Abstract
Numerous studies show efflux as a universal bacterial mechanism contributing to antibiotic resistance and also that the activity of the antibiotics subject to efflux can be enhanced by the combined use of efflux inhibitors. Nevertheless, the contribution of efflux to the overall drug resistance levels of clinical isolates of Mycobacterium tuberculosis is poorly understood and still is ignored by many. Here, we evaluated the contribution of drug efflux plus target-gene mutations to the drug resistance levels in clinical isolates of M. tuberculosis. A panel of 17 M. tuberculosis clinical strains were characterized for drug resistance associated mutations and antibiotic profiles in the presence and absence of efflux inhibitors. The correlation between the effect of the efflux inhibitors and the resistance levels was assessed by quantitative drug susceptibility testing. The bacterial growth/survival vs. growth inhibition was analyzed through the comparison between the time of growth in the presence and absence of an inhibitor. For the same mutation conferring antibiotic resistance, different MICs were observed and the different resistance levels found could be reduced by efflux inhibitors. Although susceptibility was not restored, the results demonstrate the existence of a broad-spectrum synergistic interaction between antibiotics and efflux inhibitors. The existence of efflux activity was confirmed by real-time fluorometry. Moreover, the efflux pump genes mmr, mmpL7, Rv1258c, p55, and efpA were shown to be overexpressed in the presence of antibiotics, demonstrating the contribution of these efflux pumps to the overall resistance phenotype of the M. tuberculosis clinical isolates studied, independently of the genotype of the strains. These results showed that the drug resistance levels of multi- and extensively-drug resistant M. tuberculosis clinical strains are a combination between drug efflux and the presence of target-gene mutations, a reality that is often disregarded by the tuberculosis specialists in favor of the almost undisputed importance of antibiotic target-gene mutations for the resistance in M. tuberculosis.
Keywords: tuberculosis, synergism, efflux inhibitors, TB eXIST, time to detection
Introduction
The development of mutations in the genes associated with the resistance to the antituberculosis drugs have long been considered the sole cause of resistance in tuberculosis (Zhang and Yew, 2009; da Silva and Palomino, 2011). Nevertheless, Mycobacterium tuberculosis also presents intrinsic drug resistance, mainly attributed to the unusual structure of its mycolic acid-containing cell wall combined with effective efflux mechanisms (Jarlier and Nikaido, 1994; da Silva et al., 2011). The balance between the reduced permeability of the cell wall that acts synergistically with the activity of efflux pumps and the increased expression of genes that code for those efflux pumps is believed to constitute the first step for the development and stabilization of drug resistant phenotypes (Machado et al., 2012; Schmalstieg et al., 2012; Viveiros et al., 2012; da Silva et al., 2016).
Previous studies have demonstrated the contribution of efflux mechanisms to antibiotic resistance in M. tuberculosis revealing the presence of several putative efflux pumps of different classes involved in the transport of different compounds (Viveiros et al., 2012; Black et al., 2014; da Silva et al., 2016; Supplementary Table 1). The best-represented families of efflux transporters in M. tuberculosis are the ATP-binding cassette (ABC) superfamily and the major facilitator superfamily (MFS) followed by the resistance nodulation cell division (RND) superfamily of transporters. The most well-characterized ABC transporters showed to be involved in the transport of multiple drugs are the efflux pumps DrrAB, Rv2686c-2687c-2688c, Rv1456c-Rv1457c-Rv1458c, and the Rv1217c-1218c (Choudhuri et al., 2002; Pasca et al., 2004; Balganesh et al., 2010; Hao et al., 2011; Wang et al., 2013). Among the MFS, the most studied efflux pumps are the Rv1258c (Tap-like), associated with resistance to tetracycline, rifampicin and clofazimine (Siddiqi et al., 2004; Ramón-García et al., 2012); the P55, that confer resistance to aminoglycosides, tetracycline, rifampicin and clofazimine (da Silva et al., 2001; Ramón-García et al., 2009; Bianco et al., 2011); and the EfpA efflux transporter that is associated with resistance to isoniazid, fluoroquinolones and dyes (Doran et al., 1997). The RND efflux pumps are associated with the transport of a wide variety of substrates in M. tuberculosis (Bailo et al., 2015). Among these, it was shown that the MmpL7 protein confer low-level isoniazid resistance when overexpressed (Pasca et al., 2005; Machado et al., 2012). Likewise, the overexpression of the MmpS5-MmpL5 efflux transporter was associated with the resistance of M. tuberculosis to azoles (Milano et al., 2009) and with the acquired resistance to bedaquiline, the diarylquinoline recently approved for the treatment of multidrug resistant tuberculosis (Andries et al., 2014). The Mmr efflux transporter is the only efflux pump from the small multidrug resistance (SMR) family present in the M. tuberculosis genome and is associated with the reduced susceptibility of M. tuberculosis to dyes and antibiotics such as isoniazid, erythromycin, and fluoroquinolones (De Rossi et al., 1998; Rodrigues et al., 2013).
The overexpression of these and other efflux pumps is believed to decrease the intracellular levels of the antibiotics and prevent the drug to reach its cellular target, allowing the emergence of a subpopulation presenting high-level resistance (Machado et al., 2012; Schmalstieg et al., 2012). Noteworthy, efflux-based mechanisms were recently implicated in the intrinsic resistance to bedaquiline, and the efflux system MmpS5-MmpL5 was identified as being responsible for the non-target based resistance (Andries et al., 2014). In this sense, an approach to overcome the emergence of this efflux-mediated drug resistance can be the combined use of efflux inhibitors and antibiotics.
Up to now, a number of compounds have been shown as potential inhibitors of the M. tuberculosis efflux systems (Viveiros et al., 2012; da Silva et al., 2016). Among these, the most studied are the ion channel blockers like the phenothiazines thioridazine and chlorpromazine and the phenylalkylamine verapamil (Viveiros et al., 2012; da Silva et al., 2016; Machado et al., 2016). The mechanism by which these compounds are active against M. tuberculosis is not fully understood, however, some hypothesis for their mode action have been raised and discussed (Viveiros et al., 2012; Adams et al., 2014; Rayasam and Balganesh, 2015). Verapamil, a calcium channel blocker in eukaryotic cells, is able to decrease the minimum inhibitory concentrations (MICs) of antituberculosis drugs in M. tuberculosis clinical strains (Louw et al., 2011; Machado et al., 2012; Coelho et al., 2015), accelerates the bactericidal and sterilizing activities of antituberculosis drugs in a mouse model (Gupta et al., 2013), potentiates the activity of bedaquiline (Gupta et al., 2014), and inhibits macrophage-induced antibiotic tolerance of M. tuberculosis (Adams et al., 2011, 2014). Likewise, the phenothiazines have shown similar activities in spite of their toxicity compared to verapamil and other efflux inhibitors (Ordway et al., 2003; Viveiros et al., 2012). The phenothiazines disturb the calcium-calmodulin transport and signaling pathways (Salih et al., 1991; Pluta et al., 2011), cause modifications at the level of the bacterial membrane and on nucleic acid stability (Pluta et al., 2011; Thorsing et al., 2013), inhibit the type II NADH-ubiquinone dehydrogenase and other electron-transport chain enzymes (Weinstein et al., 2005), and have significant activity against M. tuberculosis in vitro, ex vivo, and in murine models (Ordway et al., 2003; Machado et al., 2012; Dutta et al., 2014; Singh and Sharma, 2014; Coelho et al., 2015). Recently, we observed that these ion channel blockers lead to a significant decrease in the intracellular mycobacterial load as result of the inducement of phagosome acidification and activation of lysosomal hydrolases (Machado et al., 2016). The potential use of efflux inhibitors in combination with antibiotics may constitute an important alternative as adjuvants of the antituberculosis conventional therapeutic regimen. Previously, we have demonstrated that multidrug resistance and in particular the development of resistance to isoniazid develops in M. tuberculosis laboratory strains associated with the genetic and physiological activity of efflux pumps (Machado et al., 2012). Nonetheless, two questions remain answered—the role of the efflux systems in M. tuberculosis resistant strains that already carry mutations in the drug target genes and how do they respond to the drug pressure despite the presence of these mutations—a contribution to the overall resistance level poorly understood and still disregarded by many.
The main purpose of this study is to assess the role played by the efflux systems in M. tuberculosis strains that carry mutations in the drug target genes. In order to do this, we have studied the contribution of efflux to the overall resistance levels toward antituberculosis drugs in a panel of mono-, multi- and extensively drug resistant (MDR/XDR) M. tuberculosis clinical isolates from two geographical distinct areas baring well-known and characterized drug-resistance associated mutations. To quantify the role of efflux in these strains and establish clinical correlation, we performed quantitative drug susceptibility testing (qDST) and investigated the time to detection (TTD) of growth of each strain for first- and second-line antituberculosis drugs in the presence and absence of the efflux inhibitors verapamil, thioridazine, and chlorpromazine. The results obtained revealed that the addition of efflux inhibitors enhanced the effect of the antituberculosis drugs independently of the genotype of the M. tuberculosis strains. Here we have demonstrated the effectiveness of a synergistic combination of drugs with the conventional therapy despite the presence of a mutation conferring resistance that showed to be beneficial since it reduced the resistance level of the strain. Moreover, these results strongly support the relevance of these inhibitors as adjuvants in tuberculosis chemotherapy.
Materials and methods
Strains
The strains selected for this study are described in Table 1 and were isolated from Portuguese and Brazilian tuberculosis patients as part of the routine mycobacteriology laboratory services provided by Universidade NOVA de Lisboa (Lisboa, Portugal) and Universidade Federal do Rio Grande (Porto Alegre, Brazil) to the local hospitals. Since the main objective of this work was to analyze the interplay between the mutational resistance and the efflux activity, the strains were selected based on their drug resistance pattern in order to include pan-susceptible, isoniazid monoresistant, rifampicin monoresistant, ofloxacin monoresistant, poly-drug resistant (resistant to more than one first-line antituberculosis drugs other than both isoniazid and rifampicin), MDR (resistant at least to isoniazid and rifampicin), and XDR (MDR plus resistance to any fluoroquinolone and amikacin, kanamycin, or capreomycin) strains. The H37Rv ATCC27294T reference strain was obtained from the American Type Culture Collection (Virginia, USA), and used as control. Given the retrospective nature of the work involving only anonymized bacterial isolates, informed consent was not required for this study.
Table 1.
Categorization of the M. tuberculosis strains studied according to their lineage, resistance pattern, and phenotype.
| Strain | Isolation date | Lineage* | Resistance pattern | Phenotype |
|---|---|---|---|---|
| H37Rv | – | H37Rv | None | Susceptible |
| MtbPT1 | 2006 | LAM9—Orphan | None | Susceptible |
| MtbPT2 | 2008 | Unknown—SIT150 | None | Susceptible |
| MtbPT3 | 2003 | T1 | INH | Monoresistant to INH |
| MtbPT4 | 2009 | LAM1 | INH | Monoresistant to INH |
| MtbPT5 | 2003 | LAM1 | RIF | Monoresistant to RIF |
| MtbPT6 | 2005 | LAM1 | RIF | Monoresistant to RIF |
| MtbBR1 | 2009 | EAI1-SOM | OFX | Monoresistant to OFX |
| MtbBR2 | 2007 | Beijing | INH, OFX | Poly-drug resistant |
| MtbBR3 | 2010 | H3 | INH, RIF | MDR |
| MtbBR4 | 2011 | T1 | INH, RIF | MDR |
| MtbBR5 | 2011 | T2 | INH, RIF | MDR |
| MtbPT7 | 2009 | LAM1—Lisboa3& | INH; RIF | MDR |
| MtbBR6 | 2008 | Beijing | INH, RIF, AMK, CAP | MDR |
| MtbPT8 | 2011 | Beijing | INH; RIF; AMK; CAP | MDR |
| MtbPT9 | 2009 | LAM1—Lisboa3& | INH; RIF; AMK; CAP; OFX | XDR |
| MtbPT10 | 2009 | LAM4—Q1& | INH; RIF; AMK; CAP; OFX | XDR |
| MtbPT11 | 2012 | LAM4—Q1& | INH; RIF; AMK; CAP; OFX | XDR |
Spoligotype lineages/sublineages according to the international spoligotype database SITVITWEB rules. “Unknown” designates patterns with signatures that do not belong to any of the major lineages defined in the SITVITWEB database.
Lisboa3 and Q1 M. tuberculosis strains were clustered by 24-loci MIRU-VNTR analysis. AMK, amikacin; CAP, capreomycin; EAI, East-African-Indian; H, Haarlem. INH, isoniazid; LAM, Latin American-Mediterranean; MDR, multidrug resistant; OFX, ofloxacin; RIF, rifampicin; SIT, spoligotype international type; XDR, extensively drug resistant.
