Abstract
Determining the spatial relationship of individual proteins in dense assemblies remains a challenge for superresolution nanoscopy. The organization of aquaporin-4 (AQP4) into large plasma membrane assemblies provides an opportunity to image membrane-bound AQP4 antibodies (AQP4-IgG) and evaluate changes in their spatial distribution due to alterations in AQP4 isoform expression and AQP4-IgG epitope specificity. Using stimulated emission depletion nanoscopy, we imaged secondary antibody labeling of monoclonal AQP4-IgGs with differing epitope specificity bound to isolated tetramers (M1-AQP4) and large orthogonal arrays of AQP4 (M23-AQP4). Imaging secondary antibodies bound to M1-AQP4 allowed us to infer the size of individual AQP4-IgG binding events. This information was used to model the assembly of larger AQP4-IgG complexes on M23-AQP4 arrays. A scoring algorithm was generated from these models to characterize the spatial arrangement of bound AQP4-IgG antibodies, yielding multiple epitope-specific patterns of bound antibodies on M23-AQP4 arrays. Our results delineate an approach to infer spatial relationships within protein arrays using stimulated emission depletion nanoscopy, offering insight into how information on single antibody fluorescence events can be used to extract information from dense protein assemblies under a biologic context.
Introduction
Protein spatial distribution within larger assemblies is often intimately linked to protein function. Historical approaches to visualize protein distribution at high spatial resolution have been largely limited to electron microscopy, as conventional light microscopy is restricted to low spatial resolution (∼200–300 nm laterally) (1). In stimulated emission depletion (STED) nanoscopy, optical superresolution is obtained by depletion of the fluorescence emission peripheral to the excitation beam target. Depletion is elicited by a red-shifted STED beam that is shaped into a donutlike intensity distribution for two-dimensional (2D) resolution enhancement (2, 3), with the zero intensity centered over the excitation beam. The STED beam effectively switches off fluorescent molecules in the periphery of the excitation spot, but not in the zero-intensity center. As a result, STED nanoscopy increases resolution down to tens of nanometers, allowing for novel studies of protein spatial distribution and function. For example, STED localization of surface protein assemblies has provided insight into both vesicular membrane protein recycling and HIV-1 host cell infectivity (4, 5).
It remains a challenge to identify the geometric arrangement and stoichiometry of individual proteins within larger assemblies in vivo. The light emitted by a single fluorophore forms a Gaussian intensity distribution—a finite-sized spot—that will blend with light emitted by other fluorophores when packed at high densities. Recognizing individual proteins within dense assemblies often requires restrictive experimental conditions that perturb the model system away from the in vivo environment to reorganize protein assemblies into resolvable components or to observe real-time protein dynamics (6). In addition, fluorophore tags or secondary fluorescent antibodies may interfere with normal structural arrangement or biological function. Access to primary and secondary antibody epitopes and variable labeling efficiency pose further imaging challenges (7). Finally, uncertainty in the orientation of the fluorophores and the target protein itself provide additional restrictions. A better understanding of how individual fluorophores can be resolved at higher densities in a nondisruptive biologic context would allow for the development of more rigorous methods to correlate spatial protein distributions with functional outcomes.
In the central nervous system (CNS) disorder neuromyelitis optica (NMO), aquaporin-4 autoantibodies (AQP4-IgG) bind to the extracellular domains of AQP4 tetramers expressed by CNS astrocytes and initiate injury via classical complement pathway activation (8, 9, 10, 11). High-level classical pathway activation is triggered when multimeric contacts are made between the complement protein C1q and membrane-bound antibodies (12, 13). Elucidating the molecular mechanisms driving the formation of multimeric contacts between C1q and aquaporin-4-(AQP4) recombinant antibody (rAb) within the C1q–AQP4-IgG–AQP4 complex is therefore of considerable interest to combat CNS injury in NMO. AQP4-IgG will only activate C1q when AQP4-IgG binds over large arrays of AQP4 protein termed “orthogonal arrays of particles” (OAP), suggesting that high density binding of AQP4-IgG to AQP4 arrays facilitates multivalent C1q-antibody contacts and complement activation (14). We have generated a large repertoire of AQP4-specific monoclonal rAbs from NMO patient cerebrospinal fluid plasmablasts, allowing us to test this hypothesis by visualizing multiple unique antibody binding patterns both as isolated binding events and in larger clusters.
Due to its deterministic technique, STED nanoscopy is well suited to dissect blended fluorescence emission after the recognition of single fluorescence events. Here, we use STED nanoscopy to analyze these isolated and blended fluorescence patterns produced by AQP4-IgG to develop a framework for evaluating dynamic changes in protein distribution in a biologic context. The result is an algorithm that deconstructs blended fluorescence emission patterns to infer the distribution of fluorophore molecules without direct resolution. Application of this algorithm to STED images of multiple AQP4 rAbs bound to AQP4 tetramers and OAPs demonstrates an unappreciated role that target epitopes may play in organizing C1q–AQP4-IgG–AQP4 complexes.
Materials and Methods
Cell samples and AQP4 autoantibodies
Monoclonal anti-AQP4 rAbs were generated from NMO patient CSF as described in Bennett et al. (15), and bound to live CHO cells stably expressing pure M1-AQP4 or M23-AQP4. Each AQP4 rAb binds a unique extracellular epitope (16). AQP4 tetramers and OAPs were labeled with a rabbit polyclonal anti-human AQP4 antibody specific to the intracellular C terminus (Santa Cruz Biotechnology, Santa Cruz, CA) and detected with goat anti-rabbit STAR590 (Rockland Immunochemicals, Limerick, PA). Extracellular AQP4 rAb was detected with biotinylated Fab goat anti-human Fc (Novus Biologicals, Littleton, CO) and streptavidin-conjugated Atto647N (Atto-Tec, Siegen, Germany; see the Supporting Material).
Image acquisition and analysis
STED images were obtained on a noncommercial two-color STED nanoscope at the Anschutz Medical Campus Light Microscopy Core. The nanoscope is described in Meyer et al. (17); lateral resolution was calculated by imaging FluoSpheres carboxylate-modified microspheres, 0.02 μm, filled with dark-red fluorescent fluorophores (660/680, ThermoFisher Cat. No. F8783; Thermo Fisher Scientific, Waltham, MA) ± unconjugated Atto647N fluorophores (Atto-Tec). Details on acquired and simulated image acquisition, processing and quantitative analyses are described in Supporting Material.
