Abstract
In this study, we investigated the hemocytes’ immune response to white spot syndrome virus (WSSV) or Vibrio alginolyticus infection at the protein level. The differential proteomes from crab hemocytes infected with WSSV or V. alginolyticus were analyzed using the isobaric tags for relative and absolute quantitation approach immediately after infection. Using this approach, we identified 1,799 proteins by their by LC–MS/MS spectra and sequencing data. These included 157 upregulated proteins and 164 downregulated proteins after WSSV infection. Similarly, 243 proteins were determined to be differentially expressed during V. alginolyticus infection, of these, 121 were upregulated and 122 were downregulated after infection. Interestingly, among these differentially expressed proteins, 106 were up- or downregulated significantly in both WSSV and V. alginolyticus infection. Six genes, β-actin, myosin-9, anti-lipopolysaccharide factor isoform 4, anti-lipopolysaccharide factor 4, transketolase-like protein 2-like isoform 1, and sarcoplasmic calcium-binding protein 1 were chosen for further study. The expression of these genes all showed a trend of upregulation at 24 h post-WSSV or V. alginolyticus infection except for myosin-9 in response to WSSV. To confirm the protective effects of the six genes, crabs were injected with specific dsRNAs before WSSV or V. alginolyticus challenge. The results showed that the knockdown of these genes led to an increase in the morbidity and mortality (P < 0.01) rate, and a decrease in infection time in WSSV-infected crabs. During the first 84 h, knockdown of these genes also led to an increase in the morbidity rates in V. alginolyticus -infected crabs, and results of four genes showed a higher mortality rate than that of the control after they were knocked down. This is the first report of the proteome response in crab hemocytes during WSSV or V. alginolyticus infection. These findings will contribute to our understanding of the immune response to WSSV and V. alginolyticus infection in crabs.
Keywords: proteomic, hemocyte, Scylla paramamosain, white spot syndrome virus, Vibrio alginolyticus, isobaric tags for relative and absolute quantitation
Introduction
The mud crab Scylla paramamosain, one of a number of commercially important crustaceans, is widely distributed along the coast line of southern China and the broader Pacific area. Little is known about the disease burden of these animals in the juvenile or adult phases of S. paramamosain, but susceptibility to infection has been identified during the larval stages. Several bacterial and viral pathogens have been reported to infect crabs (1). At present, bacterial infections are the main cause of crab disease and constraint in the crab farming industry in China (2, 3). In aquaculture, Vibrio alginolyticus is an important bacterial pathogen that would cause huge economic losses and has caused many cases of natural infection in the Chinese mitten crab (4). White spot syndrome has become the most hazardous and devastating disease in shrimp culture (5). White spot syndrome virus (WSSV) can also infect several species of crab both in the natural and experimental setting (6–8).
As we know, crustaceans including crabs only have an innate immune system to defend against invading microbes (9). The innate immune system includes phagocytosis, encapsulation, melanization, coagulation, and the release of antimicrobial peptides (AMPs) in crustaceans (9, 10). The hemocytes are the main site in crustaceans where the immune defense is mounted. Proteomics have been widely used to investigate the molecular mechanisms underlying the immune response to WSSV infection in crustaceans but there is no research available on V. alginolyticus infection yet (11, 12). Currently an isobaric tag-based methodology for peptide relative quantification [isobaric tags for relative and absolute quantitation (iTRAQ)] coupled to multidimensional liquid chromatography and tandem mass spectrometry enables the assessment of protein levels where four samples can be compared for their common effects (13). iTRAQ analysis has been performed to reveal the effects of Spiroplasma eriocheiris infection in Chinese mitten crab (Eriocheir sinensis) hemocytes (14). Crab hemocytes have been confirmed as the major target cells in WSSV or V. alginolyticus infection. The aim of this study was to investigate the hemocytes’ immune response to WSSV or V. alginolyticus infection at the protein level.
