Abstract
Background and Purpose
Toll‐like receptor 4 (TLR4) plays a key role in the induction of inflammatory responses both in peripheral organs and the CNS. Curcumin exerts anti‐inflammatory functions by interfering with LPS‐induced dimerization of TLR4–myeloid differentiation protein‐2 (MD‐2) complex and suppressing pro‐inflammatory mediator release. However, the inhibitory mechanism of curcumin remains to be defined.
Experimental Approach
Binding of bis‐demethoxycurcumin (GG6) and its cyclized pyrazole analogue (GG9), lacking the 1,3‐dicarbonyl function, to TLR4–MD‐2 was determined using molecular docking simulations. The effects of these compounds on cytokine release and NF‐κB activation were examined by ELISA and fluorescence staining in LPS‐stimulated primary microglia. Interference with TLR4 dimerization was assessed by immunoprecipitation in Ba/F3 cells.
Key Results
Both curcumin analogues bound to the hydrophobic region of the MD‐2 pocket. However, only curcumin and GG6, both possessing the 1,3‐diketone moiety, inhibited LPS‐induced TLR4 dimerization, activation of NF‐κB and secretion of pro‐inflammatory cytokines in primary microglia. Consistent with the ability of 1,3‐diketones to coordinate divalent metal ions, LPS stimulation in a low magnesium environment decreased pro‐inflammatory cytokine release and NF‐κB p65 nuclear translocation in microglia and decreased TLR4–MD‐2 dimerization in Ba/F3 cells. Curcumin and GG6 also significantly reduced cytokine output in contrast to the pyrazole analogue GG9.
Conclusions and Implications
These results indicate that phenolic 1,3‐diketones, with a structural motif able to coordinate magnesium ions, can modulate LPS‐mediated TLR4–MD‐2 signalling. Taken together, these studies identify a previously uncharacterized mechanism involving magnesium, underlying the inflammatory responses to LPS.
Abbreviations
- CPK
Corey, Pauling and Koltun model
- Iba1
ionized calcium binding adaptor molecule 1
- MD‐2
myeloid differentiation protein‐2
- MOE
Molecular Operation Environment
- MTT
3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyltetrazolium bromide
- PDB
Protein Data Bank
- rms
root‐mean‐square
- TLRs
toll‐like receptors
Tables of Links
| TARGETS |
|---|
| Catalytic receptors |
| TLR4 |
These Tables list key protein targets and ligands in this article that are hyperlinked to corresponding entries in http://www.guidetopharmacology.org, the common portal for data from the IUPHAR/BPS Guide to PHARMACOLOGY (Southan et al., 2016), and are permanently archived in the Concise Guide to PHARMACOLOGY 2015/16 (Alexander et al., 2015).
Introduction
Neuroinflammation is a complex defence response of the CNS involved in tissue repair and induction of a protective immune response. However, if it becomes chronic, it can be destructive to the brain, leading to neuronal cell damage and, ultimately, death associated with neurodegenerative and psychiatric disorders (Frank‐Cannon et al., 2009; Amor et al., 2010). In neuroinflammation, cellular and molecular immune components such as microglia and astrocytes, cytokines, complement and pattern‐recognition receptors are the critical players (Shastri et al., 2013). Microglia, the major resident immune cells of the CNS, provide the first line of defence against invading pathogens or injury‐related products (Hanisch and Kettenmann, 2007). Microglia‐mediated defence mechanisms begin with the recognition of different pathogens through a limited number of pattern‐recognition receptors, including toll‐like receptors (TLRs) which are transmembrane glycoproteins expressed in a number of CNS cell types, including microglia (Bsibsi et al., 2002; Kawai and Akira, 2010).
LPS, a well‐characterized pathogen‐associated molecular pattern present on the outer membrane of Gram‐negative bacteria, is a potent activator of a variety of mammalian cell types (Ulevitch and Tobias, 1995). Among TLRs, TLR4 is the major LPS receptor and is a component of LPS signalling (Poltorak et al., 1998; Hoshino et al., 1999; Park and Lee, 2013). The cellular response to LPS requires specific interaction of LPS with the co‐receptor myeloid differentiation protein‐2 (MD‐2), which is associated with the extracellular domain of TLR4 (Shimazu et al., 1999; Nagai et al., 2002; Park et al., 2009). After LPS binding, the TLR4–MD‐2 complex dimerizes and a signal is then propagated by recruitment of intracellular signalling adaptor proteins leading to activation of the transcription factor NF‐κB. The latter controls the final immune response through the synthesis of inflammatory molecules, such as cytokines (TNF‐α, IL‐1β and IL‐6), chemokines, enzymes and reactive oxygen and nitrogen species (Bryant et al., 2010). As a consequence, the binding of an antagonist to the extracellular domain would prevent receptor dimerization and, consequently, intracellular signalling activation.
Identification of molecules capable of inhibiting LPS‐mediated TLR4–MD‐2 complex activation and understanding how these molecules modulate TLR4–MD‐2‐mediated signalling represents an attractive strategy for attenuating the severity of various CNS inflammatory conditions. Recently, a number of low MW compounds have been identified as TLR4 antagonists by a variety of in silico, in vitro and in vivo assays. These natural and synthetic compounds, structurally unrelated to LPS, were shown to bind directly to the MD‐2 pocket, antagonize the cellular response to LPS and improve various inflammatory conditions (Park et al., 2012). Among them is curcumin, the main bioactive component in the rhizome of the turmeric plant (Curcuma longa), a safe and a highly pleiotropic molecule endowed with a wide range of beneficial activities, including anti‐inflammatory, anti‐tumour, anti‐oxidative and cardiovascular protective effects (Hatcher et al., 2008), and which interacts with diverse molecular targets (Gupta et al., 2013). In particular, the anti‐inflammatory properties of curcumin are linked to numerous mechanisms (He et al., 2015), including its ability to bind MD‐2 (Gradišar et al., 2007) and inhibit dimerization of the TLR4–MD‐2 complex (Youn et al., 2006) which is required for inflammatory intracellular signalling. However, the inhibitory mechanism of curcumin remains to be fully understood. Despite the fact that curcumin is an electrophilic thiol‐reactive compound containing a bis‐α, β‐unsaturated 1,3‐diketone moiety which can react with cysteine residues through a Michael reaction, it binds MD‐2 in proximity of Cys133 residue, without the formation of a covalent adduct (Gradišar et al., 2007).
In the present study, two curcumin derivatives were synthesized and used to probe the anti‐inflammatory mechanism of curcumin at the molecular level. In particular, bis‐demethoxycurcumin (GG6, 1E,4Z,6E‐5‐hydroxy‐1,7‐bis‐4‐hydroxyphenylhepta‐1,4,6‐trien‐3‐one) and its pyrazole‐based analogue lacking the 1,3‐dicarbonyl function [GG9, 4,4'‐(1E,1'E)‐(1H‐pyrazole‐3,5‐diyl‐bis‐ethene‐2,1‐diyl)diphenol] were compared with curcumin in terms of their anti‐inflammatory effects and binding mechanisms to the TLR4–MD‐2 complex. Only the 1,3 diketones suppressed neuroinflammation, with the ability of the 1,3 diketone moiety to bind divalent cations such as the magnesium ion playing an important role in inhibiting LPS‐induced TLR4–MD‐2 complex activation and subsequent inflammatory response.
Methods
Synthesis of curcumin analogues
GG6 and GG9 analogues were synthesized according to previously reported procedures (Scheme 1). Spectroscopic data were consistent with their chemical structures and in good agreement with pubblished data (Ishida et al., 2002; Di Martino et al., 2016). GG6 was prepared according to the Pabon reaction (Pabon, 1964) in which 2,4‐pentandione was first reacted with B2O3 and the obtained boric complex then condensed with 4‐hydroxybenzaldehyde in the presence of n‐tributylborate. Acidic hydrolysis removed the boron complexation and provided the desired derivative. GG6 was then reacted with hydrazine hydrate to obtain the pyrazole analogue GG9.
Scheme 1.
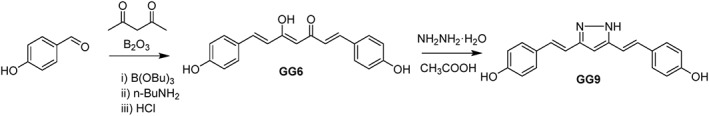
General synthetic method for GG6 and GG9.
Molecular modelling studies
Target structures
The crystallographic structure of the human TLR4‐human MD‐2‐E. coli LPS ternary complex was retrieved from the Protein Data Bank (PDB code: 3FXI) (Park et al., 2009).
