Abstract
Bacteria and archaea possess numerous defense systems to combat viral infections and other mobile genetic elements. Uniquely among these, CRISPR-Cas (clustered, regularly interspaced short palindromic repeats-CRISPR associated) provides adaptive genetic interference against foreign nucleic acids. Here we review recent advances on the CRISPR-Cas9 system in Neisseria spp, with a focus on its biological functions in genetic transfer, its mechanistic features that establish new paradigms and its technological applications in eukaryotic genome engineering.
Keywords: CRISPR-Cas9, genome engineering, RNA-guided adaptive interference, Neisseria spp
The CRISPR-Cas9 system in Neisseria spp.
INTRODUCTION
The genus Neisseria comprises many Gram-negative β-proteobacteria that interact with eukaryotic hosts, but only two organisms, the gonococcus (Gc) and its close relative the meningococcus (Mc), are human pathogens, both of which colonize mucosal surfaces (Bratcher, Bennett and Maiden 2012; Rotman and Seifert 2014). Neisseria gonorrhoeae (Gc) is the sole causative agent of the sexually transmitted disease, gonorrhea, and N. meningitidis (Mc) is an opportunistic pathogen that usually colonizes the nasopharynx asymptomatically, but can invade the blood and/or the brain and cause life-threatening septicemia and meningitis (Bratcher, Bennett and Maiden 2012). These invasive meningococcal diseases are rare but have high morbidity and mortality rates worldwide (Bratcher, Bennett and Maiden 2012). Many non-pathogenic Neisseria species also colonize the human nasopharynx, and among them N. lactamica is the most widely studied commensal (Rotman and Seifert 2014).
Neisseria species undergo frequent and extensive genetic exchange via natural transformation, which underscores their remarkable ability to generate antigenic variability and to quickly spread new traits including antibiotic resistance markers (Hamilton and Dillard 2006). Many factors such as Type IV pili, restriction-modification (R-M) systems and recombination machineries are known to affect genetic exchange in these organisms (Hamilton and Dillard 2006; Rotman and Seifert 2014). In recent years, a CRISPR-Cas9 system was discovered in meningococcus as a new player that can limit natural transformation (Zhang et al.2013).
Clustered, regularly interspaced, short palindromic repeat (CRISPR) loci and their associated cas genes constitute a small RNA-guided, adaptive immune system that helps prokaryotes to fend off invasive genetic elements (Barrangou et al.2007; Brouns et al.2008; Marraffini and Sontheimer 2008). CRISPR loci contain arrays of repeats, separated by unique short ‘spacers’ that often match sequences from plasmids or bacteriophage genomes (Bolotin et al.2005; Mojica et al.2005; Pourcel, Salvignol and Vergnaud 2005). CRISPR-Cas systems are widespread, present in about 40% of sequenced bacteria and most archaea (Makarova et al.2015). Prokaryotic genomes are shaped not only by vertical genetic transmission, but also by inheritance-independent acquisition of genetic material—a process known as horizontal gene transfer (HGT). Initially, CRISPR was discovered as a sequence-based barrier against two major routes of HGT: bacteriophage infection and plasmid conjugation (Barrangou et al.2007; Marraffini and Sontheimer 2008). A pair of studies later demonstrated that a native CRISPR system in N. meningitidis (Zhang et al.2013) and a heterologous CRISPR system in Streptococcus pneumoniae (Bikard et al.2012) can each prevent natural transformation, broadening the biological implications of CRISPR interference.
Over the past 10 years, CRISPR function and mechanism has been an area of intensive investigation, and much has been learned at the genetic, molecular, biochemical and structural levels (for comprehensive CRISPR reviews, see Marraffini 2015; Wright, Nunez and Doudna 2016). Overall, the CRISPR pathway can be divided into three phases: adaptation, CRISPR RNA (crRNA) biogenesis and target interference (Fig. 1). The first phase (adaptation) refers to the acquisition of the invasive DNA snippets—‘protospacers’—and their incorporation as new spacers into the host's CRISPR locus. These adaptive events can immunize the bacterial host to combat future attacks by similar invaders, and to maintain a genomically recorded immunological memory of past encounters (Barrangou et al.2007; Deveau et al.2008). In the second phase, crRNA precursors are produced by transcription across the CRISPR array and then further processed into small, mature crRNAs, each with sequences derived from a single spacer and one or both adjacent repeats (Brouns et al.2008; Carte et al.2008; Deltcheva et al.2011). Each crRNA assembles with Cas protein(s) into an effector complex, and during the final phase (target interference) guides the effector complex to the target nucleic acid (DNA or/and RNA) for recognition and eventual destruction. In most CRISPR systems, target interference requires not only crRNA/target complementarity, but also a short flanking sequence called a protospacer adjacent motif (PAM) (Deveau et al.2008; Mojica et al.2009).
Figure 1.
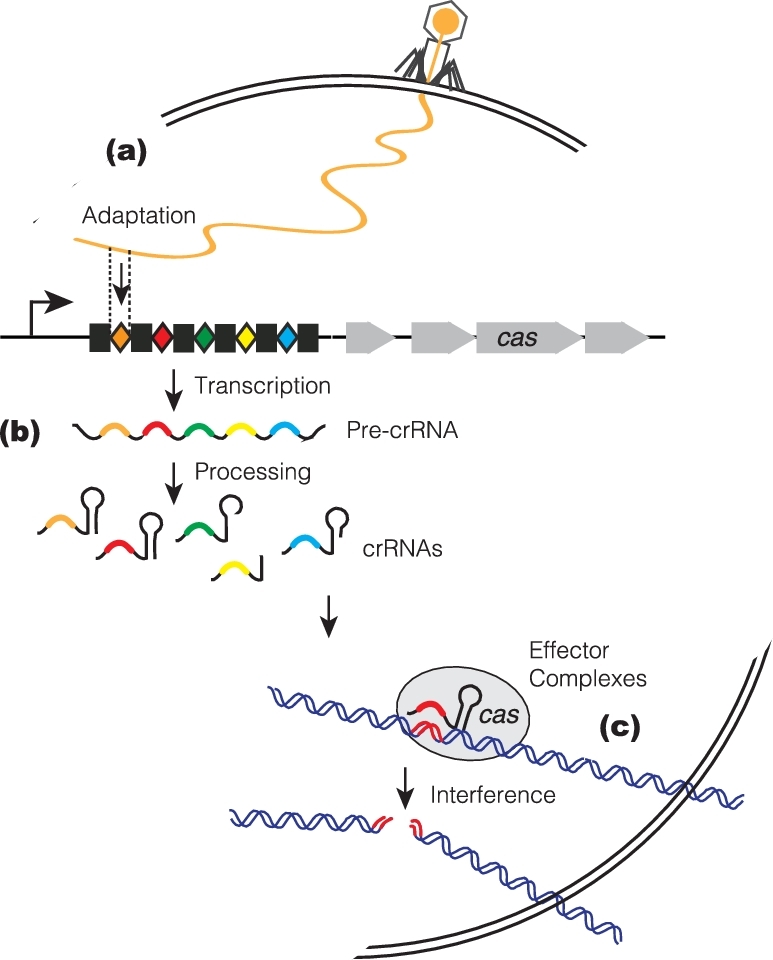
Three stages of the CRISPR-Cas interference pathway. CRISPR loci consist of arrays of short repeats (black boxes) interspaced by variable short spacers (colored diamonds), some of which are of bacteriophages or plasmids origin. CRISPR is flanked by a cluster of CRISPR-associated (cas) genes (grew arrows) that encode protein machineries for the three stages of this prokaryotic adaptive immune system. (a) Adaptation stage. Upon entering into the cells, small snippets of the invader DNAs are incorporated as new spacers into the CRISPR array of the host chromosome. (b) CrRNA biogenesis stage. Pre-crRNAs are produced by transcription across the CRISPR array, and further processed into short mature crRNAs, each with sequences derived from a single spacer and flanking repeat(s). (c) Target interference stage. The crRNAs assemble with Cas proteins into effector complexes, and serve as antisense guides for the effector complexes to locate and destroy invasive nucleic acid targets that carry cognate target sequences.
