davenport AP, Kelly E, Marrion N, Peters JA, Benson HE et al. (2015a). The Concise Guide to PHARMACOLOGY 2015/16: G protein‐coupled receptors. Br J Pharmacol 172: 5744–5861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander SPH, Fabbro D, Kelly E, Marrion N, Peters JA, Benson HE et al. (2015b). The Concise Guide to PHARMACOLOGY 2015/16: Enzymes. Br J Pharmacol 172: 6024–6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bathgate R, Oh M, Ling J, Kaas Q, Hossain A, Gooley P et al. (2013b). Elucidation of relaxin‐3 binding interactions in the extracellular loops of RXFP3. Front Endocrinol 4: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bathgate RA, Ivell R, Sanborn BM, Sherwood OD, Summers RJ (2006a). International Union of Pharmacology LVII: recommendations for the nomenclature of receptors for relaxin family peptides. Pharmacol Rev 58: 7–31. [DOI] [PubMed] [Google Scholar]
- Bathgate RA, Lin F, Hanson NF, Otvos L Jr, Guidolin A, Giannakis C et al. (2006b). Relaxin‐3: improved synthesis strategy and demonstration of its high‐affinity interaction with the relaxin receptor LGR7 both in vitro and in vivo. Biochemistry 45: 1043–1053. [DOI] [PubMed] [Google Scholar]
- Bathgate RA, Samuel CS, Burazin TC, Layfield S, Claasz AA, Reytomas IG et al. (2002). Human relaxin gene 3 (H3) and the equivalent mouse relaxin (M3) gene. Novel members of the relaxin peptide family. J Biol Chem 277: 1148–1157. [DOI] [PubMed] [Google Scholar]
- Bathgate RA, Zhang S, Hughes RA, Rosengren KJ, Wade JD (2012). The structural determinants of insulin‐like peptide 3 activity. Front Endocrinol 3: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bathgate RAD, Halls ML, van der Westhuizen ET, Callander GE, Kocan M, Summers RJ (2013a). Relaxin family peptides and their receptors. Physiol Rev 93: 405–480. [DOI] [PubMed] [Google Scholar]
- Belgi A, Bathgate RA, Kocan M, Patil N, Zhang S, Tregear GW et al. (2013a). Minimum active structure of insulin‐like peptide 5. J Med Chem 56: 9509–9516. [DOI] [PubMed] [Google Scholar]
- Belgi A, Bathgate RAD, Tregear GW, Wade JD, Akhter Hossain M (2013b). Preliminary structure–function relationship studies on insulin‐like peptide 5 (INSL5). Int J Pept Res Ther 19: 71–79. [Google Scholar]
- Belgi A, Hossain MA, Shabanpoor F, Chan L, Zhang S, Bathgate RAD et al. (2011). Structure and function relationship of murine insulin‐like peptide 5 (INSL5): free C‐terminus is essential for RXFP4 receptor binding and activation. Biochemistry 50: 8352–8361. [DOI] [PubMed] [Google Scholar]
- Büllesbach EE, Schwabe C (1986). Preparation and properties of porcine relaxin derivatives shortened at the amino terminus of the A chain. Biochemistry 25: 5998–6004. [DOI] [PubMed] [Google Scholar]
- Büllesbach EE, Schwabe C (1987). Relaxin structure. Quasi allosteric effect of the NH2‐terminal A‐chain helix. J Biol Chem 262: 12496–12501. [PubMed] [Google Scholar]
- Büllesbach EE, Schwabe C (1988). On the receptor binding site of relaxins. Int J Pept Protein Res 32: 361–367. [DOI] [PubMed] [Google Scholar]
- Büllesbach EE, Schwabe C (1991). Total synthesis of human relaxin and human relaxin derivatives by solid‐phase peptide synthesis and site‐directed chain combination. J Biol Chem 266: 10754–10761. [PubMed] [Google Scholar]
