Abstract
The last decade established that the dynamic properties of the phosphoproteome are central to function and its modulation. The temporal dimension of phosphorylation effects remains nonetheless poorly understood, particularly for intrinsically disordered proteins. Osteopontin, selected for this study due to its key role in biomineralization, is expressed in many species and tissues to play a range of distinct roles. A notable property of highly phosphorylated isoforms of osteopontin is their ability to sequester nanoclusters of calcium phosphate to form a core-shell structure, in a fluid that is supersaturated but stable. In Biology, this process enables soft and hard tissues to coexist in the same organism with relative ease. Here, we extend our understanding of the effect of phosphorylation on a disordered protein, the recombinant human-like osteopontin rOPN. The solution structures of the phosphorylated and unphosphorylated rOPN were investigated by small-angle x-ray scattering and no significant changes were detected on the radius of gyration or maximum interatomic distance. The picosecond-to-nanosecond dynamics of the hydrated powders of the two rOPN forms were further compared by elastic and quasi-elastic incoherent neutron scattering. Phosphorylation was found to block some nanosecond side-chain motions while increasing the flexibility of other side chains on the faster timescale. Phosphorylation can thus selectively change the dynamic behavior of even a highly disordered protein such as osteopontin. Through such an effect on rOPN, phosphorylation can direct allosteric mechanisms, interactions with substrates, cofactors and, in this case, amorphous or crystalline biominerals.
Introduction
Osteopontin and biomineralization
Osteopontin (OPN) is one of the main regulators of mineralization in hard and soft tissues, as well as biofluids (1, 2, 3, 4, 5). In bovine milk, the most abundant phosphoform is OPN1-149, formed by cleavage at or around residue 149 (6). In human urine, a much less phosphorylated peptide known as “uropontin” appears to be the most common form (7, 8). In mineralized tissues multiple short OPN fragments are found, as a result of the action of the protease PHEX (9). Phosphorylated OPN has been shown to mediate the intrafibrillar mineralization of collagen and activation of osteoclasts (10, 11), as well as nucleation, growth, and precipitation of calcium phosphate (12, 13, 14). The function of OPN is highly reliant on tissue-specific posttranslational processing (see for example (15, 16)), including phosphorylation by the Golgi FAM20C (17, 18) and other kinases, N- and O-glycosylation (4), and proteolysis (19). A variety of structural methods, including nuclear magnetic resonance (NMR) spectroscopy, synchrotron radiation circular dichroism, and small-angle x-ray scattering (SAXS), have been deployed to study various osteopontin peptides (20, 21, 22, 23, 24).
A critical point of regulation in physiological biomineralization is the initial formation of an amorphous hydrated phase of calcium phosphate, susceptible to control by interactions with phosphoproteins (25, 26, 27). OPN contains multiple sites of phosphorylation, which can cluster in the so-called phosphate centers (PCs): a sequence of 10 or fewer consecutive residues containing at least three sites of phosphorylation, no cysteines and fewer than three hydrophobic residues, as per the definition used in Holt et al. (22). Bovine OPN1-149 can sequester amorphous calcium phosphate through interactions involving its three PCs, forming nanostructures in which the core is surrounded by a shell of the phosphopeptides (28). Similar structures can be formed by casein phosphopeptides containing a single PC (see for example (29)).
Phosphorylation of intrinsically disordered proteins
The majority of mammalian proteins are phosphorylated (30) and phosphorylation sites are frequently found in disordered regions of protein structure (31). Phosphorylation serves dynamic cellular processes well (32, 33, 34), due to its reversibility and fast kinetics. Intrinsically disordered proteins (IDPs) can gain regular secondary structure or fold into a more compact state upon phosphorylation, with implications for their properties and function (35). Osteopontin generally shows random coil-like behavior, but quail OPN contains distinct local secondary structure elements with reduced conformational flexibility (36). Hyperphosphorylation of Tau, a known substrate of the protein kinase CK2 (37), has been associated with amyloid fibril formation in Alzheimer’s disease (38) and even subtle changes in conformational dynamics can dictate amyloidogenicity (39). In some Tau fragments, phosphorylation promotes the formation of, or stabilizes, poly-L-proline type II α-helical structures, while elsewhere in the sequence phosphorylation has no apparent structural effect (40). Furthermore, phosphorylation can also promote disorder (41): studies of kinase substrates (42) highlighted that phosphorylation by CK2 can stabilize helix unfolding.
Phosphorylation and disordered conformations are common among proteins controlling biomineralization (22, 43, 44). The modulating role of phosphorylation on the biological functions of OPN and other proteins is widely recognized, allowing for OPN to interact with prenucleation clusters (45), step-specific interactions with crystal surfaces, and by altering mineral interfacial energies (46, 47) where the extent of phosphorylation has also been shown to have an impact (48). The mapping of specific effects of phosphorylation are nevertheless poorly understood, partly due to difficulties in determining phosphorylation stoichiometry (see, for example, (49)), but mostly due to the challenges of producing phosphoproteins in high yields with quantitative and complete phosphorylation at specific sites. With significant progress in both fields—for a discussion, see, for example, Oza et al. (50)—the time is ripe for studies of phosphorylation effects at different time and length scales (51, 52, 53). Here we compare the SAXS solution structures of phosphorylated and unphosphorylated rOPN. Information on the internal dynamics of rOPN hydrated powders was collected on two neutron spectrometers to cover a broad frequency range. In the hydrated powder state, the center-of-mass diffusion of rOPN is suppressed, thus providing unambiguous access to the internal molecular fluctuations probed by the hydrogen atoms, uniformly distributed throughout the rOPN sequence. Data were obtained from elastic (EINS) and quasi-elastic (QENS) incoherent neutron scattering (54). EINS provides mean square displacements, a geometrical signature of the dynamical confinement, while QENS allows vibrational and diffusive dynamics to be distinguished.
Materials and Methods
Protein expression, purification, and preliminary characterization
Phosphorylated and unphosphorylated rOPN peptides were produced. A comparison to the bovine OPN1–149 peptide guided the design of a human-like rOPN (Fig. S1). Albeit with limited success in terms of controlling the phosphorylation sites, the rOPN sequence was chosen with the aim of decreasing the number of PCs, to produce a homogeneous functional sample. Disorder predictions and a charge-hydropathy analysis of rOPN was carried out using the PONDR VLX2 algorithm (PONDR.com), which allowed comparisons to be made with sets of folded and unfolded proteins (see Supporting Material for more details).
Unphosphorylated rOPN was produced by overexpression of a construct in Escherichia coli as described in Clegg and Holt (55). Phosphorylated rOPN was coexpressed with a constitutive serine/threonine kinase—mostly serine (42)—with increased expression and activity in numerous cancers (56). A second plasmid contained an IPTG-inducible α-subunit of casein kinase on a pACYC Duet vector. The latter includes a chloramphenicol resistance gene, allowing for dual selection for OPN and kinase clones, in the presence of their respective antibiotics. For both rOPN forms, a final purification step was carried out by size exclusion chromatography, using a 24 mL column packed with Superdex S75 resin (GE Healthcare, Little Chalfont, UK), preequilibrated with two column-volumes of buffer (50 mM phosphate pH 7.5, 300 mM NaCl and 0.02% NaN3: where M denotes molar concentrations in mol/L). Electron spray ionization mass spectrometry measurements were carried out on the two rOPN forms, at the Grenoble platform service of the Partnership for Structural Biology (Grenoble, France).
The ability of the phosphorylated rOPN peptide to stabilize calcium phosphate nanoclusters, like the native OPN, was tested using dynamic light scattering (DLS): samples of 10 mg/mL were prepared in phosphate buffer, as per the method of Holt et al. (22), and allowed to stabilize. DLS of the corresponding solution was measured on a Zetasizer unit (Malvern Instruments, Malvern, UK), with an incident wavelength of 633 nm. At least 10 consecutive measurements of the correlation time of the scattered light intensity were performed and averaged for each sample. The data were fitted using the Malvern analysis software package (Malvern Instruments).
