Abstract
Advances in computation have been enabling many recent advances in biomolecular applications of NMR. Due to the wide diversity of applications of NMR, the number and variety of software packages for processing and analyzing NMR data is quite large, with labs relying on dozens, if not hundreds of software packages. Discovery, acquisition, installation, and maintenance of all these packages is a burdensome task. Because the majority of software packages originate in academic labs, persistence of the software is compromised when developers graduate, funding ceases, or investigators turn to other projects. To simplify access to and use of biomolecular NMR software, foster persistence, and enhance reproducibility of computational workflows, we have developed NMRbox, a shared resource for NMR software and computation. NMRbox employs virtualization to provide a comprehensive software environment preconfigured with hundreds of software packages, available as a downloadable virtual machine or as a Platform-as-a-Service supported by a dedicated compute cloud. Ongoing development includes a metadata harvester to regularize, annotate, and preserve workflows and facilitate and enhance data depositions to BioMagResBank, and tools for Bayesian inference to enhance the robustness and extensibility of computational analyses. In addition to facilitating use and preservation of the rich and dynamic software environment for biomolecular NMR, NMRbox fosters the development and deployment of a new class of metasoftware packages. NMRbox is freely available to not-for-profit users.
Main Text
NMR spectroscopy is arguably the most versatile analytic method available for investigating matter. In biomedicine, NMR is employed in structural biology to determine the structures of biomolecules as well as the rates and extent of dynamic fluctuations of those structures. In the field of metabolomics it is used to determine the concentration of metabolites in biofluids and intact cells. In drug discovery it is used to screen for lead compounds that interact with a target protein and to characterize the binding site and affinity, and to confirm the active conformational state of biologics such as monoclonal antibodies.
Computation has played a seminal role in NMR spectroscopy, leading Richard Ernst to comment “without computers, no modern NMR” (1). Biomolecular applications, in particular, have been enabled in large measure by advances in computation. A simple two-dimensional Fourier transformation that required 12 h on a mainframe computer in the early 1980s now takes milliseconds on a laptop. More importantly, a rich variety of software applications for analysis of complex NMR spectra has evolved, enabling automation of many complex tasks as well as novel analyses to extract new types of information. A burgeoning area of research is the application of non-Fourier methods (2), which enables nonuniform sampling strategies (3) that greatly enhance the power of multidimensional experiments. One measure of the breadth of the software environment for bio-NMR is that more than 120 different NMR-specific software packages have been cited in the last four years in BioMagResBank (BMRB), the international data repository for biomolecular NMR data (4). Additional programs are being reported in the literature at the pace of several dozen per year. Adding the various compilers, run-time libraries, and scripting languages that are needed to support these programs, a complete software environment for NMR encompasses hundreds of software packages.
Presently there is no single source for obtaining a comprehensive set of these software packages. Individual laboratories thus face the task of discovering, acquiring, and installing all of the packages. For developers, the fragmentation of operating systems, compilers, and run-time libraries used by different laboratories greatly exacerbates the burdens of development and support. The continual evolution of operating systems also creates a substantial burden for both users and developers. This directly leads to a persistence problem, when programs cease to be supported. The persistence problem is particularly acute in bio-NMR, because many software packages are developed by academic laboratories. It is not uncommon for development and support to cease because students graduate or postdoctoral trainees move on, grants end, or research priorities change.
There are notable efforts to ease the burden of software discovery, acquisition, and use in bio-NMR. CCPN (5, 6) is a long-running European collaborative project to link new and existing NMR software via a common data standard. SBGrid (7) is a member-supported consortium of structural biologists that provides access to selected NMR software packages. WeNMR (8, 9) is another European collaborative for NMR and structural biology that provides cloud-based solutions. None of these projects, however, explicitly addresses the problems of software or platform persistence that are a raison d’etre for NMRbox. NMRbox is unique in that it provides persistence not only for NMR software titles, but also for the development and run-time environments needed to compile and run the software.
