Abstract
The strong and usually denaturing interaction between anionic surfactants (AS) and proteins/enzymes has both benefits and drawbacks: for example, it is put to good use in electrophoretic mass determinations but limits enzyme efficiency in detergent formulations. Therefore, studies of the interactions between proteins and AS as well as nonionic surfactants (NIS) are of both basic and applied relevance. The AS sodium dodecyl sulfate (SDS) denatures and unfolds globular proteins under most conditions. In contrast, NIS such as octaethylene glycol monododecyl ether (C12E8) and dodecyl maltoside (DDM) protect bovine serum albumin (BSA) from unfolding in SDS. Membrane proteins denatured in SDS can also be refolded by addition of NIS. Here, we investigate whether globular proteins unfolded by SDS can be refolded upon addition of C12E8 and DDM. Four proteins, BSA, α-lactalbumin (αLA), lysozyme, and β-lactoglobulin (βLG), were studied by small-angle x-ray scattering and both near- and far-UV circular dichroism. All proteins and their complexes with SDS were attempted to be refolded by the addition of C12E8, while DDM was additionally added to SDS-denatured αLA and βLG. Except for αLA, the proteins did not interact with NIS alone. For all proteins, the addition of NIS to the protein-SDS samples resulted in extraction of the SDS from the protein-SDS complexes and refolding of βLG, BSA, and lysozyme, while αLA changed to its NIS-bound state instead of the native state. We conclude that NIS competes with globular proteins for association with SDS, making it possible to release and refold SDS-denatured proteins by adding sufficient amounts of NIS, unless the protein also interacts with NIS alone.
Introduction
Protein-surfactant interactions play a significant role within food, washing powder detergents, personal care, and pharmaceutical industries (1). In the detergent industry, focus has been on lowering the washing temperatures to save energy wasted on heating water, and to enhance the lifetime of clothing. This is challenged by the decreased effectiveness of surfactants at lower temperatures, but it can be mitigated by adding enzymes that degrade different sources of dirt such as proteins, fats, and starch under these conditions. Unfortunately, most enzymes are denatured by anionic surfactants (AS) (2, 3, 4, 5, 6), leading to loss of activity of the enzyme. AS are included in most detergent formulations because of their ability to keep solubilized dirt away from fabrics. They are most efficient at concentrations above the critical micelle concentration where they form micelles. Detergent formulations also include nonionic surfactants (NIS), which are not precipitated by Ca2+ or Mg2+ salts and solubilize oils and organic dirt better than AS, particularly at lower temperatures. NIS do not generally bind globular proteins but can weaken the effects of anionic surfactants by forming mixed micelles. Generally, the higher the mole fraction of NIS in these mixed micelles, the lower the denaturation potency. For example, addition of the NIS dodecyl maltoside (DDM) to sodium dodecyl sulfate (SDS) strongly reduces the rate constant of unfolding of bovine serum albumin (BSA) (2), and BSA can be protected from unfolding by SDS by the presence of nonionic surfactants in the form of ethoxylated alkyls (A. Würtz, M.A. Behrens, G.V. Jensen, T.H. Callisen, J.S.P., unpublished). Likewise, the β-sheet protein TNfn3 unfolds significantly less cooperatively in micelles containing 75% SDS and 25% DDM, while higher DDM mole percentages prevent complete unfolding (7). Additionally, some membrane proteins denatured in SDS can be refolded by adding appropriate amounts of NIS (8, 9, 10) (typically >50% mole percentage is required to refold the protein). These observations suggest that addition of sufficient amounts of NIS to an SDS-denatured globular protein may in fact reverse the unfolding process, although the mechanism of renaturation may be different than that for membrane proteins. Unlike globular proteins, membrane proteins also bind NIS, so the addition of NIS to an SDS-denatured protein likely just alters the composition of the micelle encasing the protein, leading to conformational changes for the protein. In contrast, it could be conjectured that SDS-denatured globular proteins are refolded in NIS by transferring SDS from the protein surface to NIS micelles so that mixed micelles are formed and proteins are released from these micelles. Another possibility is that NIS incorporate into the SDS-protein complexes and thus weaken SDS’ denaturation potency.
Here, we investigate how the addition of NIS to AS-denatured proteins refolds these proteins. All studies are performed well above the critical micelle concentration to ensure that micelles are the dominant denaturing species. We approach the refolding aspect using circular dichroism (CD) to monitor changes in secondary and tertiary structure, and small-angle x-ray scattering (SAXS) to determine the population of different molecular species, i.e., free proteins, free micelles, and protein-surfactant complexes. As model systems we use BSA, α-lactalbumin (αLA), hen egg white lysozyme (LYZ), and β-lactoglobulin (βLG). BSA has long been the archetypal model protein for interactions with SDS; the native state can bind a small number of monomeric SDS molecules without denaturation but undergoes cooperative unfolding at higher SDS concentrations (11, 12). The Ca(II)-binding milk protein αLA is highly sensitive to unfolding by both SDS and NIS, particularly in the apo-form (5), while its Ca(II)-free homolog LYZ resists denaturation by NIS but is unfolded by SDS in several steps (13). Like BSA, βLG has hydrophobic binding sites in the native state that allow a small number of SDS molecules to bind and stabilize the protein (14) before it is eventually unfolded at higher SDS concentrations (15).
Our study confirms that in the presence of SDS, all these proteins unfold to form protein-decorated SDS micelles. In contrast, the nonionic surfactants octaethylene glycol monododecyl ether (C12E8) and DDM do not unfold BSA, βLG, and LYZ but induce some conformational changes in αLA. When the nonionic surfactants are added to the SDS-unfolded proteins, they are able to extract SDS from the complexes, thereby liberating proteins from the surfactant micelles and allowing them to fold. To the best of our knowledge, this is the first direct demonstration that NIS facilitate refolding of SDS-denatured proteins by extraction of SDS.
Materials and Methods
Materials
SDS (≥99.0%), octaethylene glycol monododecyl ether (≥98%), albumin bovine serum, α-lactalbumin (from bovine milk; 85%), lysozyme (from chicken egg white), β-lactoglobulin (from bovine milk; 90%), and NaCl were from Sigma-Aldrich (St. Louis, MO). n-Dodecyl β-maltoside (>99.5%) was purchased from Glycon Biochemicals (Luckenwalde, Germany). Sodium phosphate monobasic anhydrous (≥99.0%) and di-sodium hydrogen phosphate 2-hydrate (99.5%) were from Fluka (Sigma-Aldrich) and Merck (Kenilworth, NJ), respectively.
All experiments were carried out using a buffer of 50 mM NaCl and 10 mM phosphate (pH 7) and stock solutions of 1–2 wt % protein and 80–300 mM SDS, DDM, or C12E8. Buffer and protein stocks were stored in the fridge. Surfactant stock solutions were stored at room temperature. All samples were prepared by weighing out the appropriate amount of protein, surfactant, and buffer solutions. Initially, the SDS stock solution was added to the protein stock and left overnight. The samples that included protein were prepared to have final protein and SDS concentrations of either 5 mg/mL protein and 25 mM SDS, or 2 mg/mL protein and 10 mM SDS (corresponding to at least one SDS micelle per protein), depending on the required mole fraction. Then the appropriate amounts of buffer and C12E8 or DDM stock solutions were added to obtain the desired protein concentration. All samples were kept at room temperature for at least 1 h before measuring.
SAXS
The SAXS measurements were performed at the laboratory-based instruments at Aarhus University. One is a Bruker AXS NanoStar SAXS instrument (16, 17) with a Cu rotating anode and the other one is a Bruker AXS NanoStar (Bruker, Karlsruge, Germany) with a Ga liquid metal jet source (18). Both instruments are flux optimized and both are equipped with a homebuilt scatterless slit/pinhole (17) in front of the sample, so that a two-pinhole geometry with higher x-ray intensity is used. All measurements were carried out at 20°C and lasted 900 s at the Cu-source instrument and 600 s at the Ga-source instrument. The sample holders are homebuilt flow-through quartz capillaries glued into stainless-steel holders. The buffers were measured as background, and background subtraction and all necessary normalizations were made with the SUPERSAXS package (C.L.P. Oliveira and J.S.P., unpublished data) using standard methods (19) with the modifications described in Pedersen (16). The final intensity is displayed as a function of the modulus of the scattering vector as follows:
where λ = 1.54 and 1.34 Å is the x-ray wavelength for Cu and Ga sources, respectively, and 2θ is the angle between the incident and scattered x-rays.