CRISPR systems are remarkably diverse. Based on cas gene composition, they can be categorized into two major classes and six types, among which the molecular events underlying crRNA biogenesis and target interference differ drastically (Makarova et al.2015). Class I systems comprise Types I, III and IV that possess multisubunit effector complexes, whereas Class II comprises Types II, V and VI, each with a single, large, crRNA-bound Cas protein as their effector complex (Makarova et al.2015; Shmakov et al.2015). Members of the Cas9 family of proteins, which are the effectors of Type II CRISPR systems, function as RNA-guided DNA endonucleases that cleave double-stranded DNA targets (Gasiunas et al.2012; Jinek et al.2012), and this activity has been harnessed to enable RNA-programmable, locus-specific genome editing and genome regulation in a wide range of eukaryotic organisms (Cho et al.2013; Cong et al.2013; Hwang et al.2013; Jinek et al.2013; Mali et al.2013b). Another interesting feature of the Type II systems is that their crRNA processing is mediated by a host factor, RNase III, and a second small non-coding RNA cofactor called tracrRNA (Deltcheva et al.2011). This is in contrast to Class I systems, which use a dedicated Cas protein as the crRNA processing endonuclease, and do not employ an auxiliary tracrRNA (Brouns et al.2008; Carte et al.2008). The past year has witnessed the discovery and characterization of many novel CRISPR-Cas types and subtypes, and mechanistic distinctions among Class II systems that employ the effector proteins Cas9, Cpf1, C2c1 and C2c2 (Types II, V-A, V-B and VI, respectively) have also started to emerge (Makarova et al.2015; Shmakov et al.2015; Zetsche et al.2015; Abudayyeh et al.2016; Fonfara et al.2016; Burstein et al.2017).
CRISPR, BACTERIAL PHYSIOLOGY AND PATHOGENESIS
In recent years, hints are emerging that certain CRISPR-Cas components can have alternative roles in regulating endogenous genes and host cell physiology, beyond their canonical genome defense function (Sampson and Weiss 2013, 2014; Louwen et al.2014). However, these alternative roles are still very poorly understood. In a few species, there is strong evidence that CRISPRs are involved in bacterial stress responses. For example, a Type I-B CRISPR-Cas system in Myxococcus xanthus controls the stress-dependent development of fruiting bodies (Viswanathan et al.2007). In Escherichia coli, envelope stress induces the expression of a Type I-E CRISPR-Cas operon, which then silences a plasmid encoding a defective protein that is transported across the cell membrane (Perez-Rodriguez et al.2011). In many other organisms, direct or indirect evidence has been reported that has linked CRISPR-Cas to aspects of bacteria physiology such as biofilm formation, DNA repair, immune evasion, quorum sensing and other processes (Sampson and Weiss 2013, 2014; Louwen et al.2014).
Type II CRISPR-Cas systems, compared to other CRISPR-Cas types, are over-represented in eukaryotic host-associated pathogenic and commensal bacteria (Makarova et al.2011). The best-characterized example of Cas9's role in bacterial virulence is in the intracellular pathogen Francisella novicida, where Cas9 promotes virulence by silencing the gene encoding an endogenous bacterial lipoprotein (blp) (Sampson et al.2013). The lack of this surface Blp facilitates the pathogen's survival in host macrophages and evasion of host innate immunity. Unexpectedly, the F. novicida Cas9 ortholog (FnoCas9) employs the 3΄ tail of its tracrRNA as the ‘guide’ to recognize the mRNA target by base pairing; this interaction triggers the blp mRNA's degradation by mechanisms that are not yet known (Sampson et al.2013). Furthermore, a novel, CRISPR-Cas-associated small RNA called scaRNA (distinct from crRNA) is a necessary co-factor for this process (Sampson et al.2013). Interestingly, by analyzing cas9 knockout mutants, Sampson et al. (2013) also found that cas9 is important for the ability of Campylobacter jejuni and Neisseria meningitidis to adhere to and invade human epithelial cells, traits essential for their virulence. This corroborated a similar finding by Louwen et al. (2013) that in C. jejuni, the presence of a Cas9 ortholog (CjeCas9) correlates with enhanced virulence and reduced swarming in Guillain-Barré syndrome-inducing isolates. Although these examples of CRISPR-Cas alternative functions are extremely intriguing, they lack a unifying mechanistic theme, suggesting that different microbial hosts might have co-opted distinct CRISPR-Cas components for diverse functional outcomes. And importantly, the degree to which any such CRISPR-Cas-driven outcomes affect meningococcal virulence is underexplored and very poorly understood.
CRISPR-CAS NEW SPACER ADAPTATION
Acquisition of snippets of invader DNAs as new spacers into the host CRISPR array, a fascinating process termed adaptation, has been observed experimentally thus far in several CRISPR subtypes, with most mechanistic insights obtained from Type I-E and II-A systems (Marraffini 2015; Sternberg et al.2016; Wright, Nunez and Doudna 2016). Most new spacers are added in a highly polarized fashion at the leader-proximal end of the CRISPR array (Deveau et al.2008; Yosef, Goren and Qimron 2012), and cas1 and cas2 (the two cas genes present in nearly all CRISPR systems) play central roles in this process (Yosef, Goren and Qimron 2012). Accordingly, the spacers at the leader-distal end of the array (and therefore at the 3΄ end of the pre-crRNA transcript) tend to be the most ancient and conserved. The (Cas1)4/(Cas2)2 heterohexameric integrase complex, as revealed by structural (Nunez et al.2014, 2015a) and in vitro reconstitution studies (Nunez et al.2015b), catalyzes sequence-specific spacer incorporation at the leader-repeat junction via staggered cuts, with concurrent duplication of the first repeat. Short sequences in the foreign DNA that are flanked by a PAM are selected as new spacers and incorporated into CRISPR in a consistent orientation (Deveau et al.2008; Goren et al.2012; Heler et al.2015) to ensure that the production of functional crRNAs that can guide effective interference by the effector complexes.
In the Escherichia coli Type I-E system, cas1 and cas2 suffice for the ‘naïve’ adaptation process, where previously un-encountered invader DNAs are captured as new spacers (Yosef, Goren and Qimron 2012). A related but more complex pathway in E. coli is known as ‘primed’ adaptation. In this case, hyperactivated spacer acquisition is stimulated by a pre-existing CRISPR spacer that partially matches the invader, and this process (unlike naïve adaptation) also requires the active interference machinery, in addition to Cas1 and Cas2 (Datsenko et al.2012). It is likely that primed adaptation evolved to allow the bacterial host to rapidly adapt to predators that evaded CRISPR interference by escape mutations in the protospacer. In contrast, the Type II-A systems from Streptococcus pyogenes and S. thermophilus only have naïve adaptation, and remarkably, nearly all of the Type II-A CRISPR-Cas components—cas9, tracrRNA, csn2, cas1, cas2 and the leader-repeat junction—are required (Heler et al.2015; Wei, Terns and Terns 2015). Cas9's PAM-interacting domain defines the PAM specificity of the newly acquired spacers (Heler et al.2015), yet its HNH and RuvC nuclease activities are dispensable for Type II-A adaptation (Heler et al.2015; Wei, Terns and Terns 2015). The auxiliary gene cas4, encoded in several CRISPR subtypes (including Type II-B), is also involved in adaptation (Sternberg et al.2016; Wright, Nunez and Doudna 2016). Yet, the molecular function of Cas4 (and of the Type II-A auxiliary factor Csn2) in adaptation remains elusive.