Limited proteolysis by proteinase K
Limited proteolysis was carried out on the rOPN peptides with proteinase K (Sigma-Aldrich, St. Louis, MO), according to the method of Denning et al. (57). Equine skeletal muscle myoglobin (Sigma-Aldrich) was also lysed for comparison with a globular protein of similar molecular weight (17 kDa) to the unphosphorylated rOPN. A 5 μg sample of each protein was incubated with 100 ng of proteinase K at 37°C. Aliquots were removed after 1, 2, 5, 10, 15, 30, and 60 min and the proteolysis was stopped by boiling the aliquots dispersed in sodium dodecylsulfate (SDS) loading buffer. They were then loaded onto an SDS-polyacrylamide gel electrophoresis (PAGE) (12% polyacrylamide) gel and stained with Coomassie Blue.
1D 1H HET-SOFAST NMR
NMR spectra of phosphorylated and unphosphorylated rOPN were measured on a Varian 600 MHz spectrometer (Agilent Technologies, Santa Clara, CA) at the Institut de Biologie Structurale (Grenoble, France). The 1D 1H HET-SOFAST technique allows for a semiquantitative characterization of the polypeptide disorder in solution, by comparison to empirical thresholds defined as benchmarks for protein compactness (58) through a measurement of the 1H-1H spin diffusion or NOE effect (λNOE). All measurements were performed at room temperature, with 150 μM protein in a solution containing 200 mM NaCl and 50 mM phosphate buffer, pH 7.
SAXS
SAXS measurements were performed on the rOPN peptides, using an online HPLC system (Viscotek GPCmax; Malvern Instruments) at the BM29 Bio-SAXS beamline of the European Synchrotron Radiation Facility (Grenoble, France). A 2.4 mL Superdex 200 Increase 5/150 GL column (GE Healthcare) was used and calibrated with protein standards (Bio-Rad, Hercules, CA). Before each run, the column was equilibrated with two column-volumes of buffer (50 mM phosphate pH 7.5, 300 mM NaCl, 0.02% NaN3). A sample of 100 μL was loaded onto the column and eluted at a flow rate of 0.1 mL/min. The elution was monitored by absorption at 280 nm.
X-ray scattering data were collected on the eluate as a function of the scattered wave vector (q = 4πsinθ/λ), where 2θ is the scattering angle and the q range covered was 0.08–4 nm−1. Data were collected at a wavelength λ of 0.99 Å and at 1 frame every 2 s. Radially integrated, calibrated and normalized one-dimensional scattering profiles were obtained with the EDNA software pipeline (59). All repeat frames were compared to monitor radiation damage and exclude outliers from the averaging. Buffer frames were also averaged for background subtraction. Radii of gyration (Rg) were extracted from the x-ray data for the rOPN peptides. The Guinier equation was used to fit the data close to the zero scattering angle:
| (1) |
where I(0) is the forward scattering intensity. A Kratky plot of q2I(q)/I(0) as a function of q was also used as a qualitative estimation of protein compactness (60). The real-space pair distribution function P(r) was calculated by indirect Fourier transform using GNOM from the ATSAS package (61). The ensemble optimization method was used to produce an ensemble of structures, taking into account protein flexibility by allowing the coexistence of several different conformational isomers in the scattering solution (62). A pool of 10,000 random models was generated, based on the primary structure of the rOPN peptides. A genetic algorithm was then employed to select the 50 models that best fit the scattering curve.
Incoherent neutron scattering
Neutron spectroscopy provides the possibility to explore the hierarchy of geometrically confined, superimposed motions as a function of (reciprocal) observation length and time, and a direct verification of molecular dynamics simulations that provide space-time trajectories. Data were collected on two spectrometers: IRIS (63), at the ISIS pulsed Neutron and Muon source (Chilton, UK) and IN16B (64), at the Institut Laue Langevin (ILL, Grenoble, France). IRIS and IN16B cover similar momentum transfer q ranges of 0.3–1.85 and 0.1–1.8 Å−1, respectively, but differ in energy resolution: 17 μeV (150 ps, IRIS) and 0.8 μeV (4 ns, IN16B). Monitor normalization was performed for all data, and the detectors were calibrated by using the sample signal from the lowest-temperature data.
The rOPN samples were in the form of D2O-hydrated powders, obtained through three cycles of drying under vacuum and resuspension in D2O. Each sample contained 100 mg of protein powder, with 0.44 g of D2O per gram of protein (determined by interrupting the drying and weighting of the sealed sample to monitor when the desired weight was obtained). Fixed window elastic scans were measured while the temperature was continuously increased from 10 to 310 K at a rate of 1 K/min. The sample was sealed in a flat aluminum cell with an internal spacing of 0.3 mm, at an angle of 135° with respect to the incident beam. The so-called apparent mean square displacements (MSD; 〈u2〉) were obtained from analysis of the integrated intensity over the frequency window corresponding to the instrumental resolution around the elastic peak. The incoherent scattering from the rOPN nonlabile hydrogen atoms largely dominates the EINS signal: average MSD of the hydrogen atoms were extracted from the q-dependence of the scattered intensity (energy transfer ω), using a q4 model correction (65) to the Gaussian approximation:
| (2) |
The distinctly nonvanishing a2 q4 term illustrates the deviation of the observed motion from the Gaussian approximation, significantly improving the fit to the data and allowing for a maximum of the measured q2 information to be used in the fits: 0.19–3.4 Å−2 for IRIS data, and 0.19–1.6 Å−2 for IN16B data.
Quasi-elastic spectra at selected temperatures were also recorded at both the IRIS and IN16B spectrometers. The choppers and the Doppler drive were set to allow for an energy transfer range of ±30 μeV on IN16B, whereas the chopper settings on IRIS allowed for energy transfers of ±500 μeV. For IRIS data, the empty cell signal introduced errors (most likely due to nonmatching geometrical orientations) if subtracted. As the empty cell Debye-Waller factor of the aluminum in the q-dependence is a very small contribution, this data was not used. It is, however, represented by the elastic signal—a free parameter in the QENS fits. It may slightly affect the result for the fraction of immobile atoms (Fig. S11), but again its contribution can be assumed to be nearly constant in q.
Results and Discussion
Recombinant protein expression and levels of phosphorylation
Phosphorylated and unphosphorylated rOPN were purified to homogeneity, as determined by SDS-PAGE and Western blots. The deconvoluted electrospray ionization time-of-flight mass spectrum of the unphosphorylated rOPN produced one clear peak at 18.115 kDa, consistent with the theoretical molecular weight. This excludes the possibility of prokaryotic phosphorylation—for a review, see Macek et al. (66)—and validates the use of E. coli as a heterologous expression host for studies of CK2 phosphorylation of rOPN. The equivalent spectrum for the phosphorylated rOPN sample (Fig. S8) showed a distribution of peaks, differing by multiples of the mass of a phosphate group. With the information currently available on CK2 recognition frequencies or patterns, often biased by kinase priming of phosphorylation by another kinase, inferring a trend in the results would be speculative. The rOPN samples were designed assuming a distinct preference by a consensus sequence (see also the Supporting Material) that, by itself, is not consistent with the higher incidence of 4–9 phosphorylation sites. For the purposes of comparing phosphorylation effects on the structure and dynamics of the two rOPN isoforms, however, this is not a central issue.
The size of calcium phosphate nanoclusters formed by phosphorylated peptides is not only dependent upon phosphorylation but also the sequence and length of the chain surrounding the phosphorylated region. We observed that phosphorylated rOPN forms calcium phosphate nanoclusters (CPN) in a solution that is supersaturated with respect to the bone and tooth mineral hydroxyapatite. Notwithstanding this, the solution is stable because it is undersaturated with respect to the obligate precursor amorphous phase (22, 26). The solutions of the nanocluster complexes remained clear after storage for prolonged periods and the absence of aggregation was confirmed by the DLS results (Fig. S9). The size of the nanoclusters agrees with the 22-nm radius of gyration of CPN previously determined for bovine OPN1-149 (22), consistent with the mode of calcium phosphate binding being similar for rOPN.