Results
Characterizing STED nanoscope resolution using a biologic approach
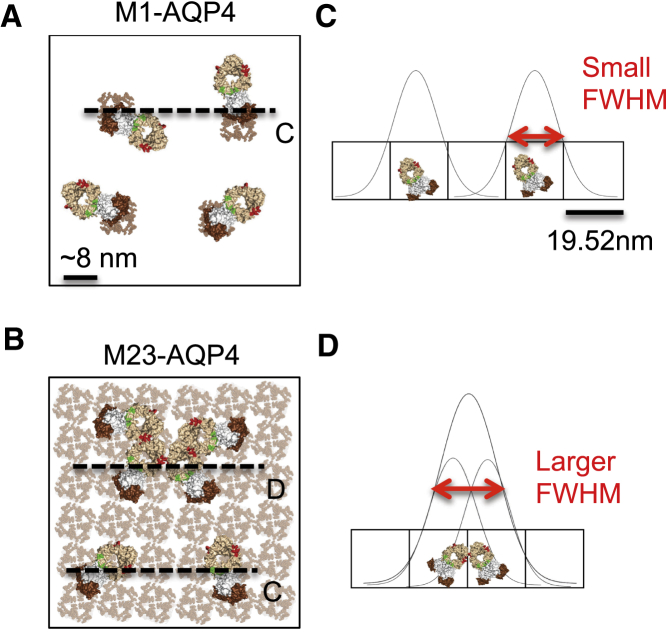
To establish a framework for evaluating dynamic changes in fluorescence emission summation patterns based on fluorophore spatial distribution, we first considered the biological variables that may influence fluorophore spatial distribution. In astrocytes, AQP4 is expressed in two isoforms: M1-AQP4 and M23-AQP4. M1- and M23-AQP4 are coassembled into tetramers with differential abilities to coalesce into larger orthogonal arrays of particles (OAPs) (18, 19). M1-AQP4 inhibits OAP assembly and, when solely expressed, assembles into isolated plasma membrane tetramers. In distinction, M23-AQP4 facilitates AQP4 tetramer assembly into larger OAPs that may span hundreds of nanometers in size. Based on the relative size of an AQP4 tetramer (8–9 nm) and IgG (12–15 nm), only a single AQP4 rAb molecule is presumed to bind to a M1-AQP4 tetramer, whereas multiple rAbs may bind to the surface of M23-AQP4 OAPs (Fig. 1, A and B) (20). In this model, single antibodies bound to plasma membrane M1-tetramers should be readily resolved because the distance between fluorophores is large compared to the resolution of the STED nanoscope (Fig. 1 C). In contrast, on M23-AQP4 OAPs, the short distance between bound AQP4 rAbs may allow fluorophores to cluster at distances smaller than the resolution of the STED nanoscope, thereby interfering with the detection of discrete binding events. As a result, on OAPs, fluorescent signal may represent multiple antibodies binding in close proximity (a cluster), rather than a single antibody molecule (Fig. 1 D).
Figure 1.
AQP4 autoantibodies have the potential to bind in multiple spatial distributions to M1-AQP4 and M23-AQP4. (A and B) Surface schematics of AQP4 rAbs bound to cell surface AQP4. (A) M1-AQP4 tetramers repel each other on the cell membrane, resulting in isolated, bound AQP4 autoantibodies. (B) M23-AQP4 organizes into large arrays on a cell membrane, offering the potential for multiple spatial distributions of bound autoantibodies depending on epitope geometry and organization. (C and D) The size of a resolvable object, defined as the FWHM of a Gaussian fit, is proportional to the size of antibody clusters given the size of each pixel relative to the size of a single antibody. The antibody distributions in (C) and (D) represent the distributions indicated by dashed lines in (A) and (B), with STED pixel coordinates and a theoretical Gaussian fit superimposed.
It is also possible that the geometric arrangement of targeted AQP4 epitopes may influence the ability for AQP4 rAb to bind in dense clusters over larger arrays (Fig. 1 B). Biochemical single amino acid epitope mapping has demonstrated that AQP4 rAb vary in both the number of extracellular loops contacted and the amino acids contacted within each loop (16). It is unclear if the epitope surface area overlays a single AQP4 monomer or spans multiple monomers within each tetramer (20); the global arrangement of all extracellular loops within the tetramer is compatible with each possibility but favors contacts spanning AQP4 monomers. Targeted epitopes may therefore differ in amino acids contacted, overall size, and geometric patterning across AQP4 tetramers and OAPs. Furthermore, steric considerations may influence epitope availability. Epitopes whose area crosses the center point of the AQP4 tetramer would have at least partially overlapping surface areas given the fourfold symmetry of an individual tetramer; consequently only one of four available epitopes could be occupied at any given time. Alternatively, a bound antibody could mask neighboring epitopes given the larger size of an antibody (∼10–15 nm) relative to the size of an AQP4 tetramer. Thus, while targeted epitopes theoretically are distributed uniformly across M23-AQP4 OAPs to permit high-density antibody binding, binding could be limited to lower densities if the accessible pool of vacant epitopes is restricted by AQP4 epitope geometry and steric considerations. We therefore hypothesized that the observed heterogeneity of target epitopes across M23-AQP4 OAPs would produce nonuniform patterns of AQP4 rAb binding with differing abilities to form dense protein assemblies: some target epitopes may be spaced to permit large numbers of AQP4 rAb binding in close proximity (Fig. 1 B, line D), whereas others may restrict rAb binding in close proximity (Fig. 1 B, line C). Consequently, some monoclonal AQP4 rAbs may appear as similar-sized objects on both AQP4 isoforms, while others may appear as larger objects on M23-AQP4 OAPs (Fig. 1, B–D).
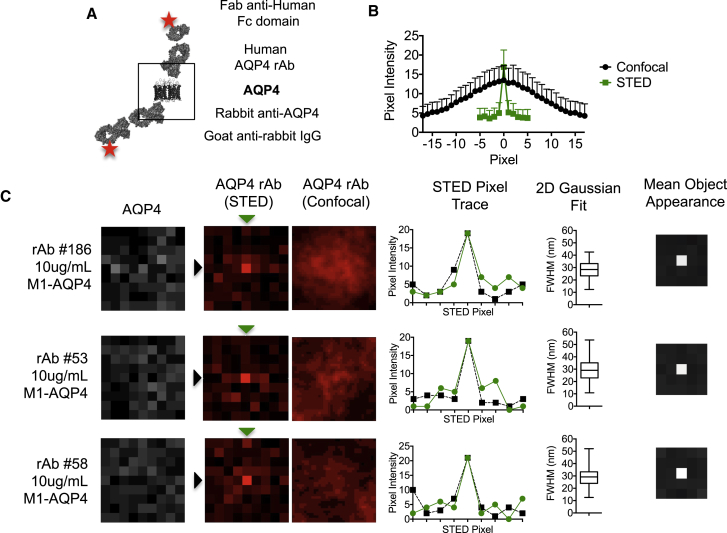
To test these hypotheses, several individual monoclonal AQP4 autoantibodies recognizing unique AQP4 extracellular epitopes were bound to live cells expressing either M1-AQP4 or M23-AQP4 and imaged via STED nanoscopy. The fluorophore labeling approach takes advantage of monoclonal antibody labels to isolate Atto647N fluorophores (Fig. 2 A), and all AQP4 tetramers are intracellularly labeled with a distinct commercial antibody to eliminate competition for extracellular rAb epitopes. AQP4 rAb bound to M1-AQP4 consistently appeared as single pixels that directly overlaid or neighbored an AQP4 tetramer; the bulk size of the intracellular antibody labeling approach has the potential to place the labeling fluorophore on a neighboring pixel (Fig. 2, A and C). Comparison to confocal images of rAb in identical fields of view reveals significant resolution improvement with STED (Fig. 2, B and C). A least-squares 2D Gaussian fit over isolated peaks quantified rAb appearance over M1-AQP4, yielding a mean peak full width at half-maximum (FWHM) of 29.1 ± 8.7, 29.1 ± 7.3, and 28.4 ± 6.5 nm for rAbs #53 (n = 102), #58 (n = 105), and #186 (n = 99), respectively, congruent with the mean appearance of rAb signal (Fig. 2 C, right). These mean peak FWHM are consistent with the appearance of Atto647N fluorophores when imaged in the same focal plane as 28 nm fluorescent beads filled with dark-red 660/680 fluorophores sampled at both 10 and 20 nm pixel sizes (Figs. S1 and S2; one-way ANOVA, p = 0.90). Single pixel localization was further verified via imaging of serial fluorophores dilutions when imaged at 20 nm pixel sampling size (Fig. S1); single pixels also fluoresced across three repetitive images in the same field of view for 19.2 ± 1.0% of events despite photobleaching (data not shown). Hence, in our imaging system, rAb fluorescence over M1-AQP4 definitively localizes to single 20 nm pixels with a mean peak FWHM of <30 nm.