Additional obstacles to reproducibility of computational analyses in bio-NMR are posed by steps that are not fully automated, or by missing metadata describing parameters used to control automated steps. A prime example is the task of peak-picking, or identifying features in spectra that correspond to nuclear resonances. Peak-picking software is indispensable for the analysis of the complex spectra of biomolecules that contain tens of thousands of peaks. However, the current generation of available software is not fool-proof, and automatically generated peak lists are manually cleaned to remove artifacts and/or add missing peaks. These cleaned peak lists are the ones typically deposited in the BMRB database. Without accompanying annotations explicitly describing the edits to the peak list, or metadata describing the parameters used to control the peak-picking software, reproducing the results of the analysis, even when the raw primary data is deposited, may be difficult if not impossible. Software that lowers the burden on the spectroscopist for annotating common manual peak list edits is one approach to overcoming the problem (10). The peak-picking task is just one example of the larger need for a robust and rigorous platform for statistical evaluation of experimental results.
Development of approaches and tools to facilitate greater reproducibility of computational workflows in bio-NMR is a prime motivation behind the recent establishment of https://NMRbox.org: National Center for Biomolecular NMR Data Processing and Analysis. Here we describe the software platform being implemented by the Center to help bio-NMR investigators to more closely approach the “gold standard” (11) of reproducibility for computational analyses, of providing the “complete software environment required to reproduce the results” (12). Although the problem of reproducibility is enjoying recent attention (13), and the problem of software persistence has been raised (14), the resource described here appears to be the only active effort to address the preservation of the functional computational environment needed to reproduce a computational analysis in a domain of biophysics.
Discovery
One of the prime motivations behind NMRbox is the abundance and diversity of NMR software. This reflects both the myriad ways that NMR is used in biomedicine, and the multitude of different approaches to the same problem. For example, we count more than a dozen software packages currently available for analyzing NMR relaxation data to characterize biomolecular dynamics. Simply discovering the existence of bio-NMR software is one of the first challenges confronting an NMR spectroscopist. To address this challenge, a principal component of NMRbox is a software registry. Searchable by keyword, developer name or institution, and related or similar packages, the registry provides active links to depositions in BMRB that cite the software, links to the primary literature citation in PubMed, links to documentation, and usage statistics collected for users of NMRbox. The University of Connecticut, the host for NMRbox, is licensed to distribute all the embedded software packages to not-for-profit or government laboratories. Where software is already available as open source, developers were nevertheless contacted as a courtesy. For non-open-source software, individual licenses were negotiated. A condition of access to NMRbox is acceptance of the terms of these software agreements.
Virtualization
“Virtualization” refers to methods for simulating a hardware and/or software environment. Virtualization was originally conceived as a means of sharing a single computer among different operating systems (OSs), and developed by IBM in the late 1960s. More recently, virtualization has enjoyed a resurgence, as modern processors are designed with virtualization technology built-in, thereby allowing guest virtual machines (VMs) to run with near-native performance on a host computer. A supervisory program called a “hypervisor” abstracts the host’s resources, such as CPUs, memory, and disks, and provides them to guest VMs. The NMRbox VMs (described below) can be downloaded and executed locally or accessed remotely as a cloud-based Platform-as-a-Service (PaaS) through a virtual network computing (VNC) client. Virtualization is a robust technology that delivers multiple benefits: 1) a zero-configuration computing platform, 2) easy access to distributed computing, 3) simplified software development and maintenance on a single target OS, 4) efficient sharing of computational resources, 5) simplified system administration, and 6) long-term persistence of software that utilize deprecated programming languages or obsolete OSs.
In NMRbox development, NMR software packages are installed within an instance of the NMRbox VM. The operating system versions, NMR software packages, and all scripts necessary to provision and configure the VM are managed via the Apache Subversion system (https://subversion.apache.org) and each NMRbox VM release incorporates the latest versions of each NMR software package. Importantly, each release of the NMRbox VM is assigned a unique version identifier, archived, and remains accessible to the community. The primary experimental data, along with the NMRbox VM version utilized for a project are thus sufficient to reconstruct the workspace of an NMR project. The challenges of reconstructing the project workflow are more complex and are the subject of ongoing development, described below.