Drug susceptibility testing and MIC determination
First- and second-line drug susceptibility testing (DST) and MIC determination were done using the MGIT 960 system using the Epicenter V5.80A software and the TB eXIST module (Becton Dickinson, Diagnostic Systems, Sparks, MD, USA). The critical concentrations used for DST were as follows: isoniazid, 0.1 μg/ml; rifampicin, 1 μg/ml; streptomycin, 1 μg/ml; ethambutol, 5 μg/ml; amikacin, 1 μg/ml; capreomycin, 2.5 μg/ml; and ofloxacin, 1 μg/ml. The results were interpreted as follows: at the time of positivity of the proportional control [Growth units (GU) = 400] if the tubes containing the drugs were GU > 100, they were considered resistant to that concentration. If the GU of the tube containing the drugs were <100 they were considered susceptible (Springer et al., 2009; Cambau et al., 2015). In these assays, the critical concentration of the antituberculosis drugs are defined as the concentration that is required to eliminate more than 99% of the population of a control strain that never has been in contact with the drug. A strain is considered susceptible to a given drug tested at their critical concentration if, among the population, the proportion of resistant cells is <1% (critical proportion). The strain is considered resistant when the number of drug-resistant bacteria present in the drug-containing tube is >1%, when compared with the drug-free proportional growth control.
MIC determination of the antibiotics and efflux inhibitors was done within the MGIT 960 system and the growth monitored with the Epicenter V5.80A software as previously described (Machado et al., 2016). The MIC was considered as the lowest concentration with GU < 100 when the drug-free proportional control tube reached a GU = 400. The lowest concentration tested corresponded to the critical concentration of each antibiotic (susceptibility breakpoint).
An absolute growth control (undiluted) was included in every assay to monitor the normal growth of each strain. In this study, the TTD for positivity of the drug-free proportional control varied between 8 and 16 days depending on the strains. DST and MIC determination were performed at least in duplicate and the final value was given as the result of two concordant values.
Quantitative drug susceptibility testing
qDST of the antibiotics was conducted using the MGIT 960 system and the growth monitored with the Epicenter V5.80A software and the TB eXIST module (Becton Dickinson). The test was performed and interpreted according to the protocol previously described (Cambau et al., 2015). Isoniazid was tested at 0.1, 1, 3, and 10 μg/ml, rifampicin and amikacin at 1, 4, and 20 μg/ml, capreomycin at 2.5, 5, and 25 μg/ml, and ofloxacin at 1, 2, and 10 μg/ml. Antibiotics for which the strains were resistant at the highest concentration tested by qDST, were further tested at higher concentrations by MIC determination. The interpretation of the results was done as described above. The antibiotic resistance levels were quantified as follows: isoniazid low-level resistance when resistant (R) at 0.1 and susceptible (S) at 1 μg/ml; isoniazid high-level resistance when R ≥ 1 μg/ml; rifampicin and amikacin low-level resistance when R at 4 and S at 20 μg/ml; rifampicin and amikacin high-level resistance when R ≥ 20 μg/ml; capreomycin low-level resistance when R at 5 and S at 25 μg/ml; capreomycin high-level resistance when R ≥ 25 μg/ml; ofloxacin low-level resistance when R at 1 and S at 2 μg/ml; ofloxacin high-level resistance when R ≥ 2 μg/ml (Cambau et al., 2015). The evaluation of bacterial growth/survival vs. growth inhibition was done by the comparison between the TTD, defined as the time from the start of incubation to the positivity signal of growth (Diacon et al., 2014; Bowness et al., 2015), of a given strain in the presence and absence of an inhibitor (ΔTTD). To obtain the degree of potentiation of each compound on the antibiotic activity, the ΔTTD was normalized against the control tube with no inhibitor. qDST was performed at least in duplicate and the final value was given as the result of two concordant values.
Detection of mutations associated with resistance
Genomic DNA was isolated using the QIAamp DNA mini kit (QIAGEN, GmbH, Hilden, Germany) according to the manufacturer's instructions. The most common mutations in inhA regulatory region, and katG, gyrA, and rrs genes were studied using the Genotype MTBDRplus and MTBDRsl (Hain Lifescience GmbH, Nehren, Germany) according to the manufacturer's instructions. Genomic analysis of the complete inhA, katG, and tlyA genes and eis promoter region was performed by PCR amplification and DNA sequencing using the primers described elsewhere (Feuerriegel et al., 2009; Machado et al., 2013; Perdigão et al., 2013). The annealing temperatures were 60°C for inhA, 62°C for katG, 56°C for eis, and 55°C for tlyA.
Evaluation of ethidium bromide accumulation and efflux
Real-time detection of efflux activity was performed with a semi-automated fluorometric method using a Rotor-Gene 3000 thermocycler (Corbett Research, Sydney, Australia) (Paixão et al., 2009; Viveiros et al., 2010; Rodrigues et al., 2015). The assays were performed as previously described (Machado et al., 2012). M. tuberculosis strains were grown in 10 ml of Middlebrook 7H9 (Difco, Madrid, Spain) supplemented with 10% OADC (oleic acid, albumin, dextrose, catalase; Becton and Dickinson) with 0.05% Tween 80 at 37°C until an OD600 nm(OD600) of 0.8. The cells were collected by centrifugation at 2940 × g for 3 min, the supernatant discarded, the pellet washed in PBS and centrifuged again in the same conditions.
Ethidium bromide accumulation assays
For the ethidium bromide accumulation assays, the pellet was resuspended in PBS and the OD600 adjusted to 0.8. To determine the concentration of ethidium bromide that establishes the equilibrium between efflux and influx, the cells were incubated with different concentrations of ethidium bromide (0.625–5 μg/ml). The assays were conducted at 37°C in a Rotor-Gene 3000 (530; 585 nm) and the equilibrium concentration determined for each strain as the concentration that promoted a plateau of no more than 10% of relative fluorescence units during the 60 min of the assay. To evaluate the effect of the efflux inhibitors on the accumulation of ethidium bromide, the assays were performed as described above with the exception that each inhibitor was added to the buffer solution at half MIC and ethidium bromide was used at the equilibrium concentration (determined for each strain). The inhibitory activity of the compounds was determined by the calculation of the relative final fluorescence (RFF; Machado et al., 2011) which is a measure of how effective the compound is on the inhibition of ethidium bromide efflux (at a given concentration) by comparison of the final fluorescence at the last time point (60 min) of the treated cells with the cells in presence of only ethidium bromide. An index of activity above zero indicated that the cells accumulate more ethidium bromide under the condition used than those of the control (non-treated cells). Negative RFF values indicated that the treated cells accumulated less ethidium bromide than those of the control condition. Values above 1 in the presence of the efflux inhibitors indicated enhanced accumulation of ethidium bromide inside the cells. The experiments were done in triplicate and the RFF values are presented as the average of three independent assays plus standard deviation (±SD).
Ethidium bromide efflux assays
For the ethidium bromide efflux assays, the pellet was resuspended in PBS and the OD600 adjusted to 0.4. The cell suspensions were exposed to ethidium bromide at the equilibrium concentration determined above, in the presence of verapamil at half MIC for 1 h at room temperature. The cells were then pelleted by centrifugation and resuspended in new buffer with and without 0.4% glucose in the presence and absence of half MIC of verapamil and the fluorescence measured as described above. Data was normalized by comparing the relative fluorescence units obtained under the conditions that promote efflux (presence of glucose and absence of verapamil) with the relative fluorescence units from the control in which there is no efflux (presence of verapamil and no glucose). The RFF corresponds to the ratio of the fluorescence that remains per unit of time, relatively to the cells in presence of the inhibitor.
RT–qPCR analysis of putative efflux pumps
Strains were grown in MGIT tubes (mycobacteria growth indicator tubes; Becton Dickinson) for the MGIT 960 system in the presence of half MIC of each antibiotic as follows: MtbPT3 was exposed to isoniazid; MtbPT5 was exposed to rifampicin; MtbBR1 was exposed to ofloxacin; H37Rv, MtbPT7, and MtbPT11 were exposed to isoniazid or rifampicin. Total RNA was extracted using a GTC/Trizol method as previously described (Machado et al., 2016). The relative expression level of the efflux pump genes mmpL7, p55, efpA, mmr, Rv1258c, and Rv2459 was analyzed by RT-qPCR. The primer sets and sequences of oligonucleotides used are described elsewhere (Machado et al., 2012). RT-qPCR assay was performed in a Rotor-Gene 3000 thermocycler and followed the protocol recommended for use with the QuantiTect SYBR Green RT-PCR Kit (QIAGEN) with the following amplification program: reverse transcription for 30 min at 50°C; initial activation step for 15 min at 95°C; 35 cycles of denaturation at 94°C for 30 s, annealing at 52°C for 30 s and extension at 72°C for 30 s; a final extension step at 72°C for 5 min; and an additional step at 50°C for 15 s followed by melt analysis (50–99°C). The determination of the relative mRNA expression level was performed using the comparative quantification cycle (Cq) method (Livak and Schmittgen, 2001). The relative expression of each target gene was determined by comparison of the relative quantity of the respective mRNA in the presence of the antibiotic to the non-exposed condition. Each strain was assayed in triplicate using total RNA obtained from three independent cultures. A relative expression level equal to 1 indicated that the expression level was identical to the unexposed strain. Genes showing expression levels above one were considered to be overexpressed. Genes showing expression levels above two were considered to be significantly overexpressed. Normalization of the mRNA levels was done using the M. tuberculosis 16S rDNA as internal control for each experiment and presented as the mean-fold change (±SD) compared with the control.
MIRU-VNTR and spoligotyping analysis
Mycobacterial Interspersed Repetitive Unit—Variable Number of Tandem Repeats (MIRU-VNTR) genotyping was performed by multiplex PCR amplification of 24-loci MIRU-VNTR, as described previously (Supply et al., 2006). Spoligotyping was performed as per Kamerbeek et al. (1997). The genotype of these strains was analyzed using the MIRU-VNTRplus web application (Allix-Béguec et al., 2008; Weniger et al., 2010) and the SITVITWEB database (Demay et al., 2012).
Data analysis
Data analysis was carried out using the Student's t-test. A *P < 0.05 was considered statistically significant and highly significant when **P < 0.01 and ***P < 0.001 (two-tailed tested).
Results
Correlation between the phenotype and mutations associated with antibiotic resistance
Firstly, to establish a correlation between the phenotypic drug resistance level and the presence of resistance associated mutations, we searched for mutations in the genes associated with drug resistance to the antibiotics studied (Table 2).
Table 2.
Mutations associated with resistance to first- and second-line drugs detected in the M. tuberculosis strains in study.
| Strain ID | Mutations | |||||||
|---|---|---|---|---|---|---|---|---|
| INH | RIF | AMK/CAP | AMK | CAP | OFX | |||
| inhA prom | inhA ORF | katG | rpoB RRDR | rrs 1400 region | eis prom | tlyA | gyrA QRDR | |
| SUSCEPTIBLE | ||||||||
| H37Rv | wt | wt | wt | wt | wt | wt | wt | wt |
| MtbPT1 | wt | wt | wt | wt | wt | wt | wt | wt |
| MtbPT2 | wt | wt | wt | wt | wt | wt | wt | wt |
| INHR | ||||||||
| MtbPT3 | wt | wt | S315T | wt | wt | nt | nt | wt |
| MtbPT4 | C-15T | wt | wt | wt | wt | nt | nt | wt |
| RIFR | ||||||||
| MtbPT5 | wt | nt | wt | S531L | wt | wt | wt | wt |
| MtbPT6 | wt | nt | wt | S531L | wt | nt | nt | wt |
| OFXR | ||||||||
| MtbBR1 | wt | nt | wt | wt | wt | nt | nt | D94N |
| Poly-DR | ||||||||
| MtbBR2 | wt | nt | S315T | D516Y | wt | nt | nt | A90V |
| MDR | ||||||||
| MtbBR3 | wt | nt | S315T | S531L | wt | wt | nt | wt |
| MtbBR4 | wt | nt | S315T | S531L | wt | wt | nt | wt |
| MtbBR5 | wt | nt | S315T | S531L | wt | wt | nt | wt |
| MtbPT7 | C-15T | S94A | wt | S531L | wt | wt | wt | wt |
| MtbBR6 | wt | nt | D735A | S531L | A1401G | wt | wt | wt |
| MtbPT8 | wt | wt | S315T | S531L | wt/A1401G* | wt | wt | wt |
| XDR | ||||||||
| MtbPT9 | C-15T | S94A | wt | S531L | wt | G-10A | ins GT at pos 755/756 | S91P |
| MtbPT10 | C-15T | I194T | wt | S531L | A1401G | wt | wt | D94A |
| MtbPT11 | C-15T | I194T | wt | S531L | A1401G | nt | nt | D94A |
Heteroresistance. AMK, amikacin; CAP, capreomycin; ID, identification; INH, isoniazid; MDR, multidrug resistant; nt, not tested; OFX, ofloxacin; ORF, open reading frame; Poly-DR, poly-drug resistant; Prom, promoter; QRDR, quinolone resistance determining region; RIF, rifampicin; RRDR, rifampicin resistance determining region; wt, wild-type sequence; XDR, extensively drug resistant.