Figure 2.
STED imaging of AQP4 rAb over M1-AQP4. (A) Depiction of labeling scheme is given. Red stars indicate position of fluorophore label. Box represents size of 19.5 × 19.5 nm STED pixel; the schematic is approximately to scale. (B) Pixel intensity traces for STED and confocal detection of binding events over M1-AQP4 are given. Data represents n = 30 events (n = 10 each for rAbs #53, #58, #186). (C) Representative STED and confocal images of antibodies binding to AQP4 are given. Pixel intensity traces for all STED rAb rows/columns labeled with an arrowhead are shown immediately adjacent. The mean peak FWHM quantifications using a least-squares 2D Gaussian fit, and the mean object appearance for rAbs #186 (n = 99), rAb #53 (n = 102), and rAb #58 (n = 105), are depicted on far right. Scale: all image pixels are 19.5 × 19.5 nm.
rAb bind in multiple arrangements over M1- and M23-AQP4
We next addressed how the appearance of rAb bound over M1-AQP4 could be exploited to test for differing spatial arrangements when the same rAb is bound over M23-AQP4. We first asked if we could detect gross changes in rAb clustering for rAb bound over M1- versus M23-AQP4 as rationale for pursuing more rigorous deconstructions of fluorescence blur. rAb clustering was analyzed by calculating the FWHM of a 2D Gaussian fit (herein called FWHMrAb spread) for the 2D array representing mean blur across an entire image, acquired using an adapted blinded deconvolution algorithm based on a maximum likelihood algorithm (21). Differences in FWHMrAb spread, arising from differing fluorescence summation patterns, indicate distinct spatial distributions of bound fluorophores given the relative size of a single antibody compared to the STED pixel dimensions (12–15 vs. 19.52 nm, respectively; Fig. 1, C and D). As anticipated by AQP4 membrane biology, the FWHMrAb spread for all three unique AQP4 rAb bound over M1-AQP4 was similar to both single fluorophore point sources and fluorescently labeled antibodies randomly immobilized on glass coverslips (Fig. 3 D, one-way ANOVA, p = 0.16). The data establishes the base FWHMrAb spread of the predominant fluorescence signal, and further supports that the majority of resolvable objects for AQP4 rAb bound to M1-AQP4 tetramers represent isolated antibodies on the cell surface.
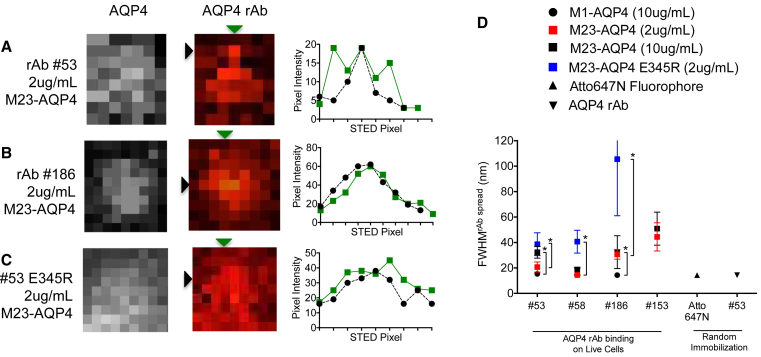
Figure 3.
Quantification of cluster spread for rAb bound over M1- and M23-AQP4. (A–C) Representative antibody images are shown for several AQP4 autoantibodies binding to M23-AQP4. An intracellular antibody (left) detects all AQP4; extracellular autoantibody binding (middle) is shown with pixel intensity traces (right) for all rows/columns labeled with an arrow. (D) Calculated mean FWHMrAb spread across an entire image. Data was compared using one-way ANOVA with Tukey’s test for multiple comparisons (∗, adjusted p < 0.05). STED image scale: 19.5 × 19.5 nm/pixel. Scale: all image pixels are 19.5 × 19.5 nm.
In contrast, the FWHMrAb spread for rAb bound to M23-AQP4 OAPs increased significantly for multiple AQP4 rAb antibodies. At 10 μg/mL, the FWHMrAb spread for rAb #58 was similar between M1-AQP4 tetramers and M23-AQP4 OAPs; however, the mean FWHMrAb spread for rAbs #53 and #186 were significantly larger (Fig. 3 D), suggesting an epitope-dependent clustering of antibody on M23-AQP4 OAPs (Fig. 3, A, B, and D). To test this conclusion, we introduced a point mutation (E345R) into the Fc domain of several AQP4 rAbs to promote the assembly of individual antibodies into larger surface clusters (22). AQP4 antibodies containing the E345R Fc mutation were detected as larger rAb clusters compared to their wild-type counterparts (Fig. 3, A, C, and D). Increasing antibody concentrations did not significantly shift the distribution of the FWHMrAb spread for rAb #58, #186, and #153, demonstrating that the increase in cluster size represented an antibody-intrinsic, epitope-driven phenomenon rather than a random juxtaposition of multiple independent binding events on AQP4 OAPs (Fig. 3 D). Interestingly, the FWHMrAb spread for rAb #53 trended toward a significant increase at higher concentrations on M23-AQP4 OAPs (21 ± 3 nm at 2 μg/mL versus 33 ± 3 nm at 10 μg/mL; Tukey’s adjusted p = 0.055), suggesting that increased occupancy of rAb #53 epitopes facilitated surface clustering over this concentration range. In summary, manipulation of AQP4 assembly (M1-AQP4 tetramers versus M23-AQP4 OAPs) and rAb interaction (E345R Fc mutation) allowed us to detect shifting spatial arrangements of AQP4 rAbs on the cell surface.