Utilizing NMRbox
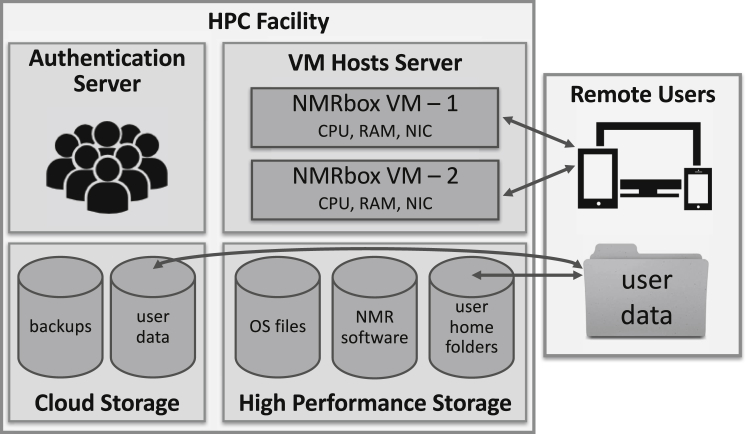
The portal to NMRbox is the website https://NMRbox.org. The first step in accessing NMRbox is to register for an account. Accounts are manually verified to ensure the user is from a nonprofit organization, and once verified the account is created. Account creation consists of multiple steps: 1) an entry is created on an LDAP authentication server, 2) a fully qualified domain name (FQDN) is created at username.nmrbox.org in a domain name server, 3) a PaaS NMRbox VM instance of the latest NMRbox version is started and network settings set to the FQDN, and 4) user home folders are created on a high performance network-attached storage server at /home/nmrbox/username and an archive folder on a cloud storage system at /nmr/archive/username. No user data is stored inside the NMRbox VM, allowing for seamless transition between different NMRbox versions. User data is transferred via a secure copy protocol (scp/sFTP), through built-in file transfer in VNC, or through Globus (https://www.globus.org) to an NMRbox endpoint. A schematic representation of the NMRBox PaaS infrastructure is shown in Fig. 1.
Figure 1.
Schematic overview of the NMRbox PaaS infrastructure. The NMRbox hardware is located in a state-of-the-art high performance computing facility with UPS-backed power, a backup generator, and redundant cooling along with a 40 GbE dark-fiber connection to an off-site disaster recovery location. The networks inside the datacenter are fully redundant with 40 GbE bandwidth with redundant 10 GbE connections to the VM host servers along with an 80 GbE-capable firewall. The HPC networks are connected to the world via a 100 GbE network to Internet2 and our ISP. There are currently five VM host servers with dual Intel Xeon E5 v4 processors (Intel, Santa Clara, CA) with a combined 196 cores and 2.3 TB of RAM and 10 TB of local RAID 10 storage. Users’ home folders and VM datastores are located on a Qumulo QC24 scale-out network-attached storage (qumulo.com) with 96 TB of capacity. Backups and additional storage for users is provided on a University-owned 3.8 PB AmpliStor on-premise cloud storage appliance (http://amplidata.com/himalaya/) with a dedicated NMRbox bucket. User data and authentication is decentralized from the NMRbox VMs allowing seamless transitions between NMRbox VM versions. Users access the NMRbox PaaS VMs through Real VNC Viewer with a full graphical user interface or via secure shell (shown by arrows). Data can be transferred between users’ local computers and their NMRbox accounts (shown by arrows) via scp/sFTP, and a Globus NMRbox institutional endpoint that allows easy and efficient transfers, including the ability to sync data, and which resolves many issues with firewalls. Expansion of the VM hosts hardware and storage is ongoing.
The NMRbox VM running as a PaaS can be accessed via a secure shell (with or without X11 graphics) or more commonly with a full graphical user interface through a VNC client (“RealVNC Viewer”, recently renamed “VNC Connect”; due to the commercial version of the VNC server that is employed by NMRbox this is the only VNC client that is supported). The commercial VNC server has several advantages including: 1) a daemon mode so users only need to connect from the VNC client without any need to manually start the VNC server session; 2) single sign-on through the NMRbox LDAP server, eliminating the need to create a separate VNC password; 3) full encryption of the entire session; 4) built-in file transfer; 5) mapping of local printers; and 6) free versions of the client for all modern platforms (available from http://www.realvnc.com). VNC sessions are persistent when the user disconnects. A tool for dynamically resetting the screen resolution is embedded in NMRbox, allowing for easy transitions between different client computers.