Experimental Procedures
Crab and Tissue Preparation
Healthy crabs were kept in a 50 L tank containing sea water with an air pump at room temperature in our laboratory. The crabs were bound with nylon rope quickly before injection and bleeding. WSSV was prepared and used immediately for the challenge according to a previous report (15). V. alginolyticus was cultured and used to challenge crabs according to the method published in a previous report (16). The hemocytes of pathogen-challenged crabs were collected for RNA isolation immediately and kept on ice to prevent RNA degradation.
Pathogen Challenge and Mortality Count
For the pathogen challenge, healthy crabs were randomly distributed into three groups (n = 10 per group). All groups received injections in the third walking foot base of crabs. The control group received injections of phosphate-buffered saline (PBS) alone, the WSSV/V. alginolyticus groups received injections of 100 µL WSSV (105 copies/mL)/V. alginolyticus (1.5 × 106/mL) in PBS, and the dsRNA + WSSV/V. alginolyticus groups received injections of dsRNA (50 µg per crab) and 100 µL WSSV (105 copies/mL)/V. alginolyticus (1.5 × 106/mL). The mortality data were arranged and analyzed in Microsoft Excel 2003.
Protein Preparation
Crab hemocytes lysates were prepared according to the previous report (14). The proteins in the supernatant were kept at −80°C for further analysis.
iTRAQ Labeling and Strong Cation Exchange (SCX) Fractionation
For iTRAQ assays, two biological replicates of each group were prepared for the proteomics experiments. Total protein (100 µg) was taken out of each sample solution, and then the protein was digested with Trypsin Gold (Promega, Madison, WI, USA) with the ratio of protein:trypsin = 30:1 at 3°C for 16 h. After trypsin digestion, peptides were dried by vacuum centrifugation. Peptides were reconstituted in 0.5 M tetraethyl ammonium bromide and processed according to the manufacture’s protocol for 8-plex iTRAQ reagent (Applied Biosystems, CA, USA). Briefly, 1 U of iTRAQ reagent was thawed and reconstituted in 24 µL isopropanol. Samples were labeled with the iTRAQ tags. The peptides were labeled with the isobaric tags, incubated at room temperature for 2 h. The labeled peptide mixtures were then pooled and dried by vacuum centrifugation. SCX chromatography was performed with an LC-20AB HPLC Pump system (Shimadzu, Kyoto, Japan). The iTRAQ-labeled peptide mixtures were reconstituted with 4 mL buffer A [25 mM NaH2PO4 in 25% acetonitrile (ACN), pH 2.7] and loaded onto a 4.6 mm × 250 mm Ultremex SCX column containing 5 µm particles (Phenomenex, Torrance, CA, USA). The peptides were eluted at a flow rate of 1 mL/min with a gradient of buffer A for 10 min, 5–60% buffer B (25 mM NaH2PO4, 1 M KCl in 25% ACN, pH 2.7) for 27 min, 60–100% buffer B for 1 min. The system was then maintained at 100% buffer B for 1 min before equilibrating with buffer A for 10 min prior to the next injection. Elution was monitored by measuring the absorbance at 214 nm, and fractions were collected every 1 min. The eluted peptides were pooled into 20 fractions, desalted with a Strata X C18 column (Phenomenex, Torrance, CA, USA) and vacuum dried.