Generation of score of antibody spatial arrangement
The spatial arrangement of antibody molecules bound over surface epitopes may have a profound impact on antibody Fc-domain mediated effector function activation (22). For example, effector complement activation requires C1q binding to multiple antibodies on a target surface (Fig. 4 A). The target surface likely organizes antibodies into clusters that facilitate multivalent C1q contacts, as isolated bound antibodies would more closely resemble circulating monomeric antibodies that have only a low affinity for C1q. Indeed, the limited ability of AQP4 rAb to bind C1q and activate the classical complement pathway over M1-AQP4 suggests that the spatial organization of bound antibodies contributes to complement activation (14). We therefore formulated a metric to quantify the C1q multivalent binding potential for antibody spatial arrangements, based on the mean appearance of rAb binding over M1-AQP4 (Fig. 2 C), to ask if fluorophore spread within variable blended fluorescence patterns could be more rigorously dissected.
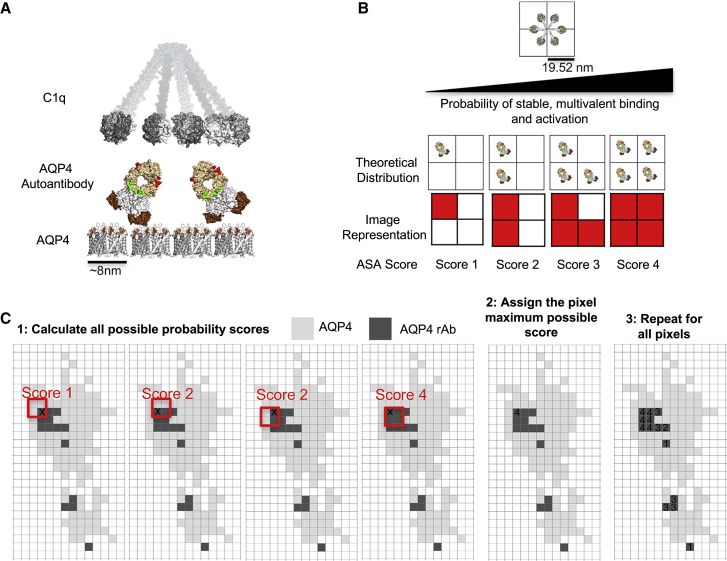
Figure 4.
A method to score rAb antibody scaffolds that interact with C1q. (A) A schematic of C1q interacting with membrane-bound AQP4 autoantibodies demonstrating the potential assembly of multivalent autoantibodies on AQP4 OAPs is shown. (B) (Top) Top-down depiction of a C1q molecule super-imposed over STED pixel coordinates is given. (Bottom) Schematic paradigm for scoring distributions of AQP4 antibodies that support multivalent C1q binding is given. (C) Sample application of the ASA scoring system to a theoretical STED image is given. The ASA score is determined in an iterative fashion using code from the software MATLAB (The MathWorks, Natick, MA). All pixels with a fluorescence signal are scored for each of the four possible ASA score arrangements (1), then the pixel is assigned the maximum possible ASA score (2), and the process is repeated for all pixels across the image (3).
C1q has a stem-and-tulips structure, with each of the six stems containing a globular head that is able to bind to an antibody Fc domain (Fig. 4 A). While there are multiple spatial distributions of AQP4 rAb that can support multivalent C1q binding, a hexamer pattern potentially represents a best-fit solution that can engage each of the globular heads (22). When superimposed over STED image pixel dimensions, multivalent C1q binding has the potential to occur across four pixels in a 2 × 2 pixel area (Fig. 4 B, top). We developed a four-point scoring system to quantify the probability of multivalent C1q binding termed the “antibody spatial arrangement” (ASA) score. Each pixel from a processed image receives a ranked integer score (1–4) based on the maximum number of neighboring pixels with signal intensity across a 2 × 2 area (Fig. 4 B, bottom). ASA score 1 represents an isolated binding event with low probability for multivalent binding, whereas ASA score 4 represents a dense cluster with the highest probability for multivalent C1q binding. The effective application of this scoring system across an entire cell (Fig. 4 C) is dependent on the ability to localize a fluorophore to each pixel, consistent with the imaging of isolated Atto647N fluorophores (Fig. S1). However, because individual fluorophores cannot theoretically be resolved on neighboring pixels (see Fig. 1, C and D), a series of data simulations were performed to reconstruct single binding events (as observed on M1-AQP4) within denser clusters (as observed on M23-AQP4) to test the possibility of developing an image processing algorithm that can assign fluorophores to any given pixel in an otherwise nonresolvable object.
Data simulations validate the ASA algorithm
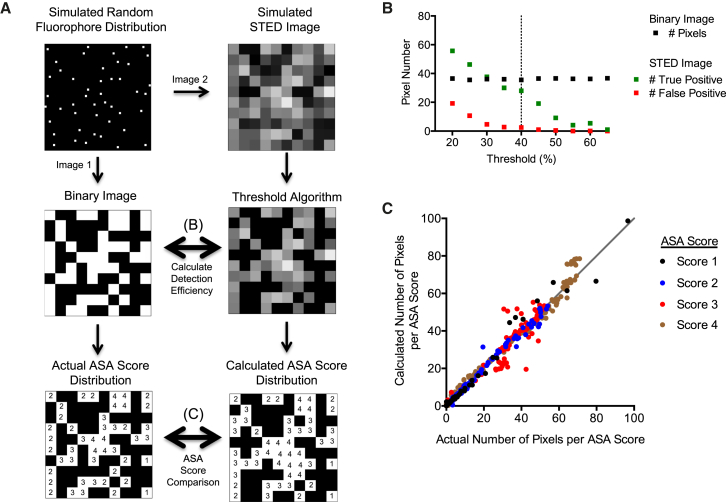
Random fluorophore distributions were generated to model antibody spatial distributions over 100 AQP4 arrays on a simulated cell (Fig. 5 A, top left). Two simulated images were generated for each random distribution. In one image, STED image pixel dimensions were immediately superimposed to create a binary image where each pixel with an intensity value represents a pixel containing at least one Gaussian fluorophore peak (Fig. 5 A, middle left). In a second image, each fluorophore was convolved based on the appearance of single rAb #58 binding events over M1-AQP4. STED image pixel dimensions were then superimposed over the second image to generate a simulated image representing a STED image for that particular fluorophore distribution (Fig. 5 A, top right).
Figure 5.