- Büllesbach EE, Schwabe C (2000). The relaxin receptor‐binding site geometry suggests a novel gripping mode of interaction. J Biol Chem 275: 35276–35280. [DOI] [PubMed] [Google Scholar]
- Büllesbach EE, Schwabe C (2002). The primary structure and the disulfide links of the bovine relaxin‐like factor (RLF). Biochemistry 41: 274–281. [DOI] [PubMed] [Google Scholar]
- Büllesbach EE, Schwabe C (2005a). LGR8 signal activation by the relaxin‐like factor. J Biol Chem 280: 14586–14590. [DOI] [PubMed] [Google Scholar]
- Büllesbach EE, Schwabe C (2005b). The trap‐like relaxin‐binding site of the leucine‐rich G‐protein‐coupled receptor 7. J Biol Chem 280: 14051–14056. [DOI] [PubMed] [Google Scholar]
- Büllesbach EE, Schwabe C (2006). The mode of interaction of the relaxin‐like factor (RLF) with the leucine‐rich repeat G protein‐activated receptor 8. J Biol Chem 281: 26136–26143. [DOI] [PubMed] [Google Scholar]
- Büllesbach EE, Schwabe C (2012). Replacement of disulfides by amide bonds in the relaxin‐like factor (RLF/INSL3) reveals a role for the A11‐B10 link in transmembrane signaling. Biochemistry 51: 4198–4205. [DOI] [PubMed] [Google Scholar]
- Büllesbach EE, Yang S, Schwabe C (1992). The receptor‐binding site of human relaxin II. A dual prong‐binding mechanism. J Biol Chem 267: 22957–22960. [PubMed] [Google Scholar]
- Burnicka‐Turek O, Shirneshan K, Paprotta I, Grzmil P, Meinhardt A, Engel W et al. (2009). Inactivation of insulin‐like factor 6 disrupts the progression of spermatogenesis at late meiotic prophase. Endocrinology 150: 4348–4357. [DOI] [PubMed] [Google Scholar]
- Chan L, Wade J, Separovic F, Bathgate RD, Hossain M (2013). The importance of tryptophan B28 in H2 relaxin for RXFP2 binding and activation. Int J Pept Res Ther 19: 55–60. [Google Scholar]
- Chan LJ, Rosengren KJ, Layfield SL, Bathgate RAD, Separovic F, Samuel CS et al. (2012). Identification of key residues essential for the structural fold and receptor selectivity within the A‐chain of human gene‐2 (H2) relaxin. J Biol Chem 287: 41152–41164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang GTG, Steenbeek M, Schippers E, Blok LJ, van Weerden WM, van Alewijk DCJG et al. (2000). Characterization of a zinc‐finger protein and its association with apoptosis in prostate cancer cells. J Natl Cancer Inst 92: 1414–1421. [DOI] [PubMed] [Google Scholar]
- Chassin D, Laurent A, Janneau JL, Berger R, Bellet D (1995). Cloning of a new member of the insulin gene superfamily (INSL4) expressed in human placenta. Genomics 29: 465–470. [DOI] [PubMed] [Google Scholar]
- Conklin D, Lofton‐Day CE, Haldeman BA, Ching A, Whitmore TE, Lok S et al. (1999). Identification of INSL5, a new member of the insulin superfamily. Genomics 60: 50–56. [DOI] [PubMed] [Google Scholar]
- Crawford RJ, Hudson P, Shine J, Niall HD, Eddy RL, Shows TB (1984). Two human relaxin genes are on chromosome 9. EMBO J 3: 2341–2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Borgo MP, Hughes RA, Bathgate RAD, Lin F, Kawamura K, Wade JD (2006). Analogs of insulin‐like peptide 3 (INSL3) B‐chain are LGR8 antagonists in vitro and in vivo. J Biol Chem 281: 13068–13074. [DOI] [PubMed] [Google Scholar]
- Del Borgo MP, Hughes RA, Wade JD (2005). Conformationally constrained single‐chain peptide mimics of relaxin B‐chain secondary structure. J Pept Sci 11: 564–571. [DOI] [PubMed] [Google Scholar]