In addition to NMRbox VM as a PaaS, the user can download an NMRbox VM version as an open virtual appliance file that may be run locally with hypervisor software such as Oracle’s VirtualBox (free) or VMware Fusion (paid), Workstation (paid), or Player (free). Unlike the PaaS version of NMRbox VM, users have full administrative privileges for the downloaded version. When running locally, the user will need to install software drivers inside the NMRbox VM to allow more seamless interaction with the host computer, for example file sharing, dynamic screen resolution changes, copy/paste, and improved graphics performance. Specific installation instructions are based on the computer platform and hypervisor software and are beyond the scope of this manuscript. The location for storage of user data should be considered carefully when utilizing ANY locally executing VM, including NMRbox VM. We strongly encourage storing user data outside the VM file system, either through shared folders or through an external hard drive or network attached storage. The VM file system is stored as a single file on the host computer and can easily be corrupted or deleted, along with user data.
From the https://NMRbox.org, website users can log in with their NMRbox account credentials to gain access to a User Dashboard. From the Dashboard the user can reset their password, edit their personal profile, download NMRbox VM versions, and select the version of the PaaS NMRbox VM that is associated with their username.nmrbox.org FQDN. When new versions of NMRbox VM are released users can choose when to upgrade and older versions of NMRbox VM can also be accessed.
The NMRbox VM
The target OS for NMRbox VM is Xubuntu LTS, which has a new release every two years and each version is supported for five years from release. Xubuntu, which is based on Ubuntu, was chosen due to Ubuntu’s large user base and the XFCE desktop environment which is clean, easy to use, and provides good performance with VNC. The current version of NMRbox is built on Xubuntu 16.04 LTS. During provisioning of NMRbox all the dependencies for each of the NMR software packages to be installed are combined and installed together. Currently there are 147 unique dependency packages needed for the software programs installed in NMRbox. In addition, NMRbox is provisioned with many common productivity tools that are part of the Ubuntu distribution such as text editors, drawing programs, office software, statistics packages, plotting tools, and many others. These are described in the NMRbox release notes at https://NMRbox.org. For each target OS version each of the NMR software packages is installed and saved as a Debian package, allowing for efficient provisioning of the NMR software into an NMRbox VM and easy customization of a VM, if desired. Currently there are more than 70 NMR software packages installed in NMRbox, covering a wide range of categories as shown in Table 1. Major releases of NMRbox will occur every two years, corresponding to a new Ubuntu LTS release. Minor releases will occur five to ten times per year with new NMR software additions and updates to existing packages.
Table 1.
NMR Software in the NMRbox 2.0 Release
| 4DSPOT | ABACUS | Adapt NMR Enhancer | Almost |
| AmberTools | ARIA | ArShift | Azara |
| BMRB CS-Rosetta | CalRW | CalRW+ | CAMERA |
| CARA | CcpNmr-Analysis | CH3Shift | Chimera |
| CNS | CNS-ARIA | COMPASS | CPMG_FIT |
| CSCDP | DASHA | DLV | DYNAMO |
| FastModelFree | GLOVE | GOAP | hmsIST |
| MD2NOE | MDD | Modelfree | Modeler |
| MOLMOL | MVAPACK | NESSY | NESTA-NMR |
| NMRDraw | NMRFAM-Sparky | NMRFx Processor | nmrglue |
| NMRmix | NMRPipe | NMRViewJ | nmr_wash |
| PALES | PLUQ | PONDEROSA | PROMEGA |
| PyMOL | RasMol | RAW | RCI |
| REDCAT | REDCRAFT | relax | rNMR |
| RNMRTK | ROSETTA | SHIFTS | SHIFTX2 |
| SMILE | SPARTA+ | Spectrum-Translator | SQLite |
| SSP | TALOS+ | TALOSN | TREND |
| VMD | XPLOR-NIH | YESWORKFLOW |
Software Development on NMRbox
Beyond simplifying discovery, acquisition, operation, and maintenance of a complex software environment for spectroscopists, NMRbox holds a number of distinct benefits for software developers. The use of a single target operating system in the VM avoids the fragmentation problem of developing, deploying, and maintaining separate versions for different operating systems. The availability of hypervisors for virtually any modern operating system (Windows, MacOS, Linux) along with NMRbox running as a PaaS provide platform independence.