LC–ESI–MS/MS Analysis Based on Triple TOF 5600
Each fraction was resuspended in buffer C [5% ACN, 0.1% formaldehyde (FA)] and centrifuged at 20,000 g for 10 min, the final concentration of peptide was about 0.5 µg/µL on average. Ten microliters of supernatant were loaded on an LC-20AD nanoHPLC (Shimadzu, Kyoto, Japan) by the autosampler onto a 2 cm C18 trap column (monolithic silica capillary column; Waters, USA). Then, the peptides were eluted onto a 10 cm analytical C18 column (monolithic silica capillary column; inner diameter 75 µm, Waters, USA) packed in-house. The samples were loaded at 8 µL/min for 4 min, then the 35 min gradient was run at 300 nL/min starting from 2 to 35% B (95% ACN, 0.1% FA), followed by 5 min linear gradient to 60%, then, followed by 2 min linear gradient to 80%, and maintenance at 80% B for 4 min, and finally return to 5% in 1 min. Data acquisition was performed with a TripleTOF 5600 System (AB SCIEX, Concord, ON, Canada) fitted with a Nanospray III source (AB SCIEX, Concord, ON, Canada) and a pulled quartz tip as the emitter (New Objectives, Woburn, MA, USA). Data were acquired using an ion spray voltage of 2.5 kV, curtain gas of 0.2 MPa, nebulizer gas of 0.1 MPa, and an interface heater temperature of 150. The MS was operated with an RP (reverse phase) of greater than or equal to 30,000 full width at half maximum for TOF MS scans. For isotope dilution analysis, survey scans were acquired in 250 ms and as many as 30 product ion scans were collected if exceeding a threshold of 120 counts per second (counts/s) and with a 2+ to 5+ charge state. Total cycle time was fixed to 3.3 s. Q2 transmission window was 100 Da for 100%. The signals from four-anode detectors were summed for each scan at a pulser frequency value of 11 kHz through monitoring of the 40 GHz multichannel TDC detector with four-anode channel detect ion. A sweeping collision energy setting of 35 ± 5 eV coupled with iTRAQ adjust rolling collision energy was applied to all precursor ions for collision-induced dissociation. Dynamic exclusion was set for 1/2 of peak width (15 s), and then the precursor was refreshed off the exclusion list.
Data Analysis
Raw data files acquired from the Orbitrap were converted into MGF files using Proteome Discoverer 1.2 (PD 1.2, Thermo Fisher Scientific, Hudson, USA), 5,600 ms converter and the MGF files were searched. Protein identification was performed using the Mascot search engine (Matrix Science, London, UK, version 2.3.02). For protein identification, a mass tolerance of 0.05 Da (ppm) was permitted for intact peptide masses and 0.1 Da for fragmented ions, with allowance for one missed cleavages in the trypsin digests. Gln → pyro-Glu (N-term Q), oxidation (M), deamidated (NQ) as the potential variable modifications, and carbamidomethyl (C), iTRAQ8plex (N-term), iTRAQ8plex (K) as fixed modifications. The charge states of peptides were set to +2 and +3. Specifically, an automatic decoy database search was performed in Mascot by choosing the decoy checkbox in which a random sequence of database is generated and tested for raw spectra as well as the real database. To reduce the probability of false peptide identification, only peptides with significance scores (geq20) at the 99% confidence interval by a Mascot probability analysis greater than “identity” were counted as identified. And each confident protein identification involves at least one unique peptide. For protein quantitation, it was required that a protein contains at least two unique peptides. The quantitative protein ratios were weighted and normalized by the median ratio in Mascot. We only used ratios with P-values < 0.05, and only fold changes of >1.2 was considered as significant. Functional annotations of the proteins were conducted using Blast 2 GO program against the non-redundant protein database (NR; NCBI). The Kyoto Encyclopedia of Genes and Genomes (KEGG) database1 and the Cluster of Orthologous Groups of proteins (COG) database2 were used to classify and group these identified proteins. Gene ontology (GO) is an international standardization of gene function classification system, and it provides a set of dynamic updating controlled vocabulary to describe genes and gene products attributes in the organism. GO has three ontologies, which can describe molecular function, cellular component, biological process, respectively. COG is the database for protein orthologous classification. Proteins in COG are grouped by common ancestry. KEGG pathway is a collection of manually drawn pathway maps representing our knowledge on the molecular interaction and reaction networks. Molecules are represented as nodes, and the biological relationship between two nodes is represented as an edge.
Expression Analysis by Real-time Polymerase Chain Reaction (PCR)
The expression levels of a gene in multiple organs of both healthy and pathogen-challenged crabs were analyzed by real-time quantitative PCR using SYBR green. The procedure was done according to the published method described previously (17). And the primers were shown in Table S1 in Supplementary Material.