- Diepenhorst NA, Petrie EJ, Chen CZ, Wang A, Hossain MA, Bathgate RAD et al. (2014). Investigation of interactions at the extracellular loops of the relaxin family peptide receptor 1 (RXFP1). J Biol Chem 289: 34938–34952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X‐J, Bathgate RAD, Samuel CS, Dart AM, Summers RJ (2010). Cardiovascular effects of relaxin: from basic science to clinical therapy. Nat Rev Cardiol 7: 48–58. [DOI] [PubMed] [Google Scholar]
- Eigenbrot C, Randal M, Quan C, Burnier J, O'Connell L, Rinderknecht E et al. (1991). X‐ray structure of human relaxin at 1.5 A. Comparison to insulin and implications for receptor binding determinants. J Mol Biol 221: 15–21. [PubMed] [Google Scholar]
- Glister C, Satchell L, Bathgate RAD, Wade JD, Dai Y, Ivell R et al. (2013). Functional link between bone morphogenetic proteins and insulin‐like peptide 3 signaling in modulating ovarian androgen production. Proc Natl Acad Sci 110: E1426–E1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosse J, Heffron H, Burling K, Akhter Hossain M, Habib AM, Rogers GJ et al. (2014). Insulin‐like peptide 5 is an orexigenic gastrointestinal hormone. Proc Natl Acad Sci 111: 11133–11138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley J, Hudson P, Scanlon D, John M, Cronk M, Shine J et al. (1982). Porcine relaxin: molecular cloning and cDNA structure. DNA 1: 155–162. [DOI] [PubMed] [Google Scholar]
- Halls ML, van der Westhuizen ET, Bathgate RA, Summers RJ (2007). Relaxin family peptide receptors‐‐former orphans reunite with their parent ligands to activate multiple signalling pathways. Br J Pharmacol 150: 677–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugaard‐Jönsson LM, Hossain MA, Daly NL, Craik DJ, Wade JD, Rosengren KJ (2009). Structure of human insulin‐like peptide 5 and characterization of conserved hydrogen bonds and electrostatic interactions within the relaxin framework. Biochem J 419: 619–627. [DOI] [PubMed] [Google Scholar]
- Haugaard‐Kedström LM, Hossain MA, Daly NL, Bathgate RAD, Rinderknecht E, Wade JD et al. (2015). Solution structure, aggregation behavior, and flexibility of human relaxin‐2. ACS Chem Biol 10: 891–900. [DOI] [PubMed] [Google Scholar]
- Haugaard‐Kedström LM, Shabanpoor F, Hossain MA, Clark RJ, Ryan PJ, Craik DJ et al. (2011). Design, synthesis, and characterization of a single‐chain peptide antagonist for the relaxin‐3 receptor RXFP3. J Am Chem Soc 133: 4965–4974. [DOI] [PubMed] [Google Scholar]
- Hojo K, Hossain MA, Tailhades J, Shabanpoor F, Wong LLL, Ong‐Palsson EEK et al. (2016). Development of a single‐chain peptide agonist of the relaxin‐3 receptor using hydrocarbon stapling. J Med Chem 59: 7445–7456. [DOI] [PubMed] [Google Scholar]
- Hossain MA, Bathgate RAD, Kong CK, Shabanpoor F, Zhang S, Haugaard‐Jönsson LM et al. (2008a). Synthesis, conformation, and activity of human insulin‐like peptide 5 (INSL5). Chembiochem 9: 1816–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain MA, Kocan M, Yao ST, Royce SG, Nair VB, Siwek C et al. (2016). A single‐chain derivative of the relaxin hormone is a functionally selective agonist of the G protein‐coupled receptor, RXFP1. Chem Sci 7: 3805–3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain MA, Rosengren KJ, Haugaard‐Jonsson LM, Zhang S, Layfield S, Ferraro T et al. (2008b). The A‐chain of human relaxin family peptides has distinct roles in the binding and activation of the different relaxin family peptide receptors. J Biol Chem 283: 17287–17297. [DOI] [PubMed] [Google Scholar]