The three pan-susceptible strains showed no mutation in any of the genes evaluated. Resistance to isoniazid was associated with mutations in the katG and inhA genes which are in accordance with published data (Cambau et al., 2015; Domínguez et al., 2016). MtbPT3 showed the S315T mutation in katG and MtbPT4 presented the C-15T substitution in the inhA promoter region. Among the nine MDR strains, five harbored mutations in katG, of which four had the S315T mutation and one has a rare mutation at codon 735. The remaining four strains harbored double mutations in inhA. The double mutations C-15T/S94A were detected in strains MtbPT7 and MtbPT9 and the pair C-15T/I194T was detected in MtbPT10 and MtbPT11.
In respect to rifampicin resistance, all MDR strains and the two rifampicin monoresistant strains showed the mutation S531L in rpoB. This is the most common mutation found in clinical isolates resistant to rifampicin (Cambau et al., 2015; Domínguez et al., 2016). Strain MtbBR2 carries the mutation D516Y in rpoB but it was found susceptible to rifampicin. Although was not our aim to evaluate susceptible strains, MtbBR2 was initially found to be resistant to rifampicin at 1 μg/ml (critical concentration). After re-testing MtbBR2 for rifampicin resistance, it was found susceptible, although with a MIC close to the breakpoint, and was reclassified as poly-drug resistant. The role of the mutation D516Y in rifampicin resistance is not clear. It has been reported by others to be associated with high-level rifampicin resistance (Cambau et al., 2015), low-level resistance (Williams et al., 1998; Zaczek et al., 2009), as well it has been found in rifampicin susceptible strains (Somoskovi et al., 2006; Andres et al., 2014), therefore a non-consensual genotype associated to resistance important to be included and analyzed in the context of this study.
Regarding second-line drugs, the MDR strains MtbBR6 and MtbPT8 were additionally resistant to one second line-injectable drug, and MtbBR2 was resistant to ofloxacin. MtbBR6 and MtbPT8 harbored a mutation in rrs, A1401G, and in strain MtbBR2 was found the A90V mutation in gyrA. MtbPT9, MtbPT10, and MtbPT11 were classified as XDR. Strains MtbPT10 and MtbPT11 harbored the rrs A1401G. This is the most common mutation found in clinical isolates resistant to second-line aminoglycosides (Cambau et al., 2015; Domínguez et al., 2016). For MtbPT9, no mutation could be found in the rrs gene. The resistance to amikacin was assigned to a mutation in eis promoter and the resistance to capreomycin was attributed to the insertion of a GT base pair at position 755/756 of the tlyA gene. The mutation G-10A in the eis promoter region was previously showed to be associated with resistance to kanamycin (Zaunbrecher et al., 2009) and with low-level resistance to amikacin (Perdigão et al., 2013). The insertion of a GT base pair at position 755/766 of tlyA was previously showed to be associated with capreomycin resistance (Perdigão et al., 2013). Resistance to ofloxacin of these three strains was associated with common mutations in gyrA at codons 91 and 94 (Table 2; Cambau et al., 2015; Domínguez et al., 2016). MtbBR1 exhibited monoresistance to ofloxacin associated with a mutation in gyrA, D94N.
Summing up, we have selected a representative and diverse sample of clinical strains of M. tuberculosis from two geographical distinct areas harboring the most representative gene mutations in known drug targets, accounting for clinical resistance to the most important first- and second-line antituberculosis drugs. As per the most commonly accepted dogma about the mechanisms accounting for drug resistance in M. tuberculosis, the sole cause of the resistance levels noticed in these strains are expected to be derived exclusively from the diminished affinity of the mutated genes toward the respective antibiotic (Zhang and Yew, 2009; Domínguez et al., 2016).
Quantification of the drug resistance levels in the presence of efflux inhibitors
Secondly, we quantified the drug resistance levels of the strains toward the antibiotics in study. For this purpose, the MICs of the antibiotics and efflux inhibitors were determined for each strain and the results are presented in Table 3. The MICs of the efflux inhibitors were found to be quite homogenous among the seven groups of strains. Conversely, the MICs of the antibiotics showed that for the same mutation different resistance levels were present. Therefore, we hypothesize that the different resistance levels presented for the same genotype are due to different levels of active efflux of the antibiotics. The existence of active efflux was evaluated indirectly through the reduction of the resistance levels of the antibiotics in the presence of the efflux inhibitors verapamil, thioridazine and chlorpromazine by qDST and MIC determination (Table 4).
Table 3.
MIC of antibiotics and efflux inhibitors for the M. tuberculosis strains studied.
| Strain ID | MIC (μg/ml) | |||||||
|---|---|---|---|---|---|---|---|---|
| Antibiotics* | Efflux inhibitors | |||||||
| INH | RIF | AMK | CAP | OFX | VP | TZ | CPZ | |
| SUSCEPTIBLE | ||||||||
| H37Rv | 0.1 | 1 | 1 | 2.5 | 1 | 256 | 15 | 30 |
| MtbPT1 | 0.1 | 1 | 1 | 2.5 | 1 | 256 | 30 | 30 |
| MtbPT2 | 0.1 | 1 | 1 | 2.5 | 1 | 256 | 15 | 30 |
| INHR | ||||||||
| MtbPT3 | 10 | 1 | 1 | 2.5 | 1 | 256 | 15 | 30 |
| MtbPT4 | 0.4 | 1 | 1 | 2.5 | 1 | 256 | 15 | 30 |
| RIFR | ||||||||
| MtbPT5 | 0.1 | 256 | 1 | 2.5 | 1 | 256 | 15 | 30 |
| MtbPT6 | 0.1 | 512 | 1 | 2.5 | 1 | 256 | 15 | 30 |
| OFXR | ||||||||
| MtbBR1 | 0.1 | 1 | 1 | 2.5 | 32 | 128 | 15 | 30 |
| Poly-DR | ||||||||
| MtbBR2 | 3 | 1 | 1 | 2.5 | 10 | 256 | 15 | 30 |
| MDR | ||||||||
| MtbBR3 | 10 | 320 | 1 | 2.5 | 1 | 256 | 15 | 15 |
| MtbBR4 | 10 | 640 | 1 | 2.5 | 1 | 256 | 15 | 15 |
| MtbBR5 | 10 | 160 | 1 | 2.5 | 1 | 128 | 15 | 15 |
| MtbPT7 | 3 | 320 | 1 | 2.5 | 1 | 256 | 15 | 30 |
| MtbBR6 | 80 | 80 | 320 | 5 | 1 | 128 | 30 | 30 |
| MtbPT8 | 20 | 320 | 40 | 25 | 1 | 256 | 15 | 30 |
| XDR | ||||||||
| MtbPT9 | 20 | 80 | 4 | 25 | 10 | 128 | 15 | 30 |
| MtbPT10 | 3 | 320 | 640 | 25 | 10 | 256 | 15 | 30 |
| MtbPT11 | 3 | 320 | 640 | 50 | 10 | 256 | 15 | 30 |
The lowest concentration tested corresponded to the critical concentration for each antibiotic (see Section Materials and Methods for details). AMK, amikacin; CAP, capreomycin; CPZ, chlorpromazine; INH, isoniazid; MDR, multidrug resistant; OFX, ofloxacin; Poly-DR, poly-drug resistant; RIF, rifampicin; TZ, thioridazine; VP, verapamil; XDR, extensively drug resistant.
Table 4.
Quantitative drug susceptibility testing for isoniazid, rifampicin, amikacin, and ofloxacin in the presence and absence of efflux inhibitors.
| Antibiotic | Quantitative susceptibility testing (μg/ml) | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Suscep. | INHR | RIFR | OFXR | Poly-DR | MDR | XDR | |||||||||||
| H37Rv | MtbPT3 | MtbPT4 | MtbPT5 | MtbPT6 | MtbBR1 | MtbBR2 | MtbBR3 | MtbBR4 | MtbBR5 | MtbPT7 | MtbBR6 | MtbPT8 | MtbPT9 | MtbPT10 | MtbPT11 | ||
| INH | No EI | 0.1 | 10 | 0.4 | – | – | – | 10 | 10 | 10 | 10 | 3 | 80 | 20 | 20 | 3 | 3 |
| +VP | 0.1 | 10 | 0.4 | – | – | – | 3 | 3 | 3 | 3 | 1 | 3 | 10 | 1 | 1 | 1 | |
| +TZ | 0.1 | 3 | 0.4 | – | – | – | 3 | 3 | 10 | 3 | 1 | 3 | 3 | 20 | 1 | 3 | |
| +CPZ | 0.1 | 3 | 0.4 | – | – | – | 1 | 10 | 10 | 3 | 1 | 3 | 3 | 1 | 1 | 1 | |
| RIF | No EI | 1 | – | – | 256 | 512 | – | 1 | 320 | 640 | 160 | 320 | 80 | 320 | 80 | 320 | 320 |
| +VP | 1 | – | – | 32 | 32 | – | <0.125 | 40 | 40 | 20 | 20 | 20 | 20 | 20 | 20 | 20 | |
| +TZ | 1 | – | – | 256 | 32 | – | <0.125 | 80 | 320 | 40 | 320 | 80 | 320 | 20 | 320 | 320 | |
| +CPZ | 1 | – | – | 32 | 20 | – | <0.125 | 80 | 320 | 40 | 320 | 20 | 20 | 4 | 20 | 20 | |
| AMK | No EI | 1 | – | – | – | – | – | – | – | – | – | – | 320 | 40 | 4 | 640 | 640 |
| +VP | 1 | – | – | – | – | – | – | – | – | – | – | 320 | 1 | 4 | 640 | 640 | |
| +TZ | 1 | – | – | – | – | – | – | – | – | – | – | 320 | 1 | 4 | 640 | 640 | |
| +CPZ | 1 | – | – | – | – | – | – | – | – | – | – | 320 | 1 | 4 | 40 | 40 | |
| CAP | No EI | 2.5 | – | – | – | – | – | – | – | – | – | – | 5 | 25 | 25 | 25 | 50 |
| +VP | 2.5 | – | – | – | – | – | – | – | – | – | – | 5 | 25 | 25 | 5 | 5 | |
| +TZ | 2.5 | – | – | – | – | – | – | – | – | – | – | 2.5 | 25 | 25 | 5 | 25 | |
| +CPZ | 2.5 | – | – | – | – | – | – | – | – | – | – | 2.5 | 25 | 25 | 5 | 5 | |
| OFX | No EI | 1 | – | – | – | – | 32 | 10 | – | – | – | – | – | – | 10 | 10 | 10 |
| +VP | 1 | – | – | – | – | 16 | 2 | – | – | – | – | – | – | 10 | 10 | 10 | |
| +TZ | 1 | – | – | – | – | 16 | <1 | – | – | – | – | – | – | 10 | 10 | 10 | |
| +CPZ | 1 | – | – | – | – | 16 | <1 | – | – | – | – | – | – | 10 | 10 | 10 | |
The lowest concentration tested corresponds to the critical concentration for each antibiotic except for rifampicin testing for strain MtbBR2 (see Section Materials and Methods for details). Synergistic interactions are in bold; –, not done. AMK, amikacin; CAP, capreomycin; CPZ, chlorpromazine; EI, efflux inhibitor; INH, isoniazid; MDR, multidrug resistant; OFX, ofloxacin; RIF, rifampicin; TZ, thioridazine; VP, verapamil; XDR, extensively drug resistant.