Assessment of crystallographic structure quality was performed with the Structure Preparation tool of the Molecular Operation Environment program (MOE, version 2014.09; Chemical Computing Group Inc., 1010 Sherbooke St. West, Suite #910, Montreal, QC, Canada, H3A 2R7, 2015). Critical structural issues (such as missing or poorly resolved atomic data, anomalous topological properties present in amino acid units and anomalous bonding patterns of non‐amino acid units) were fixed when necessary. Hydrogen atoms were added, and their appropriate protonation state fixed using the Protonate3D tool as implemented in the MOE program. To minimize contacts between hydrogen atoms, the structures were subjected to Amber99 force field (Cornell et al., 1995) minimization until the root‐mean‐square (rms) of the conjugate gradient was <0.1 kcal·mol−1·Å−1 keeping the heavy atoms fixed at their crystallographic positions.
Molecular docking protocol
Curcumin and its analogues were built using the ‘Builder’ module of MOE, and each compound was docked into the presumptive binding site (LPS binding site) using flexible MOE‐Dock methodology. The purpose of MOE‐Dock is to search for favourable binding configurations between a small, flexible ligand and a rigid macromolecular target. Searching is conducted within a user‐specified 3D docking box, using the ‘tabù search’ (Baxter et al., 1998) protocol and the MMFF94 force field (Halgren, 1996). Charges for ligands were imported from the MOPAC program (Stewart, 1993) output files. MOE‐Dock performs a user‐specified number of independent docking runs (50 in the present case) and writes the resulting conformations and their energies to a molecular database file. The resulting ligand/protein complexes were subjected to MMFF94 all‐atom energy minimization until the rms of conjugate gradient was <0.1 kcal·mol−1·Å−1. GB/SA approximation (Wojciechowski and Lesyng, 2004) was used to model the electrostatic contribution to the free energy of solvation in a continuum solvent model. Interaction energy values were calculated as the energy of the complex minus the energy of the ligand, minus the energy of the protein: ΔEinter = E(complex)−(E(L) + E(Protein)).
Cell cultures
All animal care and experimental procedures were performed in accordance with National Institutes of Health guidelines for the care and use of laboratory animals and those of the Italian Ministry of Health (D.L. 26/2014) and were approved by the Institutional Animal Care and Use Committee of the University of Padua (958/2016‐PR). Animal studies are reported in compliance with the ARRIVE guidelines (Kilkenny et al., 2010; McGrath and Lilley, 2015). One‐day‐old Sprague‐Dawley rat pups (CD strain; from our Animal Facility) of both sexes were rapidly decapitated, minimizing suffering, discomfort or stress. Primary microglial cells were isolated from mixed glial cell cultures prepared from cerebral cortex, as previously described (Skaper et al., 2012). Briefly, upon reaching confluence (7–10 days after isolation) microglia adhering to the astroglial monolayer were dislodged by shaking (200 r.p.m. for 1 h at 37°C), resuspended in high‐glucose DMEM supplemented with 2 mM L‐glutamine, 10% heat‐inactivated FCS, 100 U·mL−1 penicillin, 100 μg·mL−1 streptomycin and 50 μg·mL−1 gentamicin and plated on uncoated plastic wells at a density of 1.25 × 105 cells·cm−2. Cells were allowed to adhere for 45 min and then washed to remove non‐adhering cells. After a 24 h incubation period, the medium was replaced with either a serum‐free medium or a low‐magnesium solution containing 2.5 mM CaCl2, 4.7 mM KCl, 1.2 mM KH2PO4, 118 mM NaCl, 25 mM NaHCO3, 0.1 mM MgSO4 and 11 mM glucose. Purity of the cultures was confirmed by immunocytochemistry using a primary polyclonal antibody against ionized calcium binding adaptor molecule 1 (Iba1, 1:800, Wako Chemicals USA Inc., Richmond, VA, USA). Ninety‐seven per cent of the cells were Iba1 immunopositive. Murine Ba/F3 cells, an IL‐3 dependent pro‐B cell line, stably expressing human TLR4‐GFP, human TLR4‐Flag, human MD‐2‐Flag and human CD14 were kindly provided by Dr Kensuke Miyake (University of Tokyo, Tokyo, Japan). Cells were cultured in RPMI1640 medium, supplemented with 10% FCS, 100 μM 2‐mercaptoethanol and recombinant murine IL‐3 (~ 70 U·mL−1) (Saitoh et al., 2004). Cells were maintained at 37°C in a humidified atmosphere containing 5% CO2/95% air.
Cell viability assay
Microglial cell viability was evaluated by a quantitative colorimetric method utilizing the metabolic dye 3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyltetrazolium bromide (MTT) (Mosmann, 1983). Briefly, cells were seeded at the density of 45 000 cells per well in 96‐well plates and treated with test compounds. After 6 h incubation, the medium was removed and the cells incubated with MTT (0.18 mg·mL−1) for 4 h at 37°C. The formazan crystals were solubilized with DMSO. The plates were then analysed on a microplate reader (Victor2 Multilabel Counter, Wallac, Cambridge, MA, USA) using a test wavelength of 570 nm and a reference wavelength of 630 nm, and the results expressed as percentage viability relative to LPS‐treated cultures.
Cytokine determination
Primary microglia were pretreated for 1 h with increasing concentrations of test compounds and then stimulated with 100 ng·mL−1 Ultra‐Pure LPS‐EB for an additional 6 h. At the end of incubation, culture medium was collected, and IL‐1β and TNF‐α were assayed using commercially available ELISA kits (Antigenix America, Huntington Station, NY, USA), according to the manufacturer's instructions. Cytokine concentrations (pg·mL−1) in the medium were determined by reference to standard curves obtained with known amounts of IL‐1β or TNF‐α, and the results were expressed as percentage relative to LPS‐stimulated cultures.
Immunofluorescence
Microglia were grown on coverslips in 12‐well plates and pretreated for 1 h with 10 μM curcumin, GG6 or GG9 and then stimulated with 100 ng·mL−1 Ultra‐Pure LPS‐EB for an additional 90 min. Cells were fixed with 4% paraformaldehyde (pH 7.4, for 15 min at room temperature) and subsequently blocked with 5% normal goat serum/0.1% Triton X‐100 in PBS for 1 h (Zusso et al., 2012). Cells were incubated with a mouse anti‐p65 antibody (NF‐κB p65, 1:500; Santa Cruz Biotechnology Santa Cruz, CA, USA) followed by an Alexa Fluor 555‐conjugated anti‐mouse secondary antibody (1:1000; Invitrogen). Nuclei were stained with DAPI (Sigma), and coverslips were mounted on microscope slides with Fluoromount‐G mounting medium (Fisher Scientific, Milan, Italy). Fluorescent images for p65 staining were captured with a confocal laser‐scanning microscope (Zeiss LSM 800; Carl Zeiss AG, Oberkochen, Germany) and analysed with ZEN 2.0 imaging software (Carl Zeiss AG). ImageJ software (National Institutes of Health, Bethesda, MD, USA) was used to profile the fluorescence emission intensity of single cells.
Western blotting
Microglial cells were pretreated for 1 h with 10 μM curcumin, GG6 or GG9 and then stimulated with 100 ng·mL−1 Ultra‐Pure LPS‐EB for an additional 60 min. Cells were lysed on ice with NP40 cell lysis buffer (Life Technologies, San Giuliano Milanese, Italy), containing protease and phosphatase inhibitors (Sigma‐Aldrich), and centrifuged at 13 000 x g for 10 min at 4°C. Equal amounts of protein (30 μg) were separated by 10% SDS‐PAGE and electrotransferred onto nitrocellulose membranes. The membranes were blocked with phosphate‐buffered saline containing 0.1% Triton X‐100 and 3% non‐fat dry milk and probed overnight with rabbit anti‐IκBα (1:500; Cell Signaling Technology, Beverly, MA, USA), rabbit anti‐NF‐κB p65 phospho S536 (p‐p65; 1:2000; Abcam, Cambridge, UK) or mouse anti‐β‐actin (1:25 000; Sigma‐Aldrich). Following incubation with appropriate horseradish peroxidase‐conjugated secondary antibodies, the immunoreactive bands were visualized with enhanced chemiluminescence (ECL) substrate (#RPN2106; GE Healthcare Amersham, Buckinghamshire, UK) using the VersaDoc Imaging System (Bio‐Rad Laboratories, Hercules, CA, USA). The band intensities were quantified by densitometric analysis using ImageJ software. Band quantification was first normalized to β‐actin, and then percentage was calculated versus LPS‐stimulated cells Data obtained from densitometric analyses were normalized to account for unequal loading of samples across the lanes on a gel and for differences in transfer efficiency across a blot.