Prokaryotic Expression and Purification of dsRNA
The primers (shown in Table S1 in Supplementary Material) with specific restriction sites (HindIII in the forward primer and BamHI in the reverse primer) were designed from the cloned nucleotide sequence. PCR product digested with HindIII/BamHI was subcloned into LITMUS 38i Vector (New England Biolabs, UK) digested with the same enzymes. The constructed plasmid was verified by restriction enzyme digestion [TLP, anti-lipopolysaccharide factor isoform 4 (ALF iso4), and myosin are BamHI/HindIII; SCP, β-actin, and anti-lipopolysaccharide factor 4 (ALF 4) are EcoRI/SalI]. The recombinant plasmid was transformed into HT115 (DE3) E. coli strain. Single colonies of the above the engineering bacteria were separately inoculated to 5 mL of LB medium containing Amp (100 µg/mL), cultured at 37°C with shaking at 200 rpm for 12–16 h, and then inoculated to LB medium containing Amp by a proportion of 1%, cultured at 37°C with shaking at 200 rpm for 2–3 h (OD600 = 0.6), and added with IPTG (with a final concentration of 0.8 mM) to induce the expression for 4 h. Then the dsRNAs were extracted and purified from the bacteria.
Statistical Analysis
Quantitative data were expressed as mean ± SD. The statistical differences were estimated by one-way analysis of variance followed by least-significant differences and Duncan’s multiple range test. All statistical analyses were carried out using SPSS Statistics version 19. A probability level of 0.01 was used to indicate statistical significance (P < 0.01).
Results
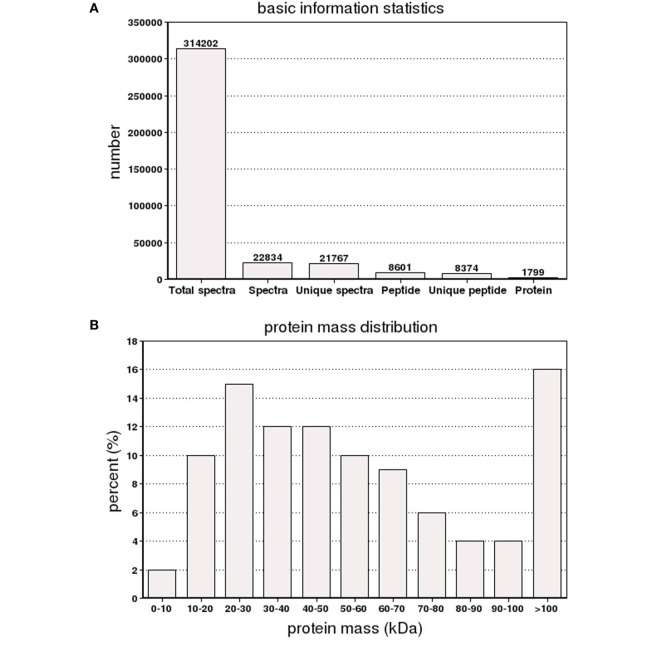
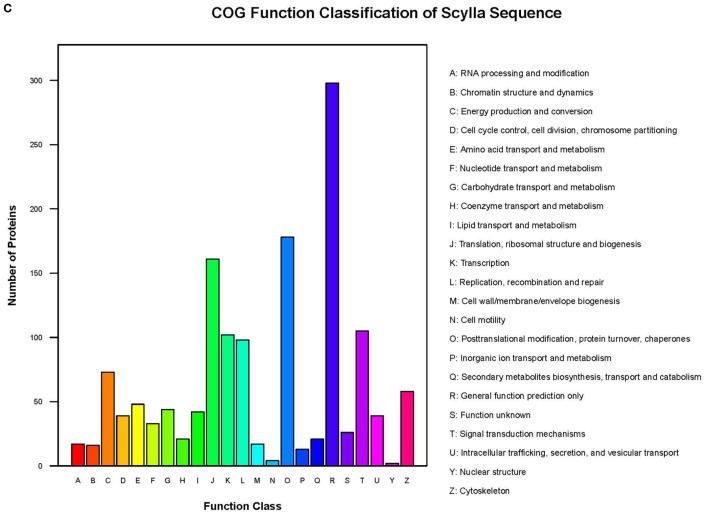
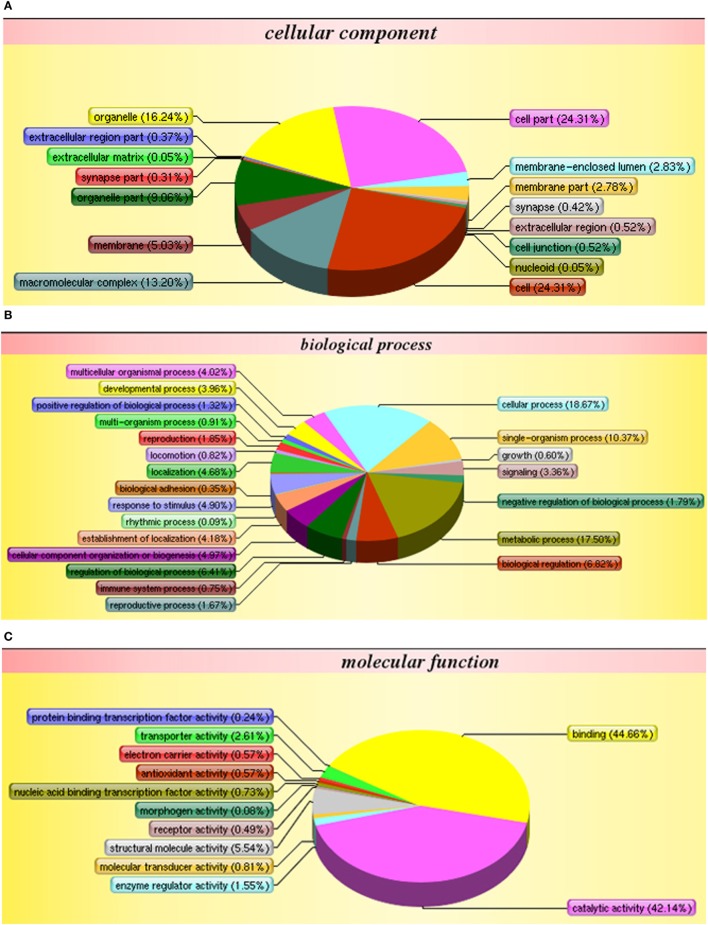
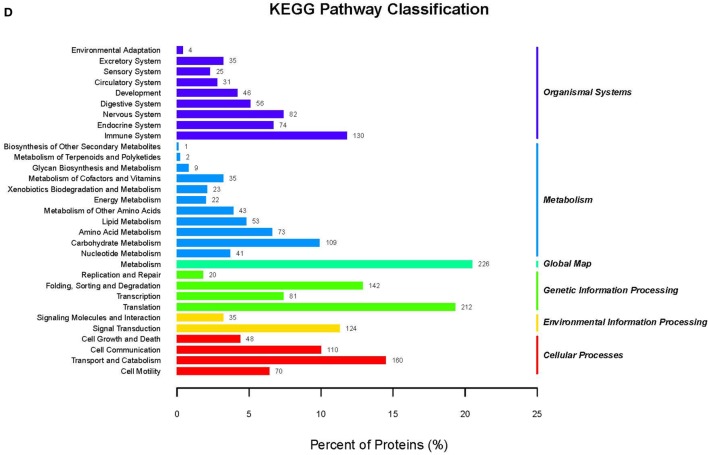