Resistance levels for isoniazid
Eleven out of 12 strains resistant to isoniazid presented high-level resistance (R ≥ 1 μg/ml). Efflux activity was detected in all except MtbPT4, the only strain that showed low-level resistance. The low-level resistance presented by this strain was attributed to the single C-15T mutation, which results in inhA overexpression and leads to titration of isoniazid and consequent mild increased resistance. For the remaining 11 strains, the resistance levels were reduced from high- to low-level (R < 1 μg/ml) in 4/11 with verapamil; 2/11 with thioridazine; and 5/11 with chlorpromazine. For the strains that do not reached low-level resistance, the MICs were reduced to 3 μg/ml. In respect to the efflux levels of the strains that harbored mutations in katG, the results showed that they have less efflux activity than those presenting mutations in inhA. Strains MtbPT7, MtbPT9, MtbPT10, and MtbPT11 have the C-15T mutation and additionally have a mutation in the inhA ORF contributing to their high-level resistance to isoniazid, as previously shown (Machado et al., 2013). The level of resistance of these strains was reduced with the efflux inhibitors to values always above that conferred by the single mutation in the inhA promoter (0.4 μg/ml), suggesting that the net resistance level conferred by inhA double mutations (promoter plus ORF) bears also a strong contribution from efflux. Noteworthy is that the strain MtbBR6 with a rare mutation in katG presents high-level resistance that could be reduced by all inhibitors tested. The difference between the levels of resistance caused by this mutation compared with those presented by the strains with the katG S315T mutation may be due to fitness cost. It is known that the mutation S315T has no fitness cost for the bacteria (Gagneux, 2009) being the most frequent and successful katG mutation accounting for isoniazid resistance worldwide (Seifert et al., 2015). Efflux activity is minimal in strains that bare this mutation since they don't need efflux to cope with the presence of isoniazid. These results confirm our previous findings that the high-levels of resistance associated with mutations in inhA have an increased component of efflux when compared with M. tuberculosis strains with high-level resistance associated with the katG S315T mutation (Machado et al., 2016).
Resistance levels for rifampicin
All the strains resistant to rifampicin presented high-level resistance (R ≥ 20 μg/ml). Efflux activity was detected in all with all the inhibitors although with different efficiencies (Table 4). The resistance levels toward rifampicin were significantly reduced in the presence of verapamil (11/11), thioridazine (4/11), and chlorpromazine (9/11). To assess the contribution of efflux activity to the borderline susceptibility toward rifampicin in MtbBR2, this strain was evaluated for susceptibility to rifampicin in the presence of efflux inhibitors. Notably, the borderline susceptibility was reversed with all inhibitors tested to concentrations well below the critical concentration (Table 4). We hypothesize that the rpoB D516Y mutation is associated with intermediate rifampicin susceptibility that has a strong association with efflux activity. For all other strains, the results demonstrated that efflux acts synergistically with mutations associated with rifampicin resistance although the rpoB S531L mutation is sufficient to produce high-levels of resistance to this antibiotic.
Resistance levels for amikacin and capreomycin
The rrs A1401G is associated with high-levels resistance to amikacin and capreomycin (Sirgel et al., 2011; Du et al., 2013). Strains MtbBR6, MtbPT8, MtbPT10, and MtbPT11 share the mutation A1401G. Efflux activity was detected in all except in MtbBR6 (Table 4). Amikacin resistance levels of MtbPT10 and MtbPT11 were reduced with chlorpromazine from 640 to 40 μg/ml, a 16-fold reduction. However, although significant, this reduction was not sufficient to reach amikacin low-level resistance. The low-level resistance to amikacin of MtbPT9 was due to a mutation in the eis promoter region and no reduction was obtained with any of the inhibitors tested. Amikacin is acetylated by Eis (Zaunbrecher et al., 2009). We hypothesize that the structural modification of the drug impairs substrate recognition and consequently its efflux by the efflux pumps but more studies are required to demonstrate this. Strain MtbPT8 showed the lowest level of resistance to amikacin (40 μg/ml) when compared with the other three amikacin resistant strains with the rrs A1401G mutation. In this strain, the resistance to amikacin was reversed with all the efflux inhibitors. This strain is heteroresistant to amikacin, i.e., the strain population is composed by an amikacin-susceptible subpopulation and an amikacin-resistant subpopulation. Heteroresistance was detected by reverse hybridization using the MTBDRsl assay. The detection of mixed genotypes (hybridization with both wild-type and resistant probes) in the same sample is dependent on the concentration of each individual genotype. Zhang et al. (2013) showed that the low-level of amikacin resistance presented by heteroresistant strains in mainly due to the presence of the drug susceptible population. This data indicates that the proportion of the amikacin-resistant subpopulation is reduced comparatively with the amikacin-susceptible subpopulation only conferring low-level resistance. Therefore, the resistant population during the acquired resistance pathway could be highly reduced in the presence of efflux inhibitors, further demonstrating the contribution of efflux to the establishment and stabilization of mutations in a M. tuberculosis population under drug selection (Machado et al., 2012; Schmalstieg et al., 2012).
Of the five strains resistant to capreomycin, four have the mutation rrs A1401G (including the heteroresistant one) and one presented a mutation in tlyA. With the exception of MtbBR6 all the others presented high-level resistance toward this antibiotic. The low-level resistance presented by MtbBR6 was reversed with thioridazine and chlorpromazine. Capreomycin resistance was reduced to low-level with verapamil (2/5), chlorpromazine (2/5), and thioridazine (1/5). Interesting, capreomycin resistance could not be reduced with any of the inhibitors tested in the heteroresistant strain (MtbPT8) for which amikacin resistance was reversed. Since cross-resistance between amikacin and capreomycin is due to the mutation rrs A1401G, this unexpected result may be explained by the existence of an unknown mechanism associated with capreomycin resistance in this strain. We search for mutations in tlyA that could co-exist with the rrs mutation but none was found and this subject will be explored in future work. It was not possible to reduce capreomycin resistance in MtbPT9 which is associated with an insertion mutation in tlyA. Capreomycin is methylated by TlyA and the loss of methyltransferase activity due to the presence of mutations in tlyA reduces susceptibility to this drug (Maus et al., 2005). Similar to that observed for Eis, we can speculate that the alteration of an essential pathway necessary for the binding of the drug to the target will hamper the substrate recognition and consequently its efflux by the mycobacterial efflux pumps.
Resistance levels for ofloxacin
Resistance to fluoroquinolones involve mainly mutations in gyrA and the most common are at positions 90, 91, and 94 which are associated with high-level resistance (Cambau et al., 2015). All the strains resistant to ofloxacin presented high-level resistance (Table 4). Resistance to ofloxacin of strains MtbPT9 to MtbPT11 could not be reduced with any of the inhibitors tested. On these strains, resistance could be solely attributed to the mutations S91P and D94A on gyrA. For MtbBR1 only basal efflux activity could be detected (Table 4). Contrary, ofloxacin resistance of strain MtbBR2 was reversed by thioridazine and chlorpromazine to levels below the critical concentration and reduced from 10 to 2 μg/ml with verapamil (Table 4). This strain harbored the gyrA A90V mutation whose role in ofloxacin resistance is controversial. The gyrA A90V mutation was recently associated with borderline levels of resistance toward ofloxacin, i.e., the level of resistance can be close or below the critical concentration of 1 μg/ml (Niward et al., 2016). In contrast, Farhat et al. (2016) reported that strains with the gyrA A90V mutation have a comparable effect on ofloxacin MIC as the mutation D94A/G (1 μg/ml>MIC<10 μg/ml). Other studies associate this mutation with ofloxacin resistance above 2 μg/ml (Poissy et al., 2010; Cambau et al., 2015). Our results showed that the A90V mutation can confer resistance up to 2 μg/ml and that the remaining resistance presented has an efflux component that can be reduced with an efflux inhibitor. This mutation have also been found in strains harboring both mutated and wild-type sequences, i.e., fluoroquinolone heteroresistant strains, or in samples harboring two different strains (van den Boogaard et al., 2011). The discrepancies noted between these studies together with the evidence that efflux in linked to this mutation may indicate that the mutation gyrA A90V is associated with intermediate resistance to ofloxacin in strong association with fluoroquinolone efflux that increases the levels of resistance, nevertheless, more studies are needed to support this hypothesis. All other gyrA mutations detected were sufficient to produce stable ofloxacin resistance demonstrating that efflux has little contribution toward ofloxacin resistance when these mutations are present.
Determination of the intrinsic efflux capacity of the M. tuberculosis strains
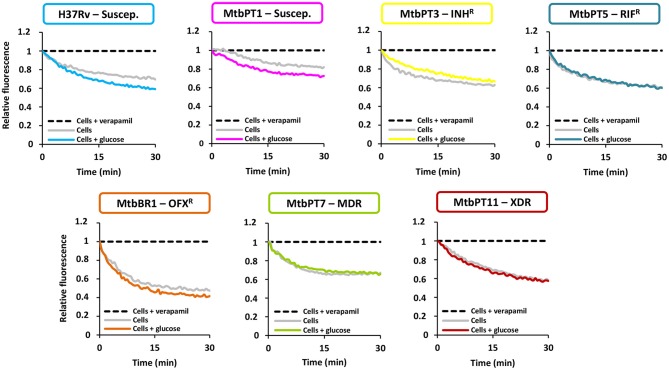
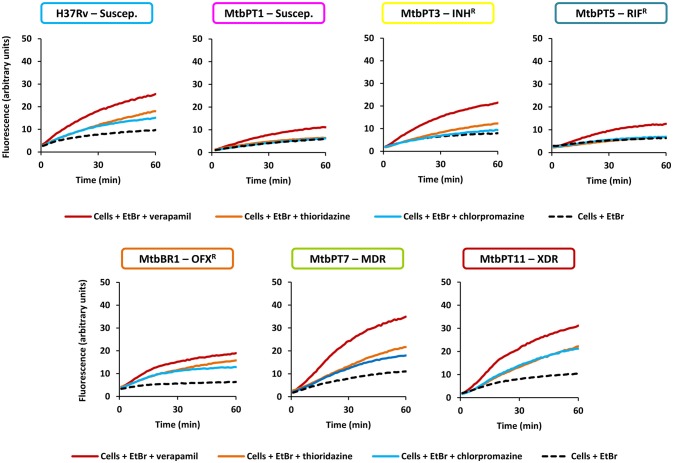
Thirdly, to confirm the existence of active efflux systems on these strains we have evaluated their ability to efflux ethidium bromide, a fluorescent efflux substrate, by real-time fluorometry (Paixão et al., 2009; Viveiros et al., 2010; Machado et al., 2012). To perform these assays, one representative of each drug resistant group was selected: MtbPT3, monoresistant to isoniazid; MtbPT5, monoresistant to rifampicin; MtbBR1, monoresistant to ofloxacin; MtbPT7, multidrug resistant, and MtbPT11, extensively drug resistant (Figure 1). M. tuberculosis H37Rv and MtbPT1 were included as representatives of drug susceptible strains. The strains were incubated with ethidium bromide at the equilibrium concentration determined for each strain and verapamil at half MIC (see Table 3 for MIC values) for 1 h. After this period, verapamil was washed out from the solution, the cells were assayed with glucose, as energy source, and without glucose, and their efflux capacity was measured by fluorometry (see Section Materials and Methods for details). As can be observed in Figure 1, efflux of ethidium bromide was detected in all these strains (colored curve), although with different efficiencies, which was inhibited in presence of verapamil at half MIC (black dashed line). It was also noted that the efflux of ethidium bromide is not affected by the presence of glucose (compare gray curve with the colored curve on each graph) which shows that the external energization of the cells is not necessary to guarantee an optimal efflux activity in M. tuberculosis, a common requirement for efficient efflux in Gram-negative bacteria (Paixão et al., 2009; Viveiros et al., 2010). The assays performed with the susceptible strains further confirmed that efflux activity in M. tuberculosis is an intrinsic characteristic of susceptible and drug resistance strains (Machado et al., 2012). The different degrees of efflux activity observed, e.g., MtbBR1 presented increased efflux activity and MtbPT1 presented reduced efflux activity compared with the other strains, may be related to the different environmental conditions to which these strains were subject in the clinical setting. To further confirm that the efflux activity of these strains could be inhibited in the presence of the efflux inhibitors, we performed ethidium bromide accumulation assays in presence of verapamil, thioridazine, and chlorpromazine (Figure 2) and determined the RFF index (Table 5). The RFF index is a measure of how efficient is the inhibitor in promoting intracellular accumulation of ethidium bromide. The results showed that the three inhibitors were capable to increase the ethidium bromide accumulation in all the tested strains (Table 5). The most efficient efflux inhibitor in these assays was verapamil (6/7), followed by thioridazine (2/7), and chlorpromazine (1/7). The results demonstrated that these compounds are able to promote intracellular accumulation of ethidium bromide on M. tuberculosis susceptible, monoresistant and MDR/XDR strains, clearly putting in evidence that active efflux is inhibited by these compounds.
Figure 1.
Assessment of efflux activity on the M. tuberculosis strains in study. Ethidium bromide was used at the equilibrium concentration for each strain as follows: 0.25 μg/ml for H37Rv, MtbPT1, MtbPT5, MtbPT7, and MtbPT11; 0.5 μg/ml, MtbPT3; and 1 μg/ml, MtbBR1; verapamil was tested at half MIC.
Figure 2.