As a first step of data analysis, an indirect Fourier transformation (20) was performed using the WIFT program (21) to get model-independent structural information in real space. The IFT procedure provides the pair distance distribution function, p(r), which is a histogram over distances between pairs of points within the particle, weighted by the excess scattering length density at the points. It thus gives information about the particle size and shape in real space, including the particles’ maximum dimension, Dmax, which can be obtained from the histogram.
The samples may contain many different components (proteins, complexes, pure and mixed micelles) and the purpose of the analysis of the SAXS data is to extract information on the amount of the various components. In the data analysis of the samples that contained protein, SDS, and NIS, we assumed that the possible structures of the constituents are the same as those that can be measured individually. This means that the protein will exist either as native protein or in complex with SDS, where both of these components have also been measured alone. Likewise, SDS must exist either in complex with the protein or in mixed micelles with NIS, which have also been measured. Finally, the NIS will exist either as pure NIS micelles (if the SDS is in complex with the protein) or in mixed micelles with SDS (if the protein refolds whereby SDS is released into the solution). Therefore, because we know the concentrations of protein, SDS, and NIS in our samples, we can calculate the concentration of all structural components (native protein, unfolded protein in complex with SDS, mixed SDS-NIS micelles, and pure NIS micelles) if the concentration of one component is specified. This fact can be exploited to constrain the fitting procedure significantly, so that only one parameter is used for fitting the data to determine the amount of all four components.
Following this, a weighted least-squares procedure was employed to fit the SAXS data as linear combinations of the measured data from the different components, as follows:
where the A values are the scale factors of the individual components, which are labeled with obvious subscripts. Using the concentration of the components in the samples, the values of all four scale factors were fitted using only one adjustable parameter. In all cases the pure mixed micelle sample with SDS mole fraction closest to the one in the fitted sample is used as input. Because concentration effects were observed for the low-q region in many samples, an overall effective hard-sphere structure factor (22) was included to account for these effects. An effective hard-sphere structure factor was multiplied on the model intensities Imodel(q). The adjustable parameters were an effective hard-sphere volume fraction and an effective hard-sphere interaction radius. Finally, an adjustable background was included to correct for slightly different background in the measurements, and 10% error on the concentrations were allowed.
The fit quality was evaluated using a reduced χ2 (i.e., goodness of fit),
where N is the number of measured points, M is the number of fit parameter, and the errors, σ, in each point included those of the individual components:
The fitting procedure was implemented in MATLAB 2015a (The MathWorks, Natick, MA).
CD
CD spectra for the samples measured by SAXS were recorded on a model No. J810 spectropolarimeter (Jasco Spectroscopic, Hachioji City, Japan) at 20°C with a band width of 2 nm, a scan speed of 100 nm/min, and an accumulation of six scans. Far-UV CD spectra were recorded in a 0.1 mm path-length cuvette (2 or 5 mg/mL protein) while near-UV CD spectra were recorded in 10 mm cuvettes (1.5 mg/mL protein). Background contributions from buffer were subtracted. The CD signal was measured in mdeg and converted to θMR for the near-UV data, as follows:
where θMR is the mean residue ellipticity given in deg cm−2 mol−1 res−1, θ is the ellipticity in mdeg, c is the concentration in g/L, l is the path length in cm, and MRW is the mean residue weight given by
where Mw is the molecular weight in g/mol, #amino acids is the number of amino acids per protein, and #amino acids −1 is the number of peptide bonds. For the near-UV data the ellipticity is shown per protein and not per mean residue.
The content of secondary structure (α-helix, β-sheet, and random coil) was estimated from the CD data using the reference spectra of Greenfield and Fasman (23) and a home-written weighted least-squares program that assigns equal weight to all data points in the CD spectrum.
Sample mixing and SAXS data fitting procedure
For all proteins, SDS and protein was mixed first, leading to a sample with SDS mole fraction χSDS of 1.0 (where χSDS = [SDS]/([NIS] + [SDS]). χSDS was then decreased to the desired values by mixing with NIS in the same buffer. The protein and SDS concentration was at 5 mg/mL and 25 mM, respectively, for the higher χSDS values and 2 mg/mL and 10 mM for the lower χSDS values (to avoid extremely high concentrations of NIS). For both protein concentrations, the amount of SDS was chosen so that the protein was completely unfolded while only insignificant amounts of free SDS micelles were present. This was determined by investigating samples with constant protein concentration and increasing amount of SDS and fitting the data by a linear combination of a core-shell model for the complexes and a contribution of free SDS micelles. The latter was important for samples with high SDS concentrations. By determining where the scale factor of the micelle contribution goes to zero, we can determine the point where free micelles start to form. All samples of protein with SDS and added NIS were studied by SAXS and far- and near-UV CD to determine changes in the secondary and tertiary structure. SAXS data provided information on the populations of proteins and surfactants in different species as described in Materials and Methods. Briefly, spectra from the mixed samples were fit by a linear combination of four basis functions (all in buffer): pure protein, pure NIS micelles, protein-SDS complexes (unfolded protein), and SDS-NIS mixed micelles. As described above, the protein/SDS concentration ratio was chosen to avoid significant contributions of SDS micelles in the data. The concentrations of all samples were included in the fitting to account for the scattering of each species on absolute scale, which constrains the results significantly. A 10% error on the concentrations was allowed. From our simple theory, it follows that the components will exist in pairs: the presence of native protein results in an equal amount of SDS-NIS mixed micelles, while the presence of protein-SDS complexes must be complemented by pure NIS micelles. Therefore, the populations of all four components are found by a single fitting parameter, and the result is simplified to two situations: either the protein is folded and free in solution or it is unfolded in association with SDS.
Results
General procedure for analyzing protein unfolding and folding in SDS and NIS
Before our refolding studies, we verified that all proteins were unfolded with SDS and not by NIS. We demonstrated this in two ways, as detailed below. Firstly, SAXS data of the protein-SDS mixtures could not be fit by a linear combination of the native protein and pure SDS micelles (Fig. S1), which shows that complexes have been formed. The SAXS data of the complexes could be described by a core-shell model (data not shown), demonstrating that the proteins have been unfolded and are situated on the SDS micelle surface as reported for several SDS-unfolded proteins (24, 25, 26, 27, 28). Contrary to the protein-SDS samples, most samples consisting of protein and NIS could be described well using linear combinations of pure protein and pure NIS micelles, demonstration that no interaction occurs (Fig. S2). Only for αLA does the poor fit quality demonstrate that αLA undergoes structural changes when DDM is added, which is in agreement with our previous observations (5). Secondly, CD spectra changed radically when the proteins were mixed with SDS (see below), but not when mixed with NIS.