Additional benefits accrue from a rich set of usage metrics: developers typically track downloads, but it is often difficult to obtain statistics for actual usage. Accounting software embedded in NMRbox provides data on the number of times each software package embedded in NMRbox is invoked. Deidentified statistics (stripped of data traceable to a specific user) are periodically sent to a central server. A Developer Dashboard provides aggregate usage statistics for individual programs and usage statistics also appear on the software registry at https://NMRbox.org.
Another significant benefit of NMRbox is that it fosters the development and deployment of metapackages, software packages that rely on the concerted action of multiple independent software packages. Prominent examples include COMPASS (15), and the NMRFAM Integrative NMR platform (16). By providing a rich set of software tools, as well as tools to facilitate interoperation, NMRbox should catalyze further developments of metasoftware for complex analyses in biomolecular NMR.
Challenges
Modern virtualization tools are highly advanced, but obstacles to persistence remain. One is the problem of virtualizing peripheral resources, such as hardware graphics accelerators. The current deployment of the NMRbox PaaS lacks graphics hardware virtualization. Software packages that require dedicated hardware graphics acceleration will function on the downloadable NMRbox VM, provided that the host has suitable hardware, but not on the NMRbox PaaS. Commercial vendors are rapidly developing methods for virtualizing and sharing hardware graphics accelerators. We are currently testing these systems and expect to have PaaS support for packages requiring hardware graphics acceleration in a future release.
Another, more fundamental challenge is that the persistence of the software embedded in NMRbox hinges on the persistence of the NMRbox project and the virtualization environment needed to execute the NMRbox VM. NMRbox is no less immune to the ephemera of research funding than any of the embedded software titles. While we cannot guarantee that the NMRbox project will survive indefinitely, we are taking steps to assure that the VMs developed by the project remain accessible to the research community, through a number of digital archives, even if active development of NMRbox is no longer supported. The software infrastructure for provisioning NMRbox VMs will be released as open source.
Case Studies
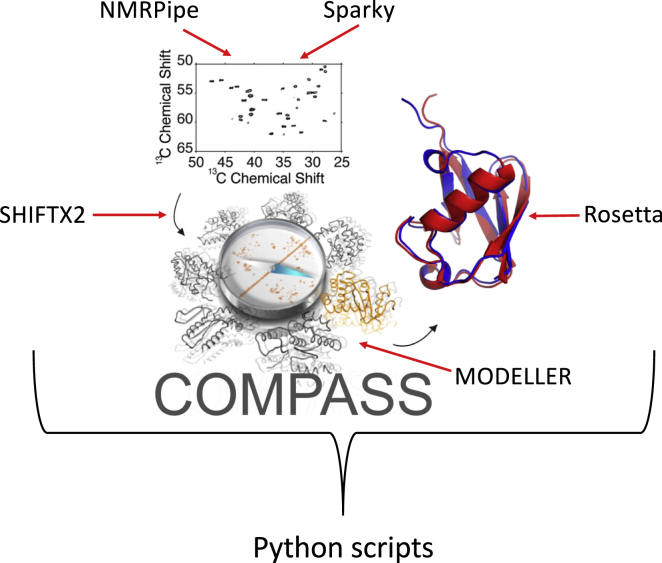
Although NMRbox is in its infancy, it has already found a number of interesting applications. The developers of COMPASS (15) (Fig. 2), a metapackage for determining protein structures from unassigned 13C chemical shifts, found that new installations of their complex software (utilizing NMRPipe (17), ROSETTA (18), MODELER (19), SPARKY (20), and SHIFTX2 (21)) to be difficult, even within their own laboratory. Installation of COMPASS in NMRbox has solved that problem. NMRbox has also found use in a number of educational settings, by providing a means for high school volunteers to instantly gain access to a rich software bio-NMR environment, and for facilitating hands-on NMR computing workshops. A study recently submitted for publication used NMRbox to facilitate comparison of 12 different non-Fourier methods for computing spectra from nonuniformly sampled NMR data. We hope and expect that the bio-NMR community will find NMRbox to be enabling in ways that we have not considered here.