Despite these and other advances in our understanding of spacer acquisition, it remains the least understood step in the CRISPR pathway, since adaptation has been established experimentally in very few organisms. There are many unanswered questions: for example, how are DNA donors selected and processed into physiological substrates for the Cas1-Cas2 integrase? What is the full scope of mechanisms that avoid the capture of self-DNAs that could otherwise lead to suicidal events? Type I-E adaptation exhibits an intrinsic preference for foreign over self-DNAs (Yosef, Goren and Qimron 2012; Diez-Villasenor et al.2013), and the RecBCD double-strand break repair complex is proposed to provide substrates for Cas1-Cas2 (Levy et al.2015). During primed adaptation in Type I-E, degradation products generated by the Cas3 effector nuclease are preferentially used as spacer donors. On the other hand, spacer acquisition in at least some Type II-A system does not appear to discriminate against self-DNAs. Catalytic inactivation of the nuclease activity of SthCas9, which abolishes its interference, results in much more robust accumulation of primarily self-DNAs derived new spacers (Wei, Terns and Terns 2015), implying that the majority of adaptation events here leads to CRISPR-mediated self-targeting and autoimmune suicide. Therefore, on a microbial population level, a subset of invaded cells may develop resistance to the invasion, at the apparent cost of occasional cell death due to adaptation-mediated autoimmunity.
MENINGOCOCCAL CRISPR-Cas9 AND GENETIC TRANSFER
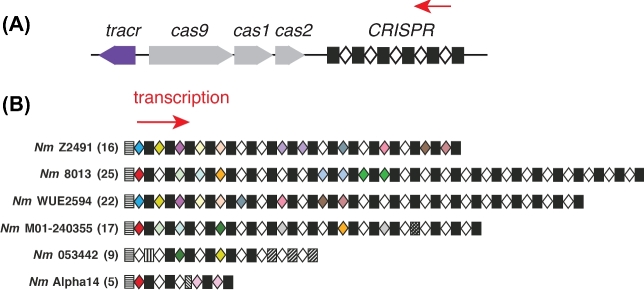
A total of 17 out of the 78 sequenced meningococcal genomes in the current NCBI nt/nr database contain a ∼6.9 kb Type II-C CRISPR-Cas locus, with three cas genes (cas1, cas2 and cas9) separating a tracrRNA locus and a CRISPR array (Fig. 2A) (Zhang et al.2013). With only three cas genes (one fewer than Type II-A and II-B), Type II-C is among the simplest CRISPR subtypes overall. As the most prevalent Type II subtype, Type II-C systems are present in many bacterial pathogens and commensals, including Neisseria meningitidis, Campylobacter jejuni and Pasteurella multocida (Fonfara et al.2014). The well-known CRISPR-Cas9s of Streptococcus pyogenes and S. thermophilus belong to Type II-A, and the system from Francisella novicida is Type II-B (Fonfara et al.2014; Makarova et al.2015).
Figure 2.
The meningococcal CRISPR-Cas9 system. (A) Schematic of the Type II-C CRISPR-Cas locus in meningcoccus. Black rectangles, CRISPR repeats; white diamonds, CRISPR spacers; in grey, three cas genes; in purple, the tracrRNA locus; red arrow, directions of CRISPR transcription. This schematic is not to scale. (B) Schematic of CRISPR loci from six meningoccal strains. Strain names are indicated. Unique spacer sequences are shown in white; non-unique spacers are in various colors. Rectangles, consensus repeats; patterned rectangles, repeat variants; red arrow, directions of CRISPR transcription.
Mc CRISPR-Cas9 confers genetic interference with natural transformation
Natural transformation is the process by which bacteria naturally take up exogenous DNAs from the environment, often incorporating them into the host genome by homologous recombination. Zhang et al. (2013) demonstrated that the native CRISPR-Cas9 of N. meningitidis strain 8013 can completely block the transformation of DNA substrates, in the forms of either integrational plasmids or meningococcal chromosomal DNAs. This blockage only occurs with substrates that carry a protospacer (that matches an existing CRISPR spacer) flanked by a suitable PAM, and the transformation frequencies, measured as antibiotic-resistant CFU/total CFU, dropped from 10−4 –10−5 to undetectable levels (< =10−8) as a result of CRISPR interference (Zhang et al.2013). Further comparisons of the wild-type, isogenic deletion or transposon insertion mutants (and their respective complementation strains) revealed that cas9, the cognate CRISPR spacer and tracrRNA are all required for this genetic interference, whereas the putative adaptation factors cas1 and cas2 are dispensable (Zhang et al.2013). This meningococcal CRISPR inference is seemingly constitutive, since interference occurs under normal conditions for liquid transformation, and abundant crRNAs and tracrRNA are constitutively expressed in mid-log and stationary phases. Zhang et al. thereby established meningococcus as a model organism to study the Type II-C CRISPR interference phenomenon as well as its underlying mechanisms.
A wealth of work has been done to understand how various Cas9 orthologs function mechanistically, and a clear picture has emerged that Cas9s are RNA-guided DNA endonucleases. Cas9 engages a crRNA and a tracrRNA that are partially duplexed, and the crRNA spacer sequence is then used to recognize double-stranded (ds) DNA targets by base pairing, leading to cleavage of both strands (Fig. 3A) (Gasiunas et al.2012; Jinek et al.2012). Stable DNA binding and cleavage in vitro, as well as genetic interference in cells, require the presence of the PAM, and crRNA/target complementarity must be nearly perfect in the 10–12 nt ‘seed’ region proximal to the PAM (Jinek et al.2012; Wright, Nunez and Doudna 2016). Cas9 employs two distinct nuclease domains, HNH and RuvC, to cleave the targeted strand and the non-complementary strand, respectively; active site mutations in both domains diminish target cleavage without affecting recognition of or binding to the DNA targets (Gasiunas et al.2012; Jinek et al.2012). Biochemical studies of N. meningitidis Cas9 (NmeCas9) revealed that these general features apply to this Type II-C Cas9 during cleavage of dsDNA targets (Zhang et al.2015). Furthermore, RuvC or HNH active site cas9 mutations in meningococcus completely abolished interference with natural transformation (Zhang et al.2013), indicating that meningococcal CRISPR-Cas9 enables genetic interference via the restriction of DNA substrates.
Figure 3.
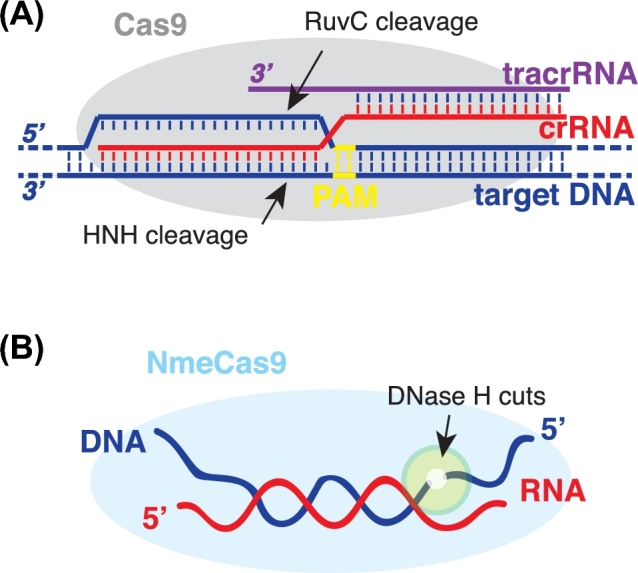
DNA targeting by the Cas9 family of RNA-guided endonucleases. (A) Schematic of dsDNA target (dark blue) recognition by Cas9s. Cas9 protein (grey) associates with two RNA partners, the crRNA (red) and the tracrRNA (purple), that are partially duplexed together. The HNH and RuvC nuclease motifs of Cas9 cleave the targeted DNA strand and the non-complementary DNA strand, respectively. Arrows denotes the cleavage sites. Recognition and cleavage of dsDNA targets require the complementarity between the DNA target and the crRNA spacer, and a short flanking protospacer-adjacent motif (PAM, yellow). (B) Schematic of NmeCas9's in vitro DNase H activity. NmeCas9 generates site-specific cuts (denoted by arrows, highlighted by green circle) in the DNA strand of a RNA-DNA hybrid substrate. Dark blue, DNA strand; red, RNA strand; light blue oval, NmeCas9.