Immunoprecipitation
Ba/F3 cells stably expressing human TLR4‐GFP, human TLR4‐Flag, human MD‐2‐Flag and human CD14 (80 × 106 cells per condition) were pretreated for 1 h with curcumin, GG6 or GG9 (50 μM) and then stimulated with 250 ng·mL−1 LPS for 30 min (Youn et al., 2006). Protein extracts were prepared as described previously (Saitoh et al., 2004). The samples were immunoprecipitated with mouse anti‐GFP antibody (#NB600–597; Novus, Littleton, CO) for 16–18 h at 4°C. The recovered immune complex was resolved using 10% SDS‐PAGE and electrotransferred to a nitrocellulose membrane. The membrane was probed overnight with either mouse anti‐Flag antibody (#F3165; Sigma‐Aldrich, St. Louis, MO) or chicken anti‐GFP antibody (#06–896; Millipore, Temecula, CA). Thereafter, the membrane was exposed to horseradish peroxidase‐conjugated secondary antibody for 1 h, and reactive bands were detected with the ECL system. Images were analysed using ImageJ software, and TLR4‐Flag expression was normalized to TLR4‐GFP levels. Results are expressed as percentage relative to LPS‐stimulated cells.
Magnesium assessment
Magnesium assessment in DMEM and in the low magnesium solution was performed using an atomic absorbance spectrophotometer (Varian AA Duo 55 b/240, Agilent Technologies, Santa Clara, CA, USA) with a Ca/Mg lamp, air/acetylene flame and detection wavelength of 285.2 nm. Absorbance readings were obtained via integrated duplicate measurement and compared with a standard, calibrated curve.
Data and statistical analysis
The data and statistical analysis comply with the recommendations on experimental design and analysis in pharmacology (Curtis et al., 2015). Data were blindly analysed using GraphPad software, version 5.01 (GraphPad Software, Inc., La Jolla, CA, USA) and expressed as mean ± SEM of at least five independent experiments. Individual data points were based on a technical duplicate to ensure the reliability of single values. As raw values varied between experiments and the variability could obscure the treatment effect, data were expressed as percentage of control cells or LPS treatment, taken as baseline of each independent experiment. Data were analysed by means of Kruskal–Wallis one‐way ANOVA followed by post hoc Dunn's test for multiple comparisons (Siegel and Castellan, 1988) against treatement with LPS alone, using P‐adjustment according to Bonferroni's method (Pohlert, 2014) or by one‐way ANOVA with Dunnett's post hoc test, as appropriate. P < 0.05 was deemed statistically significant. Post hoc tests were carried out only when the F value from ANOVA was <0.05, and there was no significant variance in homogeneity.
Materials
Unless otherwise specified, all reagents were from Sigma‐Aldrich (Milan, Italy). Tissue culture media, antibiotics and fetal calf serum (FCS) were obtained from Invitrogen (San Giuliano Milanese, Italy). LPS (Ultra‐Pure LPS‐EB from Escherichia coli, 0111:B4 strain) was purchased from InvivoGen (InvivoGen Europe, Toulouse, France). The Ultra‐Pure LPS‐EB preparation used here only activates TLR4. Falcon tissue culture plasticware was purchased from BD Biosciences (SACCO srl, Cadorago (CO), Italy).
Results
Effects of curcumin analogues on release of pro‐inflammatory cytokines from LPS‐stimulated microglia
Stimulation of microglia with TLR agonists, including LPS for TLR4, induces a pro‐inflammatory process through the increased secretion of cytokines (Lehnardt, 2009). Inhibition of LPS‐induced release of IL‐1β and TNF‐α was initially used to examine the anti‐inflammatory effects of the curcumin analogues GG6 and GG9. Cells were exposed to increasing concentrations (1–10 μM) of curcumin (positive control), GG6 or GG9 for 1 h and then stimulated with LPS for 6 h to induce an inflammatory response. Supernatants were collected and subjected to ELISA to measure IL‐1β and TNF‐α concentration. Unstimulated cells released low or undetectable amounts of IL‐1β and TNF‐α which remained unchanged after treatment with the three compounds (data not shown). LPS treatment markedly induced release of IL‐1β and TNF‐α (set to 100%; dashed lines in Figure 1A, B) that was significantly suppressed by curcumin (see also Mercanti et al., 2014) and by its 1,3‐diketone analogue GG6 (Figure 1A, B), demonstrating that GG6 had effects similar to the parent molecule. In contrast GG9, the pyrazole‐based analogue, failed to inhibit LPS‐induced release of either IL‐1β or TNF‐α (Figure 1A, B), indicating that the 1,3‐diketone moiety is crucial for anti‐inflammatory activity. None of the three compounds compromised cell viability at the concentrations tested (1–10 μM) (Figure 1C), indicating that the observed decrease in IL‐1β and TNF‐α release induced by curcumin and GG6 did not result from cytotoxicity.
Figure 1.
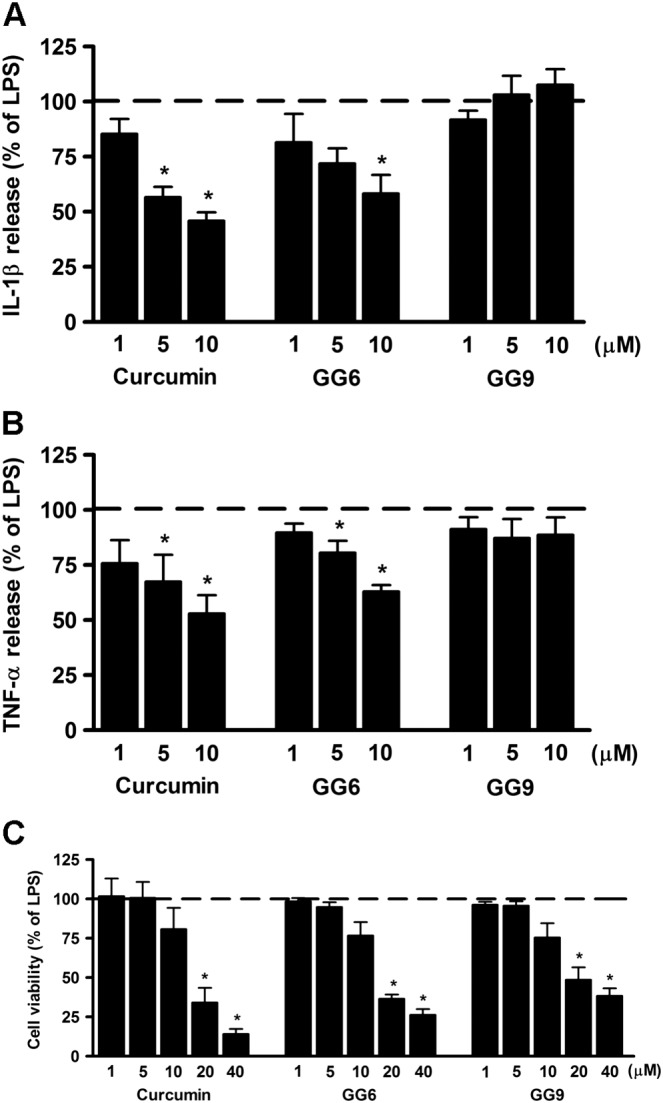
Effects of curcumin, GG6 or GG9 on cytokine release from LPS‐stimulated cortical microglia and cell viability. Microglia were cultured for 24 h in medium containing 10% serum, which was replaced with serum‐free medium before pretreatment with curcumin, GG6 or GG9 (1–10 μM) for 1 h followed by stimulation with 100 ng·mL−1 LPS for 6 h. Supernatants were collected and analysed for (A) IL‐1β and (B) TNF‐α release. (C) At the end of incubation, cell viability was also determined by MTT assay. Results are expressed as percentage of cytokine release and cell viability relative to LPS‐stimulated microglia. Data are means ± SEM (n = 5). *P < 0.05, significantly different from LPS alone (dashed line), Kruskal–Wallis followed by Dunn's post hoc test and P‐adjustment according to Bonferroni's method.
Effects of curcumin analogues on NF‐κB activation in LPS‐stimulated microglia
In an LPS‐induced inflammatory response, activation of the transcription factor NF‐κB through the translocation of its p65 subunit from the cytosol to the nucleus is considered a key event that controls the expression of a large number of cytokines and other immune response genes (Ghosh and Hayden, 2008). To investigate whether the studied compounds reduced LPS‐induced pro‐inflammatory cytokine release by preventing NF‐κB activation, the subcellular distribution of p65 subunit was studied by immunofluorescent staining in unstimulated and LPS‐stimulated microglia. In unstimulated control cells, p65 was mainly found in the cytoplasm (Figure 2A) with only minor nuclear staining, about 25% of the total p65 in the nucleus (Supporting Information Figure S1). This distribution remained unchanged after treatment with curcumin, GG6 or GG9 (10 μM) (Figure 2B–D and Supporting Information Figure S1). However, a 90 min exposure to LPS increased the proportion in the nucleus (Figure 2E), now about 75% of the total p65 (Supporting Information Figure S1), indicating NF‐κB activation. Pretreatment with curcumin and GG6 strongly attenuated this LPS‐induced nuclear localization of p65 (Figure 2F, G) to levels close to that in control cells, about 30% of total p65 (Supporting Information Figure S1). In contrast, pretreatment with GG9 did not show this effect (Figure 2H and Supporting Information Figure S1), confirming the structure–activity relationship observed in inhibition of cytokine release.