Flexibility of rOPN structure in solution
Phosphorylation of rOPN did not show a significant effect on the rate of protein degradation by proteinase K, consistent with no change in the extent of ordered structure. The susceptibility of rOPN to proteolysis is in clear contrast to that of the globular protein myoglobin (Fig. S6). Analysis of the 1H NMR chemical shift dispersion of both forms of rOPN further supports highly flexible unfolded protein structures (Fig. S7). These results are also reflected in the hydrodynamic behavior, computational predictions, and migration behavior on SDS-PAGE gels.
SAXS data of the phosphorylated and unphosphorylated rOPN are shown in Fig. 1, A and B, respectively. In Fig. 1 C, the radii obtained from the Guinier plots are found consistent with those expected for IDPs, according to the Flory equation (67), which postulates a power-law relationship between the radius of gyration Rg and the number N of residues in a polymer:
| (3) |
where R0 is a constant that varies with the persistence length of the peptide and ν is an exponential scaling factor constant. For chemically unfolded proteins (the Rg of bovine native OPN 1–149 was previously found (68) to match the predicted radius using these parameters), ν and R0 were previously determined (69) to be 0.6 ± 0.03 and 1.93 ± 0.28, respectively. Similarly, for globular proteins, accurate predictions were also made using values of ν and R0 equal to 0.39 and 0.35, respectively (used in the comparison shown in Fig. 1 C).
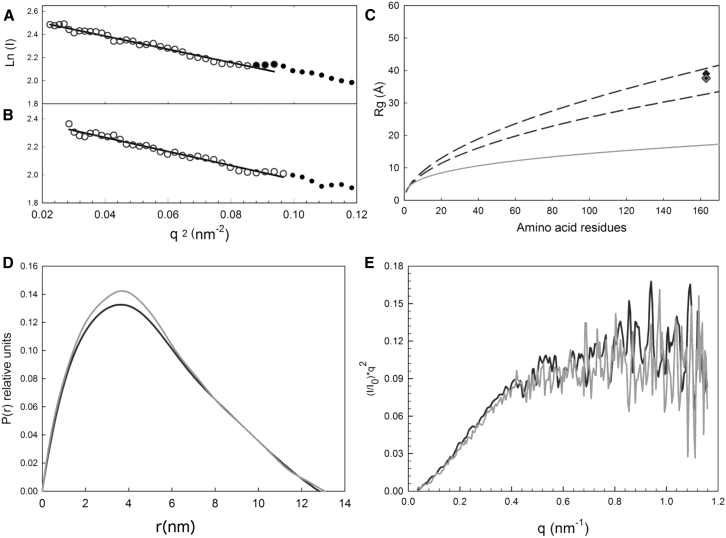
Figure 1.
Guinier plots for phosphorylated (A) and unphosphorylated (B) rOPN. Rg and I0 were recovered from the fits of the straight lines where (q × Rg) ≤ 1.3. (C) Expected Rg values for chemically denatured proteins were determined using Eq. 3, where the region between dashed lines represents the confidence interval of the Flory equation. The diamonds show experimental Rg obtained for phosphorylated (darker) and unphosphorylated rOPN (lighter). The lower continuous gray line shows the expected Rg value for globular proteins. (D) P(r) distributions of phosphorylated (black) and unphosphorylated (gray) rOPN, from which the Rg and Dmax were obtained. (E) Kratky plots are shown (same color scheme as D).
The P(r) distributions gave broadly similar results for both forms of rOPN; the asymmetrical shape of the curves is characteristic of IDPs (see Fig. 1 D; Table 1). I0 was used to produce Kratky plots (Fig. 1 E), which display a plateau at high q from Gaussian chain behavior, characteristic of highly unfolded or flexible proteins (70). Phosphorylation appears to have minimal effect on the solution scattering of rOPN, reflecting an overall unchanged conformation at the SAXS resolution. Given the distribution of phosphorylation sites, a certain degree of averaging of the structural parameters is plausible. To fully assess this effect, further studies on phosphorylated peptides are required. The ensemble-optimization method data for phosphorylated and unphosphorylated rOPN both display highly extended and flexible structures. The corresponding Rg and maximum particle size, Dmax, for the two forms of rOPN, are shown in Table 1. Fig. 2 shows the respective ensemble fits.
Table 1.
Parameters from Analysis of the SAXS Data on rOPN
| Phosphorylated | Unphosphorylated | |
|---|---|---|
| Guinier Rg (Å) | 39.0 ± 0.6 | 38.6 ± 0.6 |
| I0 | 13.4 ± 0.1 | 11.6 ± 0.1 |
| Rg (Å) from P(r) | 39.8 ± 0.2 | 39.4 ± 0.3 |
| Dmax (Å) from P(r) | 129 ± 12 | 131 ± 13 |
The confidence level for the uncertainties is 95%.
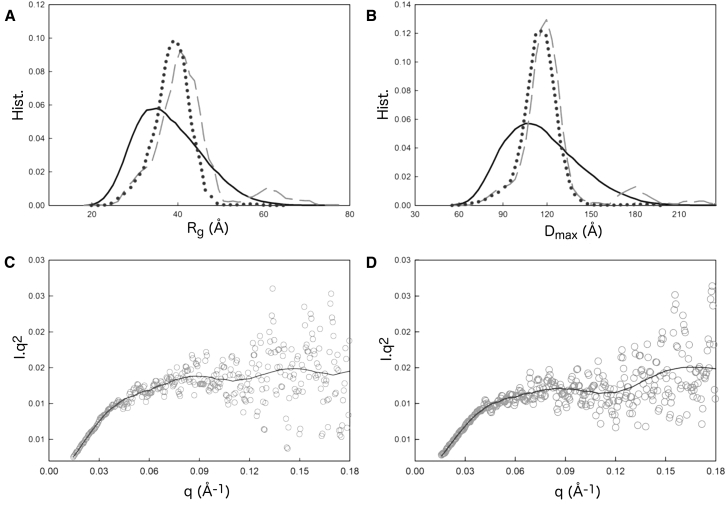
Figure 2.
Ensemble optimization analysis of the HPLC-SAXS profile measured for rOPN peptides. (A) Rg and (B) Dmax distribution for the random ensemble (solid black line), phosphorylated rOPN (dashed), and unphosphorylated rOPN (dots) is shown. The fits to the corresponding scattering curves are shown in (C) for phosphorylated rOPN and (D) unphosphorylated rOPN.