Data simulations validate the ASA scoring system on large protein assemblies. (A) Application of the ASA scoring system to a simulated OAP distribution is given. A random fluorophore distribution is created (top left) and converted into both a binary image demonstrating pixels containing a fluorophore (left middle) and a simulated STED image (top right). The simulated STED image was placed through a series of experimental thresholds to test how well a threshold may identify fluorophore-containing pixels, compared to the known distribution of fluorophores (middle row). The ASA score for both known and calculated distributions was tallied (bottom row) and compared. (B) Sample AQP4 array data demonstrating the accuracy of various detection thresholds is given. (C) Quantification of the performance of the final ASA scoring algorithm using a 40% threshold on independent simulated data set is given.
We then asked if a threshold could be reliably applied to identify pixels containing a fluorophore. The threshold represents a cutoff percentage where any pixel containing an intensity value below the threshold percentage (relative to the maximum local pixel intensity value over the AQP4 array) is reassigned a value of “0” to exclude the pixel from further analysis (see Supporting Materials and Methods). An intensity threshold effectively eliminates pixels with a low probability of containing a fluorophore, as any detected photon events from these pixels would have a low probability of falling within the FWHM of a single fluorophore event. After threshold application, each remaining pixel has a high probability of having a Gaussian peak of intensity falling somewhere within that pixel. Too low of a threshold would produce a high number of false positives (low specificity), while too high of a threshold would only detect the brightest events (low sensitivity). All simulated images were stepped through a series of thresholds at 5% increments to identify an endpoint with high sensitivity and specificity across a range of fluorophore densities (2.7–74.9% of array area, ∼100 OAPs quantified per density). The threshold accuracy was determined by comparing all pixels containing detected events (defined as all nonzero pixels postthreshold; Fig. 5 B, middle right) to the binary image to calculate the number of true- and false positive events (Fig. 5 B). With a threshold pixel intensity cutoff of 40%, 88.2% of all fluorophores were accurately localized with a 12.6% false positive rate. Although a 35% threshold yielded similar sensitivity, this threshold produced a higher number of false positives at denser concentrations (data not shown).
We subsequently used the 40% threshold to compare the calculated ASA score distribution for all positive pixels with the actual ASA score distribution on binary control images (Fig. 5 A, bottom). As anticipated from the simulation design (see Discussion), a linear relationship between calculated and actual ASA scores was not observed. However, each calculated score was readily normalized, as score distributions followed clear polynomial relationships driven by the size and complexity of blended shapes. The final algorithm was then tested with a second simulation series of random fluorophore distributions. Calculated ASA scores were highly correlated with actual ASA score distributions for ASA scores of 1, 2, and 4 (r2 = 0.98, 0.99, and 0.98, respectively). ASA score 3 showed a slightly weaker correlation (r2 = 0.83) as the algorithm was not as accurate at higher fluorophore densities (Fig. 5 C). The best fit linear regression of each score was not significantly different from y = x (f = 0.62, p = 0.65). When averaged across the entire data set, the total calculated distribution represented the theoretical distribution with 98% accuracy. We conclude that the geometric spread of dense protein clusters can be inferred at high probability from an otherwise nonresolvable object, given prior knowledge of how individual fluorophores are represented within the object.
Monoclonal antibodies display different potentials for multivaelnt C1q binding
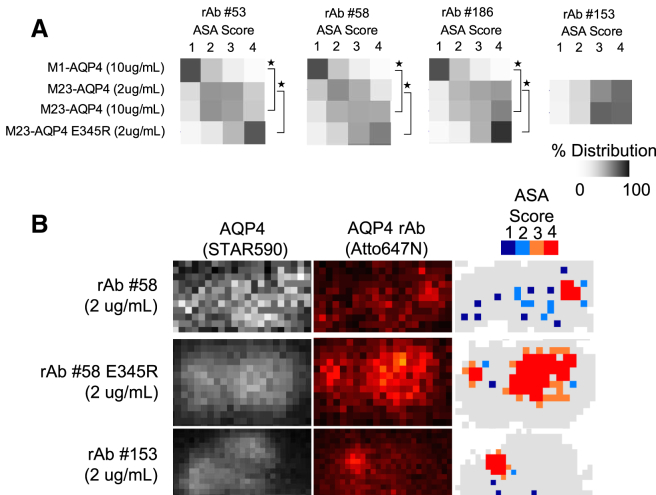
We evaluated STED images of AQP4 rAbs on M1- and M23-AQP4 to evaluate the multivalent binding potential of resolvable objects using the ASA scoring system. Consistent with the lack of C1q-mediated complement activation on M1-AQP4 tetramers, the ASA scores for images of AQP4 rAbs on M1-AQP4 differed significantly from images on M23-AQP4. On M1-AQP4, the distribution of ASA scores was heavily skewed toward “1”. For rAbs #53 and 58, ASA probability scores on M23-AQP4 were skewed toward “2” or higher, despite either no detectable increase, or only a small increase in apparent FWHMrAb spread relative to M1-AQP4 (Fig. 3 D and Fig. 6 A). An even larger skewing was observed for rAbs #186 and #153 binding to M23-AQP4 OAPs. The larger FWHMrAb spread generated by these two antibodies (Fig. 3 D) were reflected in significantly higher median ASA scores. An identical shift in the ASA scoring distribution was produced when the clustering mutation E345R was introduced into rAbs #53, #58, and #186. Interestingly, some pixels with larger ASA scores (3 and 4) were detected at significant frequencies in images of rAbs that otherwise averaged smaller FWHMrAb spread (e.g., rAbs #53 and #58). This may represent close juxtaposition of individual binding events. The optimized ASA scoring algorithm was used for the analysis for increased confidence; the overall data patterns calculated with the optimized ASA algorithm did not deviate from those that were otherwise observed using initial ASA calculations (Fig. S3). In summary, by applying a functional analysis constraint based on the dimensions of the C1q–AQP4 rAb–AQP4 complex, we were able to identify different spatial arrangements among blended fluorescence patterns produced by densely packed rAbs within a confined surface area.
Figure 6.
ASA scores of membrane-bound AQP4 rAbs. (A) The ASA score distributions for multiple AQP4 rAb are presented as a heat map representing the relative frequency of each ASA score across all OAPs (M23-AQP4) or across an entire cell (M1-AQP4). The median ASA score was significantly increased for all AQP4 rAbs on M23-AQP4 compared to M1-AQP4 and for E345R-mutated AQP4 rAbs on M23-AQP4 (∗p < 0.001, Mann-Whitney). (B) Sample STED images of AQP4 (left) and AQP4 rAb (middle) for three AQP4 rAbs are given. The corresponding ASA score (right) is shown for the AQP4 rAb image (middle). Here, gray represents an AQP4 array, and each ASA score is represented by a colored pixel (1: Dark blue, 2: light blue, 3: orange, 4: red). Scale: all image pixels are 19.5 × 19.5 nm.