Evaluation of the effect of efflux inhibitors on the accumulation of ethidium bromide on the M. tuberculosis strains in study. Ethidium bromide (EtBr) was used at the equilibrium concentration for each strain as follows: 0.25 μg/ml for H37Rv, MtbPT1, MtbPT5, MtbPT7, and MtbPT11; 0.5 μg/ml, MtbPT3; and 1 μg/ml, MtbBR1; each efflux inhibitor was tested at half MIC.
Table 5.
RFF based on the accumulation of ethidium bromide for the M. tuberculosis strains in the presence of efflux inhibitors.
| Strain | RFF of the inhibitors | ||
|---|---|---|---|
| Verapamil | Thioridazine | Chlorpromazine | |
| H37Rv | 1.69 ± 0.09*** | 0.82 ± 0.15* | 0.81 ± 0.03** |
| MtbPT1 | 0.86 ± 0.03** | 0.18 ± 0.06* | 0.13 ± 0.02* |
| MtbPT3 | 1.68 ± 0.24** | 0.78 ± 0.02* | 0.45 ± 0.02* |
| MtbPT5 | 1.17 ± 0.08** | 0.24 ± 0.09 | 0.26 ± 0.06 |
| MtbBR1 | 1.76 ± 0.21 | 1.39 ± 0.32 | 0.88 ± 0.13 |
| MtbPT7 | 2.17 ± 0.01* | 0.98 ± 0.02* | 0.67 ± 0.04* |
| MtbPT11 | 2.00 ± 0.21** | 1.14 ± 0.11* | 1.04 ± 0.03** |
Ethidium bromide was used at the equilibrium concentration for each strain as follows: 0.25 μg/ml, H37Rv, MtbPT1, MtbPT5, MtbPT7, and MtbPT11; 0.5 μg/ml, MtbPT3; and 1 μg/ml, MtbBR1; the inhibitors were tested at half MIC. The effect of the inhibitors on the accumulation of ethidium bromide was interpreted as follows: RFF values above zero indicated that the cells accumulate more ethidium bromide under the condition used than those of the control (non-treated cells). Values in boldface (above 1) indicated enhanced accumulation of ethidium bromide in the presence of the efflux inhibitors. Each assay was performed in triplicate and the results presented correspond to the average of three independent assays plus standard deviation (±SD). The results were considered significant when
P < 0.05; and highly significant when
P < 0.01 and
P < 0.001.
Effect of antibiotics on efflux pump gene expression
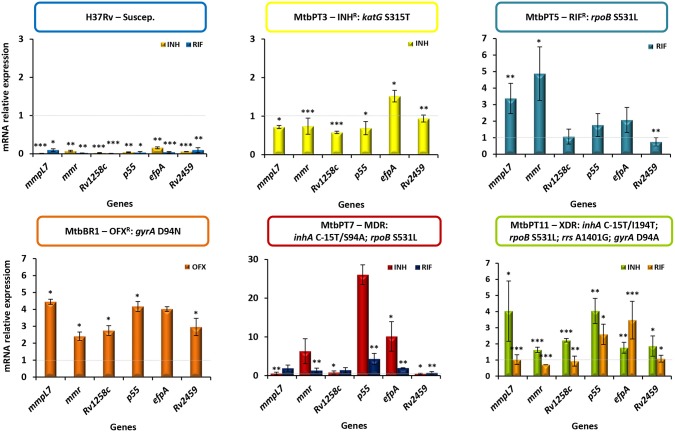
To further corroborate these findings we analyzed the transcriptional profile of putative efflux pumps described in the literature as being associated with antibiotic resistance phenotypes of M. tuberculosis strains (Viveiros et al., 2012; Black et al., 2014; da Silva et al., 2016). RT-qPCR was done for six genes previously shown to be overexpressed in response to antibiotic exposure (Machado et al., 2012, 2016). The results showed the occurrence of significant changes in the expression levels in all strains upon exposure to the antibiotics except in the susceptible H37Rv strain (Figure 3). Concerning the monoresistant strains, in MtbPT3 exposed to isoniazid only efpA was found overexpressed. In strain MtbPT5 exposed to rifampicin, from the six genes tested, four were found overexpressed namely mmpL7, mmr, p55, and efpA. The ofloxacin resistant strain MtbBR1 when exposed to ofloxacin showed significant overexpression all the genes tested (≥two-fold expression). The MDR and XDR strains were exposed to rifampicin or isoniazid and similar patterns were observed. Three to six genes were overexpressed upon exposure to isoniazid or rifampicin. P55 showed the highest level of expression on both strains and for both antibiotics. Overall, EfpA was overexpressed in all strains independently of the antibiotic to which it was exposed. Contrary, Rv2459 was found overexpressed only in the ofloxacin monoresistant strain exposed to ofloxacin. For the remaining efflux pumps analyzed we could not associate an antibiotic to the overexpression of a particular efflux pump. We have observed a general pattern of efflux pump gene expression upon the exposure to the antibiotics.
Figure 3.
Quantification of the relative mRNA expression levels of a panel of efflux pump genes. Strains were grown in MGIT tubes for the MGIT 960 system in the presence of half MIC of each antibiotic as follows: MtbPT3 was exposed to isoniazid; MtbPT5 was exposed to rifampicin; MtbBR1 was exposed to ofloxacin; H37Rv, MtbPT7, and MtbPT11 were exposed to isoniazid or rifampicin. The relative expression of the efflux pump genes was assessed by comparison of the relative quantity of the respective mRNA in the presence of an antibiotic to the non-exposed strain. A level of relative expression equal to 1 indicates that the expression level was identical to the strain that was being compared (gray dashed line in the graphs). Genes showing expression levels above 1 were considered to be expressed; genes showing expression levels above two were considered to be significant overexpressed. The results were considered significant when *P < 0.05; and highly significant when **P < 0.01 and ***P < 0.001.
M. tuberculosis growth kinetics in the presence of antibiotics and efflux inhibitors
If efflux really contributes to the overall drug resistance level seen above, the delay in the growth of each drug resistant strain, due to the stress imposed by the combination of an antibiotic plus an efflux inhibitor at subinhibitory concentrations, will render them more susceptible to the effect of the antibiotic. To test this hypothesis, we performed qDST for selected M. tuberculosis strains and investigated the TTD of growth in the presence and absence of the efflux inhibitors to attain clinical correlation (Machado et al., 2016).
The results showed a delay between the TTD of the tube containing the antibiotic plus an efflux inhibitor and the tube containing only the antibiotic ranging from 1 to 63 days depending on the antibiotic concentration and the drug combination (Supplementary Tables 2–4). Rifampicin at 1 μg/ml in combination with the efflux inhibitors showed a delay in the TTD of strain MtbPT11 of 48 h in the presence of thioridazine, corresponding to a potentiation of rifampicin activity of 63.23% when compared with rifampicin only. Similarly, in the presence of verapamil or chlorpromazine, the delay was 72 h corresponding to a potentiation of rifampicin activity of 95.81 and 96.77%, respectively. Noteworthy, this strain became susceptible to rifampicin at 20 μg/ml in the presence of verapamil (Figure 4) and chlorpromazine, with a delay in the TTD of 386 and 195 h, respectively (Supplementary Table 2). In Figure 5 is presented the qDST of isoniazid for the same strain and it can be observed the increasing of the TTD at each concentration of isoniazid in the presence of half MIC of verapamil. These results showed that the antibiotic activity is clearly potentiated by the efflux inhibitors as demonstrated by the delay on the TTD of growth although, as seen previously, they do not reach susceptibility levels due to the antibiotic-target mutations they carry.
Figure 4.
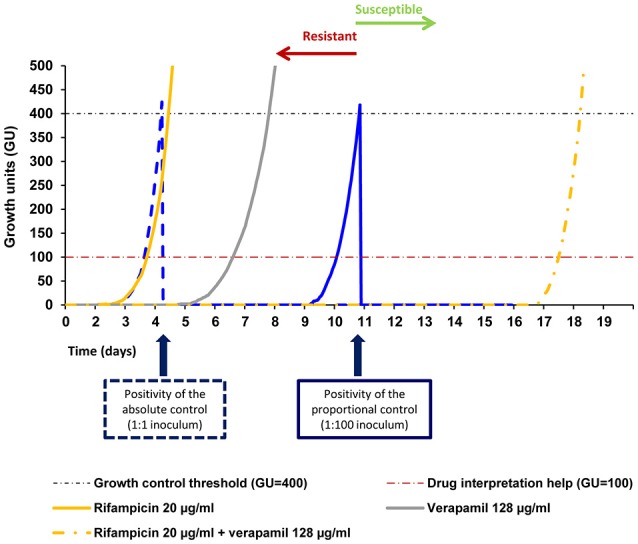
Quantitative drug susceptibility testing of rifampicin for the strain MtbPT11, in the presence or absence of verapamil. Quantitative drug susceptibility testing of isoniazid was conducted using the BACTEC 960 system and the Epicenter V5.80A software equipped with the TB eXIST module. In the Figure is presented the result of the testing for the strain MtbPT11 at 20 μg/ml of rifampicin alone (full orange curve) and rifampicin at 20 μg/ml in the presence of verapamil at half MIC (dashed orange curve). Verapamil alone does not inhibit the growth of the strain (gray curve). Absolute control is given by the dotted blue curve (undiluted strain 1:1) and the proportional control is given by the continuous blue curve (strain diluted 1:100). At the time of growth of the proportional growth control (GU = 400—black dashed line), the comparison between this tube and the tubes containing the drugs(s) was performed. If the GU of the tubes containing the drug were >100 (red dashed line), they were considered to be resistant to that concentration. If the GU of the tube containing the drug was <100 they were considered susceptible.
Figure 5.
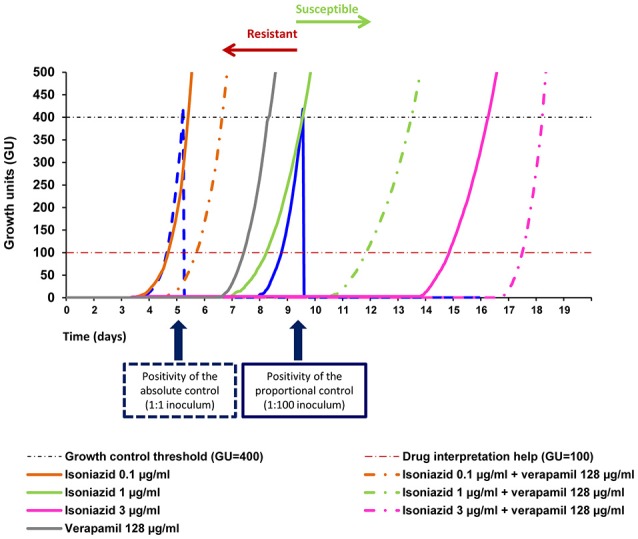
Quantitative drug susceptibility testing of isoniazid for the strain MtbPT11, in the presence or absence of verapamil. Quantitative drug susceptibility testing of isoniazid was conducted using the BACTEC 960 system and the Epicenter V5.80A software equipped with the TB eXIST module. Absolute control is given by the dotted blue curve (undiluted strain 1:1) and the proportional control is given by the continuous blue curve (strain diluted 1:100). The orange, yellow, pink, and green full curves correspond to the growth curves of the strain in the presence of only isoniazid; the respective colored dashed curves corresponded to the growth curves of the strain in the presence of isoniazid and verapamil; Verapamil alone does not inhibit the growth of the strain (gray curve). At the time of growth of the proportional growth control (GU = 400—black dashed line), the comparison between this tube and the tubes containing the drugs(s) was performed. If the GU of the tubes containing the drug were >100 (red dashed line), they were considered to be resistant to that concentration. If the GU of the tube containing the drug was <100 they were considered susceptible. Isoniazid was tested at 0.1, 1, 3, and 10 μg/ml with and without half MIC of verapamil.
Discussion
Whilst drug resistance in M. tuberculosis has long been associated with the development of mutations in the genes that code for the drug targets, efflux pump activity was only recently recognized to play a significant role in the development of drug resistant phenotypes in M. tuberculosis. Several recent studies have demonstrated the importance of the overexpression of efflux pump genes in MDR and XDR M. tuberculosis clinical strains (Calgin et al., 2013; Coelho et al., 2015; Li et al., 2015a; Yamchi et al., 2015; Kanji et al., 2016; Machado et al., 2016; Oh et al., 2017), in rifampicin monoresistant strains (Li et al., 2015b), or in the H37Rv susceptible strain after exposure to drugs (Garima et al., 2015; Caleffi-Ferracioli et al., 2016). Nevertheless, most of these studies are based on the simple assessment and evaluation of the levels of expression of M. tuberculosis efflux pump genes, and few have determined the effect of efflux inhibitors on the MICs of the antituberculosis drugs and have quantified the activity of the overexpressed efflux systems. Therefore, further studies are needed to explore the contribution of the overactivity of efflux pumps to the resistance levels in M. tuberculosis strains.