Internal protein dynamics observed by neutron scattering show a change only above the nanosecond timescale upon phosphorylation
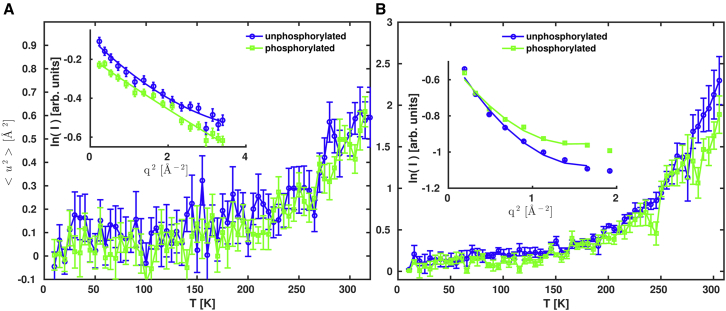
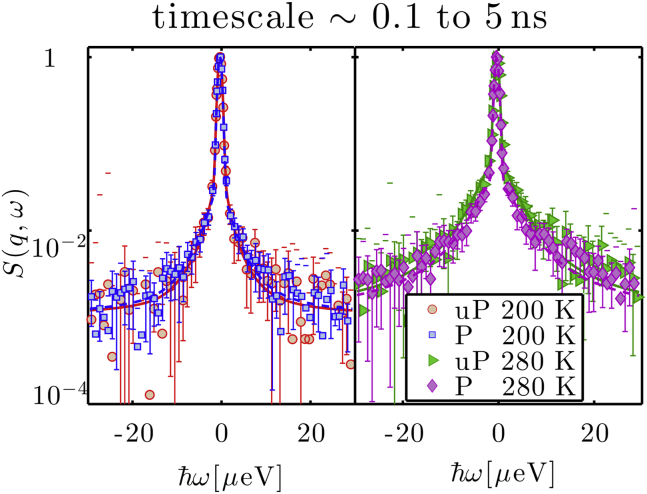
The apparent MSDs obtained for rOPN are shown in Fig. 3: the linear increase with temperature reflects the onset of motions faster than the resolution of the spectrometers used. A steeper increase at ∼200 K is typical of a dynamical transition associated with the onset of an anharmonic regime, often observed for hydrated protein powders. Within noise level, the MSDs are mostly similar for both forms of rOPN over nearly the entire temperature range measured. Above ∼280 K, a small difference in the apparent MSDs was detected for the IN16B data, and prompted the collection of full QENS spectra. These were recorded on IRIS at 300 K, and on IN16B at temperatures of 280 K and lower (Fig. 4). No differences can be observed between the two forms of rOPN at 200 K, within experimental error. Minor differences are only apparent at 280 K and for |ħω| < 10 μeV, at timescales of ∼300 ps (beyond what could be observable on the 150-ps scale of the IRIS data). At 300 K (Fig. S10), broader quasi-elastic wings are noticeable for phosphorylated rOPN. The following model was used to fit both IRIS and IN16B spectra:
| (4) |
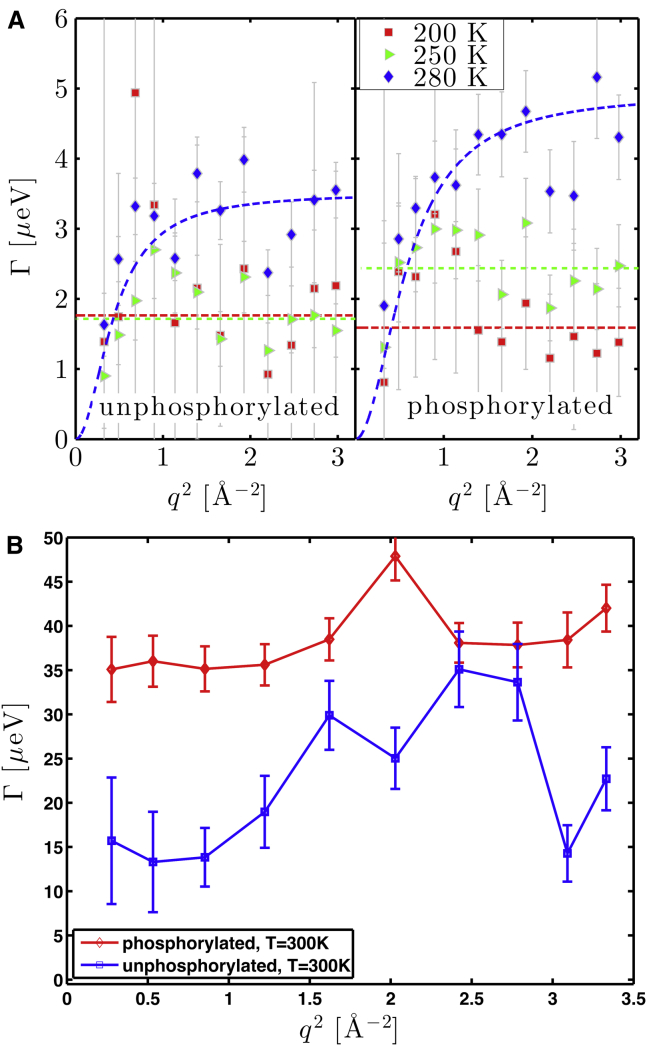
where R denotes the instrument energy resolution function, determined by fitting the spectra of a vanadium foil (IN16B), or the sample itself at the lowest temperature (10 K on IRIS), with a sum of Gaussian functions. L(Γ,ω) is a Lorentzian function with half-width at half-maximum Γ accounting for the rOPN internal molecular dynamics. Center-of-mass diffusion was ignored, given that the samples used are hydrated powders. A0(q)δ(ω) models the elastic contribution in the accessible time window, where A0(q) is the elastic incoherent structure factor (EISF) that yields information on the geometry of confinement of the atoms within the protein. The widths Γ are plotted as a function of q2 in Fig. 5. The fits to the IRIS spectra at 300 K show a trend of larger linewidths for the phosphorylated sample.
Figure 3.
Apparent MSDs of the two forms of rOPN, obtained from polynomial non-Gaussian fits to elastic fixed window temperature scans performed: (A) on the IRIS and (B) on the IN16B spectrometers. (Both insets) Elastic scattering intensities as a function of q2, at 305 K, are given. The point symbols show data binned along the temperature-axis to 5 K-intervals. The error bars correspond to 1 SD of uncertainty. Note that on the (B) inset, the error bars are smaller than the point symbols.
Figure 4.
Example QENS spectra measured on IN16B (q = 1.7 Å−1), and q-wise fit (Eq. 4) at two temperatures, for unphosphorylated (uP) and phosphorylated (P) rOPN, normalized to their maximum for a better comparison. For clarity, the data are rebinned for |ħω| > 1 μeV. The wings of the spectra—|ħω| > 15 μeV—are the same for both rOPN forms at both temperatures, within the error bars (1 SD of uncertainty).
Figure 5.
Comparison of the widths for the two rOPN forms. (A) From fits to IN16B spectra: for 200 and 250 K data, the fits (Eq. 4) are straight lines. For the 280 K data, a jump diffusion model was used (Eq. 5). The points at the two highest q values are not shown (signal-to-noise ratio too high to be included in a reliable data analysis). (B) IRIS spectra and corresponding fits (Eq. 4) at 300 K are given. The lines are guides to the eye. The error bars correspond to 1 SD of uncertainty.
For the IRIS data at 300 K (Fig. S10), as well as for the IN16B data at 200 and 250 K (Fig. 5), no particular q dependence was observed on the range. A constant can be fitted to the QENS data with an average relaxation time τ. The observation of a q-independent width may indicate that side-chain motions are restricted to localized rotations at low temperature, at the accessible timescale. At 300 K, we obtain relaxation times of 17.1 ± 3.8 ps and 29.6 ± 1.7 ps for phosphorylated and unphosphorylated rOPN, respectively. This confirms that the phosphorylation state influences the internal dynamics of rOPN on the subnanosecond timescale (see also Fig. S11) and that, on average, a shorter time elapses between two side-chain jumps for phosphorylated rOPN, when compared to the unphosphorylated form.
At 280 K, a more marked q dependence is observed, which can be fitted by a jump-diffusion model (71):
| (5) |
where D is the jump-diffusion coefficient. Such a model was used in previous studies (72, 73) to describe the dynamics of protein side chains on the nanosecond and picosecond timescales. For 200 and 250 K data, the linewidth was fitted as a global parameter. After the observations on IN16B at 280 K, a fit was performed for all q simultaneously to keep the error on the parameters as low as possible, with the scattering function:
| (6) |
where D and τ are global parameters for all scattered wave vectors. Jump-diffusion coefficients of (4.3 ± 2.9) × 10−6 and (2.3 ± 0.9) × 10−6 cm2/s were obtained from the fits for the unphosphorylated and phosphorylated rOPN, respectively. The confidence of the fit is insufficient for quantitative conclusions on a possible variation of D with phosphorylation. It is, however, noteworthy that both D coefficients do show observed internal dynamics approximately one order-of-magnitude faster than the center-of-mass motion of typical proteins (74), and one order-of-magnitude slower than the diffusion of water (75).