Much remains to be learned about the coexistence of, and interplay between, systems that either drive or oppose HGT in Neisseriae. On one hand, Neisseria species are famous for their natural competency, and they do not regulate their competency like many other naturally competent bacteria such as Bacillus subtilis and S. pneumonia do; instead they are naturally competent for DNA uptake in all growth phases (Johnston et al.2014). On the other hand, Neisseriae also limit the transformation of foreign DNAs by multiple R-M systems, and by the preference for a species-specific, non-palindromic, 10–12 nt DNA update sequences (DUS) (Hamilton and Dillard 2006; Rotman and Seifert 2014). Unlike R-M, CRISPR is an adaptive and therefore sequence-selective barrier against the transformation of loci containing specific protospacer sequences that may provide beneficial or detrimental traits under the right circumstance. Given that bacteriophages and plasmids are relatively rare in this genus, and that natural transformation is the prevalent way that Neisseriae mobilize their chromosomal DNAs for genomic plasticity (Rotman and Seifert 2014), the meningococcal CRISPR-Cas9 can be a key player that helps shape the evolution of the physiology and pathogenicity of this important human commensal/pathogen.
Spacer content and targeting rules
BLASTN searches for all 74 unique spacers from six CRISPR-containing N. meningitidis strains (Fig. 2B) revealed that most (41/74) lack known potential targets (meaning those that are perfectly matched or that have a single mismatch outside the ‘seed’) in the NCBI database, and the remaining 33 meningococcal spacers match hundreds of distinct potential targets (Zhang et al.2013). Only ∼19% of these targets are within known prophages or in predicted phage-related genes, whereas the remaining appear to be chromosomal sequences from other strains or species of Neisseria (Zhang et al.2013). This is consistent with the idea that transformation of Neisserial chromosomal DNA is the most prevalent means for Neisseria genetic exchange, but may also reflect to some extent the paucity of known bacteriophage or mobile elements sequences of meningococcus. The distribution of potential targets within the genomes of Neisseriae has no obvious strand bias or preference for gene-coding versus intergenic regions. An alignment of a large number of potential natural targets bioinformatically deduced a clear consensus PAM, N4G(A/C)TT, in the 3΄ flanking sequence (relative to the crRNA-non-complementary strand) of target regions specified by Neisseria CRISPR spacers (Zhang et al.2013).
Despite that spacers from the six CRISPR-plus meningococcal strains primarily match to genomic sequences of other Neisseria strains or species, five ‘self-targeting’ spacers exist that have perfect complementarity to self-chromosome regions outside the host CRISPR array (Zhang et al.2013). This is not surprising given that a noticeable number of ‘self-targeting’ CRISPR spacers have been reported in numerous organisms and are thought have diverse biological consequences (Stern et al.2010; Heussler and O’Toole 2016). For example, spacers that target self-chromosomal regions flanked by functional PAMs will likely lead to CRISPR-mediated self-killing that can reshape the microbial population by selecting for cells that have lost either the targeted genomic region or the functional CRISPR-Cas system (Heussler and O’Toole 2016). Partially self-matching Type I-F spacers in Pseudomonas aeruginosa have also been shown to influence group behavior such as biofilm formation and swarming motility, by inducing cell death in surface-associated bacteria but not the planktonic population (Heussler and O’Toole 2016). In meningoccus, the five ‘self-targeting’ spacers all match to self-chromosomal regions that include 3nt deviations from the N4G(A/C)TT PAM consensus; therefore, CRISPR lethality is likely abrogated due to the lack of functional PAMs (Zhang et al.2013). Whether these ‘self-targeting’ spacers have biological implications in gene regulation or bacterial physiology remain elusive.
The initial in silico target analyses were based on the assumption that targeting by CRISPR-Cas9 requires near-perfect complementary and a consensus PAM. However, subsequent experiments have revealed that CRISPR interference in meningococcus has more relaxed targeting requirements, as numerous seed mismatches and PAM deviations were well tolerated in transformation interference assays (Zhang et al.2015). For example, all single nt and many 2 nt mismatches in crRNA/target complementarity have minimal interference defects in Neisseria. And interestingly, NmeCas9 strictly requires the G residue in the N4G(A/C)TT PAM, but has relaxed and complex dependence on the other three non-G positions in the PAM; at least 18 PAM variants with 1- or 2 nt deviations from the consensus at non-G positions appear to be functional (Zhang et al.2015). These in vivo findings likely reflect intrinsic properties of the NmeCas9 enzyme, as they are corroborated by results from in vitro biochemical analysis (Zhang et al.2015). Collectively, the targeting potential for natural Neisseria spacers may be larger than previously thought. In the future, the accumulation of data for the identity of Neisseria spacers from larger collections of isolates, along with their repertoire of potential natural targets, should provide insights into the acquisition and loss of Neisseria spacers, as well as CRISPR-Cas9's effects on meningococcal evolution.
CRISPRs and Neisseria prophages
Despite their paucity, naturally occurring spacers do exist that match known Neisseria filamentous (Nf) prophages, including the MDA pathogenicity island (Zhang et al.2013). Neisseria meningitidis strain WUE2594, which lacks many Nf prophages in its genome (Joseph et al.2011), has a CRISPR with nearly a dozen spacers with extensive targeting potential against known Nf prophages, reflecting a likely role for CRISPR interference in shaping prophage content in meningococcus (Zhang et al.2013). Conversely, Neisseria Mu-like prophages (which have much larger genomes than Nf prophages) are ‘targeted’ by far fewer (if any) CRISPR spacers, and the reasons behind this apparent discrepancy are unclear.
Putative CRISPRs in other Neisseria species
Strain 020-06 of N. lactamica, the best-studied commensal Neisseria species, contains a Type II-C CRISPR-cas locus with very similar cas genes and CRISPR repeats (but nine different spacers) as those of meningococcus (Zhang et al.2013). BLASTP search using NmeCas9 protein sequences as queries can identify putative Cas9s in other Neisseria species including N. cinerea, N. mucosa, N. flavescens, N. bacilliformis and N. wadsworthii, implying that cas9, either alone or together with crispr, cas1 and cas2, might have been horizontally transferred among pathogenic and commensal species that co-inhabit the human nasopharynx. Several N. gonorrhoeae strains possess a very questionable CRISPR with just one spacer and a few Type I-C cas genes containing frameshift mutations (Zhang et al.2013). This locus is likely reminiscent of a degenerate, non-functional type I-C system and the absence of functional CRISPRs in gonococcus may have contributed to its ability to repeatedly develop and spread antibiotic resistance. The sexually transmitted disease gonorrhea, with 78 million new infections every year, is now dangerously close to being untreatable due to multidrug resistance (WHO 2016). It is also noteworthy that a putative Type I-C and/or a putative Type III-B CRISPR system also exist in certain strains of species such as N. lactamica, N. sicca, N. weaveri, N. cinerea and N. mucosa (Louwen et al.2014). Nonetheless, the functionality of the putative Type I-C and Type III-B CRISPR-Cas systems in commensal Neisseria remains to be established.