β-lactoglobulin refolds below χSDS = 0.5 and shows a conformational overshoot at lower χSDS values
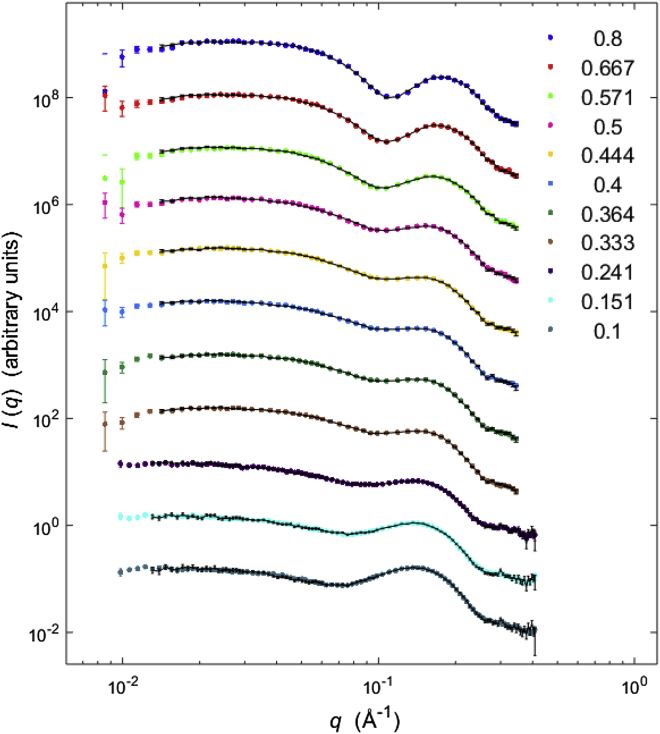
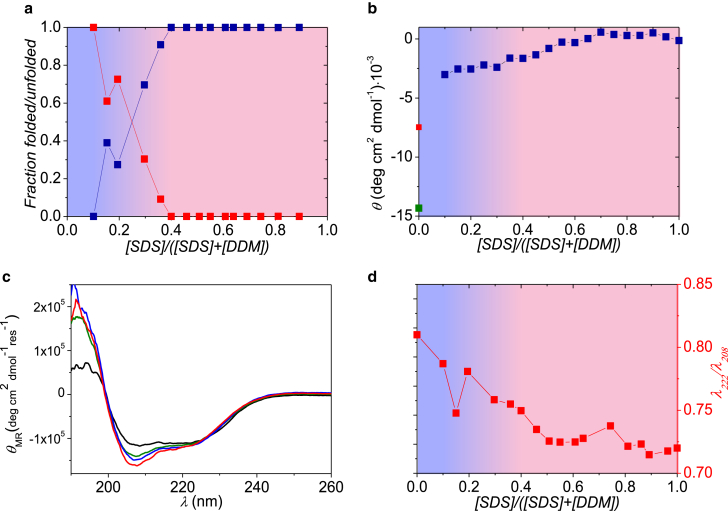
βLG is a small, globular protein, which consists mainly of β-strands. For both types of NIS (C12E8 and DDM), the fitting method described in Materials and Methods was able to fit the data very well. SAXS data of mixtures of βLG and NIS can be described by linear combinations of data from pure NIS and pure protein, confirming that NIS does not interact with βLG. In the presence of SDS, however, it is clear that the protein forms complexes with micelles. Examples of the fits from this procedure of SDS-unfolded βLG and C12E8 are provided in Fig. 1, demonstrating the good fit quality for all χSDS. The results from the analysis are plotted in Fig. 2 a, providing a comprehensive overview of the protein state (either unfolded in micelles, or folded and dissociated from micelles) at all measured χSDS values. It is evident from this figure that there is a preferred state of the protein at each χSDS, and that the transition between these states is quite sharp around χSDS = 0.5. At higher χSDS values the protein is almost entirely unfolded, while lower χSDS values result in refolding of βLG. Fig. 1 shows a gradual and characteristic change of the q dependence of the SAXS data. At high χSDS there is a pronounced minimum in the intensity close to q = 0.1 Å−1, which is due to the near-spherical shape of the core-structure of the complex with opposite excess scattering length in core and shell. At intermediate χSDS, where the SAXS data are the sum of that from refolded protein and a relatively small amount of free mixed micelles, the minimum is filled out and is more like a plateau, despite the scattering from the mixed micelles having a minimum close to 0.1 Å−1. At small χSDS there is again an observable minimum, but the scattering looks more like that of free mixed micelles due to their presence in excess.
Figure 1.
Fits of βLG and different χSDS in C12E8. Data were fit with a linear combination of folded protein in presence of mixed micelles and SDS-unfolded protein in presence of pure NIS micelles, as described in Materials and Methods. Decreasing mole fraction from top to bottom with values given in the figure. To see this figure in color, go online.
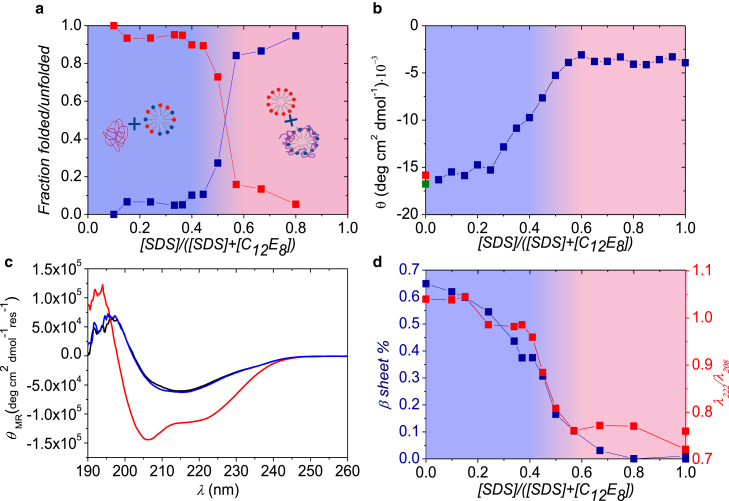
Figure 2.
Results for βLG and SDS samples mixed with C12E8. (a) Part of the protein that is either folded (upper data points at low mole fractions) or unfolded around SDS micelles (lower data points at low mole fractions) is shown. (b) Near-UV CD data at 293 nm of βLG in micelles (data as a function of mole fraction) buffer (upper point at zero mole fraction), and in 25 mM C12E8 (lower point at zero mole fraction) are shown. (c) Shown here are CD spectra for pure βLG (black), βLG mixed with SDS and C12E8 at SDS mole fractions of 0.8 (curve with deepest minimum) and 0.37 (nearly coinciding with black curve), respectively. (d) The fraction of β-sheet (points going to zero at a mole fraction of 0.8), and λ222/λ208 ratio (upper points at high mole fractions) is given. Background shading in (a), (b), and (d) indicates whether the protein is mainly free in solution (right part) or bound to micelles (left part). To see this figure in color, go online.
To corroborate these changes in protein structure, we turned to CD to monitor the protein structure directly. Near-UV CD data at 293 nm at different χSDS (Fig. 2 b) confirm a transition in the protein tertiary structure around χSDS = 0.5. Similarly, far-UV CD data in Fig. 2 c demonstrate that the secondary structure of βLG (black curve) is clearly nonnative at higher χSDS values (0.8, red curve), but that the protein returns to its native state at lower χSDS values (0.37, blue curve). Near-UV CD data at these values of χSDS show the same behavior (Fig. S3). The λ222/λ208 ratio, which is often used as a convenient measure of changes in the arrangement of helices under situations where the protein retains helical structure but is nonnative (29, 30), displays the same transition at ∼χSDS = 0.5 (Fig. 2 d, red squares) as observed with SAXS and near-UV CD data. Additionally, fitting of the far-UV CD data shows that the β-sheet content is absent for χSDS > 0.5 (Fig. 2 d, blue squares), underlining that the native structure of βLG is disrupted in this region. On the other hand, the amount of α-helix in βLG seems to increase slightly when the protein is unfolded by SDS (from ∼40 to ∼50%), in agreement with earlier observations (31).