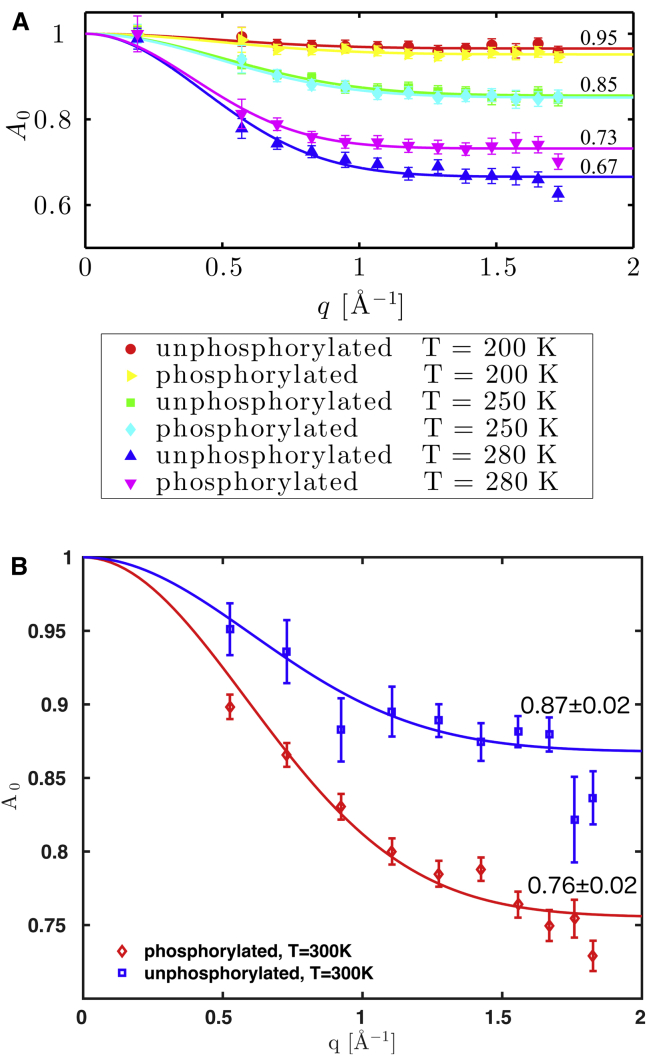
The EISF from Eq. 5 is plotted in Fig. 6 for both rOPN forms. It describes an atom in a Gaussian radial energy landscape (76) with effective radius a:
| (7) |
where p is the fraction of immobile atoms (immobile will be used, hereon, not in an absolute sense but in the context of the instrumental timescales used). We obtain an average value of 3.5 Å for the radius a (see Fig. S11), regardless of temperature and phosphorylation state. The fraction of immobile atoms can be easily deduced from the high-q value of A0, and is shown in Fig. 6. For both rOPN forms, p is closer to 1 at the lowest temperature and decreases toward higher temperatures at the nanosecond resolution. At 280 K, where the difference between EISFs of the two rOPN forms becomes clear, the phosphorylated protein has a larger number of immobile atoms on the resolution of IN16B. On the much shorter IRIS resolution timescale (at 300 K), upon phosphorylation, the number of immobile rOPN side chains decreases. The difference in absolute values between the apparent MSD data from IRIS and IN16B is then due to the different populations of motions sampled.
Figure 6.
EISF, A0, for the two forms of rOPN from fits (lines) by Eq. 7. The fraction of immobile atoms, at the resolution times of each spectrometer, is shown for each plateau: (A) IN16B QENS spectra (∼4 ns); see also Fig. S11 for more information on the fit parameters. (B) IRIS spectra (∼15 ps). The effective radius a is 2.63 ± 0.86 Å and 2.71 ± 0.36 Å for unphosphorylated and phosphorylated rOPN, respectively. The data point error bars correspond to 1 SD of uncertainty.
Overall, for phosphorylated rOPN, before applying any model we observe a larger broadening of the QENS than for the unphosphorylated rOPN, recorded on both IRIS at 300 K and IN16B at 280 K (Figs. S4 and S10). This observation is consistent with the fits of the Lorentzian Γ in Eq. 4, which show a larger Γ for the phosphorylated samples at the aforementioned temperatures. The increased flexibility is further supported by the shorter residence times at 250 and 280 K for the phosphorylated rOPN (Fig. S12). The results obtained provide further insights into the motional landscape of rOPN upon phosphorylation:
-
1)
The geometrical confinement (MSDs and p values) and relaxation of dynamics are unchanged at lower temperatures upon phosphorylation. At 280 K, phosphorylation appears to induce slightly more extensive confinement of dynamics. This could be an effect of the larger immobile fraction (with effectively very small confinement).
-
2)
We see a dynamically heterogeneous sample, as reflected by the q4 term from EINS fits and the separation into immobile and mobile atoms at 75- and 4-ns timescales.
-
3)
Slower dynamics: for immobile atoms, a large part of the hydrogen atoms is not moving considerably even on nanosecond timescales (>67% at 280 K), and it increases by ∼7% (0.73–0.67) at 280 K upon phosphorylation. The difference likely comes from the serine side chains directly affected. For mobile atoms, phosphorylation speeds up the dynamics of mobile atoms, presumably to compensate for the entropic cost of immobilizing atoms. The q signature identifies these motions as jump-like below 280 K, and jump-diffusion-like for 280 K.
-
4)
Faster dynamics: at 300 K, phosphorylation unblocks some immobile atoms (11% = 0.87–0.76). Mobile atoms appear to have faster dynamics on the picosecond scale, and the q signature identifies the motions as mainly jump-like. Possible explanations for this are only speculative without further data, but at 300 K—closer to physiological conditions—the entropic cost of immobilizing residues upon phosphorylation may become too high for a range of rOPN conformations. Specific, preferred conformations—not sufficiently different from the ensemble for the change to be reflected on the Rg—may become more populated, where faster, albeit more confined movements, are allowed to further compensate for the rigidity of the PCs. The faster picosecond motions can plausibly enhance ability to interact with crystal surfaces and provide a potential advantage in interactions with prenucleation clusters—picosecond-lived species present in supersaturated solutions of calcium phosphates and other salts (45).
Conclusions
Phosphorylation effects at various length- and time-scales are paramount to IDPs, which often use allosteric mechanisms requiring somewhat long-range intramolecular communication (77). Proteins can use fast picosecond kinetics to drive allosteric changes by strategically increasing some and decreasing other side-chain mobilities (78, 79). The relationship between function and the hierarchy of timescales in protein dynamics is not well understood and there are still few studies in the literature investigating this in sufficient level of detail.
In this study, no conformational changes were detected by small angle scattering on the solution structure of rOPN upon phosphorylation. This was previously observed for OPN peptides (48), as well as other phosphorylated proteins (80). Our studies here show that, overall, phosphorylation makes side-chain motions faster, but blocks some of the slower moving residues and releases some of the faster moving ones. Similar phosphorylation effects were previously found by NMR relaxation studies (80), where dynamic changes at the active site were seemingly opposite to those observed elsewhere in the sequence.
Phosphorylation of rOPN caused a rigidification of some residues on the accessible timescale. By selectively slowing nanosecond dynamics of PCs, phosphorylation affords sufficient free energy for sequestration of calcium phosphate and formation of CPN. Motions of side chains outside the PCs, on the other hand, can provide an entropic compensation for the arrest of the phosphorylated residues. The core-shell structure of CPN is highly dependent on the sequestering power of the phosphopeptide (caseins, for example, are known to form nanoclusters with a calcium phosphate core structure four times smaller than OPN).
Our results are also consistent with higher (closer to physiological) temperatures being a trigger to an allosteric effect of phosphorylation, but further evidence is needed to unequivocally establish this. We have shown here that rOPN displays heterogeneous, complex internal molecular dynamics, and these are known to modulate tertiary and quaternary contacts. Effects from the extent and mapping of phosphorylation events, as well as the corresponding motional landscape, will require further studies.
Author Contributions
All authors contributed to the writing of the manuscript. S.L. prepared the recombinant peptides and characterized their structure, under the supervision of M.H., T.N., and S.C.M.T. Data were collected by S.L., T.N., and S.C.M.T., with T.S. and V.G.S. assisting. M.G., F.R.-R., T.S., V.G.S., and F.S. analyzed the incoherent neutron scattering data. R.C., C.H., T.N., M.H., and S.C.M.T. designed the samples. S.C.M.T. coordinated the research.