Discussion
We quantified multiple spatial arrangements for membrane-bound AQP4 autoantibodies with STED nanoscopy using two distinct biologic contexts. Using relatively dispersed M1-AQP4 tetramers, we were able to visualize single antibody binding events in our STED images and subsequently use that information to develop methods to recognize differences in the spatial organization of antibodies in close proximity on M23-AQP4 arrays. The recognition of an individual binding event on M1-AQP4 also allowed us to more rigorously dissect differing spatial arrangements of dense protein clusters without the direct ability to absolutely resolve events between neighboring pixels. Distinct spatial organizations were observed for some, but not all, monoclonal rAbs when comparing images on M1- and M23-AQP4. These differences correlated with changes in spatial distribution induced by a Fc-mutation that facilitates antibody clustering, suggesting that target epitopes may be spaced and oriented on M23-AQP4 OAPs to facilitate AQP4 rAb interaction, and subsequent C1q binding in vivo. Characterizing the relationship between individual fluorophore distributions and blended fluorophore fluorescence summation patterns provides insight into the fields of both superresolution image analysis and neuromyelitis optica.
Superresolution imaging of protein clusters
The advent of superresolution imaging has introduced many new challenges in resolving individual proteins in vivo. Ideally, images would be obtained in a manner that allows for the resolution of individual molecular targets without compromising specificity or function. The relative size of many proteins compared to the size of the detecting fluorophore has the potential to confound precise localization and disrupt the local cellular environment. For example, our attempts to produce AQP4 rAb fusion proteins containing autofluorescent proteins resulted in disruption of both antibody binding and complement-activation (unpublished data). Labeling efficiency may also impact accuracy in quantifying protein assemblies. In our model, the C-terminal intracellular AQP4-specific commercial antibody labels entire arrays, whereas extracellular-targeted rAbs label only portions of arrays. Therefore, the two antibodies would produce different results when used to calculate the size and number of AQP4 OAPs. The impact of such labeling artifacts on image resolution was recently investigated in detail by Lau et al. (7).
Alternative superresolution imaging approaches may offer novel solutions. For example, innovative single-molecule fluorophore labeling approaches and STORM imaging have been used to investigate both the mobility of M1- versus M23-AQP4 tetramers on plasma membranes and the organization of M1- versus M23-AQP4 tetramers within OAPs (23, 24, 25). Atomic force microscopy has been used to demonstrate the potential for IgG antibodies to form hexameric assemblies (26) and for antibodies to move along repetitive surfaces and form transient antibody clusters (27). In the future, atomic force microscopy could be adapted to examine antibody hexameric assemblies on AQP4 OAPs. Regardless, overcoming the stringent lateral resolution demands imposed by protein size for resolving single proteins remains a universal challenge in constructing models of protein assemblies. For example, despite outstanding resolution, STORM imaging of AQP4 tetramers has not resolved individual AQP4 tetramers within larger arrays (23, 24, 25, 28). Modeling protein crystal structures over nonatomic images may represent novel strategies (7, 22).
Imaging environment, sample preparation, nanoscope design, and the efficiency of fluorophore depletion may influence STED nanoscope resolution. Deriving a PSF from larger beads is further influenced by object sampling, assumption of PSF and bead shapes, accuracy of bead size, bead integrity, and the need for deconvolution (4, 29, 30, 31, 32, 33, 34). Some groups have reported STED resolutions near 20 nm (29, 32, 35, 36). In our study, we calculated a mean peak FWHM <30 nm for single fluorophores despite encountering multiple challenges attributable to pixel sampling size (Figs. 2 and S2). Both sampling at a smaller 10 nm pixel size within Nyquist criterion (Fig. S2 H) and further fluorophore manipulation of 20 nm pixels (Figs. S1 and S2) improved confidence in the calculation. To our knowledge, this is the first study reporting a FWHM calculation derived using serial fluorophore dilutions imaged in the same focal plane as larger beads (Figs. S1 and S2). The experiment provides a priori information regarding fluorophore appearance. As described by Westphal et al. (30), the application of a priori information reduces the signal demand required to identify individual objects, which, in conjunction with differing fluorophore photoproperties, likely accounts for our improved ability to localize the Atto647N fluorophore compared to the larger beads filled with a distinct fluorophore (17). Future adaptations and refinements may provide further insight into how best to characterize objects potentially smaller than nanoscope PSF. Nevertheless, multiple experimental approaches consistently localized Atto647N fluorophores to single 20 nm pixels. Regardless of sampling and quantification approach for FWHM, the nanoscope resolution is more than sufficient for downstream analyses of fluorophore spread because these analyses apply distinct image transformations to detect gross changes in fluorescence summation. FWHM calculation error arising from the encountered challenges would therefore not impact algorithm performance or data interpretation, which instead relies on relative comparisons to intrinsic biologic controls.
Despite the reliance on single pixel intensities to capture fluorescence emission signal, our ASA analysis indicates that the geometric spread of protein spatial arrangements can be accurately inferred with high probability at larger pixel sampling sizes (Fig. 5). Although a multivalent IgG platform is a known requirement for C1q binding, inadequate understanding of C1q binding site geometries, combined with uncertainty regarding the position of the secondary fluorophores, precludes any further benefit from higher sampling resolutions. Furthermore, regardless of pixel size, an ASA analysis would require additional understanding of the summation of fluorescence emission distributions as the optical resolution needed to resolve single antibodies is unlikely to be achieved given the small size of a single antibody independent of superresolution imaging approach.
In our biologic model, multiple solutions for each ASA score are possible as each rAb has the potential to bind one of multiple vacant epitopes. Consequently, a range of fluorescence summation patterns are expected for each ASA score and the relationship between the calculated and actual ASA scores will be dependent on the overall complexity of the summation patterns across the entire array. As a result, linearity will be lost with more complex summation patterns; however, a basic understanding of these patterns allows for normalization. Importantly, the consistency of algorithm performance at variable levels of array saturation demonstrates that the detection of all pixels containing a fluorophore is not a prerequisite for accurate detection of AQP4 rAb protein cluster formation.
The relationship between threshold levels and raw ASA score distributions (Fig. S3) demonstrate that the observed fluorophore spatial arrangements are primarily driven by the target epitope and not the labeling approach. As a result, a small sampling of events should be sufficient to provide a robust quantification, potentially failing when the threshold becomes biased to capture only the most complex spatial arrangements (Fig. S3). Furthermore, the magnitude between the number of false positives and false negatives was relatively consistent until these higher threshold intensities were reached (Fig. 5 B). The observations suggest that successful algorithm application is not necessarily limited to a small range of threshold values, despite the inability to achieve 100% sensitivity. Further testing in other experimental environments is required to understand how generalizable the algorithm may be and to identify additional algorithms to dissect fluorophore summation patterns. Additional approaches were not pursued in this study given our well-defined imaging environment and our initial success with the ASA scoring system.