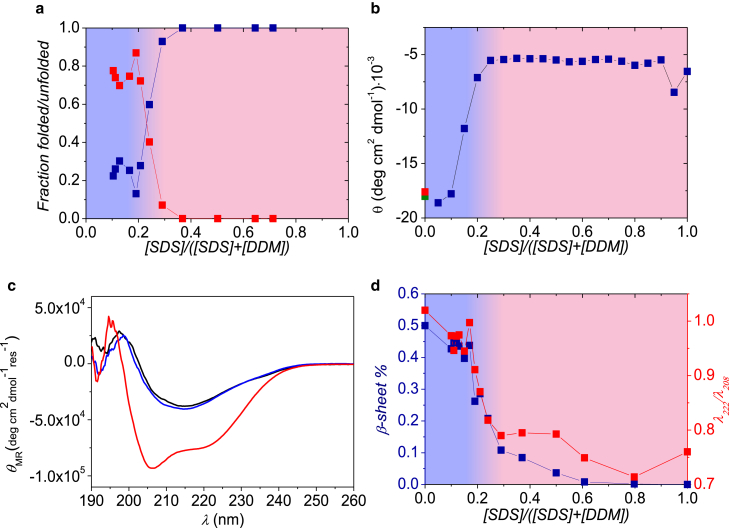
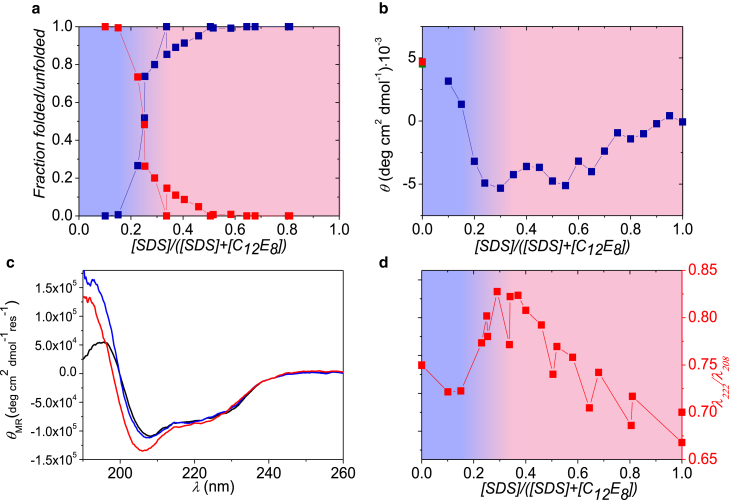
We additionally investigated samples with DDM instead of C12E8 to study whether different nonionic surfactants have different influence on protein refolding. The data on βLG unfolded in SDS and mixed with an increasing amount of DDM are displayed in Fig. 3 (fits to the SAXS data can be found in Fig. S4). Both the SAXS results (Fig. 3 a) and the near- (Fig. 3 b) and far-UV CD data (Fig. 3, c and d) show very similar results to for C12E8, except that the transition between folded and unfolded protein occurs at χSDS values of ∼0.25 in DDM and ∼0.5 in C12E8.
Figure 3.
Results for βLG and SDS samples mixed with DDM. (a) Part of the protein that is either folded (upper data points at low mole fractions) or unfolded around SDS micelles (lower data points at low mole fractions) is shown. (b) Near-UV CD data at 293 nm of βLG in micelles (data as a function of mole fraction), buffer (lowest point at zero mole fraction), and in 25 mM DDM (upper data point at zero mole fraction) are shown. (c) Shown here are CD spectra for pure βLG (black), βLG mixed SDS, and DDM with SDS mole fraction of 0.5 (curve with deepest minimum) and 0.1 (nearly coinciding with black curve), respectively. (d) The fraction of β-sheet (points going to zero at a mole fraction of 0.6), and λ222/λ208 ratio (upper points at high mole fractions) is given. To see this figure in color, go online.
αLA binds NIS, SDS, and mixed SDS-NIS micelles with different structural features
For the αLA-SDS-C12E8 data set it was observed that the fit quality throughout the series improved a little if the αLA-SDS basis function was replaced by the sample with αLA-SDS and a very small amount of C12E8, indicating that the structure of the αLA-SDS complexes changes slightly upon addition of C12E8. No additional changes were observed upon further addition of C12E8. Therefore, the SDS-unfolded αLA basis function in the fitting was replaced by αLA-SDS with a small amount of C12E8.
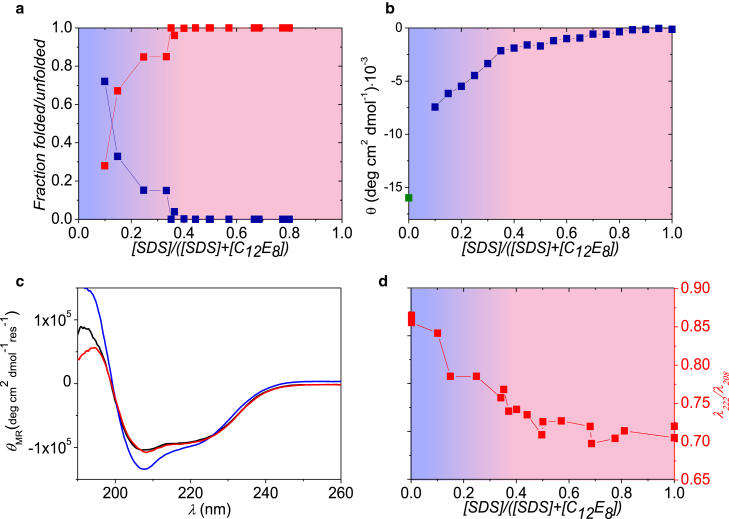
From the SAXS results in Fig. 4 a (fits in Fig. S6), a gradual transition from unfolded (at higher χSDS values) to a nativelike protein starts to occur below χSDS = 0.4. However, the folded, micelle-free state only becomes the most abundant species at the very last data point at χSDS = 0.1. The near-UV CD data indicate a small, continuous change as χSDS is decreased to 0.4, followed by a slightly steeper decrease below 0.4. Even so, there remains a large difference in signal between the value of the lowest χSDS of 0.1 and the values from pure αLA (green square) or αLA with only C12E8 micelles (data point not shown), which indicates that the tertiary structure of αLA with C12E8 is not the same as in pure buffer, consistent with previous data on DDM (5). Nevertheless, the far-UV CD data (Fig. 4 c) of the lowest χSDS (red) is almost identical to that of pure protein (black), demonstrating that under these conditions, αLA has nativelike secondary structure despite the lack of nativelike tertiary structure. CD spectra at χSDS = 0.5 (Fig. 4 c) show a marked increase in ellipticity at the 209 nm minimum, indicating a conformational change that can be interpreted as the formation of isolated helices. Near-UV data for the same values of χSDS (Fig. S7) show a complete loss of tertiary structure at χSDS = 0.5, which is then partly recovered at χSDS = 0.1. The CD data are thus in reasonable agreement with the SAXS results. As an indication of changes in secondary structure, the ratio λ222/λ208 from the far-UV CD data (Fig. 4 d) increases slowly below χSDS = 0.4, which may be caused by the transition from SDS-denatured to NIS-refolded observed from the SAXS analysis. Therefore, we conclude that αLA undergoes a conformational change from the SDS-denatured state when C12E8 is added to a globular state similar to that of the native protein, but with differences in tertiary structure.
Figure 4.
Results for αLA and SDS samples mixed with C12E8. (a) Part of the protein that is either folded (data with highest data point at low mole fraction) or unfolded around SDS micelles (goes to unity at high mole fraction) from SAXS analysis is shown. (b) Near-UV CD data at 296 nm of αLA in micelles (data as a function of mole fraction) and buffer (point at zero mole fraction) are shown. (c) Shown here are CD spectra for pure αLA (black), and αLA mixed SDS and C12E8 with SDS mole fractions of 0.5 (curve with deepest minimum) and 0.1 (nearly coinciding with black curve), respectively. (d) λ222/λ208 ratio. To see this figure in color, go online.
As in the case of βLG, we additionally investigated how DDM influences the αLA-SDS complexes. Both SAXS and CD data from the samples with αLA and DDM (with no SDS) indicated that DDM itself induces structural changes in αLA. This was observed by changes in the far-UV CD data of αLA upon addition of DDM, and from the fact that the fits of the αLA-DDM samples with a linear combination of pure αLA and DDM were poor. On the contrary, the fits improved significantly if the pure protein basis function was replaced by protein with a small amount of DDM, demonstrating an interaction of αLA with a small amount of DDM, while higher concentrations of DDM merely resulted in free micelles. It should be noted that calcium-depleted αLA was used in this study, and this apo-state state (32, 33) is more loosely packed than αLA with calcium and therefore more sensitive to different surfactants. The interaction between DDM and αLA, leading to a change in its tertiary structure, is consistent with our earlier reports on the ability of NIS to denature αLA (5, 34, 35).