Figure 2.
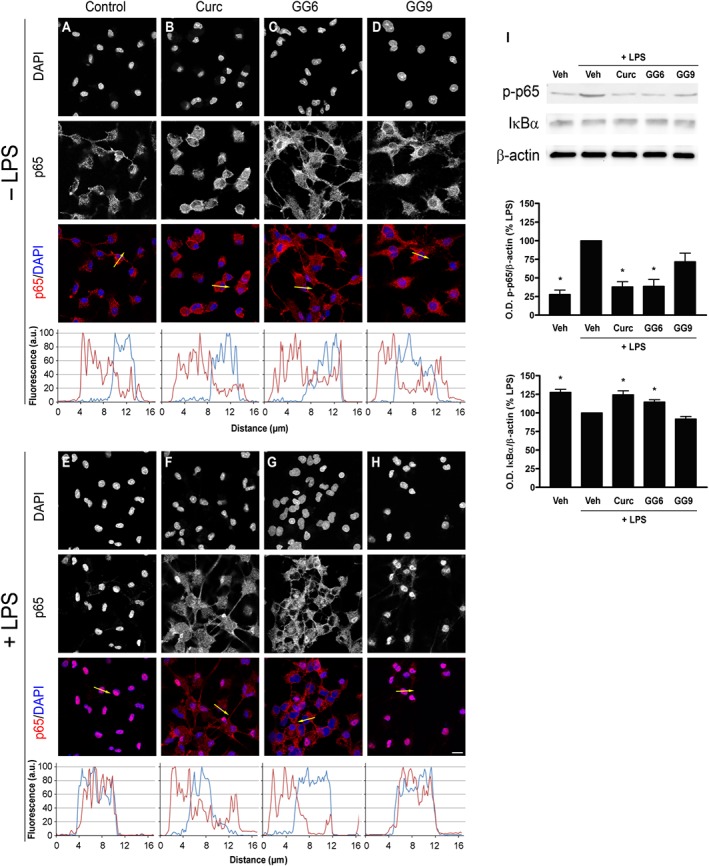
Effects of curcumin, GG6 or GG9 on NF‐κB activation in unstimulated and LPS‐stimulated cortical microglia. Microglia were subcultured for 24 h in medium containing 10% serum, which was replaced with serum‐free medium before stimulation with curcumin (Curc), GG6 or GG9 (10 μM) ± 100 ng·mL−1 LPS. Cells were then processed for NF‐κB p65 immunostaining. Fluorescence intensity profiles along an ideal oriented line across a representative cell (yellow arrow in the third line) are shown in the fourth line. Experiments were performed five times, and representative confocal images showing subcellular localization of p65 in unstimulated and LPS‐stimulated microglia are shown in (A–D) and (E–H) respectively. Scale bar, 10 μm. (I) Protein levels of IκBα and phosphorylated p65 (p‐p65) were examined by Western blot. Experiments were performed five times, and representative images are shown. Densitometric analysis of Western blots relative to β‐actin is expressed as percentage of LPS‐stimulated cells. Data are means ± SEM (n = 5). *P < 0.05, significantly different from LPS alone; Kruskal–Wallis followed by Dunn's post hoc test and P‐adjustment according to Bonferroni's method. Veh, vehicle.
To further examine the effect of curcumin and GG6 on NF‐κB activation, IκBα degradation and phosphorylation of p65, both required for NF‐κB transcriptional activity (Christian et al., 2016), were evaluated. Both curcumin and GG6 significantly decreased IκBα degradation and phosphorylation of p65 induced by a 60 min challenge with LPS (Figure 2I).
Effects of curcumin analogues on TLR4 dimerization
TLR4 dimerization is one of the initial steps involved in activation of the TLR4‐mediated signalling pathway. To investigate whether curcumin analogues could inhibit this step, Ba/F3 cells stably transfected with human TLR4‐GFP, TLR4‐Flag, MD‐2‐Flag and CD14 (Saitoh et al., 2004) were used. Cells were pre‐incubated with curcumin, GG6, GG9 or vehicle (untreated control) for 1 h prior to LPS stimulation for 30 min. To detect TLR4 dimerization, TLR4‐GFP was immunoprecipitated, and co‐precipitation of TLR4‐Flag was probed with anti‐Flag antibody (Figure 3). TLR4 dimerization increased in response to LPS stimulation, and, as previously demonstrated in this cell line (Youn et al., 2006), curcumin reduced LPS‐induced dimerization (Figure 3A, B). Importantly, the 1,3‐diketone derivative GG6 also markedly decreased this effect. In contrast, and in agreement with the effects on inhibition of cytokine release and NF‐κB activation, the pyrazole analogue GG9 did not affect dimerization of TLR4, confirming a key role for the 1,3‐diketone moiety in the first step of the inflammatory cascade (Figure 3A, B).
Figure 3.
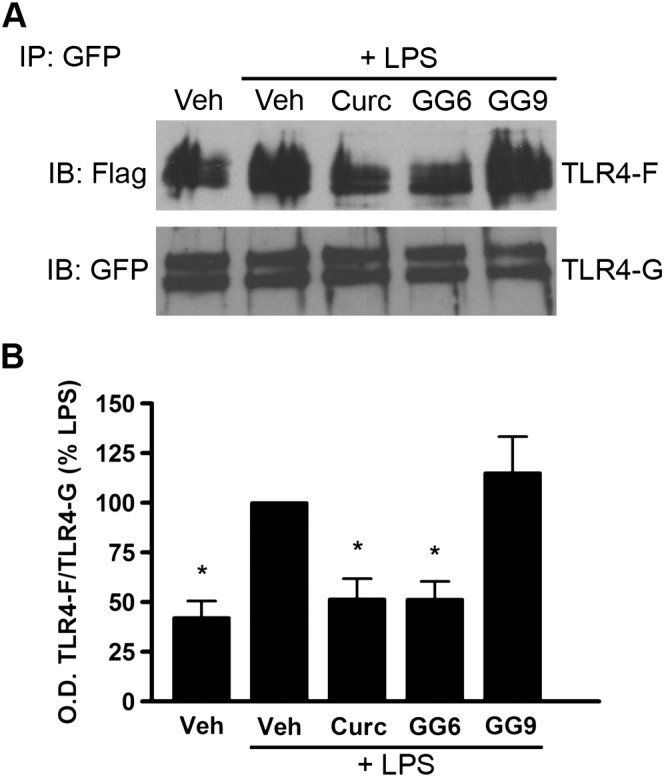
Effects of curcumin, GG6 and GG9 on LPS‐induced TLR4 dimerization. (A) Ba/F3 cells expressing TLR4‐Flag (TLR4F), TLR4‐GFP (TLR4G), MD2‐Flag and CD14 were pretreated with 50 μM curcumin (Curc), GG6 or GG9 for 1 h and then stimulated with 250 ng·mL−1 LPS for 30 min. Cell lysates were immunoprecipitated with mouse anti‐GFP antibody and immunoblotted with mouse anti‐Flag (upper) or chicken anti‐GFP (lower) antibody. Experiments were performed at least five times, and representative images are shown. (B) Histograms showing the ratio TLR4‐Flag/TLR4‐GFP, expressed as percentage of LPS‐stimulated cells. Data are means ± SEM (n = 5). *P < 0.05, significantly different from LPS alone; Kruskal–Wallis followed by Dunn's post hoc test and P‐adjustment according to Bonferroni's method. IB, immunoblot; IP, immunoprecipitation; Veh, vehicle.
The binding mode of curcumin analogues to MD‐2
To explore the observed structure–activity relationship of curcumin and its analogues in their ant‐inflammatory effects, molecular docking simulations were carried out to characterize the putative binding mode of these molecules to the LPS binding site of MD‐2. In agreement with previous studies, curcumin (Wang et al., 2015), as well as GG6 and GG9, could be accommodated into the large hydrophobic binding pocket of MD‐2 occupying a relevant portion of the LPS binding site (Figure 4) while assuming different binding modes. Among all binding modes generated for each ligand, the energetically more stable were those that showed important interactions with MD‐2, such as hydrogen bonding, engaging residues Arg90, Glu92 and Tyr102. These binding modes are well conserved in both curcumin analogues. As shown in Figure 5, curcumin (panel a), GG6 (panel b) and GG9 (panel c) are located in the LPS binding site of MD‐2, guaranteeing the same cavity occupancy and the same interaction networks.
Figure 4.