- Hossain MA, Rosengren KJ, Samuel CS, Shabanpoor F, Chan LJ, Bathgate RA et al. (2011). The minimal active structure of human relaxin‐2. J Biol Chem 286: 37555–37565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain MA, Samuel CS, Binder C, Hewitson TD, Tregear GW, Wade JD et al. (2010). The chemically synthesized human relaxin‐2 analog, B‐R13/17K H2, is an RXFP1 antagonist. Amino Acids 39: 409–416. [DOI] [PubMed] [Google Scholar]
- Hossain MA, Wade JD, Bathgate RA (2012). Chimeric relaxin peptides highlight the role of the A‐chain in the function of H2 relaxin. Peptides 35: 102–106. [DOI] [PubMed] [Google Scholar]
- Hsu SY (2003). New insights into the evolution of the relaxin‐LGR signaling system. Trends Endocrinol Metab 14: 303–309. [DOI] [PubMed] [Google Scholar]
- Hsu SY, Nakabayashi K, Nishi S, Kumagai J, Kudo M, Sherwood OD et al. (2002). Activation of orphan receptors by the hormone relaxin. Science 295: 671–674. [DOI] [PubMed] [Google Scholar]
- Hu M‐J, Shao X‐X, Wang J‐H, Wei D, Guo Y‐Q, Liu Y‐L et al. (2016a). Mechanism for insulin‐like peptide 5 distinguishing the homologous relaxin family peptide receptor 3 and 4. Sci Rep 6: 29648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M‐J, Shao X‐X, Wang J‐H, Wei D, Liu Y‐L, Xu Z‐G et al. (2016b). Identification of hydrophobic interactions between relaxin‐3 and its receptor RXFP3: implication for a conformational change in the B‐chain C‐terminus during receptor binding. Amino Acids : 1–10. [DOI] [PubMed] [Google Scholar]
- Hudson P, Haley J, Cronk M, Shine J, Niall H (1981). Molecular cloning and characterization of cDNA sequences coding for rat relaxin. Nature 291: 127–131. [DOI] [PubMed] [Google Scholar]
- Hudson P, Haley J, John M, Cronk M, Crawford R, Haralambidis J et al. (1983). Structure of a genomic clone encoding biologically active human relaxin. Nature 301: 628–631. [DOI] [PubMed] [Google Scholar]
- Kuei C, Sutton S, Bonaventure P, Pudiak C, Shelton J, Zhu J et al. (2007). R3(BDelta23 27)R/I5 chimeric peptide, a selective antagonist for GPCR135 and GPCR142 over relaxin receptor LGR7: in vitro and in vivo characterization. J Biol Chem 282: 25425–25435. [DOI] [PubMed] [Google Scholar]
- Kumagai J, Hsu SY, Matsumi H, Roh JS, Fu P, Wade JD et al. (2002). INSL3/Leydig insulin‐like peptide activates the LGR8 receptor important in testis descent. J Biol Chem 277: 31283–31286. [DOI] [PubMed] [Google Scholar]
- Laurent A, Rouillac C, Delezoide AL, Giovangrandi Y, Vekemans M, Bellet D et al. (1998). Insulin‐like 4 (INSL4) gene expression in human embryonic and trophoblastic tissues. Mol Reprod Dev 51: 123–129. [DOI] [PubMed] [Google Scholar]
- Liu C, Chen J, Kuei C, Sutton S, Nepomuceno D, Bonaventure P et al. (2005a). Relaxin‐3/insulin‐like peptide 5 chimeric peptide, a selective ligand for G protein‐coupled receptor (GPCR)135 and GPCR142 over leucine‐rich repeat‐containing G protein‐coupled receptor 7. Mol Pharmacol 67: 231–240. [DOI] [PubMed] [Google Scholar]
- Liu C, Eriste E, Sutton S, Chen J, Roland B, Kuei C et al. (2003). Identification of relaxin‐3/INSL7 as an endogenous ligand for the orphan G‐protein‐coupled receptor GPCR135. J Biol Chem 278: 50754–50764. [DOI] [PubMed] [Google Scholar]