AQP4 autoantibodies in neuromyelitis optica
Molecular models describing how AQP4 autoantibodies initiate pathologic immune activation may identify novel therapeutic targets in NMO. As anticipated by the work of Phuan et al. (14), larger M23-AQP4 arrays are able to bind multiple AQP4 rAb at high density in spatial arrangements that would support multivalent contacts with the complement protein C1q. However, some AQP4 rAb appear to have increased abilities to form multivalent contacts with C1q, indicated by a larger mean object size and a higher frequency of ASA scores 3 and 4. The epitope-dependent variation in antibody cluster size and distribution establishes that antibody-specificity, in addition to AQP4 membrane organization, impacts complement activation in NMO.
A better understanding of the molecular mechanisms driving rAb cluster assembly is needed to determine the functional significance of the observed rAb binding patterns. Indeed, some rAb formed consistently larger clusters independent of antibody concentration. The observation indicates that an unappreciated molecular mechanism likely stabilizes the organization of some, but not all, AQP4 rAb clusters and that some epitopes are able to orient bound antibodies to engage this mechanism (12, 20). An unanticipated implication of this finding is that identical ASA scores between rAb may actually reflect organized and unique molecular antibody assemblies that drive stable multivalent C1q contact and activation.
In conclusion, we were able to image two unique plasma membrane structures at high resolution using STED nanoscopy and model variable patterns of antibody clustering in relation to epitope specificity and AQP4 array assembly. Future investigations that correlate these spatial arrangements with additional functional studies of C1q activation may provide a framework to understand how AQP4-IgG promotes tissue destruction in neuromyelitis optica. Expanding this approach to additional model systems may facilitate the development of novel algorithms to dissect larger protein assemblies at lower lateral resolutions and advance our understanding of the organization and function of protein clusters.
Author Contributions
J.L.B. conceived the project. J.N.S. performed experiments. H.S. produced all rAb. J.N.S. and S.A.M. performed STED imaging and analyzed data. J.N.S., S.A.M., E.A.G., D.R., and J.L.B. contributed to analysis design, wrote, and edited the manuscript.
Acknowledgments
This study was funded by the Guthy-Jackson Charitable Foundation, a National Multiple Sclerosis Society collaborative grant, the NIH (grants No. NEI EY022936, No. NIAID UM1AI110498, No. NINDS NS048154 P30 Nanoscopy Core, and No. S10 RR023381) and the National Science Foundation (grant No. DBI-1337573). J.N.S. is a trainee of and has received support from the Medical Scientist Training Program at the University of Colorado Anschutz Medical Campus.
Editor: Laurent Blanchoin.
Footnotes
Supporting Materials and Methods and three figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)30331-4.
Supporting Citations
References (37, 38, 39, 40, 41, 42, 43, 44) appear in the Supporting Material.
Supporting Material
References
- 1.Abbe E. Archiv für Mikroskopische Anatomie. University of Michigan; Ann Arbor, MI: 1873. Beiträge zur theorie des mikroskops und der mikroskopischen wahrnehmung; pp. 413–418. [Google Scholar]
- 2.Hell S.W. Far-field optical nanoscopy. Science. 2007;316:1153–1158. doi: 10.1126/science.1137395. [DOI] [PubMed] [Google Scholar]
- 3.Hell S.W., Wichmann J. Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt. Lett. 1994;19:780–782. doi: 10.1364/ol.19.000780. [DOI] [PubMed] [Google Scholar]
- 4.Willig K.I., Rizzoli S.O., Hell S.W. STED microscopy reveals that synaptotagmin remains clustered after synaptic vesicle exocytosis. Nature. 2006;440:935–939. doi: 10.1038/nature04592. [DOI] [PubMed] [Google Scholar]
- 5.Chojnacki J., Staudt T., Kräusslich H.G. Maturation-dependent HIV-1 surface protein redistribution revealed by fluorescence nanoscopy. Science. 2012;338:524–528. doi: 10.1126/science.1226359. [DOI] [PubMed] [Google Scholar]
- 6.Heller I., Sitters G., Wuite G.J. STED nanoscopy combined with optical tweezers reveals protein dynamics on densely covered DNA. Nat. Methods. 2013;10:910–916. doi: 10.1038/nmeth.2599. [DOI] [PubMed] [Google Scholar]
- 7.Lau L., Lee Y.L., Moerner W.E. STED microscopy with optimized labeling density reveals 9-fold arrangement of a centriole protein. Biophys. J. 2012;102:2926–2935. doi: 10.1016/j.bpj.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Papadopoulos M.C., Bennett J.L., Verkman A.S. Treatment of neuromyelitis optica: state-of-the-art and emerging therapies. Nat. Rev. Neurol. 2014;10:493–506. doi: 10.1038/nrneurol.2014.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ratelade J., Asavapanumas N., Verkman A.S. Involvement of antibody-dependent cell-mediated cytotoxicity in inflammatory demyelination in a mouse model of neuromyelitis optica. Acta Neuropathol. 2013;126:699–709. doi: 10.1007/s00401-013-1172-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ratelade J., Zhang H., Verkman A.S. Neuromyelitis optica IgG and natural killer cells produce NMO lesions in mice without myelin loss. Acta Neuropathol. 2012;123:861–872. doi: 10.1007/s00401-012-0986-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Phuan P.-W., Zhang H., Verkman A.S. C1q-targeted monoclonal antibody prevents complement-dependent cytotoxicity and neuropathology in in vitro and mouse models of neuromyelitis optica. Acta Neuropathol. 2013;125:829–840. doi: 10.1007/s00401-013-1128-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaboriaud C., Ling W.L., Rossi V. Deciphering the fine details of c1 assembly and activation mechanisms: “mission impossible”? Front. Immunol. 2014;5:565. doi: 10.3389/fimmu.2014.00565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holers V.M. Complement and its receptors: new insights into human disease. Annu. Rev. Immunol. 2014;32:433–459. doi: 10.1146/annurev-immunol-032713-120154. [DOI] [PubMed] [Google Scholar]