In a previous work, we presented a model on how multidrug resistance develops in tuberculosis patients, the role of efflux pumps on the development of isoniazid resistance once exposed to the critical concentration of this antituberculosis drug and how these events determine the basis of acquired multidrug resistance (Machado et al., 2012). From that work two questions were raised: (i) what is the role played by the efflux systems in M. tuberculosis strains that already carry mutations in the drug target genes? and (ii) do they respond in the same way to the drug pressure despite the presence of a mutation? In this work, we selected a panel of M. tuberculosis strains presenting different phenotypes and genotypes to study the relationship between the effects of efflux inhibitors on the levels of resistance of the main antituberculosis drugs, the overexpression of efflux pumps upon drug pressure, and the presence of active efflux systems. This study was designed and executed to provide a simple and comprehensive demonstration that drug resistance in M. tuberculosis is a combination between efflux and the presence of a mutation in the drug target gene, a controversial subject in debate for many years (Jarlier and Nikaido, 1994; Louw et al., 2009; da Silva et al., 2011, 2016; Machado et al., 2012; Schmalstieg et al., 2012; Viveiros et al., 2012).
First we analyzed the effect of verapamil, thioridazine and chlorpromazine, known inhibitors of M. tuberculosis efflux pumps, on the resistance levels of first- and second-line drugs against a panel of monoresistant, MDR and XDR M. tuberculosis strains. The results obtained demonstrated that the resistance levels of the M. tuberculosis clinical strains toward the first- and second-line antibiotics studied could be reduced by an efflux inhibitor, independently of the genotype of the strains, demonstrating that the efflux pump inhibitors could be useful in combination with the standard antituberculosis therapy. These results showed that the drug resistant level observed on these strains is a combination between the efflux activity and the mutation in a drug resistance target. With the concentrations of the efflux inhibitors that we tested (half MIC), we can only aim to reduce the efflux component but the resistance component due to the mutation remains untouched. It is not plausible that a compound, whose main function is the inhibition of efflux activity, could reverse the antibiotic resistance in the presence of a mutation associated with high-level resistance. Since all these strains presented mutations associated with high-level drug resistance, these antibiotics are useless in these cases. Although the degree of synergism does not reach full susceptibility, the reduction of the MIC value of the antibiotics to values close or within the serum concentration can improve therapy outcomes demonstrating the possible usefulness of the efflux inhibitors as helper-compounds of antibiotic activity in tuberculosis treatment (Böttger, 2011; Machado et al., 2012; Viveiros et al., 2012; da Silva et al., 2016). Furthermore, it is described that the antibiotics can penetrate and concentrate within macrophages (Prokesch and Hand, 1982) increasing, by this manner, their effective concentrations.
We also demonstrated that the efflux inhibitors verapamil, thioridazine, and chlorpromazine are able to promote the intracellular accumulation of ethidium bromide, a broad range substrate for efflux pumps (Blair and Piddock, 2016), on the M. tuberculosis strains, clearly putting in evidence that active efflux is inhibited by these compounds. At the present, we do not know the exact mechanism by which these compounds inhibit the efflux activity in M. tuberculosis. It has been hypothesized in several studies that these efflux inhibitors affect M. tuberculosis membrane energetics and that they inhibit efflux by inhibiting the energy required for the pumps function (Black et al., 2014; Pule et al., 2016; da Silva et al., 2016). Due to the paucity of new antituberculosis drugs in the tuberculosis drug discovery pipeline and the increasing of drug resistant tuberculosis there is an urgent need for the development of new drugs and the implementation of new therapeutic strategies. The use of compounds that have the ability to inhibit mycobacterial efflux pumps promoting the retention of the co-administered antibiotics that are subject to efflux will obviously improve the efficacy and will extend the clinical utility of the existing antibiotics. In this sense, in the last years, various compounds have been developed and described as putative efflux pump inhibitors, e.g., verapamil analogs were shown to be effective antituberculosis inhibitors being able to reduce the level of resistance to rifampicin by binding to the efflux pump Rv1258c (Singh et al., 2014); thioridazine derivatives have demonstrated to potentiate the antituberculosis drug activity against M. tuberculosis in vitro and ex vivo (Pieroni et al., 2015), and hybrid efflux pump inhibitors, combining the activities of verapamil and thioridazine, were also shown to possess in vitro antituberculosis activity and intracellular activity in macrophages (Kumar et al., 2016).
Next, we analyzed the expression levels of five putative efflux pumps upon exposure to the antibiotics in the H37Rv susceptible strain, and in the monoresistant, MDR and XDR M. tuberculosis clinical isolates. The efflux pump genes mmr, mmpL7, Rv1258c, p55, and efpA were shown to be overexpressed in presence of antibiotics demonstrating the contribution of these efflux pumps to the resistance phenotype of the strains studied, with the exception of the H37Rv strain. We have noticed a general overexpression of almost all efflux genes studied upon exposure to the antibiotics in the drug resistant strains independently on the genotype of the strains. Indeed, when we look to the substrate specificity of each of the M. tuberculosis efflux pumps described in the literature we cannot find a correlation between a given substrate and a specific efflux pump (Supplementary Table 1). These results indicated that M. tuberculosis efflux pumps are promiscuous in their activity as we cannot associate the extrusion of drugs to a specific gene. Moreover, the RT-qPCR data combined with the real-time fluorometry results showed that the drug resistant clinical strains are more prompt to respond, via an efflux-mediated response, to the antibiotics, whereas the susceptible H37Rv strain shows a less prompt detoxifying response to the drugs to which it was exposed. Altogether, the results obtained demonstrate that M. tuberculosis clinical strains are primed to efflux noxious compounds and showed that efflux pump induction is involved in the antibiotic resistance pattern of the strains in study, reinforcing the validity of the hypotheses that efflux contributes to the resistance level of these strains, contributing also to their MDR phenotype.
We also showed that the TTD of M. tuberculosis growth in the MGIT 960 culture system in combination with qDST is a valuable methodology to evaluate M. tuberculosis drug response and to characterize the potency of drug combinations for personalized treatment, especially for MDR/XDR-TB patient regimens. Since the TTD and the inoculum concentration are inversely correlated (Diacon et al., 2010; Bowness et al., 2015), this synergistic activity can be highly beneficial during tuberculosis therapy, either in drug susceptible or drug-resistant strains, allowing the antibiotic, combined with the host's immune system, to tackle in the most effective way the infecting strain. This method allowed the monitoring, on a real-time basis, of bacterial growth/survival vs. growth inhibition by the comparison between the TTD of growth of a given strain in the presence and absence of an efflux inhibitor. The methodology used proved to be useful to study the contribution of the efflux mechanisms to M. tuberculosis drug resistance and simultaneously evaluate the efficacy of an efflux inhibitor to decrease the level of resistance of a strain to a given drug in the perspective of the development of new therapeutic strategies for susceptible and drug resistant tuberculosis, incorporating efflux inhibitors in the control and prevention of drug resistance during therapy.
In conclusion, this study allowed us to show that the main mechanisms associated with drug resistance in M. tuberculosis correlates mutations in target genes with increased efflux and that compounds that inhibit efflux activity can significantly reduce the phenotypic level of such resistance. The level of drug resistance in M. tuberculosis is a combination between the presence of a mutation in the drug target genes and a general stress response to the presence of noxious compounds that regulates the intracellular level of a drug. The data obtained in the presented study corroborated our previous findings (Machado et al., 2012; Coelho et al., 2015) now tested on a larger and diverse panel of M. tuberculosis clinical strains. The demonstration that the efflux activity modulates the levels of antibiotic resistance by complementing the resistance due to target-gene mutations, is a very relevant finding in the context of the ongoing discussion on the ability and clinical reliability of sole molecular based detection of the target-gene mutations as the future routine DST for M. tuberculosis (Böttger, 2011; Domínguez et al., 2016; Pankhurst et al., 2016; Schön et al., 2016). The use of efflux inhibitors as adjuvants of the antituberculosis therapy may be a promise for the development of new and shorter therapeutic strategies as they may potentiate the activity of the current antituberculosis drugs shortening the recommended 6 month treatment for cure, they can increase the activity of drugs that are no longer used due to the emergence of resistance, and they may also be used to protect the activity and usefulness of the new antituberculosis drugs from the development of drug resistance.
Author contributions
DM and MV conceived and designed the study. DM, TC, JP, and CP performed the experiments. DM, TC, JP, IC, IP, RM, DR, AV, MR, PS, MV analyzed the data. IP, IC, and MV contributed with reagents/materials. DM and MV wrote the manuscript. All authors reviewed and approved the final version of the manuscript.
Funding
This work was partially supported by projects PTDC/BIA-MIC/121859/2010 and UID/Multi/04413/2013 from Fundação para a Ciência e a Tecnologia (FCT), Portugal, and project “Ciência sem Fronteiras/Professor Visitante Especial” (Ref. 88881.064961/2014-01) from CAPES/MEC/Brazil.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
DM and JP were supported by post-doctoral grants SFRH/BPD/100688/2014 and SFRH/BPD/95406/2013, respectively, from Fundação para a Ciência e a Tecnologia (FCT), Portugal. TC and DR were supported by doctoral and post-doctoral grants, respectively, from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior/Ministério da Educação (CAPES/MEC), Brazil. PEA has a Research Productivity Fellowship of National Council for Scientific and Technological Development (CNPq)/MCT/Brazil.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00711/full#supplementary-material
References
- Adams K., Szumowski J., Ramakrishnan L. (2014). Verapamil, and its metabolite norverapamil, inhibit macrophage-induced, bacterial efflux pump-mediated to multiple anti-tubercular drugs. J. Infect. Dis. 210, 456–466. 10.1093/infdis/jiu095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams K., Takaki K., Connolly L., Wiedenhoft H., Winglee K., Humbert O., et al. (2011). Drug tolerance in replicating mycobacteria mediated by a macrophage-induced efflux mechanism. Cell 145, 39–53. 10.1016/j.cell.2011.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allix-Béguec C., Harmsen D., Weniger T., Supply P., Niemann S. (2008). Evaluation and strategy for use of MIRU-VNTRplus, a multifunctional database for online analysis of genotyping data and phylogenetic identification of Mycobacterium tuberculosis complex isolates. J. Clin. Microbiol. 46, 2692–2699. 10.1128/JCM.00540-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres S., Hillemann D., Rüsch-Gerdes S., Richter E. (2014). Occurrence of rpoB mutations in isoniazid-resistant but rifampicin-susceptible Mycobacterium tuberculosis isolates from Germany. Antimicrob. Agents Chemother. 58, 590–592. 10.1128/AAC.01752-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andries K., Villellas C., Coeck N., Thys K., Gevers T., Vranckx L., et al. (2014). Acquired resistance of Mycobacterium tuberculosis to bedaquiline. PLoS ONE 9:e102135. 10.1371/journal.pone.0102135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailo R., Bhatt A., Aínsa J. A. (2015). Lipid transport in Mycobacterium tuberculosis and its implications in virulence and drug development. Biochem. Pharmacol. 96, 159–167. 10.1016/j.bcp.2015.05.001 [DOI] [PubMed] [Google Scholar]