Because αLA changes structure slightly in the presence of DDM, it is inappropriate to use the data for αLA in buffer as a basis function in the refolding studies, because this species will not exist when the solution contains DDM. Therefore, data from the pure protein basis function were replaced by a sample with αLA and a low concentration of DDM. The overall results were comparable to those obtained for C12E8 (Fig. 5; SAXS fits in Fig. S8). The SAXS results show a well-defined transition below χSDS = 0.4, while the near-UV CD data (Fig. 5 b) reveal a modest increase in the absolute value of the CD signal (i.e., an increase in structure) starting at ∼χSDS = 0.7 and stabilizing at ∼χSDS = 0.4. The large difference in molar ellipticity at 296 nm between protein with DDM and pure protein also suggests an interaction and binding of DDM to the protein as described above. Fig. 5 c demonstrates that the CD spectra for αLA at χSDS 0.1 (blue line) is significantly different from pure αLA (black line), but similar to αLA with only DDM (no SDS, green line). This shows that the αLA species at χSDS = 0.1 is just as folded as when only DDM is present. On the other hand, the CD data at χSDS = 0.5 (red line) are significantly different, revealing that αLA has a more unfolded structure at this χSDS. Similar conclusions can be drawn from the near-UV CD data (Fig. S9): addition of DDM to pure αLA decreases the signal significantly, the structure is completely lost at χSDS = 0.5, and at χSDS = 0.1 it is partly restored but still less than for the αLA-DDM data. The relatively small variations in λ222/λ208 ratio (Fig. 5 d) as a function of χSDS determined from the CD data indicate relatively small changes in secondary structure in the different samples. Nevertheless, there is an increase in λ222/λ208 ratio below χSDS = 0.4, just as seen for C12E8. All results considered, it is most likely that there is a transition from SDS-unfolded to DDM-modified αLA at very low χSDS values, similar to the case for αLA and C12E8.
Figure 5.
Results for αLA and SDS samples mixed with DDM. (a) Part of the protein that is either folded (highest data points at low mole fraction) or unfolded around SDS micelles (goes to unity at high mole fraction) from SAXS analysis is shown. (b) Near-UV CD data at 296 nm of αLA in micelles (data as a function of mole fraction), buffer (lowest point at zero mole fraction), and in DDM (highest point at zero mole fraction) are given. (c) Shown here are CD spectra for pure αLA (black), αLA mixed with DDM (second curve from top at 207 nm), and αLA mixed with SDS and DDM to SDS mole fractions of 0.5 (deepest minimum) and 0.1 (second curve from below at 207 nm), respectively. (d) λ222/λ208 ratio. To see this figure in color, go online.
To get more insight into the structure of the DDM-αLA complexes, we performed modeling, on an absolute scale, of SAXS data from a series with constant protein concentration and increasing amounts of DDM using a program originally developed for analysis of detergent-solubilized proteins (36). At DDM concentrations of 6 mM DDM and higher, the SAXS data could be described by a model with a dimer of αLA with 2–3 mM DDM bound between two partly unfolded αLA molecules with the rest of the DDM present as free micelles (Fig. S9). The unfolded αLA was manually constructed by opening the structure in the middle. The amount of bound DDM corresponds to 13–20 DDM molecules per dimer complex. Although such a modeling is not unique with respect to the protein structure, it gives a good impression for a likely structure of the DDM-αLA complex.
Lysozyme refolds to a micelle-free state in the presence of NIS
Given the special interactions between αLA and NIS, we turned to the homologous (but much more stable) protein lysozyme to determine if it behaves differently when exposed to SDS and NIS. As C12E8 and DDM appear to have similar effect on the protein-refolding capabilities of αLA and βLG, only C12E8 was included in these following refolding studies. In contrast to αLA, LYZ is not influenced by addition of C12E8 alone (data not shown). However, as was also observed for αLA, the fit quality of the data series improved significantly if the LYZ-SDS complexes were replaced by the LYZ-SDS sample with a small amount of C12E8, which points to a change in structure of the LYZ-SDS complexes in presence of C12E8. Therefore, this sample with LYZ, SDS, and a small concentration of C12E8 was used as a basis function in the fitting instead of the pure LYZ-SDS complexes.
The results from the refolding study of SDS-unfolded LYZ with C12E8 are summarized in Fig. 6 (SAXS fits in Fig. S10). The SAXS analysis (Fig. 6 a) indicates a transition from unfolded complexes to folded protein at ∼χSDS = 0.25, and the rather steep changes in both the near-UV CD data (Fig. 6 b) and the λ222/λ208 ratio (Fig. 6 d) around this χSDS support this conclusion.
Figure 6.
Results for LYZ and SDS samples mixed with C12E8. (a) Part of the protein that is either folded (upper data points at low mole fraction) or unfolded around SDS micelles (unity at high mole fraction) from SAXS analysis is shown. (b) Near-UV CD data at 296 nm of LYZ in micelles (data as function of mole fraction), buffer (lowest point, nearly covered, at zero mole fraction), and in C12E8 (upper point at zero mole fraction) are shown. (c) CD spectra for pure LYZ (black), LYZ-mixed SDS, and C12E8 with SDS mole fractions of 1 (with deepest minimum) and 0.1 (nearly coinciding with black curve above 202 nm), respectively, are given. (d) λ222/λ208 ratio. To see this figure in color, go online.
Analysis of the CD data (Fig. 6 c) show that the secondary state of pure LYZ (black line), SDS-unfolded LYZ (red line), and samples with χSDS = 0.1 (blue line) is mostly α-helical. This is expected because native LYZ contains a high degree of α-helices, and SDS is known to induce α-helices when unfolding proteins (37). Even though all three CD data sets reflect mainly α-helical structure, there is still a clear difference between SDS-unfolded LYZ compared to pure LYZ and the sample with χSDS = 0.1, which are very similar. Additionally, the far-UV data (Fig. S11) demonstrate very clearly that the tertiary structure of LYZ is lost when mixed with SDS, while it is completely restored when C12E8 is present at χSDS = 0.1. Therefore, we conclude that LYZ is refolded by C12E8 at low χSDS values.
BSA can be folded to the native state by NIS at low χSDS
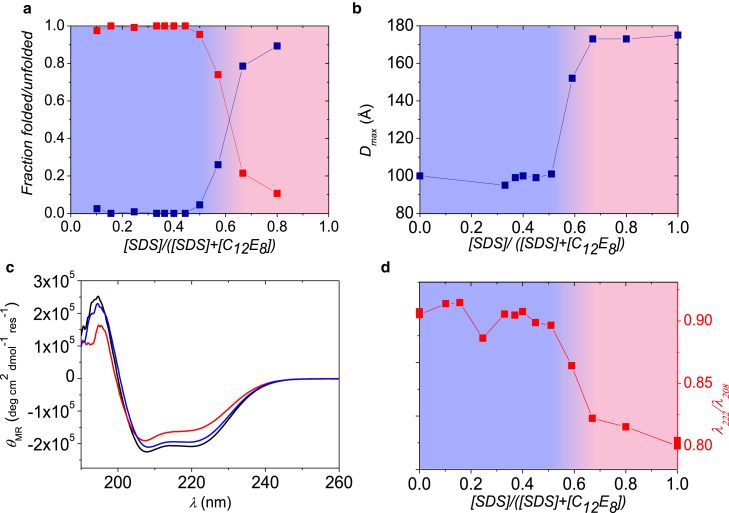
Finally, to examine whether the observations from the above refolding studies apply to larger proteins as well, we examined whether C12E8 could refold SDS-unfolded BSA. An overview of these results is presented in Fig. 7, with the SAXS fits again included in Fig. S12. Addition of C12E8 alone did not influence the protein structure. BSA is known to be able to bind a broad array of amphipathic and hydrophobic molecules in the native state without any major conformational changes (38). The results from the SAXS analysis (Fig. 7 a) clearly indicate that free micelles and folded protein are present below χSDS = 0.6, while protein-micelle complexes are formed at higher χSDS values. Additional evidence for formation of larger complexes is demonstrated in Fig. 7 b, which shows the maximum particle diameter estimated from the pair distance distribution function, p(r), obtained from the SAXS data through the IFT procedure. The data show that the maximum particle diameter almost doubles when χSDS is >0.6. This supports the other results—namely, that the protein is unfolded and forms complexes with the surfactant in this range. Modeling of the complexes using a model with protein-decorated core-shell micelles shows that the complexes contained two SDS micelles and this explains the larger maximum size of the complexes compared to the folded protein.