FEATURES OF Mc CRISPR-Cas9 THAT ESTABLISH NEW PARADIGMS
Genetically encoded inhibitors of NmeCas9
As a new twist on the co-evolutionary arms race between bacteria and their phages, Pawluk et al. (2016) recently reported the discovery of three families of anti-CRISPR (Acr) genes (acrIIC1Nme, acrIIC2Nme, and acrIIC3Nme) with Neisseria homologs that can evade CRISPR interference by binding to and disarming the NmeCas9 machinery. The acrIIC1Nme gene exists in one Neisseria sequence contig displaying mobile genetic element (MGE) properties, and acrIIC2Nme and acrIIC3Nme coexist in a putative prophage within a few meningococcal strains. These three genes encode the first natural Cas9 inhibitors to be discovered, and their utility as ‘off switches’ for NmeCas9 genome editing in mammalian cells has also been validated (Pawluk et al.2016). Pawluk et al. provided a proof of principle that anti-CRISPRs, previously reported only for Type I-E and I-F systems, now extend into Type II-C CRISPR-Cas systems. These results suggested that MGE-encoded inhibitor genes for other Class II CRISPR effectors (including the widely used SpyCas9) might also exist. Indeed, this possibility has since been confirmed by the discovery of Type II-A anti-CRISPRs widespread in Listeria monocytogenes (Rauch et al.2017). Rauch et al. found four acr genes (acrIIA1, acrIIA2 acrIIA3, and acrIIA4) encoded by L. monocytogenes prophages that can prevent the Type II-A LmoCas9's function in CRISPR interference. Two of these inhibitors cross-react with SpyCas9 (another Type II-A Cas9), and inhibit SpyCas9-enabled gene editing in human cells (Rauch et al.2017). Further mechanistic and structural studies are needed to understand how the newfound Cas9 inhibitors exert their effects. Continued exploration of novel NmeCas9 inhibitors may also provide new insights into Neisseria phage biology, as well as the co-evolution of bacteria and their parasitic mobile elements. Anti-CRISPR genes may have profound impacts on CRISPRs and HGT across prokaryotes, and it is likely that an onslaught of anti-CRISPR gene discovery and validation is now underway.
An expanded range of DNA cleavage properties for NmeCas9
Members of the divergent Cas9 family of endonucleases employ the HNH and RuvC nuclease domains to cut the crRNA-complementary and non-complementary strands of the dsDNA substrate, respectively (Fig. 3A), and these activities strictly require the presence of both small RNA partners (crRNA and tracrRNA) (Jinek et al.2012; Fonfara et al.2014). The best-studied Type II-A Cas9 from Streptococcus pyogenes cleaves ssDNA targets inefficiently (compared to duplexed DNA targets) in vitro (Jinek et al.2012). Strikingly, the Type II-C NmeCas9 can direct robust in vitro cleavage of ssDNA targets, and the cleavages can occur in the presence or absence of the tracrRNA co-factor, and in the presence or absence of a PAM (Zhang et al.2015). Further biochemical studies revealed that this tracrRNA-independent activity requires an active HNH domain (but not RuvC active site residues), divalent metal ions and a reprogrammable RNA guide with a minimum of 17–18 contiguous nts base paired with its ssDNA target (Zhang et al.2015). In essence, this is a ‘DNase H’ activity, reminiscent of the RNase H activity that degrades the RNA strand of a RNA-DNA hybrid duplex, except that NmeCas9's ‘DNase H’ activity cleaves the DNA rather than the RNA strand of the hybrid duplex (Fig. 3B) (Zhang et al.2015). The cuts occur at very specific positions of the duplex, with the sites determined by an apparent ruler mechanism that measures from the 5΄ end of the ssDNA's RNA-paired region; furthermore, this activity can occur with hybrid duplexes that are preformed passively in solution, i.e. without NmeCas9 first engaging an ssRNA guide (Zhang et al.2015). This DNase H activity could provide the basis for new biotechnological tools in the future. Collectively, NmeCas9 has fundamental distinctions from SpyCas9, despite its capability for the conventional dual RNA-mediated dsDNA targeting. Crystal and cryo-EM structures of SpyCas9 in multiple functional states (with and without its RNA cofactors, and with single- and double-stranded target DNA bound) have been solved (Anders et al.2014; Jinek et al.2014; Nishimasu et al.2014; Jiang et al.2015); as for Type II-C, the apo structure of a separate Cas9 ortholog (AnaCas9) is known (Jinek et al.2014), but no guide-loaded (let alone DNA-bound) Type II-C structures have been reported. Future structural insights are needed to understand how NmeCas9 engages different small RNA partners and accommodates different types of nucleic acid substrates.
Natural transformation occurs in multiple stages, and it is not yet known whether all stages are susceptible to NmeCas9-based interference. Exogenous DNAs are thought to internalize in single-stranded form (accompanied by the degradation of the opposite strand), with the imported ssDNAs then loaded with RecA recombinase and ssDNA-binding proteins (SSB) and usually integrate into the host chromosome by homologous recombination (Johnston et al.2014). The strand of the external dsDNA that is ultimately internalized is selected randomly (Johnston et al.2014), yet a crRNA that only pairs with one target strand can block the formation of all transformants. This argues for a model where some, if not all, targeting events must occur after the double strandedness of DNA is restored by integration and chromosome replication (Johnston et al.2013). This model also ensures that those bacterial cells that have integrated unwanted CRISPR targets into their genomes, due to a failure or escape of interference during uptake and recombination, could still be cleared from the population. In light of the recent finding that NmeCas9 can efficiently cleave ssDNA targets (including those pre-loaded with RecA or SSB) in vitro (Zhang et al.2015), it is tempting to speculate that CRISPRs may also target earlier ssDNA stages of Neisseria transformation. Furthermore, it was also reported that several other Type II-C Cas9s have also robust ssDNA cleaving activities in vitro (Ma et al.2015a), hinting that Type II-C systems, which are thought to be phylogenetically ancestral to Types II-A and II-B, might have evolved initial functions such as targeting transforming ssDNAs or ssDNA bacteriophages (Ma et al.2015a; Zhang et al.2015). The physiological meaning of DNase H activity by NmeCas9 remains to be established.
A distinct crRNA biogenesis pathway
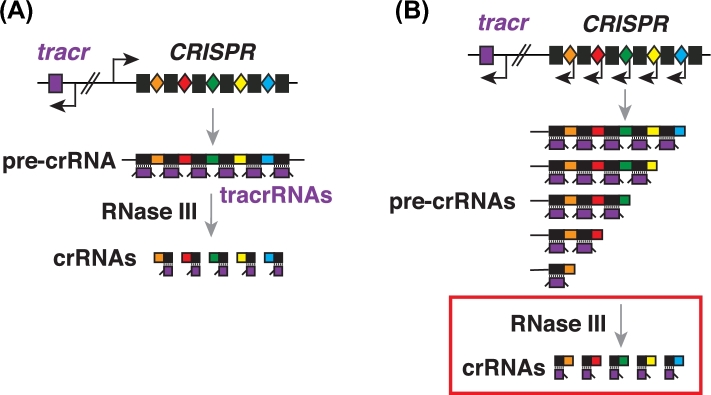
A long crRNA precursor (containing multiple repeat-spacer units) is usually derived from the CRISPR array, and its transcription is driven from an external promoter within the A/T-rich ‘leader’ region preceding the CRISPRs (Pougach et al.2010; Deltcheva et al.2011). Correct processing of this multimeric crRNA precursor into mature monomers has been defined as an essential step for interference in multiple CRISPR-Cas systems (Brouns et al.2008; Deltcheva et al.2011; Wright, Nunez and Doudna 2016). In Types I and III, the pre-crRNAs are processed by the Cas6 (and occasionally Cas5) family of ribonucleases (Brouns et al.2008; Carte et al.2008; Charpentier et al.2015), whereas in Type II, processing relies on a host factor (RNase III) and the tracrRNA co-factor (Deltcheva et al.2011). TracrRNA has an ‘anti-repeat’ region that base pairs with the repeats in the pre-crRNA, resulting in RNA duplexes that are then cleaved by RNase III and that associate with Cas9 to direct DNA targeting (Fig. 4A) (Deltcheva et al.2011). Despite the divergence in lengths and sequences of tracrRNA orthologs across Type II systems, the ‘anti-repeat’ region of the tracrRNA and the repeat regions of the pre-crRNA co-evolved to maintain their base-pairing potential (Deltcheva et al.2011; Chylinski et al.2014).