The iTRAQ analysis of S. paramamosain hemocytes proteome showed 22, 834 queries in the database (314,202 sequences) and 1,799 proteins were identified from LC–MS/MS spectra and peptides (Figure 1A). The protein mass distribution showed that protein mass above 100 kDa occupied 16% and protein mass between 10 and 60 kDa occupied 59% (Figure 1B) of the identified proteome. In an effort to analyze the functional distribution of the identified proteins, the known proteins from S. paramamosain hemocytes were used as a reference in the COG database (Figure 1C). The functional classification of the S. paramamosain sequences showed that the most common function was R: general function prediction only, which indicated that many sequences had an unknown function. The second function was O: posttranslational modification, protein turnover, chaperones, the third function was J: translation, ribosomal structure, and biogenesis. KEGG pathway classification showed that the major organismal system was the metabolism that contained 226 identified proteins, and the major metabolic pathway was carbohydrate metabolism (109 proteins). The second pathway was translation (212 proteins), which represents genetic information processing, and the third pathway was transport and catabolism (160 proteins), which represents cellular processes. The cellular component identified by GO analysis is depicted in Figure 2A. GO analyses showed that the peptides can be categorized into several biological process, i.e., cellular process (18.67%), metabolic process (17.50%), single-organism process (10.37%), and the following two regulatory stages: biological regulation (6.82%) and regulation of biology processes (6.41%) (Figure 2B). The major molecular functions identified by GO analysis of the peptides were binding (44.66%) and catalytic activity (42.14%) (Figure 2C). The KEGG pathway classification of proteins in hemocytes from healthy crabs showed that metabolic pathways and translation are very important (Figure 2D).
Figure 1.
The basic information and statistics for the isobaric tags for relative and absolute quantitation analysis of Scylla paramamosain hemocyte proteome. (A) Coverage of proteins by the identified peptides. (B) Distribution of identified proteins among different molecular weights (in kilodaltons). (C) Distribution of proteins containing different number of identified peptides.