Figure 7.
Results from measurements and analysis of BSA and SDS samples mixed with C12E8. (a) Part of the protein that is either folded (data at unity at low mole fraction) or unfolded around SDS micelles (data at zero at low mole fraction) from SAXS analysis is shown. (b) Estimate of maximum particle diameter determined using IFT is given. (c) CD spectra for pure BSA (black) and BSA in mixed SDS-C12E8 micelles with an SDS mole fraction of 0.8 (upper curve near the minima) and 0.51 (middle curve), respectively, are given. (d) λ222/λ208 ratio. To see this figure in color, go online.
The CD spectra for pure BSA and BSA with χSDS = 0.8 are distinctly different (Fig. 7 c). When χSDS is decreased to 0.51, the CD data approach the shape of the CD spectra for pure BSA, verifying that SDS-unfolded BSA refolds upon addition of C12E8. The ratio λ222/λ208 of the signal for pure BSA and BSA mixed with surfactants (Fig. 7 d) is similar at χSDS < 0.6, whereas it is reduced markedly at χSDS > 0.6. This also shows that BSA indeed does refold when the nonionic surfactant is present at high concentrations. Estimation of the secondary structure elements supports that the secondary structure of BSA changes with χSDS.
We also attempted to follow BSA’s conformational changes using near-UV CD as for the other proteins. However, the large number of aromatic residues in BSA tend to cancel each other out, and therefore BSA does not have a very well-defined near-UV CD spectrum. As a consequence, it is not possible to measure a distinct transition from the native to the SDS-denatured state this way (data not shown).
Discussion
The results from this study show that all investigated proteins are able to refold from their SDS-unfolded complexes when mixed with the nonionic surfactants C12E8 and DDM. Nevertheless, for αLA the refolded state in C12E8 had a different fold than the native one, and in the presence of DDM the refolded state was complexed with DDM, though not in the micellar state of DDM. The two tested NIS had almost identical influence on the refolding capabilities of αLA, while for βLG the transition between folded and unfolded protein shifted from χSDS of ∼0.5 for C12E8 to ∼0.25 for DDM. Because both surfactants were able to refold αLA (to some extent) and βLG, only C12E8 was used in the studies LYZ and BSA.
The use of both SAXS and far- and near-UV CD in these protein refolding experiments allowed us to probe both the overall sample composition and the overall features of protein secondary and tertiary structure, respectively. This made it possible to compare results from different techniques to strengthen our conclusions. In general, the transitions were easy to follow, and there was a good correlation between the results from SAXS and those from CD. Only the results from αLA were less obvious, because the transition found using SAXS was accompanied by only subtle changes in the CD data. However, the combined results can be interpreted as refolding of αLA beginning at χSDS = 0.4 and below. It should be kept in mind that this refolding for DDM was only to the DDM-altered state of αLA and not the native protein state, because addition of DDM surfactants alone influenced the structure of the calcium-depleted αLA used in this study, reflecting the highly dynamic nature of the unstable apo-state of αLA and its known ability to undergo conformational changes in the presence of different NIS (5).
We find our data to be noteworthy on two accounts, namely the reversibility of protein-SDS interactions and the NIS’ ability to completely remove micellar SDS from their complex with proteins. Because all protein samples were initially mixed with SDS with complete unfolding as a result, the ability of the proteins to refold must mean that the proteins are not trapped in complex with SDS but form equilibrium structures. If the complexes were kinetically trapped states, the protein would not refold upon addition of nonionic surfactants. Furthermore, isothermal titration calorimetry studies show coexistence of free and protein-bound SDS molecules (5, 7, 28, 39, 40, 41) although usually with a lower free SDS concentration than the CMC of pure SDS. Likewise, the proteins’ ability to refold at low χSDS values to micelle-free structures demonstrate that the advantage of transferring SDS molecules into mixed micelles, with a very favorable mixing entropy, outweighs the electrostatic interactions of SDS with the protein at low χSDS. This redistribution of SDS into NIS micelles makes it possible to withdraw sufficient amounts of SDS from the complexes to allow the proteins to refold. Such an entropically driven transfer is less efficient, but also gentler, than chelatelike systems that strip SDS from proteins through very strong binding interactions. Probably the best example is provided by the SDS-cyclodextrin pair, which has been used by Rozema and Gellman (42, 43) to develop artificial chaperone folding systems and has been expanded to include cationic surfactants (44, 45). In this system, the protein, initially denatured in a chemical denaturant such as urea, is first mixed with an ionic surfactant, allowing the denaturant to be removed by dilution or dialysis, while keeping the protein soluble but still denatured. Subsequently, the detergent can be rapidly stripped off the protein by addition of excess equivalents of cyclodextrin, allowing the protein to fold. The high affinity of the SDS-cyclodextrin pair combined with high association rate constants allows stripping to occur and equilibrate within milliseconds, and provides an alternative approach to monitor the kinetics of folding of, e.g., the protein S6 (46). (An analogous approach is to remove SDS by cooling (47, 48) or by chromatography (49), leading to protein refolding, although this removal obviously happens much less rapidly than with cyclodextrin stripping.) In contrast, transfer of SDS to NIS micelles is at least an order-of-magnitude slower, so that folding is coupled to SDS transfer and therefore the measured refolding rate constant of S6 is reduced by at least a factor of 10 compared to the SDS-cyclodextrin system (D.E.O., data not shown). A slower transfer of SDS may be advantageous, provided the SDS that remains transiently bound to the protein during the equilibration process helps to reduce aggregation while allowing the protein to refold. More systematic studies will be required to ascertain whether this is favorable in practice. Other approaches to refold SDS-denatured proteins include the use of alcohols such as methyl-pentanediol as cosolvents that dissociate SDS micelles by screening the sulfate headgroup and shielding the alkyl chain’s solvent accessibility. This leads to a shift from harsh to gentle surfactant, reducing interactions with proteins and thus allowing them to refold (50, 51, 52). Similarly, dilution of SDS into NIS micelles will dilute out the otherwise repulsive effects of the sulfate headgroup and also sequester the alkyl chain from potentially disruptive hydrophobic interactions with proteins.
From separate studies it is known that SDS can bind at low concentrations to the native state of proteins such as BSA (38), βLG (15), and LYZ (53), but this interaction can be treated formally as a simple ligand-binding equilibrium that stabilizes the native state and does not lead to major conformational changes. Consequently, this state cannot be distinguished from the SDS-free native state and will go undetected in our SAXS analysis.
The existence of detergent-free/detergent-poor species observed in our studies is fundamentally different from the folding and unfolding of membrane proteins in mixed-micelle systems (8). Numerous membrane proteins have been shown to undergo reversible unfolding as the χSDS value varies between 0 and 1 (54), but the membrane protein intrinsically requires itself to be embedded in a membranelike environment to avoid aggregation. This embedding can occur either via nonionic micelles clustering around the transmembrane region in the folded structure, as demonstrated by recent SAXS studies of the β-barrel protein OmpA (36) and the α-helical aquaporin (55), or by more conventional dissolution of the protein in SDS micelles, leading to greatly increased interhelical distances in, e.g., bacteriorhodopsin, but no major change in the distribution of protein end-to-end distances (56).
The methodology presented here will also be fully transferable to unfolding-refolding studies of membrane proteins as a function of χSDS. Despite the larger similarity between SAXS data from folded and unfolded membrane proteins, the internal constraints of the SAXS analysis (together with complementary information on the secondary and tertiary structure from CD and fluorescence measurements) will surely make it possible to distinguish the two species from each other.