- Balganesh M., Kuruppath S., Marcel N., Sharma S., Nair A., Sharma U. (2010). Rv1218c, an ABC transporter of Mycobacterium tuberculosis with implications in drug discovery. Antimicrob. Agents Chemother. 54, 5167–5172. 10.1128/AAC.00610-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco M. V., Blanco F. C., Imperiale B., Forrellad M. A., Rocha R. V., Klepp L. A., et al. (2011). Role of P27-P55 operon from Mycobacterium tuberculosis in the resistance to toxic compounds. BMC Infect. Dis. 11:195. 10.1186/1471-2334-11-195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black P., Warren R., Louw G., van Helden P., Victor T., Kana B. (2014). Energy metabolism and drug efflux in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 58, 2491–2503. 10.1128/AAC.02293-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair J. M., Piddock L. J. (2016). How to measure export via bacterial multidrug resistance efflux pumps. MBio 7:e00840-16. 10.1128/mBio.00840-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttger E. (2011). The ins and outs of Mycobacterium tuberculosis drug susceptibility testing. Clin. Microbiol. Infect. 17, 1128–1134. 10.1111/j.1469-0691.2011.03551.x [DOI] [PubMed] [Google Scholar]
- Bowness R., Boeree M., Aarnoutse R., Dawson R., Diacon A., Mangu C., et al. (2015). The relationship between Mycobacterium tuberculosis MGIT time to positivity and CFU in sputum samples demonstrates changing bacterial phenotypes potentially reflecting the impact of chemotherapy on critical sub-populations. J. Antimicrob. Chemother. 70, 448–455. 10.1093/jac/dku415 [DOI] [PubMed] [Google Scholar]
- Caleffi-Ferracioli K. R., Amaral R. C. R., Demitto F. O., Maltempe F. G., Canezin P. H., Scodro R. B., et al. (2016). Morphological changes and differentially expressed efflux pump genes in Mycobacterium tuberculosis exposed to a rifampicin and verapamil combination. Tuberculosis 97, 65–72. 10.1016/j.tube.2015.12.010 [DOI] [PubMed] [Google Scholar]
- Calgin M. K., Sahin F., Turegun B., Gerceker D., Atasever M., Koksal D., et al. (2013). Expression analysis of efflux pump genes among drug-susceptible and multidrug-resistant Mycobacterium tuberculosis clinical isolates and reference strains. Diagn. Microbiol. Infect. Dis. 76, 291–297. 10.1016/j.diagmicrobio.2013.02.033 [DOI] [PubMed] [Google Scholar]
- Cambau E., Viveiros M., Machado D., Raskine L., Ritter C., Tortoli E., et al. (2015). Revisiting susceptibility testing in MDR-TB by a standardized quantitative phenotypic assessment in a European multicentre study. J. Antimicrob. Chemother. 70, 686–696. 10.1093/jac/dku438 [DOI] [PubMed] [Google Scholar]
- Choudhuri B., Bhakta S., Barik R., Basu J., Kundu M., Chakrabarti P. (2002). Overexpression and functional characterization of an ABC (ATP-binding cassette) transporter encoded by the genes drrA and drrB of Mycobacterium tuberculosis. Biochem. J. 367, 279–285. 10.1042/bj20020615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho T., Machado D., Couto I., Maschmann R., Ramos D., von Groll A., et al. (2015). Enhancement of antibiotic activity by efflux inhibitors against multidrug resistant Mycobacterium tuberculosis clinical isolates from Brazil. Front. Microbiol. 6:330. 10.3389/fmicb.2015.00330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva P. E. A., Palomino J. C. (2011). Molecular basis and mechanisms of drug resistance in Mycobacterium tuberculosis: classical and new drugs. J. Antimicrob. Chemother. 66, 1417–1430. 10.1093/jac/dkr173 [DOI] [PubMed] [Google Scholar]
- da Silva P. E. A., Machado D., Ramos D., Couto I., von Groll A., Viveiros M. (2016). Efflux pumps in mycobacteria: antimicrobial resistance, physiological functions, and role in pathogenicity, in Efflux-Mediated Antimicrobial Resistance in Bacteria, eds Li X-Z., Elkins C. A., Zgurskaya H. I. (Cham: Springer International Publishing AG; ), 527–559. [Google Scholar]
- da Silva P. E. A., von Groll A., Martin A., Palomino J. C. (2011). Efflux as a mechanism for drug resistance in Mycobacterium tuberculosis. FEMS Immunol. Med. Microbiol. 63, 1–9. 10.1111/j.1574-695X.2011.00831.x [DOI] [PubMed] [Google Scholar]
- da Silva P., Bigi F., Santangelo M., Romano M., Martín C., Cataldi A., et al. (2001). Characterization of P55, a multidrug efflux pump in Mycobacterium bovis and Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 45, 800–804. 10.1128/AAC.45.3.800-804.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rossi E., Branzoni M., Cantoni R., Milano A., Riccardi G., Ciferri O. (1998). mmr, a Mycobacterium tuberculosis gene conferring resistance to small cationic dyes and inhibitors. J. Bacteriol. 180, 6068–6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demay C., Liens B., Burguiere T., Hill V., Couvin D., Millet J., et al. (2012). SITVITWEB - a publicly available international multimarker database for studying Mycobacterium tuberculosis genetic diversity and molecular epidemiology. Infect. Genet. Evol. 12, 755–766. 10.1016/j.meegid.2012.02.004 [DOI] [PubMed] [Google Scholar]
- Diacon A., Maritz J., Venter A., van Helden P., Andries K., McNeeley D., et al. (2010). Time to detection of the growth of Mycobacterium tuberculosis in MGIT 960 for determining the early bactericidal activity of antituberculosis agents. Eur. J. Microbiol. Infect. Dis. 29, 1561–1565. 10.1007/s10096-010-1043-7 [DOI] [PubMed] [Google Scholar]
- Diacon A., van der Merwe L., Demers A. M., von Groote-Bidlingmaier F., Venter A., Donald P. R. (2014). Time to positivity in liquid culture predicts colony forming unit counts of Mycobacterium tuberculosis in sputum specimens. Tuberculosis 94, 148–151. 10.1016/j.tube.2013.12.002 [DOI] [PubMed] [Google Scholar]
- Domínguez J., Boettger E. C., Cirillo D., Cobelens F., Eisenach K. D., Gagneux S., et al. (2016). Clinical implications of molecular drug resistance testing for Mycobacterium tuberculosis: a TBNET/RESIST-TB consensus statement. Int. J. Tuberc. Lung Dis. 20, 24–42. 10.5588/ijtld.15.0221 [DOI] [PubMed] [Google Scholar]
- Doran J., Pang Y., Mdluli K., Moran A., Victor T., Stokes R., et al. (1997). Mycobacterium tuberculosis efpA encodes an efflux protein of the QacA transporter family. Clin. Diagn. Lab. Immunol. 4, 23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Q., Dai G., Long Q., Yu X., Dong L., Huang H., et al. (2013). Mycobacterium tuberculosis rrs A1401G mutation correlates with high-level resistance to kanamycin, amikacin, and capreomycin in clinical isolates from mainland China. Diagn. Microbiol. Infect. Dis. 77, 138–142. 10.1016/j.diagmicrobio.2013.06.031 [DOI] [PubMed] [Google Scholar]
- Dutta N. K., Pinn M. L., Karakousis P. C. (2014). Reduced emergence of isoniazid resistance with concurrent use of thioridazine against acute murine tuberculosis. Antimicrob. Agents Chemother. 58, 4048–4053. 10.1128/AAC.02981-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhat M., Jacobson K. R., Franke M. F., Kaur D., Sloutsky A., Mitnick C. D., et al. (2016). Gyrase mutations are associated with variable levels of fluoroquinolone resistance in Mycobacterium tuberculosis. J. Clin. Microbiol. 54, 727–733. 10.1128/JCM.02775-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuerriegel S., Cox H., Zarkua N., Karimovich H. A., Braker K., Rüsch-Gerdes S., et al. (2009). Sequence analyses of just four genes to detect extensively drug-resistant Mycobacterium tuberculosis strains in multidrug-resistant tuberculosis patients undergoing treatment. Antimicrob. Agents Chemother. 53, 3353–3356. 10.1128/AAC.00050-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagneux S. (2009). Fitness cost of drug resistance in Mycobacterium tuberculosis. Clin. Microbiol. Infect. 15, 66–68. 10.1111/j.1469-0691.2008.02685.x [DOI] [PubMed] [Google Scholar]
- Garima K., Pathak R., Tandon R., Rathor N., Sinha R., Bose M., et al. (2015). Differential expression of efflux pump genes of Mycobacterium tuberculosis in response to varied subinhibitory concentrations of antituberculosis agents. Tuberculosis 95, 155–161. 10.1016/j.tube.2015.01.005 [DOI] [PubMed] [Google Scholar]
- Gupta S., Cohen K., Winglee K., Maiga M., Diarra B., Bishai W. (2014). Efflux inhibition with verapamil potentiates bedaquiline in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 58, 574–576. 10.1128/AAC.01462-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S., Tyagi S., Almeida D., Maiga M., Ammerman N., Bishai W. (2013). Acceleration of tuberculosis treatment by adjunctive therapy with verapamil as an efflux inhibitor. Am. J. Respir. Crit. Care Med. 188, 600–607. 10.1164/rccm.201304-0650OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao P., Shi-Liang Z., Ju L., Ya-Xin D., Biao H., Xu W., et al. (2011). The role of ABC efflux pump, Rv1456c-Rv1457c-Rv1458c, from Mycobacterium tuberculosis clinical isolates in China. Folia Microbiol. 56, 549–553. 10.1007/s12223-011-0080-7 [DOI] [PubMed] [Google Scholar]
- Jarlier V., Nikaido H. (1994). Mycobacterial cell wall: structure and role in natural resistance to antibiotics. FEMS Microbiol. Lett. 123, 11–18. 10.1111/j.1574-6968.1994.tb07194.x [DOI] [PubMed] [Google Scholar]
- Kamerbeek J., Schouls L., Kolk A., van Agterveld M., van Soolingen D., Kuijper S., et al. (1997). Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35, 907–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanji A., Hasan R., Zhang Y., Shi W., Imtiaz K., Iqbal K., et al. (2016). Increased expression of efflux pump genes in extensively drug-resistant isolates of Mycobacterium tuberculosis. Int. J. Mycobacteriol. 5:S150. 10.1016/j.ijmyco.2016.09.067 [DOI] [PubMed] [Google Scholar]
- Kumar M., Singh K., Naran K., Hamzabegovic F., Hoft D. F., Warner D. F., et al. (2016). Design, synthesis, and evaluation of novel hybrid efflux pump inhibitors for use against Mycobacterium tuberculosis. ACS Infect. Dis. 2, 714–725. 10.1021/acsinfecdis.6b00111 [DOI] [PubMed] [Google Scholar]
- Li G., Zhang J., Guo Q., Jiang Y., Wei J., Zhao L. L., et al. (2015a). Efflux pump gene expression in multidrug-resistant Mycobacterium tuberculosis clinical isolates. PLoS ONE 10:e0119013. 10.1371/journal.pone.0119013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Zhang J., Guo Q., Wei J., Jiang Y., Zhao X., et al. (2015b). Study of efflux pump gene expression in rifampicin-monoresistant Mycobacterium tuberculosis clinical isolates. J. Antibiot. 68, 431–435. 10.1038/ja.2015.9 [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25, 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Louw G. E., Warren R. M., van Pittius N. C., Leon R., Jimenez A., Hernandez-Pando R., et al. (2011). Rifampicin reduces susceptibility to ofloxacin in rifampicin-resistant Mycobacterium tuberculosis through efflux. Am. J. Respir. Crit. Care Med. 184, 269–276. 10.1164/rccm.201011-1924OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louw G. E., Warren R. M., van Pittius N. C., McEvoy C. R. E., Van Helden P. D., Victor T. C. (2009). A balancing act: efflux/influx in mycobacterial drug resistance. Antimicrob. Agents Chemother. 53, 3181–3189. 10.1128/AAC.01577-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado D., Couto I., Perdigão J., Rodrigues L., Portugal I., Baptista P., et al. (2012). Contribution of efflux to the emergence of isoniazid and multidrug resistance in Mycobacterium tuberculosis. PLoS ONE 7:e34538. 10.1371/journal.pone.0034538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado D., Perdigão J., Ramos J., Couto I., Portugal I., Ritter C., et al. (2013). High-level resistance to isoniazid and ethionamide in multidrug resistant Mycobacterium tuberculosis of the Lisboa family is associated with inhA double mutations. J. Antimicrob. Chemother. 68, 1728–1732. 10.1093/jac/dkt090 [DOI] [PubMed] [Google Scholar]
- Machado D., Pires D., Perdigão P., Couto I., Portugal I., Martins M., et al. (2016). Ion channel blockers as antimicrobial agents, efflux inhibitors, and enhancers of macrophage killing activity against drug resistant Mycobacterium tuberculosis. PLoS ONE 11:e0149326. 10.1371/journal.pone.0149326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado L., Spengler G., Evaristo M., Handzlik J., Molnar J., Viveiros M., et al. (2011). Biological activity of twenty-three hydantoin derivatives on intrinsic efflux pump system of Salmonella enterica serovar Enteritidis NCTC 13349. In vivo 25, 769–772. [PubMed] [Google Scholar]