Figure 2.
Gene ontology analysis of proteins in hemocytes of healthy crabs based on cellular component (A), biological process (B), molecular function (C), and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway classification (D) of proteins in the hemocytes of healthy crabs.
The crabs were challenged with WSSV or V. alginolyticus, and then the samples were used for iTRAQ analysis and compared with the control group. We applied a 1.2-fold (P < 0.05) increase or decrease as a threshold for significant differences in protein expression to qualify a physiologically important change, 321 proteins were shown to meet this criteria for differential expression, including 157 upregulated proteins and 164 downregulated proteins after WSSV infection (Figures 3A,B). A further 243 proteins showed a 1.2-fold (P < 0.05) increase or decrease in protein expression during V. alginolyticus infection including, 121 upregulated proteins and 122 downregulated proteins. WSSV infection affected more protein expression in crab hemocytes than V. alginolyticus infection. Interestingly, 106 differentially expressed proteins were common to both infections (Figure 3B). Of these, copine-8, troponin C isoform 2b, and sarcoplasmic calcium-binding protein 1 were upregulated significantly, and Sacsin, ALF 4, ALF iso4, and Pseudomonas aeruginosa pathogenicity island (PAPI) I were downregulated significantly (Tables 1–3). The data indicate that these proteins may participate in the immune response to WSSV infection and V. alginolyticus infection. These 106 proteins can be categorized into some important pathways including carbon metabolism, ribosome, mitogen-activated protein kinase (MAPK) signaling pathway, Ras signaling pathway, phagosome, hippo signaling pathway among others (data not shown). The major molecular functions determined by GO analysis for these proteins were binding, catalytic activity, and organelle components (Figure 3C).
Figure 3.