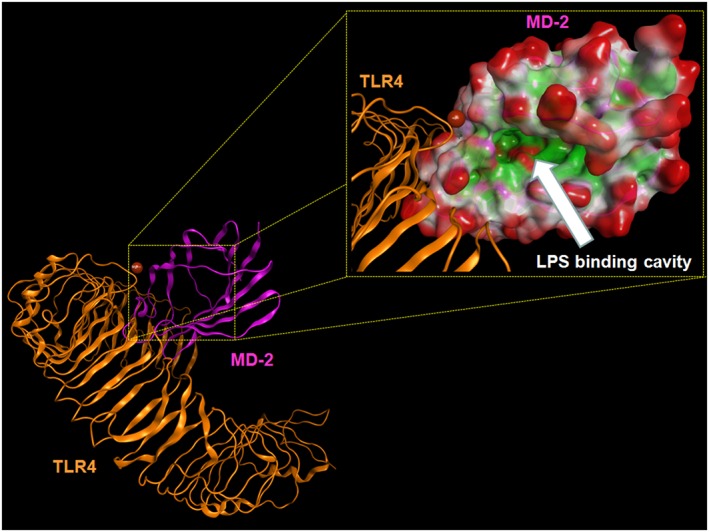
Overall structure of the human TLR4–M‐D2 complex derived by the crystallographic structure 3FXI. Some atoms have been voluntarily omitted. Bottom left: TLR4 (orange) and MD‐2 (magenta) are represented showing their secondary structure. Top right: the LPS binding cavity of MD‐2 is enlarged and shown as the Connolly surface. Hydrophobic regions of the Connolly surface are coloured in green, polar in magenta and those exposed to solvent in red. Mg2+ is represented by Corey, Pauling and Koltun Model (CPK) (bronze).
Figure 5.
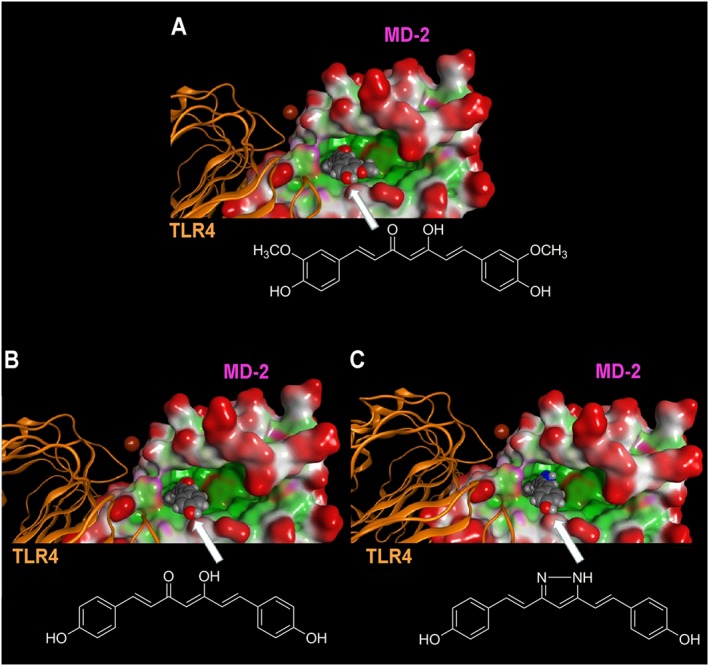
Molecular docking analysis of the energetically most stable binding poses between (A) curcumin, (B) GG6, (C) GG9 and MD‐2. As described in Figure 4, the LPS binding cavity of MD‐2 is enlarged and shown as the Connolly surface. Hydrophobic regions of the Connolly surface are green, polar in magenta and those exposed to solvent in red. Mg2+ is represented by CPK (bronze).
Interestingly, the crystallographic structure coded by PDB as 3FXI, in proximity to entrance of the LPS binding site of MD‐2, showed a metal ion at the interface between the MD‐2‐LPS complex to TLR4, either directly or indirectly through water molecules, as shown in (Park et al., 2009). The original crystallographic study suggested this to be Mg2+, whose role at that time was thought to be non‐essential for LPS binding and receptor dimerization (Park et al., 2009). New molecular docking simulations were performed by using a sampling grid large enough to include the metal ion. These studies showed that the presence of a 1,3‐diketone moiety, as well as its enolate form, allows the appearance of an alternative binding mode close to magnesium ion (Figure 6). On the contrary, the pyrazole analogue GG9 lacked this behaviour, thus confirming its inability to coordinate metal ions.
Figure 6.
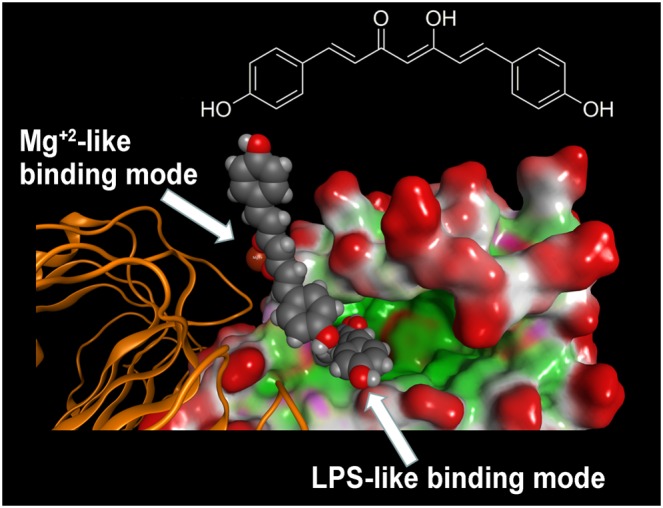
Molecular docking analysis of the two alternative binding modes between GG6 and MD‐2. As described in Figure 4, the LPS binding cavity of MD‐2 is enlarged and shown as the Connolly surface. Hydrophobic regions of the Connolly surface are green, polar in magenta and those exposed to solvent in red. Mg2+ ion is represented by CPK (bronze).
Effect of a low magnesium environment on inflammatory response
To examine the effect of magnesium on the inflammatory response, microglial cells cultured in a low magnesium environment (0.1 ± 0.05 mM, determined by atomic absorbance spectroscopy) were exposed to curcumin, GG6 or GG9 for 1 h and then stimulated with LPS for 6 h. Cell viability and low basal levels of IL‐1β and TNF‐α were not significantly affected by a low extracellular magnesium concentration (Figure 7A–C). Under these conditions, however, LPS‐induced release of both cytokines was significantly decreased compared with that under control conditions (DMEM; Mg2+, 0.9 ± 0.1 mM), as shown in Figure 7D, E, (white bars). Treatment with curcumin and GG6 brought about a marked concentration‐dependent reduction in IL‐1β and TNF‐α release – unlike GG9 (Figure 7D, E, grey bars). Furthermore, the diminished cytokine release observed in a low magnesium environment correlated well with decreased NF‐κB p65 nuclear translocation in microglia (Figure 8A, B) and suppression of TLR4 dimerization in Ba/F3 cells (Figure 8C).
Figure 7.
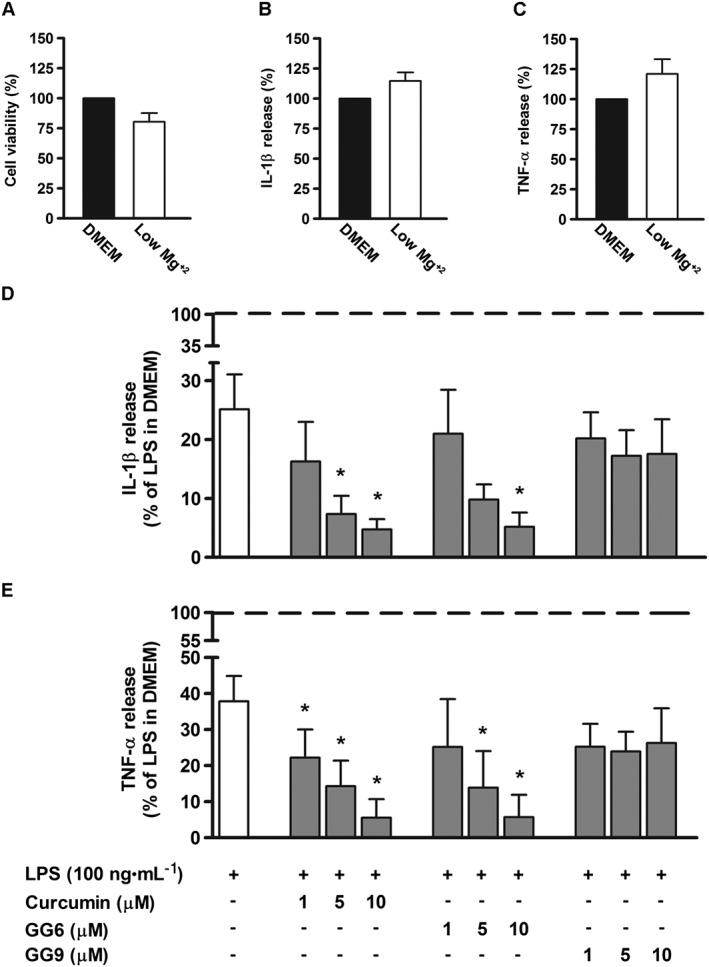
Effect of curcumin, GG6 or GG9 in a low magnesium environment on cytokine release from LPS‐stimulated cortical microglia. Microglia were cultured for 24 h in medium containing 10% serum, which was then replaced with a solution containing a low magnesium concentration before pretreatment with curcumin (Curc), GG6 or GG9 (1–10 μM) for 1 h and further stimulation with 100 ng·mL−1 LPS for 6 h. (A) Microglial cell viability was determined by MTT assay after exposing cells to a low magnesium environment. Supernatants were collected and analysed for (B, D) IL‐1β and (C, E) TNF‐α release. Results are expressed as percentage of cell viability and cytokine release relative to control (DMEM) or LPS‐stimulated microglia. Data are means ± SEM (n = 5). *P < 0.05, significantly different from LPS alone (dashed line), one‐way ANOVA followed by Dunnett's post hoc test.