Acknowledgments
We are grateful to the European Synchrotron Radiation Facility for beamtime at the BM29 Bio-SAXS, and to M. Brennich for assistance with the data collection; to the Institut Laue-Langevin (ILL) for beamtime at IN16B (data DOI:10.5291/ILL-DATA.8-04-719); and the ISIS pulsed Neutron and Muon source (Rutherford Appleton Laboratory, Oxfordshire, UK) for beamtime at the IRIS instrument (data DOI:10.5286/ISIS.E.47624569). S.C.M.T. is very grateful to G. Zaccai for guidance and useful discussions, to the ILL Large Scale Structures and Life Sciences groups, as well as to the NIST Neutron Research Centre (National Institute of Standards and Technology) and the University of Delaware for general support. We thank L. Signor for the electron spray ionization mass spectrometry measurements and Adrien Favier for the NMR measurements (IBS, Grenoble, France).
This work used the platforms of the Grenoble Instruct Centre (ISBG; UMS 3518 CNRS-CEA-UJF-EMBL), with support from FRISBI (grant No. ANR-10-INSB-05-02) and GRAL (grant No. ANR-10-LABX-49-01) within the Grenoble Partnership for Structural Biology. S.C.M.T., T.N., and S.L. are grateful for the Ph.D. studentship cofunded by the ILL, Keele University (EPSAM, UK), and Lund University (Sweden). M.G. is grateful for the Ph.D. studentship cofunded by the ILL and University of Tübingen (Germany).
Editor: Jill Trewhella.
Footnotes
Supporting Materials and Methods and twelve figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)30292-8.
Supporting Citations
References (81, 82, 83, 84, 85, 86, 87, 88, 89, 90) appear in the Supporting Material.
Supporting Material
References
- 1.Mazzali M., Kipari T., Hughes J. Osteopontin—a molecule for all seasons. QJM. 2002;95:3–13. doi: 10.1093/qjmed/95.1.3. [DOI] [PubMed] [Google Scholar]
- 2.O’Neill W.C., Hamano T., Rakugi H. Mineral complexes and vascular calcification. Kidney Int. 2009;76:915. doi: 10.1038/ki.2009.273. [DOI] [PubMed] [Google Scholar]
- 3.Heiss A., Pipich V., Schwahn D. Fetuin-A is a mineral carrier protein: small angle neutron scattering provides new insight on Fetuin-A controlled calcification inhibition. Biophys. J. 2010;99:3986–3995. doi: 10.1016/j.bpj.2010.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boskey A.L., Christensen B., Sørensen E.S. Post-translational modification of osteopontin: effects on in vitro hydroxyapatite formation and growth. Biochem. Biophys. Res. Commun. 2012;419:333–338. doi: 10.1016/j.bbrc.2012.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho H.-J., Cho H.-J., Kim H.-S. Osteopontin: a multifunctional protein at the crossroads of inflammation, atherosclerosis, and vascular calcification. Curr. Atheroscler. Rep. 2009;11:206–213. doi: 10.1007/s11883-009-0032-8. [DOI] [PubMed] [Google Scholar]
- 6.Sørensen E.S., Højrup P., Petersen T.E. Posttranslational modifications of bovine osteopontin: identification of twenty-eight phosphorylation and three O-glycosylation sites. Protein Sci. 1995;4:2040–2049. doi: 10.1002/pro.5560041009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christensen B., Petersen T.E., Sørensen E.S. Post-translational modification and proteolytic processing of urinary osteopontin. Biochem. J. 2008;411:53–61. doi: 10.1042/BJ20071021. [DOI] [PubMed] [Google Scholar]
- 8.Min W., Shiraga H., Hoyer J.R. Quantitative studies of human urinary excretion of uropontin. Kidney Int. 1998;53:189–193. doi: 10.1046/j.1523-1755.1998.00745.x. [DOI] [PubMed] [Google Scholar]
- 9.Barros N.M., Hoac B., McKee M.D. Proteolytic processing of osteopontin by PHEX and accumulation of osteopontin fragments in Hyp mouse bone, the murine model of X-linked hypophosphatemia. J. Bone Miner. Res. 2013;28:688–699. doi: 10.1002/jbmr.1766. [DOI] [PubMed] [Google Scholar]
- 10.Rodriguez D.E., Thula-Mata T., Gower L.B. Multifunctional role of osteopontin in directing intrafibrillar mineralization of collagen and activation of osteoclasts. Acta Biomater. 2014;10:494–507. doi: 10.1016/j.actbio.2013.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qin C., Baba O., Butler W.T. Post-translational modifications of sibling proteins and their roles in osteogenesis and dentinogenesis. Crit. Rev. Oral Biol. Med. 2004;15:126–136. doi: 10.1177/154411130401500302. [DOI] [PubMed] [Google Scholar]
- 12.Pampena D.A., Robertson K.A., Hunter G.K. Inhibition of hydroxyapatite formation by osteopontin phosphopeptides. Biochem. J. 2004;378:1083–1087. doi: 10.1042/BJ20031150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schlieper G., Westenfeld R., Ketteler M. Inhibitors of calcification in blood and urine. Semin. Dial. 2007;20:113–121. doi: 10.1111/j.1525-139X.2007.00257.x. [DOI] [PubMed] [Google Scholar]
- 14.Ito S., Saito T., Amano K. In vitro apatite induction by osteopontin: interfacial energy for hydroxyapatite nucleation on osteopontin. J. Biomed. Mater. Res. A. 2004;69:11–16. doi: 10.1002/jbm.a.20066. [DOI] [PubMed] [Google Scholar]
- 15.Razzouk S., Brunn J.C., Butler W.T. Osteopontin posttranslational modifications, possibly phosphorylation, are required for in vitro bone resorption but not osteoclast adhesion. Bone. 2002;30:40–47. doi: 10.1016/s8756-3282(01)00637-8. [DOI] [PubMed] [Google Scholar]
- 16.Kariya Y., Kanno M., Hashimoto Y. Osteopontin O-glycosylation contributes to its phosphorylation and cell-adhesion properties. Biochem. J. 2014;463:93–102. doi: 10.1042/BJ20140060. [DOI] [PubMed] [Google Scholar]
- 17.Tagliabracci V.S., Engel J.L., Dixon J.E. Secreted kinase phosphorylates extracellular proteins that regulate biomineralization. Science. 2012;336:1150–1153. doi: 10.1126/science.1217817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang X., Yan W., Wang X. Family with sequence similarity member 20C is the primary but not the only kinase for the small-integrin-binding ligand N-linked glycoproteins in bone. FASEB J. 2016;30:121–128. doi: 10.1096/fj.15-273607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christensen B., Schack L., Sørensen E.S. Osteopontin is cleaved at multiple sites close to its integrin-binding motifs in milk and is a novel substrate for plasmin and cathepsin D. J. Biol. Chem. 2010;285:7929–7937. doi: 10.1074/jbc.M109.075010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fisher L.W., Torchia D.A., Fedarko N.S. Flexible structures of SIBLING proteins, bone sialoprotein, and osteopontin. Biochem. Biophys. Res. Commun. 2001;280:460–465. doi: 10.1006/bbrc.2000.4146. [DOI] [PubMed] [Google Scholar]
- 21.Schedlbauer A., Ozdowy P., Konrat R. Backbone assignment of osteopontin, a cytokine and cell attachment protein implicated in tumorigenesis. Biomol. NMR Assign. 2008;2:29–31. doi: 10.1007/s12104-007-9076-2. [DOI] [PubMed] [Google Scholar]
- 22.Holt C., Sørensen E.S., Clegg R.A. Role of calcium phosphate nanoclusters in the control of calcification. FEBS J. 2009;276:2308–2323. doi: 10.1111/j.1742-4658.2009.06958.x. [DOI] [PubMed] [Google Scholar]