- Maus C., Plikaytis B., Shinnick T. (2005). Mutation of tlyA confers capreomycin resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 49, 571–577. 10.1128/AAC.49.2.571-577.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milano A., Pasca M., Provvedi R., Lucarelli A., Manina G., Ribeiro A., et al. (2009). Azole resistance in Mycobacterium tuberculosis is mediated by the MmpS5–MmpL5 efflux system. Tuberculosis 89, 84–90. 10.1016/j.tube.2008.08.003 [DOI] [PubMed] [Google Scholar]
- Niward K., Angeby K., Chryssanthou E., Paues J., Bruchfeld J., Jureen P., et al. (2016). Susceptibility testing breakpoints for Mycobacterium tuberculosis categorize isolates with resistance mutations in gyrA as susceptible to fluoroquinolones: implications for MDR-TB treatment and the definition of XDR-TB. J. Antimicrob. Chemother. 71, 333–338. 10.1093/jac/dkv353 [DOI] [PubMed] [Google Scholar]
- Oh T. S., Kim Y. J., Kang H. Y., Kim C. K., Cho S. Y., Lee H. J. (2017). RNA expression analysis of efflux pump genes in clinical isolates of multidrug-resistant and extensively drug-resistant Mycobacterium tuberculosis in South Korea. Infect. Genet. Evol. 49, 111–115. 10.1016/j.meegid.2017.01.002 [DOI] [PubMed] [Google Scholar]
- Ordway D., Viveiros M., Leandro C., Bettencourt R., Almeida J., Martins M., et al. (2003). Clinical concentrations of thioridazine kill intracellular multidrug-resistant Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 47, 917–922. 10.1128/AAC.47.3.917-922.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paixão L., Rodrigues L., Couto I., Martins M., Fernandes P., de Carvalho C., et al. (2009). Fluorometric determination of ethidium bromide efflux kinetics in Escherichia coli. J. Biol. Eng. 3:18. 10.1186/1754-1611-3-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankhurst L. J., del Ojo Elias C., Votintseva A. A., Walker T. M., Cole K., Davies J., et al. (2016). Rapid, comprehensive, and affordable mycobacterial diagnosis with whole-genome sequencing: a prospective study. Lancet Respir. Med. 4, 49–58. 10.1016/S2213-2600(15)00466-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasca M., Guglierame P., Arcesi F., Bellinzoni M., De Rossi E., Riccardi G. (2004). Rv2686c-Rv2687c-Rv2688c, an ABC fluoroquinolone efflux pump in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 48, 3175–3178. 10.1128/AAC.48.8.3175-3178.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasca M., Guglierame P., De Rossi E., Zara F., Riccardi G. (2005). mmpL7 gene of Mycobacterium tuberculosis is responsible for isoniazid efflux in Mycobacterium smegmatis. Antimicrob. Agents Chemother. 49, 4775–4777. 10.1128/AAC.49.11.4775-4777.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdigão J., Macedo R., Silva C., Machado D., Couto I., Viveiros M., et al. (2013). From multidrug-resistant to extensively drug-resistant tuberculosis in Lisbon, Portugal: the stepwise mode of resistance acquisition. J. Antimicrob. Chemother. 68, 23–33. 10.1093/jac/dks371 [DOI] [PubMed] [Google Scholar]
- Pieroni M., Machado D., Azzali E., Santos Costa S., Couto I., Costantino G., et al. (2015). Rational design and synthesis of thioridazine analogues as enhancers of the antituberculosis therapy. J. Med. Chem. 58, 5842–5853. 10.1021/acs.jmedchem.5b00428 [DOI] [PubMed] [Google Scholar]
- Pluta K., Morak-Młodawska B., Jeleń M. (2011). Recent progress in biological activities of synthesized phenothiazines. Eur. J. Med Chem. 46, 3179–3189. 10.1016/j.ejmech.2011.05.013 [DOI] [PubMed] [Google Scholar]
- Poissy J., Aubry A., Fernandez C., Lott M. C., Chauffour A., Jarlier V., et al. (2010). Should moxifloxacin be used for the treatment of extensively drug-resistant tuberculosis? An answer from a murine model. Antimicrob. Agents Chemother. 54, 4765–4771. 10.1128/AAC.00968-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokesch R. C., Hand W. L. (1982). Antibiotic entry into human polymorphonuclear leukocytes. Antimicrob. Agents Chemother. 21, 373–380. 10.1128/AAC.21.3.373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pule C. M., Sampson S. L., Warren R. M., Black P. A., van Helden P. D., Victor T. C., et al. (2016). Efflux pump inhibitors: targeting mycobacterial efflux systems to enhance TB therapy. J. Antimicrob. Chemother. 71, 17–26. 10.1093/jac/dkv316 [DOI] [PubMed] [Google Scholar]
- Ramón-García S., Martín C., Thompson C. J., Aínsa J. A. (2009). Role of the Mycobacterium tuberculosis P55 efflux pump in intrinsic drug resistance, oxidative stress responses, and growth. Antimicrob. Agents Chemother. 53, 3675–3682. 10.1128/AAC.00550-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramón-García S., Mick V., Dainese E., Martín C., Thompson C., De Rossi E., et al. (2012). Functional and genetic characterization of the Tap efflux pump in Mycobacterium bovis BCG. Antimicrob. Agents Chemother. 56, 2074–2083. 10.1128/AAC.05946-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayasam G. V., Balganesh T. S. (2015). Exploring the potential of adjunct therapy in tuberculosis. Trends Pharmacol. Sci. 36, 506–513. 10.1016/j.tips.2015.05.005 [DOI] [PubMed] [Google Scholar]
- Rodrigues L., Villellas C., Bailo R., Viveiros M., Aínsa J. A. (2013). Role of the Mmr efflux pump in drug resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 57, 751–757. 10.1128/AAC.01482-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues L., Viveiros M., Aínsa J. A. (2015). Measuring efflux and permeability in mycobacteria, in Mycobacteria Protocols, eds Parish T., Roberts D. M. (New York, NY: Springer Science+Business Media; ), 227–239. [Google Scholar]
- Salih F. A., Kaushik N. K., Sharma P., Choudary G. V., Murthy P. S., Venkitasubramanian T. A. (1991). Calmodulin-like activity in mycobacteria. Indian J. Biochem. Biophys. 28, 491–495. [PubMed] [Google Scholar]
- Schmalstieg A. M., Srivastava S., Belkaya S., Deshpande D., Meek C., Leff R., et al. (2012). The antibiotic resistance arrow of time: efflux pump induction is a general first step in the evolution of mycobacterial drug resistance. Antimicrob. Agents Chemother. 56, 4806–4815. 10.1128/AAC.05546-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schön T., Miotto P., Köser C. U., Viveiros M., Böttger E., Cambau E. (2016). Mycobacterium tuberculosis drug-resistance testing: challenges, recent developments and perspectives. Clin. Microbiol. Infect. 23, 154–160. 10.1016/j.cmi.2016.10.022 [DOI] [PubMed] [Google Scholar]
- Seifert M., Catanzaro D., Catanzaro A., Rodwell T. C. (2015). Genetic mutations associated with isoniazid resistance in Mycobacterium tuberculosis: a systematic review. PLoS ONE 10:e0119628. 10.1371/journal.pone.0119628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqi N., Das R., Pathak N., Banerjee S., Ahmed N., Katoch V., et al. (2004). Mycobacterium tuberculosis isolate with a distinct genomic identity overexpresses a tap-like efflux pump. Infection 32, 109–111. 10.1007/s15010-004-3097-x [DOI] [PubMed] [Google Scholar]
- Singh A., Sharma S. (2014). Chemotherapeutic efficacy of thioridazine as an adjunct drug in a murine model of latent tuberculosis. Tuberculosis 94, 695–700. 10.1016/j.tube.2014.08.016 [DOI] [PubMed] [Google Scholar]
- Singh K., Kumar M., Pavadai E., Naran K., Warner D. F., Ruminski P. G., et al. (2014). Synthesis of new verapamil analogues and their evaluation in combination with rifampicin against Mycobacterium tuberculosis and molecular docking studies in the binding site of efflux protein Rv1258c. Bioorg. Med. Chem. Lett. 24, 2985–2990. 10.1016/j.bmcl.2014.05.022 [DOI] [PubMed] [Google Scholar]
- Sirgel F., Tait M., Warren R., Streicher E. M., Böttger E. C., van Helden P. D., et al. (2011). Mutations in the rrs A1401G gene and phenotypic resistance to amikacin and capreomycin in Mycobacterium tuberculosis. Microb. Drug Resist. 18, 193–197. 10.1089/mdr.2011.0063 [DOI] [PubMed] [Google Scholar]
- Somoskovi A., Dormandy J., Mitsani D., Rivenburg J., Salfinger M. (2006). Use of smear-positive samples to assess the PCR-based genotype MTBDR assay for rapid, direct detection of the Mycobacterium tuberculosis complex as well as its resistance to isoniazid and rifampin. J. Clin. Microbiol. 44, 4459–4463. 10.1128/JCM.01506-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer B., Lucke K., Calligaris-Maibach R., Ritter C., Böttger E. C. (2009). Quantitative drug susceptibility testing of Mycobacterium tuberculosis using MGIT960 and the EpiCenter instrumentation. J. Clin. Microbiol. 47, 1773–1780. 10.1128/JCM.02501-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supply P., Allix C., Lesjean S., Cardoso-Oelemann M., Rüsch-Gerdes S., Willery E., et al. (2006). Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 44, 4498–4510. 10.1128/JCM.01392-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsing M., Klitgaard J. K., Atilano M. L., Skov M. N., Kolmos H. J., Filipe S. R., et al. (2013). Thioridazine induces major changes in global gene expression and cell wall composition in methicillin-resistant Staphylococcus aureus USA300. PLoS ONE 8:e64518. 10.1371/journal.pone.0064518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Boogaard J., Semvua H. H., van Ingen J., Mwaigwisya S., van der Laan T., van Soolingen D., et al. (2011). Low rate of fluoroquinolone resistance in Mycobacterium tuberculosis isolates from northern Tanzania. J. Antimicrob. Chemother. 66, 1810–1814. 10.1093/jac/dkr205 [DOI] [PubMed] [Google Scholar]
- Viveiros M., Martins M., Rodrigues L., Machado D., Couto I., Ainsa J., et al. (2012). Inhibitors of mycobacterial efflux pumps as potential boosters for anti-tubercular drugs. Expert Rev. Anti. Infect. Ther. 10, 983–998. 10.1586/eri.12.89 [DOI] [PubMed] [Google Scholar]
- Viveiros M., Rodrigues L., Martins M., Couto I., Spengler G., Martins A., et al. (2010). Evaluation of efflux activity of bacteria by a semi-automated fluorometric system, in Antibiotic Resistance Protocols, 2nd Edn., eds Gillespie S. H., McHugh T. D. (New York, NY: Springer Science+Business Media LLC; Humana Press; ), 159–172. [DOI] [PubMed] [Google Scholar]
- Wang K., Pei H., Huang B., Zhu X., Zhang J., Zhou B., et al. (2013). The expression of ABC efflux pump, Rv1217c–Rv1218c, and its association with multidrug resistance of Mycobacterium tuberculosis in China. Curr. Microbiol. 66, 222–226. 10.1007/s00284-012-0215-3 [DOI] [PubMed] [Google Scholar]
- Weinstein E., Yano T., Li L., Avarbock D., Avarbock A., Helm D., et al. (2005). Inhibitors of type II NADH: menaquinone oxidoreductase represent a class of antitubercular drugs. Proc. Natl. Acad. Sci. U.S.A. 122, 4548–4553. 10.1073/pnas.0500469102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weniger T., Krawczyk J., Supply P., Niemann S., Harmsen D. (2010). MIRU-VNTRplus: a web tool for polyphasic genotyping of Mycobacterium tuberculosis complex bacteria. Nucleic Acids Res. 38, W326–W331. 10.1093/nar/gkq351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams D. L., Spring L., Collins L., Miller L. P., Heifets L. B., Gangadharam P. R. J., et al. (1998). Contribution of rpoB mutations to development of rifamycin cross-resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 42, 1853–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamchi J. K., Haeili M., Feyisa S. G., Kazemian H., Shahraki A. H., Zahednamazi F., et al. (2015). Evaluation of efflux pump gene expression among drug susceptible and drug resistant strains of Mycobacterium tuberculosis from Iran. Infect. Genet. Evol. 36, 23–26. 10.1016/j.meegid.2015.08.036 [DOI] [PubMed] [Google Scholar]
- Zaczek A., Brzostek A., Augustynowicz-Kopec E., Zwolska Z., Dziadek J. (2009). Genetic evaluation of relationship between mutations in rpoB and resistance of Mycobacterium tuberculosis to rifampin. BMC Microbiol. 9:10. 10.1186/1471-2180-9-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaunbrecher M., Sikes R., Metchock B., Shinnick T. M., Posey J. E. (2009). Overexpression of the chromosomally encoded aminoglycoside acetyltransferase eis confers kanamycin resistance in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 106, 20004–20009. 10.1073/pnas.0907925106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Zhao B., Huang H., Zhu Y., Peng J., Dai G., et al. (2013). Co-occurrence of amikacin-resistant and -susceptible Mycobacterium tuberculosis isolates in clinical samples from Beijing, China. J. Antimicrob. Chemother. 68, 1537–1542. 10.1093/jac/dkt082 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Yew W. (2009). Mechanisms of drug resistance in Mycobacterium tuberculosis. Int. J. Tuberc. Lung Dis. 13, 1320–1330. 10.5588/ijtld.15.0389 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.