Figure 8.
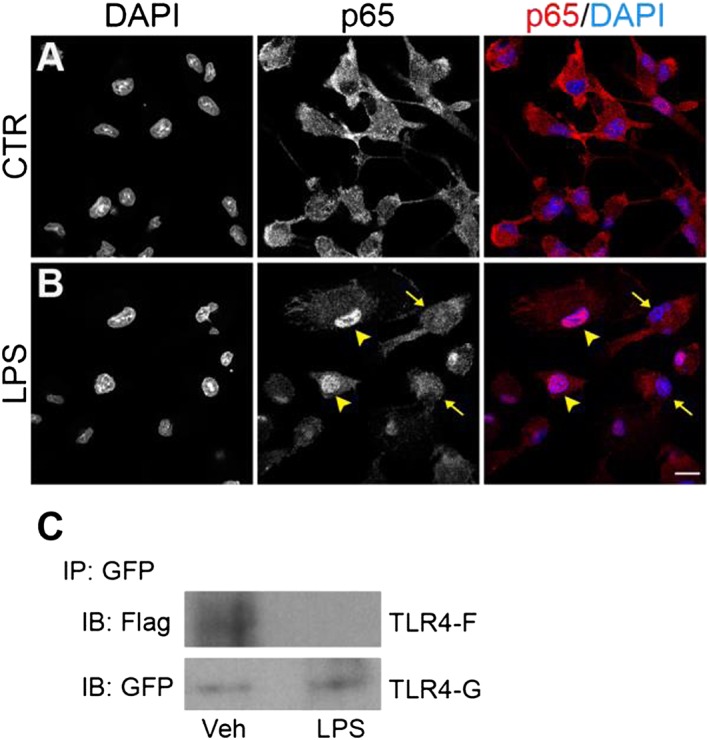
Effect of a low magnesium environment on NF‐κB p65 nuclear translocation and TLR4 dimerization. (A, B) Microglial cells, exposed to a low magnesium environment, were stimulated with 100 ng·mL−1 LPS for 90 min and further processed for NF‐κB p65 immunostaining. DAPI counterstaining is shown in the images in the left column and in the merged images in the right column. Experiments were performed at least five times, and representative confocal images showing subcellular localization of p65 in unstimulated and LPS‐stimulated microglia are shown in (A) and (B) respectively. Arrowheads point to examples of p65 nuclear localization, while arrows point to examples of cytoplasmic distribution. Scale bar, 10 μm. (C) Ba/F3 cells expressing TLR4‐Flag (TLR4F), TLR4‐GFP (TLR4G), MD2‐Flag and CD14, exposed to a low magnesium environment, were stimulated with 250 ng·mL−1 LPS for 30 min. Cell lysates were then immunoprecipitated with mouse anti‐GFP antibody and immunoblotted with mouse anti‐Flag (upper) or chicken anti‐GFP (lower) antibody. Experiments were performed five times, and representative results are shown. IB, immunoblot; IP, immunoprecipitation; Veh, vehicle.
Discussion
Curcumin possesses well‐known anti‐inflammatory activities, and many studies have suggested that MD‐2 may be the direct target of curcumin in its inhibition of LPS signalling (Youn et al., 2006; Gradišar et al., 2007; He et al., 2015). However, the exact interaction between curcumin and MD‐2 is not completely understood. Several curcumin analogues have recently been synthesized with the aim to learn more about curcumin mechanism of action and to establish structure–activity relationships. Although the 1,3‐diketone moiety in the curcumin structure has been considered responsible for its chemical instability (Oliveira et al., 2015) and some mono‐carbonyl analogues of curcumin displayed anti‐inflammatory effects in vitro and in vivo (Zhang et al., 2014; Zhang et al., 2015), this structural motif seems to be important for inhibition of LPS‐mediated TLR4–MD‐2 activation/dimerization (Youn et al., 2006). Here, we approached the question of this chemical moiety's involvement in the microglial anti‐inflammatory response by using two curcumin derivatives: bis‐demethoxycurcumin (GG6) and its cyclized pyrazole analogue (GG9), lacking the 1,3‐dicarbonyl function. Non‐cytotoxic concentrations of curcumin and GG6 suppressed, in a similar manner, LPS‐induced inflammatory response in primary cortical microglia. Previous studies have suggested that prevention of the inflammatory cascade by curcumin may be linked to its ability to inhibit LPS‐induced dimerization of TLR4 (Youn et al., 2006). GG6, sharing with curcumin the 1,3‐diketone moiety, suppressed this effect, whereas the pyrazole‐based analogue GG9 was unable to inhibit LPS‐induced pro‐inflammatory response in microglia and TLR4 dimerization.
These observations highlight the fundamental role of the 1,3 diketone moiety in the anti‐inflammatory effects of curcumin and GG6, by directly suppressing LPS‐induced initiation of receptor signalling. This structural information prompted us to characterize the binding mode of GG6 and GG9 to the LPS binding site on MD‐2. Molecular modelling studies showed that, as for curcumin (Wang et al., 2015), both analogues could bind the hydrophobic region of the MD‐2 pocket, leading to a stable complex via interactions with Arg90, Glu92 and Tyr102, all of which are conserved between human, mouse and rat (Shimazu et al., 1999; Akashi et al., 2000; Winnall et al., 2011). Considering the size of the LPS binding cavity, it is also not surprising that these compounds may assume different poses inside this binding site. Based on the above considerations, however, it is not possible to explain the remarkable decrease in anti‐inflammatory activity of the pyrazole analogue GG9, compared with the 1,3‐diketone analogue GG6.
As previously reported, a metal ion, probably Mg2+, is present at the interface between the MD‐2–LPS complex and TLR4. As EDTA was unable to block LPS binding and receptor dimerization (Park et al., 2009), the presence of Mg2+ ions has been considered to be non‐essential. Diketones efficiently coordinate divalent metal ions (including copper, nickel, cobalt, zinc and magnesium) (Vyas et al., 2013). Conceivably, the presence of a metal ion like Mg2+ could affect docking simulations facilitating ligand movement from inside the LPS binding cavity of MD‐2 closer to the metal coordination sphere, where possible in terms of complex stability. Molecular docking simulations including Mg2+ confirmed that only the presence of a 1,3‐diketone moiety allowed the appearance of poses close to the metal ion, confirming the inability of the pyrazole analogue GG9 to coordinate metal ions. This computational evidence led us to hypothesize that the anti‐inflammatory activity of 1,3‐diketone derivatives is associated with their ability to coordinate Mg2+ which could in turn affect proper assembly of the TLR4–MD‐2–LPS ternary complex. Exposing microglial to a low magnesium environment significantly decreased LPS‐induced IL‐1β and TNF‐α release and NF‐κB nuclear translocation, compared with normal magnesium concentrations. Moreover, curcumin and GG6 markedly reduced cytokine output in contrast to GG9, suggesting further that the capability of the 1,3 diketone moiety to coordinate divalent cations, such as Mg2+, may play a critical role in anti‐inflammatory responses. These findings contrast with previous studies in both rodent and human cells, which suggested a link between magnesium deficiency and inflammation and, conversely, showed that elevated magnesium limited the inflammatory response by inhibiting NF‐κB activation and pro‐inflammatory cytokine production (Lin et al., 2010; Lee et al., 2011; Sugimoto et al., 2012; Gao et al., 2013). This discrepancy may reflect differences in experimental conditions, such as the length of exposure to a low magnesium environment or LPS concentration. Sugimoto and colleagues showed a decreased cytokine production induced by magnesium supplementation with LPS concentration (50 pg·mL−1) too low to activate all cells. In contrast, high LPS concentrations, similar to those used here, abolished the effects of magnesium (Sugimoto et al., 2012).