Conclusions
This work shows that the nonionic surfactants C12E8 and DDM are able to refold four different proteins into their native or NIS-altered structure after the proteins have been unfolded by SDS. The proteins chosen included both small and large proteins, as well as proteins consisting of mainly either α-helices or β-sheets. Therefore, because all proteins were able to refold, we suggest that refolding of SDS-unfolded proteins by nonionic surfactants may be a general feature for water-soluble globular proteins. This suggestion remains to be consolidated by more extensive future studies involving additional representatives of the diverse class of NIS as well as multimeric proteins.
Author Contributions
J.S.P. and D.E.O. designed the experiments. J.D.K., A.S., and D.J.M. performed experiments and analyzed data. D.E.O., J.S.P., J.D.K., and A.S. wrote the manuscript.
Acknowledgments
We are grateful to Jannik Nedergaard Pedersen for help in collecting SAXS spectra.
This work was supported by the Danish Research Council for Independent Research, Natural Sciences through grant No. 0602-01878B.
Editor: Jill Trewhella.
Footnotes
Jørn Døvling Kaspersen and Anne Søndergaard contributed equally to this work.
Twelve figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)30332-6.
Contributor Information
Daniel E. Otzen, Email: dao@inano.au.dk.
Jan Skov Pedersen, Email: jsp@chem.au.dk.
Supporting Material
References
- 1.Kirk O., Borchert T.V., Fuglsang C.C. Industrial enzyme applications. Curr. Opin. Biotechnol. 2002;13:345–351. doi: 10.1016/s0958-1669(02)00328-2. [DOI] [PubMed] [Google Scholar]
- 2.Otzen D. Protein-surfactant interactions: a tale of many states. Biochim. Biophys. Acta. 2011;1814:562–591. doi: 10.1016/j.bbapap.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 3.Jones M.N. Surfactant interactions with biomembranes and proteins. Chem. Soc. Rev. 1992;21:127–136. [Google Scholar]
- 4.Santos S.F., Zanette D., Itri R. A systematic study of bovine serum albumin (BSA) and sodium dodecyl sulfate (SDS) interactions by surface tension and small angle x-ray scattering. J. Colloid Interface Sci. 2003;262:400–408. doi: 10.1016/S0021-9797(03)00109-7. [DOI] [PubMed] [Google Scholar]
- 5.Otzen D.E., Sehgal P., Westh P. α-lactalbumin is unfolded by all classes of detergents but with different mechanisms. J. Coll. Int. Sci. 2009;329:273–283. doi: 10.1016/j.jcis.2008.10.021. [DOI] [PubMed] [Google Scholar]
- 6.Otzen D.E. Proteins in a brave new surfactant world. Curr. Opin. Colloid Interface Sci. 2015;20:161–169. [Google Scholar]
- 7.Nielsen M.M., Andersen K.K., Otzen D.E. Unfolding of β-sheet proteins in SDS. Biophys. J. 2007;92:3674–3685. doi: 10.1529/biophysj.106.101238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lau F.W., Bowie J.U. A method for assessing the stability of a membrane protein. Biochemistry. 1997;36:5884–5892. doi: 10.1021/bi963095j. [DOI] [PubMed] [Google Scholar]
- 9.Otzen D.E. Folding of DsbB in mixed micelles: a kinetic analysis of the stability of a bacterial membrane protein. J. Mol. Biol. 2003;330:641–649. doi: 10.1016/s0022-2836(03)00624-7. [DOI] [PubMed] [Google Scholar]
- 10.Paslawski W., Lillelund O.K., Otzen D.E. Cooperative folding of a polytopic α-helical membrane protein involves a compact N-terminal nucleus and nonnative loops. Proc. Natl. Acad. Sci. USA. 2015;112:7978–7983. doi: 10.1073/pnas.1424751112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanford C. Formation of Micelles and Biological Membranes. 2nd Ed. Wiley & Sons; New York: 1980. The hydrophobic effect. [Google Scholar]
- 12.Anand U., Ray S., Mukherjee S. Structural aspects of a protein-surfactant assembly: native and reduced states of human serum albumin. Protein J. 2015;34:147–157. doi: 10.1007/s10930-015-9606-1. [DOI] [PubMed] [Google Scholar]
- 13.Lad M.D., Ledger V.M., Frazier R.A. Analysis of the SDS-lysozyme binding isotherm. Langmuir. 2003;19:5098–5103. [Google Scholar]
- 14.Waninge R., Paulson M., Sellers P. Binding of sodium dodecyl sulphate and dodecyl trimethyl ammonium chloride to β-lactoglobulin: a calorimetric study. Int. Dairy J. 1998;8:141–148. [Google Scholar]
- 15.Hansted J.G., Wejse P.L., Otzen D.E. Effect of protein-surfactant interactions on aggregation of β-lactoglobulin. Biochim. Biophys. Acta. 2011;1814:713–723. doi: 10.1016/j.bbapap.2011.03.011. [DOI] [PubMed] [Google Scholar]
- 16.Pedersen J.S. A flux- and background-optimized version of the NanoSTAR small-angle x-ray scattering camera for solution scattering. J. Appl. Cryst. 2004;37:369–380. [Google Scholar]
- 17.Li Y.L., Beck R., Divinagracia M. Scatterless hybrid metal-single-crystal slit for small-angle x-ray scattering and high-resolution x-ray diffraction. J. Appl. Cryst. 2008;41:1134–1139. [Google Scholar]
- 18.Schwamberger A., De Roo B., Locquet J.P. Combining SAXS and DLS for simultaneous measurements and time-resolved monitoring of nanoparticle synthesis. Nuclear Inst. Methods Phys. Res. B. 2015;343:116–122. [Google Scholar]
- 19.Lindner P. Scattering experiments: experimental aspects, initial data reduction and absolute calibration. In: Zemb T., Lindner P., editors. Neutron, X-rays and Light. Scattering Methods Applied to Soft Condensed Matter. 1st Ed. North-Holland; Amsterdam, the Netherlands: 2002. [Google Scholar]
- 20.Glatter O. A new method for the evaluation of small-angle scattering data. J. Appl. Cryst. 1977;10:415–421. [Google Scholar]
- 21.Pedersen J.S., Hansen S., Bauer R. The aggregation behavior of zinc-free insulin studied by small-angle neutron scattering. Eur. Biophys. J. 1994;22:379–389. doi: 10.1007/BF00180159. [DOI] [PubMed] [Google Scholar]
- 22.Kinning D.J., Thomas E.L. Hard-sphere Interactions between spherical domains in diblock copolymers. Macromolecules. 1984;17:1712–1718. [Google Scholar]
- 23.Greenfield N., Fasman G.D. Computed circular dichroism spectra for the evaluation of protein conformation. Biochemistry. 1969;8:4108–4116. doi: 10.1021/bi00838a031. [DOI] [PubMed] [Google Scholar]
- 24.Sun Y., Filho P.L.O., Oliveira C.L.P. Unfolding and folding pathway of lysozyme induced by sodium dodecyl sulfate. Soft Matter. 2015;11:7769–7777. doi: 10.1039/c5sm01231g. [DOI] [PubMed] [Google Scholar]
- 25.Narayanan J., Abdul Rasheed A.S., Bellare J.R. A small-angle x-ray scattering study of the structure of lysozyme-sodium dodecyl sulfate complexes. J. Colloid Interface Sci. 2008;328:67–72. doi: 10.1016/j.jcis.2008.09.012. [DOI] [PubMed] [Google Scholar]
- 26.Abelein A., Kaspersen J.D., Gräslund A. Formation of dynamic soluble surfactant-induced amyloid β peptide aggregation intermediates. J. Biol. Chem. 2013;288:23518–23528. doi: 10.1074/jbc.M113.470450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giehm L., Oliveira C.L.P., Otzen D.E. SDS-induced fibrillation of α-synuclein: an alternative fibrillation pathway. J. Mol. Biol. 2010;401:115–133. doi: 10.1016/j.jmb.2010.05.060. [DOI] [PubMed] [Google Scholar]