Figure 4.
CrRNA biogenesis in Type II CRISPR-Cas9 systems. (A) A typical Type II-A CRISPR loci express a long pre-crRNA, and a tracrRNA cofactor that hybridizes to all the pre-crRNA repeats. The host factor RNase III cleaves the pre-crRNA/tracrRNA duplexes, yielding pairs of matured crRNA and tracrRNA species. Black rectangles, repeats; colored diamonds, spacers; purple, tracrRNA locus; black arrows, transcriptions. (B) A unique crRNA biogenesis pathway in meningoccus. Transcriptions from repeat-embedded internal promoters result in nested set of pre-crRNA transcripts of varying lengths. RNase III- and tracrRNA-mediated RNA processing (in red box) occurs in cells but is dispensable for the interference function.
Meningococcus was the first Type II-C system to have its crRNA biogenesis pathway examined, revealing an unusual crRNA biogenesis pathway distinct from that of previously characterized Type II CRISPR-Cas systems (Fig. 4B) (Zhang et al.2013). A differential RNA-seq technique that can distinguish primary versus processed transcripts enabled the discovery of an extended –10 box (TGn) promoter embedded within every repeat of the meningococcal CRISPR (Zhang et al.2013). These internal promoters initiate transcription starting within every spacer, leading to a nested set of RNA precursors that are further processed by RNase III and tracrRNA into mature species (Fig. 4B) (Zhang et al.2013). It is unclear why the meningococcal system (as well as some if not all other Type II-C CRISPRs; Dugar et al.2013) evolved repeat-embedded promoters as a general feature, as opposed to the external promoter within the A/T-rich ‘leader’. Is this a way to ensure independent production of crRNAs from each spacer? Is this a way to maximize crRNA expression from the distal end (as defined by the direction of transcription) of the CRISPR array? In addition, the promoter portion of the repeats may have additional functions in processes like new spacer acquisition. These possibilities await exploration.
Another distinctive feature of the meningococcal pathway is that post-transcriptional pre-crRNA processing, though it clearly occurs, is dispensable for interference function (Zhang et al.2013). This was demonstrated by deletion of the RNase III-encoding gene (rnc) in Mc, which disrupted pre-crRNA processing but did not prevent interference (Fig. 4B) (Zhang et al.2013). This is in stark contrast to other diverse CRISPRs characterized thus far, in which pre-crRNAs are not targeting competent until processed into mature species. The molecular basis for this tolerance to pre-crRNA processing defects is not yet understood.
Type II-C CRISPR adaptation
Spacer acquisition has not been experimentally established for the meningococcal Type II-C CRISPR system, yet some distinguishing features of this pathway point to an adaptation mechanism that exhibits differences from those of Type II-A and II-B CRISPRs. First, the polarity of spacer conservation is opposite to that observed elsewhere, with the most conserved spacers clustered at the upstream end of CRISPR (relative to the direction of transcription), and the more strain-specific spacers (likely acquired more recently) clustered at the downstream end (Zhang et al.2013). Second, the upstream-most repeat within meningococcal CRISPRs is the most likely to deviate from the consensus, whereas in other CRISPRs the downstream-most repeat is the repeat variant (Zhang et al.2013). Third, an obvious A/T-rich leader is not evident in the vicinity of Mc CRISPRs. These observations suggest that meningococcal spacer adaption could occur predominantly at the downstream end of the CRISPR via a leader-independent mechanism. This possibility is corroborated by recent findings from a different Type II-C system from Campylobacter jejuni: new spacers were indeed acquired at the downstream end of the CRISPR locus during a bacteriophage carrier state infection (Hooton and Connerton 2014). Interestingly, the C. jejuni phages used in this study encode a Cas4-like protein whose genetic requirement in adaptation has not been tested, and furthermore, the newly acquired spacers are exclusively of host rather than phage origin (Hooton and Connerton 2014). It is not yet clear whether Type II-C CRISPR-Cas systems in general, which notably lack the fourth cas gene observed in Type II-A and II-B systems (csn2 and cas4, respectively), require any additional factors (e.g. phage-encoded Cas4) for spacer acquisition. For meningococcus, neither bacteriophage carrier state nor prophage-encoded cas4-like genes has been reported, to the best of our knowledge. It is also worthy examining if and how the NmeCas9 inhibitors would affect Type II-C spacer aquisition, given the likely role of NmeCas9 in this process. If one extrapolate the findings from S. pyogenes Type II-A adaption that Cas9 defines the PAM specificity of newly acquired spacers, then potential inhibitors of NmeCas9 that disrupt its PAM recognition will likely interfere with the Type II-C adaptation process. On the other hand, it is also tempting to speculate that potential NmeCas9 inhibitors that only inactivate its nuclease activities without affecting its ability to recognize PAMs or to cooperate with the Cas1-Cas2 integrase may result in robust accumulation of new spacers acquired from self-DNAs.
Given that natural transformation with chromosomal DNAs from neighboring cells happens frequently in Neisseria, it is of particular importance to set up a genetically tractable system to address fundamental questions concerning the meningococcal Type II-C adaptation pathway. Is acquisition constitutive, or is it a tightly regulated process that is only stimulated under certain conditions (e.g. during stress response, phage attack or MGE mobilization)? Are there mechanisms that bias new spacer acquisition towards foreign rather than host DNA? Where do the substrates for adaptation come from and how are they processed? Does NmeCas9 interact with Cas1-Cas2 during acquisition, genetically and physically? Intriguingly, more than a dozen functional PAM variants exist (without a clear consensus) for NmeCas9 interference (Hou et al.2013; Zhang et al.2015; Lee, Cradick and Bao 2016), yet the N4G(A/C)TT consensus PAM was deduced from the analysis of natural targets for Neisseria spacers (Zhang et al.2013). This disparity awaits explanation. Is it possible that the adaptation PAMs are more stringent than, and only partially overlap with, the interference PAMs?
CRISPR applications
Long before the biological roles of CRISPRs in phage defense and HGT limitation were demonstrated in 2007 and 2008, the hypervariable sequences of CRISPRs already served as typing tools for the evolutionary, diagnostic and epidemiologic analyses of microbes. To date, CRISPRs have been used for typing species including Yersinia pestis, Salmonella spp., Mycobacterium tuberculosis, C. jejuni, Corynebacterium diphtheria and Legionella pneumophila (Louwen et al.2014).