- Liu C, Kuei C, Sutton S, Chen J, Bonaventure P, Wu J et al. (2005b). INSL5 is a high affinity specific agonist for GPCR142 (GPR100). J Biol Chem 280: 292–300. [DOI] [PubMed] [Google Scholar]
- Liu C, Kuei C, Sutton S, Shelton J, Zhu J, Nepomuceno D et al. (2009). Probing the functional domains of relaxin‐3 and the creation of a selective antagonist for RXFP3/GPCR135 over relaxin receptor RXFP1/LGR7. Ann N Y Acad Sci 1160: 31–37. [DOI] [PubMed] [Google Scholar]
- Lok S, Johnston DS, Conklin D, Lofton‐Day CE, Adams RL, Jelmberg AC et al. (2000). Identification of INSL6, a new member of the insulin family that is expressed in the testis of the human and rat. Biol Reprod 62: 1593–1599. [DOI] [PubMed] [Google Scholar]
- Luo X, Li T, Zhu Y, Dai Y, Zhao J, Guo Z‐Y et al. (2015). The insulinotrophic effect of insulin‐like peptide 5 in vitro and in vivo. Biochem J 466: 467–473. [DOI] [PubMed] [Google Scholar]
- Nef S, Parada LF (1999). Cryptorchidism in mice mutant for Insl3. Nat Genet 22: 295–299. [DOI] [PubMed] [Google Scholar]
- Patil NA, Bathgate RAD, Kocan M, Ang SY, Tailhades J, Separovic F et al. (2016a). The C‐terminus of the B‐chain of human insulin‐like peptide 5 is critical for cognate RXFP4 receptor activity. Amino Acids 48: 987–992. [DOI] [PubMed] [Google Scholar]
- Patil NA, Hughes RA, Rosengren KJ, Kocan M, Ang SY, Tailhades J et al. (2016b). Engineering of a novel simplified human insulin‐like peptide 5 agonist. J Med Chem 59: 2118–2125. [DOI] [PubMed] [Google Scholar]
- Rosengren KJ, Lin F, Bathgate RA, Tregear GW, Daly NL, Wade JD et al. (2006a). Solution structure and novel insights into the determinants of the receptor specificity of human relaxin‐3. J Biol Chem 281: 5845–5851. [DOI] [PubMed] [Google Scholar]
- Rosengren KJ, Zhang S, Lin F, Daly NL, Scott DJ, Hughes RA et al. (2006b). Solution structure and characterization of the LGR8 receptor binding surface of insulin‐like peptide 3. J Biol Chem 281: 28287–28295. [DOI] [PubMed] [Google Scholar]
- Ryan PJ, Büchler E, Shabanpoor F, Hossain MA, Wade JD, Lawrence AJ et al. (2013a). Central relaxin‐3 receptor (RXFP3) activation decreases anxiety‐ and depressive‐like behaviours in the rat. Behav Brain Res 244: 142–151. [DOI] [PubMed] [Google Scholar]
- Ryan PJ, Kastman HE, Krstew EV, Rosengren KJ, Hossain MA, Churilov L et al. (2013b). Relaxin‐3/RXFP3 system regulates alcohol‐seeking. Proc Natl Acad Sci 110: 20789–20794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel CS, Cendrawan S, Gao X‐M, Ming Z, Zhao C, Kiriazis H et al. (2011). Relaxin remodels fibrotic healing following myocardial infarction. Lab Invest 91: 675–690. [DOI] [PubMed] [Google Scholar]
- Schwabe C, McDonald JK (1977). Relaxin: a disulfide homolog of insulin. Science 197: 914–915. [DOI] [PubMed] [Google Scholar]
- Scott DJ, Tregear GW, Bathgate RA (2009). Modeling the primary hormone‐binding site of RXFP1 and RXFP2. Ann N Y Acad Sci 1160: 74–77. [DOI] [PubMed] [Google Scholar]
- Scott DJ, Wilkinson TN, Zhang S, Ferraro T, Wade JD, Tregear GW et al. (2007). Defining the LGR8 residues involved in binding insulin‐like peptide 3. Mol Endocrinol 21: 1699–1712. [DOI] [PubMed] [Google Scholar]