- 28.Andersen K.K., Oliveira C.L.P., Otzen D. The role of decorated SDS micelles in sub-CMC protein denaturation and association. J. Mol. Biol. 2009;391:207–226. doi: 10.1016/j.jmb.2009.06.019. [DOI] [PubMed] [Google Scholar]
- 29.Lau S.Y., Taneja A.K., Hodges R.S. Synthesis of a model protein of defined secondary and quaternary structure. Effect of chain length on the stabilization and formation of two-stranded α-helical coiled-coils. J. Biol. Chem. 1984;259:13253–13261. [PubMed] [Google Scholar]
- 30.Zhou N.E., Kay C.M., Hodges R.S. Synthetic model proteins. Positional effects of interchain hydrophobic interactions on stability of two-stranded α-helical coiled-coils. J. Biol. Chem. 1992;267:2664–2670. [PubMed] [Google Scholar]
- 31.Liu X., Shang L., Wang E. Conformational changes of β-lactoglobulin induced by anionic phospholipid. Biophys. Chem. 2006;121:218–223. doi: 10.1016/j.bpc.2005.12.015. [DOI] [PubMed] [Google Scholar]
- 32.Kataoka M., Kuwajima K., Goto Y. Structural characterization of the molten globule of alpha-lactalbumin by solution x-ray scattering. Protein Sci. 1997;6:422–430. doi: 10.1002/pro.5560060219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuwajima K. The molten globule state of α-lactalbumin. FASEB J. 1996;10:102–109. doi: 10.1096/fasebj.10.1.8566530. [DOI] [PubMed] [Google Scholar]
- 34.Andersen K.K., Otzen D.E. Denaturation of α-lactalbumin and myoglobin by the anionic biosurfactant rhamnolipid. Biochim. Biophys. Acta. 2014;1844:2338–2345. doi: 10.1016/j.bbapap.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 35.Malmendal A., Underhaug J., Nielsen N.C. Fast mapping of global protein folding states by multivariate NMR: a GPS for proteins. PLoS One. 2010;5:e10262. doi: 10.1371/journal.pone.0010262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Døvling Kaspersen J., Moestrup Jessen C., Pedersen J.S. Low-resolution structures of OmpA⋅DDM protein-detergent complexes. ChemBioChem. 2014;15:2113–2124. doi: 10.1002/cbic.201402162. [DOI] [PubMed] [Google Scholar]
- 37.Takeda K., Sasaoka H., Moriyama Y. Size and mobility of sodium dodecyl-sulfate bovine serum-albumin complex as studied by dynamic light-scattering and electrophoretic light-scattering. J. Coll. Int. Sci. 1992;154:385–392. [Google Scholar]
- 38.Kragh-Hansen U., Chuang V.T.M., Otagiri M. Practical aspects of the ligand-binding and enzymatic properties of human serum albumin. Biol. Pharm. Bull. 2002;25:695–704. doi: 10.1248/bpb.25.695. [DOI] [PubMed] [Google Scholar]
- 39.Nielsen A.D., Arleth L., Westh P. Interactions of Humicola insolens cutinase with an anionic surfactant studied by small-angle neutron scattering and isothermal titration calorimetry. Langmuir. 2005;21:4299–4307. doi: 10.1021/la047299+. [DOI] [PubMed] [Google Scholar]
- 40.Andersen K.K., Westh P., Otzen D.E. Global study of myoglobin-surfactant interactions. Langmuir. 2008;24:399–407. doi: 10.1021/la702890y. [DOI] [PubMed] [Google Scholar]
- 41.Wang H., Andersen K.K., Otzen D.E. pH regulation of the kinetic stability of the lipase from Thermomyces lanuginosus. Biochemistry. 2013;52:264–276. doi: 10.1021/bi301258e. [DOI] [PubMed] [Google Scholar]
- 42.Rozema D., Gellman S.H. Artificial chaperone-assisted refolding of carbonic anhydrase B. J. Biol. Chem. 1996;271:3478–3487. doi: 10.1074/jbc.271.7.3478. [DOI] [PubMed] [Google Scholar]
- 43.Rozema D., Gellman S.H. Artificial chaperone-assisted refolding of denatured-reduced lysozyme: modulation of the competition between renaturation and aggregation. Biochemistry. 1996;35:15760–15771. doi: 10.1021/bi961638j. [DOI] [PubMed] [Google Scholar]
- 44.Couthon F., Clottes E., Vial C. Refolding of SDS- and thermally denatured MM-creatine kinase using cyclodextrins. Biochem. Biophys. Res. Commun. 1996;227:854–860. doi: 10.1006/bbrc.1996.1596. [DOI] [PubMed] [Google Scholar]
- 45.Lanckriet H., Middelberg A.P. Operational regimes for a simplified one-step artificial chaperone refolding method. Biotechnol. Prog. 2004;20:1861–1867. doi: 10.1021/bp049897k. [DOI] [PubMed] [Google Scholar]
- 46.Otzen D.E., Oliveberg M. A simple way to measure protein refolding rates in water. J. Mol. Biol. 2001;313:479–483. doi: 10.1006/jmbi.2001.5039. [DOI] [PubMed] [Google Scholar]
- 47.Schlager B., Straessle A., Hafen E. Use of anionic denaturing detergents to purify insoluble proteins after overexpression. BMC Biotechnol. 2012;12:95. doi: 10.1186/1472-6750-12-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Putnam F., Neurath H. The precipitation of proteins by synthetic detergents. J. Am. Chem. Soc. 1944;66:692–697. [Google Scholar]
- 49.Weber K., Kuter D.J. Reversible denaturation of enzymes by sodium dodecyl sulfate. J. Biol. Chem. 1971;246:4504–4509. [PubMed] [Google Scholar]
- 50.Michaux C., Roussel G., Perpete E.A. Unravelling the mechanisms of a protein refolding process based on the association of detergents and co-solvents. J. Pept. Sci. 2016;22:485–491. doi: 10.1002/psc.2893. [DOI] [PubMed] [Google Scholar]
- 51.Michaux C., Pomroy N.C., Privé G.G. Refolding SDS-denatured proteins by the addition of amphipathic cosolvents. J. Mol. Biol. 2008;375:1477–1488. doi: 10.1016/j.jmb.2007.11.026. [DOI] [PubMed] [Google Scholar]
- 52.Roussel G., Rouse S.L., Perpète E.A. The role of 2-methyl-2, 4-pentanediol in sodium dodecyl sulfate micelle dissociation unveiled by dynamic light scattering and molecular dynamics simulations. Colloids Surf. B Biointerfaces. 2014;114:357–362. doi: 10.1016/j.colsurfb.2013.10.023. [DOI] [PubMed] [Google Scholar]
- 53.Yonath A., Podjarny A., Traub W. Crystallographic studies of protein denaturation and renaturation. 2. Sodium dodecyl sulfate induced structural changes in triclinic lysozyme. Biochemistry. 1977;16:1418–1424. doi: 10.1021/bi00626a028. [DOI] [PubMed] [Google Scholar]
- 54.Neumann J., Klein N., Schneider D. Folding energetics and oligomerization of polytopic α-helical transmembrane proteins. Arch. Biochem. Biophys. 2014;564:281–296. doi: 10.1016/j.abb.2014.07.017. [DOI] [PubMed] [Google Scholar]
- 55.Berthaud A., Manzi J., Mangenot S. Modeling detergent organization around aquaporin-0 using small-angle x-ray scattering. J. Am. Chem. Soc. 2012;134:10080–10088. doi: 10.1021/ja301667n. [DOI] [PubMed] [Google Scholar]
- 56.Krishnamani V., Hegde B.G., Lanyi J.K. Secondary and tertiary structure of bacteriorhodopsin in the SDS denatured state. Biochemistry. 2012;51:1051–1060. doi: 10.1021/bi201769z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.