The mechanism(s) by which magnesium exerts its effect on inflammatory responses is poorly understood. As a known antagonist of some calcium ion channels (Sonna et al., 1996), magnesium may counteract the toxic consequences of excessive calcium entry into immune cells, including microglia (Lin et al., 2010; Gao et al., 2013). Other potential cellular mechanisms concerning the relationship between magnesium status and inflammation have been considered, such as effects on NMDA receptors, release of substance P and activation of NF‐κB (Mazur et al., 2007). Furthermore, whether magnesium functions via an intra‐ or extracellular mechanism has been debated (Mazur et al., 2007; Gao et al., 2013). The present findings reveal a previous unrecognized direct link between a low magnesium environment and LPS‐mediated TLR4 activation, by showing inhibition of TLR4–MD‐2 complex dimerization in LPS‐stimulated Ba/F3 cells exposed to a low Mg2+ concentration. These results provide evidence for a new and alternative mechanism for the LPS‐mediated activation of TLR4, suggesting that molecules like 1,3‐diketones, which possess a structural motif able to coordinate Mg2+, could modulate LPS‐mediated TLR4–MD‐2 signalling, an important component of immune responses in the nervous system (Kigerl et al., 2014). In a broader framework, neuroinflammation and microglial activation may also involve activation of multiple TLRs by exogenous and endogenous ligands in a context‐dependent manner (Li et al., 2016). Furthermore, the TLR4–MD‐2 complex and its dimerization process may not be the exclusive targets through which curcumin and its 1,3‐diketone analogue may modulate neuroinflammation. Even so, the mechanism proposed in this study can provide a starting point for the design and development of novel agents targeting the MD‐2 protein, applied to neuropathologies in which prolonged TLR4–MD‐2 activation is known to exacerbate disease conditions.
Author contributions
M.Z., P.G. and S.M. designed the research; M.Z., G.M., F.B., R.D.M., C.M., P. B., R.L. and S.M. performed the research; M.Z., A.P., E.R., S.S., P.G. and S.M. analysed the data; M.Z. and S.M. wrote the paper; and all authors read and approved the final manuscript.
Conflict of interest
The authors declare no conflicts of interest.
Declaration of transparency and scientific rigour
This Declaration acknowledges that this paper adheres to the principles for transparent reporting and scientific rigour of preclinical research recommended by funding agencies, publishers and other organisations engaged with supporting research.
Supporting information
Figure S1 Effects of curcumin, GG6 and GG9 on NF‐κB p65 subcellular distribution in unstimulated and LPS‐stimulated cortical microglia. The fluorescence intensity of cytoplasmic and nuclear p65 subunit was calculated using ImageJ software. Results, as nuclear (dark bars) and cytoplasmic NF‐κB p65 (grey bars) are presented as a percentage of the total fluorescence. Data are mean ± SEM from at least three random fields of five separate experiments.
Figure S2 Supporting info item
Acknowledgements
This work was supported by ‘Progetto di Ateneo’, University of Padua, Italy (CPDA144389/14 to M.Z.); by a PRIN (20103W4779) project grant from MIUR, Italy; and by the Canadian Institute of Health Research (grants MOP‐123270 and MOP‐123500 to S.S.). The computational work coordinated by S.M. received support from the University of Padua. We thank Dr Carla Argentini for technical assistance, Dr Chiara Calore for assistance with atomic absorption spectroscopy and Dr Kensuke Miyake for providing the Ba/F3 cells. We are also very grateful to the Chemical Computing Group for the scientific and technical partnership. Finally, the authors would like to thank Dr Stephen D. Skaper for critical reading of the manuscript.
Zusso, M. , Mercanti, G. , Belluti, F. , Di Martino, R. M. C. , Pagetta, A. , Marinelli, C. , Brun, P. , Ragazzi, E. , Lo, R. , Stifani, S. , Giusti, P. , and Moro, S. (2017) Phenolic 1,3‐diketones attenuate lipopolysaccharide‐induced inflammatory response by an alternative magnesium‐mediated mechanism. British Journal of Pharmacology, 174: 1090–1103. doi: 10.1111/bph.13746.
References
- Akashi S, Shimazu R, Ogata H, Nagai Y, Takeda K, Kimoto M et al. (2000). Cutting edge: cell surface expression and lipopolysaccharide signaling via the toll‐like receptor 4‐MD‐2 complex on mouse peritoneal macrophages. J Immunol 164: 3471–3475. [DOI] [PubMed] [Google Scholar]
- Alexander SPH, Fabbro D, Kelly E, Marrion N, Peters JA, Benson HE et al. (2015). The Concise Guide to PHARMACOLOGY 2015/16: Catalytic receptors. Br J Pharmacol 172: 5979–6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amor S, Puentes F, Baker D, van der Valk P (2010). Inflammation in neurodegenerative diseases. Immunology 129: 154–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter CA, Murray CW, Clark DE, Westhead DR, Eldridge MD (1998). Flexible docking using Tabu search and an empirical estimate of binding affinity. Proteins Struct Funct Genet 33: 367–382. [PubMed] [Google Scholar]
- Bryant CE, Spring DR, Gangloff M, Gay NJ (2010). The molecular basis of the host response to lipopolysaccharide. Nat Rev Microbiol 8: 8–14. [DOI] [PubMed] [Google Scholar]
- Bsibsi M, Ravid R, Gveric D, van Noort JM (2002). Broad expression of toll‐like receptors in the human central nervous system. J Neuropathol Exp Neurol 61: 1013–1021. [DOI] [PubMed] [Google Scholar]
- Christian F, Smith EL, Carmody RJ (2016). The regulation of NF‐κB subunits by phosphorylation. Cells 5: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM Jr, Ferguson DM et al. (1995). A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J Am Chem Soc 117: 5179–5197. [Google Scholar]
- Curtis MJ, Bond RA, Spina D, Ahluwalia A, Alexander SP, Giembycz MA et al. (2015). Experimental design and analysis and their reporting: new guidance for publication in BJP. Br J Pharmacol 172: 3461–3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Martino RM, De Simone A, Andrisano V, Bisignano P, Bisi A, Gobbi S et al. (2016). Versatility of the curcumin scaffold: discovery of potent and balanced dual BACE‐1 and GSK‐3β inhibitors. J Med Chem 59: 531–544. [DOI] [PubMed] [Google Scholar]
- Frank‐Cannon TC, Alto LT, McAlpine FE, Tansey MG (2009). Does neuroinflammation fan the flame in neurodegenerative diseases? Mol Neurodegener 4: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Ding B, Zhou L, Gao X, Guo H, Xu H (2013). Magnesium sulfate provides neuroprotection in lipopolysaccharide‐activated primary microglia by inhibiting NF‐κB pathway. J Surg Res 184: 944–950. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Hayden MS (2008). New regulators of NF‐kappaB in inflammation. Nat Rev Immunol 8: 837–848. [DOI] [PubMed] [Google Scholar]
- Gradišar H, Keber MM, Pristovsek P, Jerala R (2007). MD‐2 as the target of curcumin in the inhibition of response to LPS. J Leukoc Biol 82: 968–974. [DOI] [PubMed] [Google Scholar]
- Gupta SC, Patchva S, Aggarwal BB (2013). Therapeutic roles of curcumin: lessons learned from clinical trials. AAPS J 15: 195–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halgren TA (1996). Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. J Comput Chem 17: 490–519. [Google Scholar]
- Hanisch UK, Kettenmann H (2007). Microglia: active sensor and versatile effector cells in the normal and pathologic brain. Nat Neurosci 10: 1387–1394. [DOI] [PubMed] [Google Scholar]
- Hatcher H, Planalp R, Cho J, Torti FM, Torti SV (2008). Curcumin: from ancient medicine to current clinical trials. Cell Mol Life Sci 65: 1631–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Yue Y, Zheng X, Zhang K, Chen S, Du Z (2015). Curcumin, inflammation, and chronic diseases: how are they linked? Molecules 20: 9183–9213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino K, Takeuchi O, Kawai T, Sanjo H, Ogawa T, Takeda Y et al. (1999). Cutting edge: toll‐like receptor 4 (TLR4)‐deficient mice are hyporesponsive to lipopolysaccharide: evidence for TLR4 as the Lps gene product. J Immunol 162: 3749–3752. [PubMed] [Google Scholar]