- 23.Kläning E., Christensen B., Vorup-Jensen T. Multiple low-affinity interactions support binding of human osteopontin to integrin αXβ2. Biochim. Biophys. Acta. 2015;1854:930–938. doi: 10.1016/j.bbapap.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 24.Platzer G., Żerko S., Konrat R. 1H, 15N, 13C resonance assignment of human osteopontin. Biomol. NMR Assign. 2015;9:289–292. doi: 10.1007/s12104-014-9594-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gericke A., Qin C., Boskey A.L. Importance of phosphorylation for osteopontin regulation of biomineralization. Calcif. Tissue Int. 2005;77:45–54. doi: 10.1007/s00223-004-1288-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holt C. Unfolded phosphopolypeptides enable soft and hard tissues to coexist in the same organism with relative ease. Curr. Opin. Struct. Biol. 2013;23:420–425. doi: 10.1016/j.sbi.2013.02.010. [DOI] [PubMed] [Google Scholar]
- 27.Holt C., Lenton S., Teixeira S.C. Mineralisation of soft and hard tissues and the stability of biofluids. J. Struct. Biol. 2014;185:383–396. doi: 10.1016/j.jsb.2013.11.009. [DOI] [PubMed] [Google Scholar]
- 28.Lenton S., Nylander T., Teixeira S.C.M. Structural studies of hydrated samples of amorphous calcium phosphate and phosphoprotein nanoclusters. Eur. Biophys. J. 2016;45:405–412. doi: 10.1007/s00249-015-1109-7. [DOI] [PubMed] [Google Scholar]
- 29.Little E.M., Holt C. An equilibrium thermodynamic model of the sequestration of calcium phosphate by casein phosphopeptides. Eur. Biophys. J. 2004;33:435–447. doi: 10.1007/s00249-003-0376-x. [DOI] [PubMed] [Google Scholar]
- 30.Olsen J.V., Blagoev B., Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 31.Iakoucheva L.M., Radivojac P., Dunker A.K. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004;32:1037–1049. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson L.N., Lewis R.J. Structural basis for control by phosphorylation. Chem. Rev. 2001;101:2209–2242. doi: 10.1021/cr000225s. [DOI] [PubMed] [Google Scholar]
- 33.Elbaum M.B., Zondlo N.J. OGlcNAcylation and phosphorylation have similar structural effects in α-helices: post-translational modifications as inducible start and stop signals in α-helices, with greater structural effects on threonine modification. Biochemistry. 2014;53:2242–2260. doi: 10.1021/bi500117c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosenlöw J., Isaksson L., Orekhov V.Y. Tyrosine phosphorylation within the intrinsically disordered cytosolic domains of the B-cell receptor: an NMR-based structural analysis. PLoS One. 2014;9:e96199. doi: 10.1371/journal.pone.0096199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bah A., Vernon R., Forman-Kay J. Folding of an intrinsically disordered protein by phosphorylation as a regulatory switch. Nature. 2015;519:106–109. doi: 10.1038/nature13999. [DOI] [PubMed] [Google Scholar]
- 36.Platzer G., Schedlbauer A., Konrat R. The metastasis-associated extracellular matrix protein osteopontin forms transient structure in ligand interaction sites. Biochemistry. 2011;50:6113–6124. doi: 10.1021/bi200291e. [DOI] [PubMed] [Google Scholar]
- 37.Pinna L.A., Meggio F. Protein kinase CK2 (“casein kinase-2”) and its implication in cell division and proliferation. Prog. Cell Cycle Res. 1997;3:77–97. doi: 10.1007/978-1-4615-5371-7_7. [DOI] [PubMed] [Google Scholar]
- 38.Buée L., Bussière T., Hof P.R. Tau protein isoforms, phosphorylation and role in neurodegenerative disorders. Brain Res. Brain Res. Rev. 2000;33:95–130. doi: 10.1016/s0165-0173(00)00019-9. [DOI] [PubMed] [Google Scholar]
- 39.Karamanos T.K., Pashley C.L., Radford S.E. A population shift between sparsely populated folding intermediates determines amyloidogenicity. J. Am. Chem. Soc. 2016;138:6271–6280. doi: 10.1021/jacs.6b02464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sibille N., Huvent I., Landrieu I. Structural characterization by nuclear magnetic resonance of the impact of phosphorylation in the proline-rich region of the disordered Tau protein. Proteins. 2012;80:454–462. doi: 10.1002/prot.23210. [DOI] [PubMed] [Google Scholar]
- 41.Nishi H., Fong J.H., Panchenko A.R. Regulation of protein-protein binding by coupling between phosphorylation and intrinsic disorder: analysis of human protein complexes. Mol. Biosyst. 2013;9:1620–1626. doi: 10.1039/c3mb25514j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meggio F., Pinna L.A. One-thousand-and-one substrates of protein kinase CK2? FASEB J. 2003;17:349–368. doi: 10.1096/fj.02-0473rev. [DOI] [PubMed] [Google Scholar]
- 43.Hunter G.K., O’Young J., Goldberg H.A. The flexible polyelectrolyte hypothesis of protein-biomineral interaction. Langmuir. 2010;26:18639–18646. doi: 10.1021/la100401r. [DOI] [PubMed] [Google Scholar]
- 44.Kalmar L., Homola D., Tompa P. Structural disorder in proteins brings order to crystal growth in biomineralization. Bone. 2012;51:528–534. doi: 10.1016/j.bone.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 45.Gebauer D., Kellermeier M., Cölfen H. Pre-nucleation clusters as solute precursors in crystallisation. Chem. Soc. Rev. 2014;43:2348–2371. doi: 10.1039/c3cs60451a. [DOI] [PubMed] [Google Scholar]
- 46.Li S., Zhang W., Wang L. Direct nanoscale imaging of calcium oxalate crystallization on brushite reveals the mechanisms underlying stone formation. Cryst. Growth Des. 2015;15:3038–3045. [Google Scholar]
- 47.Li S., Wu S., Wang L. Inhibition of pathological mineralization of calcium phosphate by phosphorylated osteopontin peptides through step-specific interactions. Chem. Mater. 2014;26:5605–5612. [Google Scholar]
- 48.Wang L., Guan X., Nancollas G.H. Phosphorylation of osteopontin is required for inhibition of calcium oxalate crystallization. J. Phys. Chem. B. 2008;112:9151–9157. doi: 10.1021/jp804282u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsai S.Y., Pokrass M.J., Su T.P. Sigma-1 receptor regulates Tau phosphorylation and axon extension by shaping p35 turnover via myristic acid. Proc. Natl. Acad. Sci. USA. 2015;112:6742–6747. doi: 10.1073/pnas.1422001112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oza J.P., Aerni H.R., Jewett M.C. Robust production of recombinant phosphoproteins using cell-free protein synthesis. Nat. Commun. 2015;6:8168. doi: 10.1038/ncomms9168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gabel F., Bicout D., Zaccai G. Protein dynamics studied by neutron scattering. Q. Rev. Biophys. 2002;35:327–367. doi: 10.1017/s0033583502003840. [DOI] [PubMed] [Google Scholar]
- 52.Stadler A.M., Garvey C.J., Zaccai G. Picosecond dynamics in haemoglobin from different species: a quasielastic neutron scattering study. Biochim. Biophys. Acta. 2014;1840:2989–2999. doi: 10.1016/j.bbagen.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 53.Grimaldo M., Roosen-Runge F., Seydel T. Hierarchical molecular dynamics of bovine serum albumin in concentrated aqueous solution below and above thermal denaturation. Phys. Chem. Chem. Phys. 2015;17:4645–4655. doi: 10.1039/c4cp04944f. [DOI] [PubMed] [Google Scholar]
- 54.Zaccai G. Neutron scattering perspectives for protein dynamics. J. Non-Cryst. Solids. 2011;357:615–621. [Google Scholar]
- 55.Clegg R.A., Holt C. An E. coli over-expression system for multiply-phosphorylated proteins and its use in a study of calcium phosphate sequestration by novel recombinant phosphopeptides. Protein Expr. Purif. 2009;67:23–34. doi: 10.1016/j.pep.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 56.Nitta R.T., Gholamin S., Li G. Casein kinase 2α regulates glioblastoma brain tumor-initiating cell growth through the β-catenin pathway. Oncogene. 2015;34:3688–3699. doi: 10.1038/onc.2014.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Denning D.P., Patel S.S., Rexach M. Disorder in the nuclear pore complex: the FG repeat regions of nucleoporins are natively unfolded. Proc. Natl. Acad. Sci. USA. 2003;100:2450–2455. doi: 10.1073/pnas.0437902100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schanda P., Forge V., Brutscher B. HET-SOFAST NMR for fast detection of structural compactness and heterogeneity along polypeptide chains. Magn. Reson. Chem. 2006;44:S177–S184. doi: 10.1002/mrc.1825. [DOI] [PubMed] [Google Scholar]