Figure 2.
Component software making up the metapackage COMPASS (15). COMPASS requires interaction among multiple software packages, mediated by Python scripts (https://www.python.org). NMRbox greatly simplifies deployment of COMPASS in all its complexity by providing all the programs preinstalled and configured. Figure was adapted from Fenwick et al. (10), with permission.
Ongoing NMRbox Development
The NMRbox VM and PaaS are a first step toward addressing some of the obstacles to reproducibility of computational workflows in bio-NMR, but NMRbox also provides a platform for developments that address additional obstacles to reproducibility. One is the development and implementation of metadata harvesting and other workflow management tools. These will facilitate regularization and annotation of workflows (including manual interventions) so that metadata describing a workflow deposited in BMRB can recapitulate the workflow. These build on the existing CONNJUR-WB system (22) and extensions to NMRFAM-SPARKY (10, 23) for annotating manual edits to peak lists. A second area of development is tools for Bayesian inference (24) to improve robustness and extensibility (for example, incorporating new types of experimental data into prior analyses). An applications programming interface being developed will enable other developers to deploy Bayesian inference, and facilitate the development of consensus approaches that combine the results of multiple software packages. These extensions to the NMbox platform will be described in forthcoming publications.
Conclusions
NMRbox provides a complete software environment for bio-NMR data. Modern virtualization tools provide the means for capturing this environment in a way that is persistent, even in the face of hardware and software obsolescence. The VM approach to capturing and preserving a complete and functional computational environment should be easily applied to other scientific disciplines. In addition to simplifying discovery and use of NMR software, NMRbox facilitates software development, deployment, and support. NMRbox also complements BMRB by simplifying and enriching BMRB depositions, and thus provides multiple tools for achieving greater reproducibility of biomolecular NMR studies.
Author Contributions
M.W.M., A.D.S., M.R.G., I.I.M., E.L.U., H.R.E., and J.C.H. formulated the concept for the resource and developed the broad outlines for implementation. M.L. advised on distributed computing and security. M.W.M. designed the system for provisioning and building VMs. I.I.M. designed the hardware and networking infrastructure. A.D.S managed software accession and licensing agreements. F.D. developed software to support interoperation. E.L.U. and P.R.R. provided interfaces between BMRB and NMRbox. J.C.H. wrote the paper with contributions from M.W.M., A.D.S., H.R.E., M.R.G., I.I.M., M.L., and F.D.
Acknowledgments
We thank David Donoho and Hatef Monajemi for useful discussions, and the many software developers who have embraced the NMRbox project by contributing their software. We thank Mosrur Chowdhury, Oksana Gorbatyuk, Dillon Jones, Gerard Weatherby, Michael Wilson, Terry Wright, Kumaran Baskaran, Vincent Chen, Hesam Dashti, Dmitry Maziuk, and Jonathan Wedell for technical support.
This work was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award No. P41GM111135.
Editor: Daniel Raleigh.