- Sethi A, Bruell S, Patil N, Hossain MA, Scott DJ, Petrie EJ et al. (2016). The complex binding mode of the peptide hormone H2 relaxin to its receptor RXFP1. Nat Commun 7: 11344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabanpoor F, Akhter Hossain M, Ryan PJ, Belgi A, Layfield S, Kocan M et al. (2012). Minimization of human relaxin‐3 leading to high‐affinity analogues with increased selectivity for relaxin‐family peptide 3 receptor (RXFP3) over RXFP1. J Med Chem 55: 1671–1681. [DOI] [PubMed] [Google Scholar]
- Shabanpoor F, Bathgate RAD, Wade JD, Hossain MA (2013). C‐Terminus of the B‐chain of relaxin‐3 is important for receptor activity. PLoS One 8: e82567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabanpoor F, Zhang S, Hughes RA, Hossain MA, Layfield S, Ferraro T et al. (2011). Design and development of analogues of dimers of insulin‐like peptide 3 B‐chain as high‐affinity antagonists of the RXFP2 receptor. Biopolymers 96: 81–87. [DOI] [PubMed] [Google Scholar]
- Silvertown JD, Symes JC, Neschadim A, Nonaka T, Kao JC, Summerlee AJ et al. (2007). Analog of H2 relaxin exhibits antagonistic properties and impairs prostate tumor growth. FASEB J 21: 754–765. [DOI] [PubMed] [Google Scholar]
- Smith CM, Walker AW, Hosken IT, Chua BE, Zhang C, Haidar M et al. (2014). Relaxin‐3/RXFP3 networks: an emerging target for the treatment of depression and other neuropsychiatric diseases? Front Pharmacol 5: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southan C, Sharman JL, Benson HE, Faccenda E, Pawson AJ, Alexander SP et al. (2016). The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. Nucleic Acids Res 44: D1054–D1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanel‐Borowski K, Schäfer I, Zimmermann S, Engel W, Adham IM (2001). Increase in final stages of follicular atresia and premature decay of corpora lutea in Insl3‐deficient mice. Mol Reprod Dev 58: 281–286. [DOI] [PubMed] [Google Scholar]
- Sudo S, Kumagai J, Nishi S, Layfield S, Ferraro T, Bathgate RA et al. (2003). H3 relaxin is a specific ligand for LGR7 and activates the receptor by interacting with both the ectodomain and the exoloop 2. J Biol Chem 278: 7855–7862. [DOI] [PubMed] [Google Scholar]
- Tan YY, Wade JD, Tregear GW, Summers RJ (1998). Comparison of relaxin receptors in rat isolated atria and uterus by use of synthetic and native relaxin analogues. Br J Pharmacol 123: 762–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teerlink JR, Cotter G, Davison BA, Felker GM, Filippatos G, Greenberg BH et al. (2013). Serelaxin, recombinant human relaxin‐2, for treatment of acute heart failure (RELAX‐AHF): a randomised, placebo‐controlled trial. Lancet 381: 29–39. [DOI] [PubMed] [Google Scholar]
- Teerlink JR, Metra M, Felker GM, Ponikowski P, Voors AA, Weatherley BD et al. (2009). Relaxin for the treatment of patients with acute heart failure (Pre‐RELAX‐AHF): a multicentre, randomised, placebo‐controlled, parallel‐group, dose‐finding phase IIb study. Lancet 373: 1429–1439. [DOI] [PubMed] [Google Scholar]
- Wang X‐Y, Guo Y‐Q, Shao X‐X, Liu Y‐L, Xu Z‐G, Guo Z‐Y (2014). Identification of important residues of insulin‐like peptide 5 and its receptor RXFP4 for ligand–receptor interactions. Arch Biochem Biophys 558: 127–132. [DOI] [PubMed] [Google Scholar]
- Wilkinson TN, Bathgate RA (2007). The evolution of the relaxin peptide family and their receptors. Adv Exp Med Biol 612: 1–13. [DOI] [PubMed] [Google Scholar]
- Wilkinson TN, Speed TP, Tregear GW, Bathgate RA (2005). Evolution of the relaxin‐like peptide family. BMC Evol Biol 5: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]