- 59.Incardona M.F., Bourenkov G.P., Svensson O. EDNA: a framework for plugin-based applications applied to x-ray experiment online data analysis. J. Synchrotron Radiat. 2009;16:872–879. doi: 10.1107/S0909049509036681. [DOI] [PubMed] [Google Scholar]
- 60.Receveur-Bréchot V., Durand D. How random are intrinsically disordered proteins? A small angle scattering perspective. Curr. Protein Pept. Sci. 2012;13:55–75. doi: 10.2174/138920312799277901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Semenyuk A., Svergun D. GNOM—a program package for small-angle scattering data processing. J. Appl. Cryst. 1991;24:537–540. [Google Scholar]
- 62.Bernadó P., Mylonas E., Svergun D.I. Structural characterization of flexible proteins using small-angle x-ray scattering. J. Am. Chem. Soc. 2007;129:5656–5664. doi: 10.1021/ja069124n. [DOI] [PubMed] [Google Scholar]
- 63.Carlile C.J., Adams M.A. The design of the IRIS inelastic neutron spectrometer and improvements to its analysers. Physica B. 1992;182:431–440. [Google Scholar]
- 64.Frick B., Mamontov E., Seydel T. Recent backscattering instrument developments at the ILL and SNS. Z. Phys. Chem. 2010;224:33–60. [Google Scholar]
- 65.Hennig P. Advances in Neural Information Processing Systems (NIPS) The MIT Press; Cambridge, MA: 2011. Optimal reinforcement learning for Gaussian systems. [Google Scholar]
- 66.Macek B., Gnad F., Mann M. Phosphoproteome analysis of E. coli reveals evolutionary conservation of bacterial Ser/Thr/Tyr phosphorylation. Mol. Cell. Proteomics. 2008;7:299–307. doi: 10.1074/mcp.M700311-MCP200. [DOI] [PubMed] [Google Scholar]
- 67.Flory P. Cornell University Press; New York: 1953. Principles of Polymer Chemistry. [Google Scholar]
- 68.Lenton S., Seydel T., Zaccai G. Dynamic footprint of sequestration in the molecular fluctuations of osteopontin. J. R. Soc. Interface. 2015;12:0506. doi: 10.1098/rsif.2015.0506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kohn J., Millet E., Dothager R. Random-coil behaviour and the dimensions of chemically unfolded proteins. Proc. Natl. Acad. Sci. USA. 2004;101:12491–12496. doi: 10.1073/pnas.0403643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Beaucage G., Rane S., Sukumaran S. Persistence length of isotactic poly(hydroxy butyrate) Macromolecules. 1997;30:4158–4162. [Google Scholar]
- 71.Rahman A., Singwi K., Sjölander A. Theory of slow neutron scattering by liquids. I. Phys. Rev. 1962;126:986. [Google Scholar]
- 72.Stadler A.M., Digel I., Artmann G.M. From powder to solution: hydration dependence of human hemoglobin dynamics correlated to body temperature. Biophys. J. 2009;96:5073–5081. doi: 10.1016/j.bpj.2009.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Grimaldo M., Roosen-Runge F., Schreiber F. Diffusion and dynamics of γ-globulin in crowded aqueous solutions. J. Phys. Chem. B. 2014;118:7203–7209. doi: 10.1021/jp504135z. [DOI] [PubMed] [Google Scholar]
- 74.Roosen-Runge F., Hennig M., Schreiber F. Protein self-diffusion in crowded solutions. Proc. Natl. Acad. Sci. USA. 2011;108:11815–11820. doi: 10.1073/pnas.1107287108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Qvist J., Schober H., Halle B. Structural dynamics of supercooled water from quasielastic neutron scattering and molecular simulations. J. Chem. Phys. 2011;134:144508. doi: 10.1063/1.3578472. [DOI] [PubMed] [Google Scholar]
- 76.Volino F., Perrin J.-C., Lyonnard S. Gaussian model for localized translational motion: application to incoherent neutron scattering. J. Phys. Chem. B. 2006;110:11217–11223. doi: 10.1021/jp061103s. [DOI] [PubMed] [Google Scholar]
- 77.Hoofnagle A.N., Stoner J.W., Ahn N.G. Phosphorylation-dependent changes in structure and dynamics in ERK2 detected by SDSL and EPR. Biophys. J. 2004;86:395–403. doi: 10.1016/S0006-3495(04)74115-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang Y., Madl T., Sattler M. Structure, phosphorylation and U2AF65 binding of the N-terminal domain of splicing factor 1 during 3′-splice site recognition. Nucleic Acids Res. 2013;41:1343–1354. doi: 10.1093/nar/gks1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stadler A.M., Knieps-Grünhagen E., Krauss U. Photoactivation reduces side-chain dynamics of a LOV photoreceptor. Biophys. J. 2016;110:1064–1074. doi: 10.1016/j.bpj.2016.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McDonald L.R., Whitley M.J., Lee A.L. Colocalization of fast and slow timescale dynamics in the allosteric signaling protein CheY. J. Mol. Biol. 2013;425:2372–2381. doi: 10.1016/j.jmb.2013.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Christensen B., Sørensen E.S. Osteopontin is highly susceptible to cleavage in bovine milk and the proteolytic fragments bind the αVβ3-integrin receptor. J. Dairy Sci. 2014;97:136–146. doi: 10.3168/jds.2013-7223. [DOI] [PubMed] [Google Scholar]
- 82.Edelson-Averbukh M., Pipkorn R., Lehmann W.D. Analysis of protein phosphorylation in the regions of consecutive serine/threonine residues by negative ion electrospray collision-induced dissociation. Approach to pinpointing of phosphorylation sites. Anal. Chem. 2007;79:3476–3486. doi: 10.1021/ac0623991. [DOI] [PubMed] [Google Scholar]
- 83.Larkin M.A., Blackshields G., Higgins D.G. ClustalW and ClustalX version 2. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 84.Goujon M., McWilliam H., Lopez R. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 2010;38(Web Server Issue):W695–W699. doi: 10.1093/nar/gkq313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kurzbach D., Schwarz T.C., Konrat R. Compensatory adaptations of structural dynamics in an intrinsically disordered protein complex. Angew. Chem. Int. Ed. Engl. 2014;53:3840–3843. doi: 10.1002/anie.201308389. [DOI] [PubMed] [Google Scholar]
- 86.Xue B., Dunbrack R.L., Uversky V.N. PONDR-FIT: a meta-predictor of intrinsically disordered amino acids. Biochim. Biophys. Acta. 2010;1804:996–1010. doi: 10.1016/j.bbapap.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Romero P., Obradovic Z., Dunker A.K. Sequence complexity of disordered protein. Proteins. 2001;42:38–48. doi: 10.1002/1097-0134(20010101)42:1<38::aid-prot50>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 88.Uversky V.N. Unusual biophysics of intrinsically disordered proteins. Biochim. Biophys. Acta. 2013;1834:932–951. doi: 10.1016/j.bbapap.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 89.Tompa P. Intrinsically unstructured proteins. Trends Biochem. Sci. 2002;27:527–533. doi: 10.1016/s0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- 90.Csizmók V., Szollosi E., Tompa P. A novel two-dimensional electrophoresis technique for the identification of intrinsically unstructured proteins. Mol. Cell. Proteomics. 2006;5:265–273. doi: 10.1074/mcp.M500181-MCP200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.