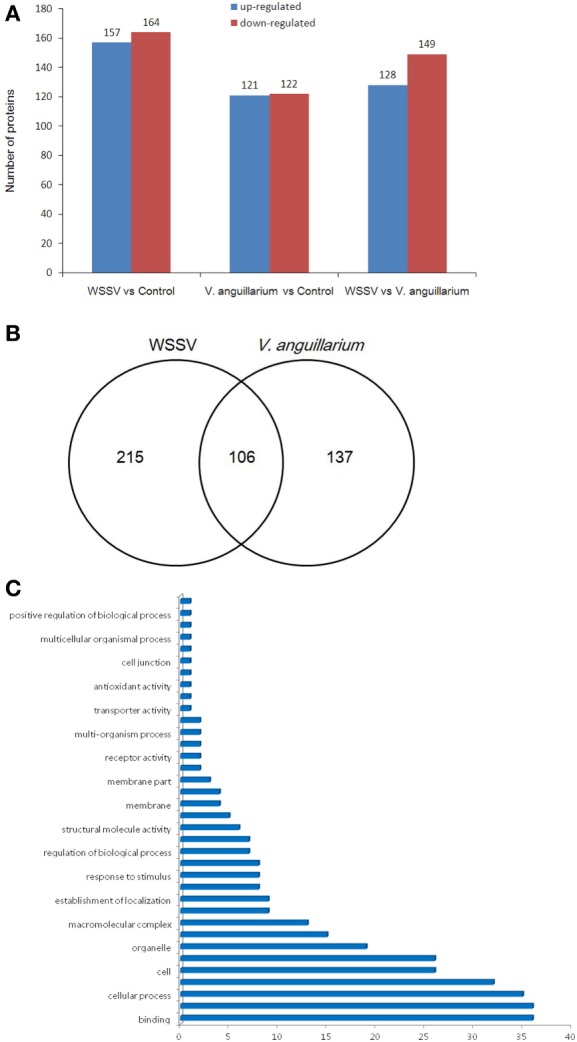
The differentially expressed proteins were evaluated by isobaric tags for relative and absolute quantitation analysis. The numbers represent the proteins that were upregulated or downregulated (more than 1.2-fold) compared with the control or Vibrio alginolyticus group (A). Venn diagram of differentially expressed genes (B). The numbers represented the proteins upregulated or downregulated (more than 1.2-fold) compared with the control. The statistics of gene ontology enrichment of the 106 proteins up- or downregulated significantly in white spot syndrome virus (WSSV) and V. alginolyticus infection (C).
Table 1.
The differential expressed proteins in Scylla paramamosain hemocytes with above two-fold change post injection with white spot syndrome virus.
| Protein name | Accession | Score | Coverage | Peptide | Fold change |
|---|---|---|---|---|---|
| Upregulated proteins | |||||
| Copine-8 | comp20718_c0_seq2_No.1 | 841 | 30.9 | 14 | 6.29 |
| Troponin C isoform 2b | comp22592_c0_seq1_No.1 | 69 | 26.3 | 3 | 5.35 |
| Sarcoplasmic calcium-binding protein 1 | comp40442_c0_seq1_No.1 | 83 | 16.1 | 3 | 4.78 |
| β-Actin | comp12583_c0_seq1_No.1 | 1,377 | 67.2 | 4 | 4.02 |
| Myosin light chain 2 | comp21132_c0_seq1_No.1 | 765 | 34.1 | 9 | 3.58 |
| Slow muscle myosin S1 heavy chain | comp20546_c0_seq16_No.1 | 2,058 | 43.7 | 26 | 2.69 |
| clathrin light chain | comp17823_c1_seq1_No.1 | 147 | 22.5 | 4 | 2.67 |
| Ubiquitin carboxyl-terminal esterase L3 | comp16837_c1_seq1_No.1 | 396 | 31.6 | 6 | 2.65 |
| 2-Phospho-d-glycerate hydrolase | comp20783_c0_seq1_No.1 | 1,685 | 50.7 | 18 | 2.65 |
| Membrane-associated protein | comp51893_c0_seq1_No.1 | 281 | 15.4 | 2 | 2.64 |
| Alpha actin | comp12775_c1_seq1_No.1 | 1,274 | 57 | 5 | 2.56 |
| Spermatogonial stem-cell renewal factor | comp72221_c0_seq1_No.1 | 240 | 41.9 | 9 | 2.48 |
| Inorganic pyrophosphatase-like protein | comp12817_c0_seq1_No.1 | 216 | 25.3 | 5 | 2.44 |
| Putative phosphoglycerate kinase | comp12041_c0_seq1_No.1 | 897 | 33.5 | 13 | 2.4 |
| Protein kinase c | comp18689_c0_seq1_No.1 | 185 | 13.4 | 8 | 2.36 |