- Ishida J, Ohtsu H, Tachibana Y, Nakanishi Y, Bastow KF, Nagai M et al. (2002). Antitumor agents. Part 214: synthesis and evaluation of curcumin analogues as cytotoxic agents. Bioorg Med Chem 10: 3481–3487. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S (2010). The role of pattern‐recognition receptors in innate immunity: update on toll‐like receptors. Nat Immunol 11: 373–384. [DOI] [PubMed] [Google Scholar]
- Kigerl KA, de Rivero Vaccari JP, Dietrich WD, Popovich PG, Keane RW (2014). Pattern recognition receptors and central nervous system repair. Exp Neurol 258: 5–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilkenny C, Browne W, Cuthill IC, Emerson M, Altman DG (2010). Animal research: reporting in vivo experiments: the ARRIVE guidelines. Br J Pharmacol 160: 1577–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Jantaratnotai N, McGeer E, McLarnon JG, McGeer PL (2011). Mg2+ ions reduce microglial and THP‐1 cell neurotoxicity by inhibiting Ca2+ entry through purinergic channels. Brain Res 1369: 21–35. [DOI] [PubMed] [Google Scholar]
- Lehnardt S (2009). Innate immunity and neuroinflammation in the CNS: the role of microglia in toll‐like receptor‐mediated neuronal injury. Glia 58: 253–263. [DOI] [PubMed] [Google Scholar]
- Li J, Csakai A, Jin J, Zhang F, Yin H (2016). Therapeutic developments targeting toll‐like receptor‐4‐mediated neuroinflammation. Chem Med Chem 11: 154–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Tsai PS, Hung YC, Huang CJ (2010). L‐type calcium channels are involved in mediating the anti‐inflammatory effects of magnesium sulphate. Br J Anaesth 104: 44–51. [DOI] [PubMed] [Google Scholar]
- Mazur A, Maier JA, Rock E, Gueux E, Nowacki W, Rayssiguier Y (2007). Magnesium and the inflammatory response: potential physiopathological implications. Arch Biochem Biophys 458: 48–56. [DOI] [PubMed] [Google Scholar]
- McGrath JC, Lilley E (2015). Implementing guidelines on reporting research using animals (ARRIVE etc.): new requirements for publication in BJP. Br J Pharmacol 172: 3189–3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercanti G, Ragazzi E, Toffano G, Giusti P, Zusso M (2014). Phosphatidylserine and curcumin act synergistically to down‐regulate release of interleukin‐1β from lipopolysaccharide‐stimulated cortical primary microglial cells. CNS Neurol Disord Drug Targets 13: 792–800. [DOI] [PubMed] [Google Scholar]
- Mosmann T (1983). Rapid colorimetric assay for cellular growth and survival: application to proliferation and citotoxicity assays. J Immunol Methods 65: 55–63. [DOI] [PubMed] [Google Scholar]
- Nagai Y, Akashi S, Nagafuku M, Ogata M, Iwakura Y, Akira S et al. (2002). Essential role of MD‐2 in LPS responsiveness and TLR4 distribution. Nat Immunol 3: 667–672. [DOI] [PubMed] [Google Scholar]
- Oliveira AS, Sousa E, Vasconcelos MH, Pinto M (2015). Curcumin: a natural lead for potential new drug candidates. Curr Med Chem 22: 4196–4232. [DOI] [PubMed] [Google Scholar]
- Pabon HJ (1964). A synthesis of curcumin and related compounds. Recl Trav Chim Pays‐Bas 83: 379–386. [Google Scholar]
- Park BS, Lee JO (2013). Recognition of lipopolysaccharide pattern by TLR4 complexes. Exp Mol Med 45: e66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park BS, Song DH, Kim HM, Choi BS, Lee H, Lee JO (2009). The structural basis of lipopolysaccharide recognition by the TLR4‐MD‐2 complex. Nature 458: 1191–1195. [DOI] [PubMed] [Google Scholar]
- Park SH, Kim ND, Jung JK, Lee CK, Han SB, Kim Y (2012). Myeloid differentiation 2 as a therapeutic target of inflammatory disorders. Pharmacol Ther 133: 291–298. [DOI] [PubMed] [Google Scholar]
- Pohlert T (2014). The pairwise multiple comparison of mean ranks package (PMCMR). R package Available at: http://CRAN.R‐project.org/package=PMCMR.
- Poltorak A, He X, Smirnova I, Liu MY, Van Huffel C, Du X et al. (1998). Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science 282: 2085–2088. [DOI] [PubMed] [Google Scholar]
- Saitoh S, Akashi S, Yamada T, Tanimura N, Kobayashi M, Konno K et al. (2004). Lipid A antagonist, lipid IVa, is distinct from lipid A in interaction with toll‐like receptor 4 (TLR4)‐MD‐2 and ligand‐induced TLR4 oligomerization. Int Immunol 16: 961–969. [DOI] [PubMed] [Google Scholar]
- Shastri A, Bonifati DM, Kishore U (2013). Innate immunity and neuroinflammation. Mediators Inflamm 2013: 342931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimazu R, Akashi S, Ogata H, Nagai Y, Fukudome K, Miyake K et al. (1999). MD‐2, a molecule that confers lipopolysaccharide responsiveness on toll‐like receptor 4. J Exp Med 189: 1777–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel S, Castellan NJ Jr (1988). Nonparametric Statistics for the Behavioral Sciences, 2nd edn. McGraw‐Hill: New York. [Google Scholar]
- Skaper SD, Argentini C, Barbierato M (2012). Culture of neonatal rodent microglia, astrocytes, and oligodendrocytes from cortex and spinal cord. Methods Mol Biol 846: 67–77. [DOI] [PubMed] [Google Scholar]
- Sonna LA, Hirshman CA, Croxton TL (1996). Role of calcium channel blockade in relaxation of tracheal smooth muscle by extracellular Mg2+ . Am J Physiol 271: L251–L257. [DOI] [PubMed] [Google Scholar]
- Southan C, Sharman JL, Benson HE, Faccenda E, Pawson AJ, Alexander SPH et al. (2016). The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. Nucl Acids Res 44 (Database Issue): D1054–D1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart JJP (1993). MOPAC 7. Fujitsu Limited: Tokyo, Japan. [Google Scholar]
- Sugimoto J, Romani AM, Valentin‐Torres AM, Luciano AA, Ramirez Kitchen CM, Funderburg N et al. (2012). Magnesium decreases inflammatory cytokine production: a novel innate immunomodulatory mechanism. J Immunol 188: 6338–6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulevitch RJ, Tobias PS (1995). Receptor‐dependent mechanisms of cell stimulation by bacterial endotoxin. Annu Rev Immunol 13: 437–457. [DOI] [PubMed] [Google Scholar]
- Vyas A, Dandawate P, Padhye S, Ahmad A, Sarkar F (2013). Perspectives on new synthetic curcumin analogs and their potential anticancer properties. Curr Pharm Des 19: 2047–2069. [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Chen G, Chen L, Liu X, Fu W, Zhang Y et al. (2015). Insights into the binding mode of curcumin to MD‐2: studies from molecular docking, molecular dynamics simulations and experimental assessments. Mol Biosyst 11: 1933–1938. [DOI] [PubMed] [Google Scholar]
- Winnall WR, Muir JA, Hedger MP (2011). Differential responses of epithelial Sertoli cells of the rat testis to toll‐like receptor 2 and 4 ligands: implications for studies of testicular inflammation using bacterial lipopolysaccharides. Innate Immun 17: 123–136. [DOI] [PubMed] [Google Scholar]
- Wojciechowski M, Lesyng B (2004). Generalized born model: analysis, refinement, and applications to proteins. J Phys Chem B 108: 18368–18376. [Google Scholar]
- Youn HS, Saitoh SI, Miyake K, Hwang DH (2006). Inhibition of homodimerization of toll‐like receptor 4 by curcumin. Biochem Pharmacol 72: 62–69. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Jiang X, Peng K, Chen C, Fu L, Wang Z et al. (2014). Discovery and evaluation of novel anti‐inflammatory derivatives of natural bioactive curcumin. Drug Des Devel Ther 8: 2161–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liang D, Dong L, Ge X, Xu F, Chen W et al. (2015). Anti‐inflammatory effects of novel curcumin analogs in experimental acute lung injury. Respir Res 16: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zusso M, Methot L, Lo R, Greenhalgh AD, David S, Stifani S (2012). Regulation of postnatal forebrain amoeboid microglial cell proliferation and development by the transcription factor Runx1. J Neurosci 32: 11285–11298. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Effects of curcumin, GG6 and GG9 on NF‐κB p65 subcellular distribution in unstimulated and LPS‐stimulated cortical microglia. The fluorescence intensity of cytoplasmic and nuclear p65 subunit was calculated using ImageJ software. Results, as nuclear (dark bars) and cytoplasmic NF‐κB p65 (grey bars) are presented as a percentage of the total fluorescence. Data are mean ± SEM from at least three random fields of five separate experiments.
Figure S2 Supporting info item