- 14.Phuan P.-W., Ratelade J., Verkman A.S. Complement-dependent cytotoxicity in neuromyelitis optica requires aquaporin-4 protein assembly in orthogonal arrays. J. Biol. Chem. 2012;287:13829–13839. doi: 10.1074/jbc.M112.344325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bennett J.L., Lam C., Hemmer B. Intrathecal pathogenic anti-aquaporin-4 antibodies in early neuromyelitis optica. Ann. Neurol. 2009;66:617–629. doi: 10.1002/ana.21802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Owens G.P., Ritchie A., Bennett J.L. Mutagenesis of the aquaporin 4 extracellular domains defines restricted binding patterns of pathogenic neuromyelitis optica IgG. J. Biol. Chem. 2015;290:12123–12134. doi: 10.1074/jbc.M115.647149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meyer S.A., Ozbay B.N., Gibson E.A. Super-resolution imaging of ciliary microdomains in isolated olfactory sensory neurons using a custom two-color stimulated emission depletion microscope. J. Biomed. Opt. 2016;21:66017. doi: 10.1117/1.JBO.21.6.066017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Furman C.S., Gorelick-Feldman D.A., Rash J.E. Aquaporin-4 square array assembly: opposing actions of M1 and M23 isoforms. Proc. Natl. Acad. Sci. USA. 2003;100:13609–13614. doi: 10.1073/pnas.2235843100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Verkman A.S., Phuan P.W., Tradtrantip L. Biology of AQP4 and anti-AQP4 antibody: therapeutic implications for NMO. Brain Pathol. 2013;23:684–695. doi: 10.1111/bpa.12085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crane J.M., Lam C., Verkman A.S. Binding affinity and specificity of neuromyelitis optica autoantibodies to aquaporin-4 M1/M23 isoforms and orthogonal arrays. J. Biol. Chem. 2011;286:16516–16524. doi: 10.1074/jbc.M111.227298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fahmy M.F., Abdel Raheem G.M., Fahmy O.F. A new fast iterative blind deconvolution algorithm. J. Signal Inf. Process. 2012;3:98–108. [Google Scholar]
- 22.Diebolder C.A., Beurskens F.J., Parren P.W. Complement is activated by IgG hexamers assembled at the cell surface. Science. 2014;343:1260–1263. doi: 10.1126/science.1248943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crane J.M., Verkman A.S. Determinants of aquaporin-4 assembly in orthogonal arrays revealed by live-cell single-molecule fluorescence imaging. J. Cell Sci. 2009;122:813–821. doi: 10.1242/jcs.042341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crane J.M., Bennett J.L., Verkman A.S. Live cell analysis of aquaporin-4 m1/m23 interactions and regulated orthogonal array assembly in glial cells. J. Biol. Chem. 2009;284:35850–35860. doi: 10.1074/jbc.M109.071670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rossi A., Moritz T.J., Verkman A.S. Super-resolution imaging of aquaporin-4 orthogonal arrays of particles in cell membranes. J. Cell Sci. 2012;125:4405–4412. doi: 10.1242/jcs.109603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ido S., Kimiya H., Yamada H. Immunoactive two-dimensional self-assembly of monoclonal antibodies in aqueous solution revealed by atomic force microscopy. Nat. Mater. 2014;13:264–270. doi: 10.1038/nmat3847. [DOI] [PubMed] [Google Scholar]
- 27.Preiner J., Kodera N., Hinterdorfer P. IgGs are made for walking on bacterial and viral surfaces. Nat. Commun. 2014;5:4394. doi: 10.1038/ncomms5394. [DOI] [PubMed] [Google Scholar]
- 28.Smith A.J., Verkman A.S. Superresolution imaging of aquaporin-4 cluster size in antibody-stained paraffin brain sections. Biophys. J. 2015;109:2511–2522. doi: 10.1016/j.bpj.2015.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Donnert G., Keller J., Hell S.W. Macromolecular-scale resolution in biological fluorescence microscopy. Proc. Natl. Acad. Sci. USA. 2006;103:11440–11445. doi: 10.1073/pnas.0604965103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Westphal V., Lauterbach M.A., Hell S.W. Dynamic far-field fluorescence nanoscopy. New J. Phys. 2007;435:1–10. [Google Scholar]
- 31.Galiani S., Harke B., Bianchini P. Strategies to maximize the performance of a STED microscope. Opt. Express. 2012;20:7362–7374. doi: 10.1364/OE.20.007362. [DOI] [PubMed] [Google Scholar]
- 32.Harke B., Keller J., Hel S.W. Resolution scaling in STED microscopy. Opt. Express. 2008;16:4154–4162. doi: 10.1364/oe.16.004154. [DOI] [PubMed] [Google Scholar]
- 33.Gould T.J., Pellett P.A., Bewersdorf J. Fluorescence Microscopy: From Principles to Biological Applications. John Wiley & Sons; Hoboken, NJ: 2013. STED microscopy. [Google Scholar]
- 34.Farahani J.N., Schibler M.J., Bentolila L.A. Stimulated emission depletion (STED) microscopy: from theory to practice. In: Mendez-Vilas A., Diaz J., editors. Microscopy: Science, Technology, Applications and Education. Formatex Research Center; Badajoz, Spain: 2010. [Google Scholar]
- 35.Göttfert F., Wurm C.A., Hell S.W. Coaligned dual-channel STED nanoscopy and molecular diffusion analysis at 20 nm resolution. Biophys. J. 2013;105:L01–L03. doi: 10.1016/j.bpj.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Westphal V., Hell S.W. Nanoscale resolution in the focal plane of an optical microscope. Phys. Rev. Lett. 2005;94:143903. doi: 10.1103/PhysRevLett.94.143903. [DOI] [PubMed] [Google Scholar]
- 37.Saphire E.O., Parren P.W., Wilson I.A. Crystal structure of a neutralizing human IGG against HIV-1: a template for vaccine design. Science. 2001;293:1155–1159. doi: 10.1126/science.1061692. [DOI] [PubMed] [Google Scholar]
- 38.Ho J.D., Yeh R., Stroud R.M. Crystal structure of human aquaporin 4 at 1.8 Å and its mechanism of conductance. Proc. Natl. Acad. Sci. USA. 2009;106:7437–7442. doi: 10.1073/pnas.0902725106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gaboriaud C., Juanhuix J., Arlaud G.J. The crystal structure of the globular head of complement protein C1q provides a basis for its versatile recognition properties. J. Biol. Chem. 2003;278:46974–46982. doi: 10.1074/jbc.M307764200. [DOI] [PubMed] [Google Scholar]
- 40.Okuyama K., Hongo C., Bächinger H.P. High-resolution structures of collagen-like peptides [(Pro-Pro-Gly) 4-Xaa-Yaa-Gly-(Pro-Pro-Gly) 4]: implications for triple-helix hydration and Hyp(X) puckering. Biopolymers. 2009;91:361–372. doi: 10.1002/bip.21138. [DOI] [PubMed] [Google Scholar]
- 41.Rainey J.K., Goh M.C. A statistically derived parameterization for the collagen triple-helix. Protein Sci. 2002;11:2748–2754. doi: 10.1110/ps.0218502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wildanger D., Medda R., Hell S.W. A compact STED microscope providing 3D nanoscale resolution. J. Microsc. 2009;236:35–43. doi: 10.1111/j.1365-2818.2009.03188.x. [DOI] [PubMed] [Google Scholar]
- 43.Fish D.A., Brincombe A.M., Walker J.G. Blind deconvolution by means of the Richardson-Lucy algorithm. J. Opt. Soc. Am. A. 1995;12:58–65. [Google Scholar]
- 44.Pinteric L., Painter R.H., Connell G.E. Ultrastructure of the Fc fragment of human immunoglobulin G. Immunochemistry. 1971;8:1041–1045. doi: 10.1016/0019-2791(71)90492-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.