The most notable application of CRISPRs, however, has been the development of CRISPR-Cas9 into facile, RNA-guided, user-programmable DNA targeting tools that have revolutionized genome-engineering technologies in a broad range of species, including humans and other mammals (Wright, Nunez and Doudna 2016; Komor, Badran and Liu 2017). Nowadays, for each desired genomic locus we only need to re-design an RNA guide for the Cas9 enzyme to introduce double-stranded breaks (DSB), which would greatly stimulate gene-editing efficiencies. In eukaryotic cells, these DSBs are processed primarily by two endogenous cellular repair pathways, either the error-prone non-homologous end joining pathway that leads to insertions or deletions and therefore gene disruptions or the homology-directed repair pathway (in the presence of a repair template) that allows precise editing (Komor, Badran and Liu 2017). Catalytically inactivated Cas9 (‘dead’ or dCas9) has also proven to be an effective platform for locus-specific, programmable genome binding. Tethering to dCas9 a wide range of effector domains, including transcriptional regulators (Mali et al.2013a; Chavez et al.2015), fluorescent proteins (Chen et al.2013; Ma et al.2015b), epigenetic modulators (Hilton et al.2015; Kearns et al.2015) and base editing enzymes (Komor et al.2016), has been shown to be useful for the regulation, or the imaging, of the target chromosomal loci. These and other Cas9-based tools have had transformational impacts on biomedical research, regenerative medicine and gene therapy.
The most commonly used Cas9, the Type II-A SpyCas9, is 1368 amino acids long and recognizes 20-bp protospacers with NGG (and, less efficiently, NAG and NGA) PAMs (Hsu et al.2013; Jiang et al.2013). Several natural Cas9 orthologs and other Class II CRISPR effectors that vary in sizes, PAM requirements, and the lengths of RNA guides, have also been adapted for eukaryotic genome engineering (Komor, Badran and Liu 2017); promising many benefits such as multiplexed applications, increased targeting fidelity, larger repertoire of targetable sites and expanded delivery options. In addition, a variety of strategies, including engineered Cas9 variants, have been developed to improve targeting specificities (i.e. mitigate off-targets) (Komor, Badran and Liu 2017).
NmeCas9 is an effective tool for genome engineering in eukaryotic cells including human pluripotent stem cells (Esvelt et al.2013; Hou et al.2013; Lee, Cradick and Bao 2016). Fusions of the catalytically inactivated dNmeCas9 to GFP or the effector domain of a transcription activator, acetyltransferase or histone demethylase have all been developed and shown to enable fluorescent labeling or the transcriptional modulation of the targeted chromosomal loci (Hilton et al.2015; Kearns et al.2015; Ma et al.2015b). NmeCas9 and its guide RNAs are orthogonal to those of SpyCas9, thereby allowing multiplexed tasks to be achieved simultaneously and independently (Esvelt et al.2013; Fonfara et al.2014; Ma et al.2015b). NmeCas9 is only 1082 residues (∼21% smaller than SpyCas9), well within range of adeno-associated virus-based delivery into cells and animals, and it is also less prone to off-target effects (Lee, Cradick and Bao 2016). Most recently, the first naturally occurring inhibitors for Type II-C and Type II-A Cas9s were discovered, and their utility as ‘off-switches’ for NmeCas9-, and SpyCas9-mediated mammalian gene editing applications have been established (Pawluk et al.2016; Rauch et al.2017). These inhibitor proteins could potentially provide handy tools to enable greater temporal, spatial or tissue-specific control of Cas9 gene editing, or to control the spread of Cas9 ‘gene drives’.
CONCLUDING REMARKS
The discovery of CRISPR-Cas systems in microbial genomes has resulted in fansicnating research areas of CRISPR biology and CRISPR-based genome engineering that have undergone true explosions in the past decade. The functional CRISPR-Cas9 system in meningoccus provided the context in which native CRISPR's role in limiting natural transformation was established, in which the first Type II-C system was examined in great mechanistic detail, and in which prophage-encoded Cas9 inhibitors were discovered. Continued investigation of the CRISPR-Cas systems in pathogenic and non-pathogenic Neisseria species promises to yield further unforeseen discoveries and exciting new technologies.
Acknowledgments
The author would like to thank Erik Sontheimer (UMass Medical School) for constructive comments and graphical help. The author is also grateful to Zhonggang Hou for critical reading of this manuscript and valuable inputs.
FUNDING
YZ is supported by National Institute of Health (NIH) grant K99 GM117268 and funds from University of Michigan.
Conflict of interest. None declared.
REFERENCES
- Abudayyeh OO, Gootenberg JS, Konermann S et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 2016;353:aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders C, Niewoehner O, Duerst A et al. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 2014;513:569–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science 2007;315:1709–12. [DOI] [PubMed] [Google Scholar]
- Bikard D, Hatoum-Aslan A, Mucida D et al. CRISPR interference can prevent natural transformation and virulence acquisition during in vivo bacterial infection. Cell Host Microbe 2012;12:177–86. [DOI] [PubMed] [Google Scholar]
- Bolotin A, Quinquis B, Sorokin A et al. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 2005;151:2551–61. [DOI] [PubMed] [Google Scholar]
- Bratcher HB, Bennett JS, Maiden MC. Evolutionary and genomic insights into meningococcal biology. Future Microbiol 2012;7:873–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouns SJ, Jore MM, Lundgren M et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 2008;321:960–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstein D, Harrington LB, Strutt SC et al. New CRISPR-Cas systems from uncultivated microbes. Nature 2017;542:237–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carte J, Wang R, Li H et al. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Gene Dev 2008;22:3489–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier E, Richter H, van der Oost J et al. Biogenesis pathways of RNA guides in archaeal and bacterial CRISPR-Cas adaptive immunity. FEMS Microbiol Rev 2015;39:428–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez A, Scheiman J, Vora S et al. Highly efficient Cas9-mediated transcriptional programming. Nat Methods 2015;12:326–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Gilbert LA, Cimini BA et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 2013;155:1479–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim JM et al. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol 2013;31:230–2. [DOI] [PubMed] [Google Scholar]
- Chylinski K, Makarova KS, Charpentier E et al. Classification and evolution of type II CRISPR-Cas systems. Nucleic Acids Res 2014;42:6091–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013;339:819–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Pougach K, Tikhonov A et al. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat Commun 2012;3:945. [DOI] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 2011;471:602–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveau H, Barrangou R, Garneau JE et al. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol 2008;190:1390–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez-Villasenor C, Guzman NM, Almendros C et al. CRISPR-spacer integration reporter plasmids reveal distinct genuine acquisition specificities among CRISPR-Cas I-E variants of Escherichia coli. RNA Biol 2013;10:792–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugar G, Herbig A, Forstner KU et al. High-resolution transcriptome maps reveal strain-specific regulatory features of multiple Campylobacter jejuni isolates. PLoS Genet 2013;9:e1003495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esvelt KM, Mali P, Braff JL et al. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods 2013;10:1116–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonfara I, Le Rhun A, Chylinski K et al. Phylogeny of Cas9 determines functional exchangeability of dual-RNA and Cas9 among orthologous type II CRISPR-Cas systems. Nucleic Acids Res 2014;42:2577–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonfara I, Richter H, Bratovic M et al. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature 2016;532:517–21. [DOI] [PubMed] [Google Scholar]
- Gasiunas G, Barrangou R, Horvath P et al. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. P Natl Acad Sci USA 2012;109:E2579–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goren MG, Yosef I, Auster O et al. Experimental definition of a clustered regularly interspaced short palindromic duplicon in Escherichia coli. J Mol Biol 2012;423:14–6. [DOI] [PubMed] [Google Scholar]
- Hamilton HL, Dillard JP. Natural transformation of Neisseria gonorrhoeae: from DNA donation to homologous recombination. Mol Microbiol 2006;59:376–85. [DOI] [PubMed] [Google Scholar]