References
- 1.Ernst R.R. Without computers—no modern NMR. In: Hoch J.C., Poulsen F.M., Redfield C., editors. Computational Aspects of the Study of Biological Macromolecules by Nuclear Magnetic Resonance Spectroscopy. Plenum Press; New York: 1991. pp. 1–25. [Google Scholar]
- 2.Hoch J.C. Modern spectrum analysis in nuclear magnetic resonance: alternatives to the Fourier transform. Methods Enzymol. 1989;176:216–241. doi: 10.1016/0076-6879(89)76014-6. [DOI] [PubMed] [Google Scholar]
- 3.Mobli M., Hoch J.C. Nonuniform sampling and non-Fourier signal processing methods in multidimensional NMR. Prog. Nucl. Magn. Reson. Spectrosc. 2014;83:21–41. doi: 10.1016/j.pnmrs.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ulrich E.L., Akutsu H., Markley J.L. BioMagResBank. Nucleic Acids Res. 2008;36(Database issue):D402–D408. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Skinner S.P., Fogh R.H., Vuister G.W. CcpNmr AnalysisAssign: a flexible platform for integrated NMR analysis. J. Biomol. NMR. 2016;66:111–124. doi: 10.1007/s10858-016-0060-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vranken W.F., Boucher W., Laue E.D. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 7.Morin A., Eisenbraun B., Sliz P. Collaboration gets the most out of software. eLife. 2013;2:e01456. doi: 10.7554/eLife.01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van der Schot G., Bonvin A.M. Performance of the WeNMR CS-Rosetta3 web server in CASD-NMR. J. Biomol. NMR. 2015;62:497–502. doi: 10.1007/s10858-015-9942-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bertini I., Case D.A., Rosato A. A Grid-enabled web portal for NMR structure refinement with AMBER. Bioinformatics. 2011;27:2384–2390. doi: 10.1093/bioinformatics/btr415. [DOI] [PubMed] [Google Scholar]
- 10.Fenwick M., Hoch J.C., Gryk M.R. CONNJUR R: an annotation strategy for fostering reproducibility in bio-NMR-protein spectral assignment. J. Biomol. NMR. 2015;63:141–150. doi: 10.1007/s10858-015-9964-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peng R.D. Reproducible research in computational science. Science. 2011;334:1226–1227. doi: 10.1126/science.1213847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donoho D.L. An invitation to reproducible computational research. Biostatistics. 2010;11:385–388. doi: 10.1093/biostatistics/kxq028. [DOI] [PubMed] [Google Scholar]
- 13.Stodden V., McNutt M., Taufer M. Enhancing reproducibility for computational methods. Science. 2016;354:1240–1241. doi: 10.1126/science.aah6168. [DOI] [PubMed] [Google Scholar]
- 14.Afgan E., Chapman B., Taylor J. Using cloud computing infrastructure with CloudBioLinux, CloudMan, and Galaxy. Curr. Protoc. Bioinformatics. 2012;Chapter 11 doi: 10.1002/0471250953.bi1109s38. Unit11.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Courtney J.M., Ye Q., Rienstra C.M. Experimental protein structure verification by scoring with a single, unassigned NMR spectrum. Structure. 2015;23:1958–1966. doi: 10.1016/j.str.2015.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee W., Cornilescu G., Markley J.L. Integrative NMR for biomolecular research. J. Biomol. NMR. 2016;64:307–332. doi: 10.1007/s10858-016-0029-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delaglio F., Grzesiek S., Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 18.Shen Y., Lange O., Bax A. Consistent blind protein structure generation from NMR chemical shift data. Proc. Natl. Acad. Sci. USA. 2008;105:4685–4690. doi: 10.1073/pnas.0800256105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Webb B., Sali A. Protein structure modeling with MODELLER. Methods Mol. Biol. 2014;1137:1–15. doi: 10.1007/978-1-4939-0366-5_1. [DOI] [PubMed] [Google Scholar]
- 20.Goddard T.D., Kneller D.G. University of California, San Francisco; San Francisco, CA: 2008. SPARKY 3. [Google Scholar]
- 21.Han B., Liu Y., Wishart D.S. SHIFTX2: significantly improved protein chemical shift prediction. J. Biomol. NMR. 2011;50:43–57. doi: 10.1007/s10858-011-9478-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fenwick M., Weatherby G., Gryk M.R. CONNJUR Workflow Builder: a software integration environment for spectral reconstruction. J. Biomol. NMR. 2015;62:313–326. doi: 10.1007/s10858-015-9946-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee W., Tonelli M., Markley J.L. NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics. 2015;31:1325–1327. doi: 10.1093/bioinformatics/btu830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bahrami A., Assadi A.H., Eghbalnia H.R. Probabilistic interaction network of evidence algorithm and its application to complete labeling of peak lists from protein NMR spectroscopy. PLOS Comput. Biol. 2009;5:e1000307. doi: 10.1371/journal.pcbi.1000307. [DOI] [PMC free article] [PubMed] [Google Scholar]