- Heler R, Samai P, Modell JW et al. Cas9 specifies functional viral targets during CRISPR-Cas adaptation. Nature 2015;519:199–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heussler GE, O’Toole GA. Friendly fire: biological functions and consequences of chromosomal targeting by CRISPR-Cas systems. J Bacteriol 2016;198:1481–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton IB, D’Ippolito AM, Vockley CM et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol 2015;33:510–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooton SP, Connerton IF. Campylobacter jejuni acquire new host-derived CRISPR spacers when in association with bacteriophages harboring a CRISPR-like Cas4 protein. Front Microbiol 2014;5:744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Z, Zhang Y, Propson NE et al. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. P Natl Acad Sci USA 2013;110:15644–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 2013;31:827–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol 2013;31:227–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Zhou K, Ma L et al. STRUCTURAL BIOLOGY. A Cas9-guide RNA complex preorganized for target DNA recognition. Science 2015;348:1477–81. [DOI] [PubMed] [Google Scholar]
- Jiang W, Bikard D, Cox D et al. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol 2013;31:233–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012;337:816–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, East A, Cheng A et al. RNA-programmed genome editing in human cells. eLife 2013;2:e00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Jiang F, Taylor DW et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science 2014;343:1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston C, Martin B, Fichant G et al. Bacterial transformation: distribution, shared mechanisms and divergent control. Nat Rev Microbiol 2014;12:181–96. [DOI] [PubMed] [Google Scholar]
- Johnston C, Martin B, Polard P et al. Postreplication targeting of transformants by bacterial immune systems? Trends Microbiol 2013;21:516–21. [DOI] [PubMed] [Google Scholar]
- Joseph B, Schwarz RF, Linke B et al. Virulence evolution of the human pathogen Neisseria meningitidis by recombination in the core and accessory genome. PLoS One 2011;6:e18441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns NA, Pham H, Tabak B et al. Functional annotation of native enhancers with a Cas9-histone demethylase fusion. Nat Methods 2015;12:401–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor AC, Badran AH, Liu DR. CRISPR-based technologies for the manipulation of eukaryotic genomes. Cell 2017;168:20–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor AC, Kim YB, Packer MS et al. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016;533:420–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CM, Cradick TJ, Bao G. The Neisseria meningitidis CRISPR-Cas9 system enables specific genome editing in mammalian cells. Mol Ther 2016;24:645–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy A, Goren MG, Yosef I et al. CRISPR adaptation biases explain preference for acquisition of foreign DNA. Nature 2015;520:505–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louwen R, Horst-Kreft D, de Boer AG et al. A novel link between Campylobacter jejuni bacteriophage defence, virulence and Guillain-Barre syndrome. Eur J Clin Microbiol 2013;32:207–26. [DOI] [PubMed] [Google Scholar]
- Louwen R, Staals RH, Endtz HP et al. The role of CRISPR-Cas systems in virulence of pathogenic bacteria. Microbiol Mol Biol Rev 2014;78:74–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma E, Harrington LB, O’Connell MR et al. Single-stranded DNA cleavage by divergent CRISPR-Cas9 enzymes. Mol Cell 2015a;60:398–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Naseri A, Reyes-Gutierrez P et al. Multicolor CRISPR labeling of chromosomal loci in human cells. P Natl Acad Sci USA 2015b;112:3002–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Haft DH, Barrangou R et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol 2011;9:467–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Alkhnbashi OS et al. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol 2015;13:722–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Aach J, Stranges PB et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol 2013a;31:833–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM et al. RNA-guided human genome engineering via Cas9. Science 2013b;339:823–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA. CRISPR-Cas immunity in prokaryotes. Nature 2015;526:55–61. [DOI] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 2008;322:1843–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica FJ, Diez-Villasenor C, Garcia-Martinez J et al. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol 2005;60:174–82. [DOI] [PubMed] [Google Scholar]
- Mojica FJ, Diez-Villasenor C, Garcia-Martinez J et al. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 2009;155:733–40. [DOI] [PubMed] [Google Scholar]
- Nishimasu H, Ran FA, Hsu PD et al. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 2014;156:935–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez JK, Harrington LB, Kranzusch PJ et al. Foreign DNA capture during CRISPR-Cas adaptive immunity. Nature 2015a;527:535–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez JK, Kranzusch PJ, Noeske J et al. Cas1-Cas2 complex formation mediates spacer acquisition during CRISPR-Cas adaptive immunity. Nat StructMol Biol 2014;21:528–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez JK, Lee AS, Engelman A et al. Integrase-mediated spacer acquisition during CRISPR-Cas adaptive immunity. Nature 2015b;519:193–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawluk A, Amrani N, Zhang Y et al. Naturally occurring off-switches for CRISPR-Cas9. Cell 2016;167:1829–38.e1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Rodriguez R, Haitjema C, Huang Q et al. Envelope stress is a trigger of CRISPR RNA-mediated DNA silencing in Escherichia coli. Mol Microbiol 2011;79:584–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pougach K, Semenova E, Bogdanova E et al. Transcription, processing and function of CRISPR cassettes in Escherichia coli. Mol Microbiol 2010;77:1367–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 2005;151:653–63. [DOI] [PubMed] [Google Scholar]
- Rauch BJ, Silvis MR, Hultquist JF et al. Inhibition of CRISPR-Cas9 with bacteriophage proteins. Cell 2017;168:150–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotman E, Seifert HS. The genetics of Neisseria species. Annu Rev Genet 2014;48:405–31. [DOI] [PubMed] [Google Scholar]
- Sampson TR, Saroj SD, Llewellyn AC et al. A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature 2013;497:254–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson TR, Weiss DS. Alternative roles for CRISPR/Cas systems in bacterial pathogenesis. PLoS Pathog 2013;9:e1003621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson TR, Weiss DS. CRISPR-Cas systems: new players in gene regulation and bacterial physiology. Front Cell Infect Microbiol 2014;4:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmakov S, Abudayyeh OO, Makarova KS et al. Discovery and Functional Characterization of Diverse Class 2 CRISPR-Cas Systems. Mol Cell 2015;60:385–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern A, Keren L, Wurtzel O et al. Self-targeting by CRISPR: gene regulation or autoimmunity? Trends Genet 2010;26:335–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg SH, Richter H, Charpentier E et al. Adaptation in CRISPR-Cas systems. Mol Cell 2016;61:797–808. [DOI] [PubMed] [Google Scholar]
- Viswanathan P, Murphy K, Julien B et al. Regulation of dev, an operon that includes genes essential for Myxococcus xanthus development and CRISPR-associated genes and repeats. J Bacteriol 2007;189:3738–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Terns RM, Terns MP. Cas9 function and host genome sampling in Type II-A CRISPR-Cas adaptation. Gene Dev 2015;29:356–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. WHO Guidelines for the Treatment of Neisseria gonorrhoeae. Geneva, 2016. [PubMed] [Google Scholar]
- Wright AV, Nunez JK, Doudna JA. Biology and applications of CRISPR systems: harnessing nature's toolbox for genome engineering. Cell 2016;164:29–44. [DOI] [PubMed] [Google Scholar]
- Yosef I, Goren MG, Qimron U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res 2012;40:5569–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Gootenberg JS, Abudayyeh OO et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 2015;163:759–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Heidrich N, Ampattu BJ et al. Processing-independent CRISPR RNAs limit natural transformation in Neisseria meningitidis. Mol Cell 2013;50:488–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Rajan R, Seifert HS et al. DNase H Activity of Neisseria meningitidis Cas9. Mol Cell 2015;60:242–55. [DOI] [PMC free article] [PubMed] [Google Scholar]