| Glutathione S-transferase | comp15053_c0_seq1_No.1 | 1,634 | 50.5 | 10 | 2.19 |
| Myosin-9 | comp12936_c0_seq1_No.1 | 382 | 18.3 | 13 | 2.1 |
| Myosin heavy chain type 1 | comp20546_c0_seq7_No.1 | 186 | 38.8 | 2 | 2.04 |
| Downregulated proteins | |||||
| Sacsin | comp19443_c0_seq1_No.1 | 42 | 0.2 | 1 | 8.76 |
| Pacifastin-like serine protease inhibitor | comp3991_c0_seq1_No.1 | 82 | 15.1 | 2 | 5.45 |
| Male reproductive tract-specific Kazal-type proteinase inhibitor | comp19421_c0_seq1_No.1 | 164 | 11.7 | 1 | 3.77 |
| Pseudomonas aeruginosa pathogenicity island I | comp20408_c0_seq1_No.1 | 54 | 7.6 | 1 | 3.1 |
| Anti-lipopolysaccharide factor isoform 5 | comp12264_c0_seq1_No.1 | 179 | 27.6 | 2 | 2.59 |
| Putative protein phosphatase | comp18826_c0_seq1_No.1 | 117 | 16.4 | 2 | 2.52 |
| Crustin antimicrobial peptide | comp12292_c0_seq1_No.1 | 462 | 35 | 2 | 2.47 |
| Glutathione-dependent prostaglandin D synthase | comp16103_c0_seq1_No.1 | 133 | 25.1 | 5 | 2.42 |
| Cuticular protein analogous to peritrophins 3 | comp12594_c0_seq1_No.1 | 147 | 9.3 | 2 | 2.4 |
| 60S acidic ribosomal protein P2-like protein | comp21230_c0_seq1_No.1 | 292 | 66.7 | 5 | 2.2 |
| Glutathione peroxidase 6 precursor | comp10211_c0_seq1_No.1 | 157 | 17.7 | 4 | 2.18 |
| Chain A, crystal structure of Chmp4b hairpin | comp21559_c0_seq1_No.1 | 356 | 35.3 | 5 | 2.15 |
| Calreticulin precursor | comp14413_c0_seq1_No.1 | 194 | 15.8 | 6 | 2.12 |
| RIKEN | comp2503_c0_seq1_No.1 | 47 | 11.3 | 2 | 2.08 |
| Lactate dehydrogenase | comp19912_c0_seq3_No.1 | 87 | 16.9 | 5 | 2.03 |
| Heat shock protein 90 | comp10311_c0_seq1_No.1 | 332 | 18.5 | 12 | 2 |
Table 3.
The selected proteins in Scylla paramamosain hemocytes with over two-fold change post infection.
| Protein name | Accession | Score | Coverage | Peptide | Fold change |
|
|---|---|---|---|---|---|---|
| White spot syndrome virus | VA | |||||
| Cytoskeleton/extracellular proteins | ||||||
| β-Actin | comp12583_c0_seq1_No.1 | 1,377 | 67.2 | 4 | +4.02 | –1.23 |
| Myosin-9 | comp12936_c0_seq1_No.1 | 382 | 18.3 | 13 | +2.11 | +1.96 |
| Immunologic proteins | ||||||
| Anti-lipopolysaccharide factor 4 | comp12264_c0_seq1_No.1 | 179 | 27.6 | 2 | –2.59 | –2.82 |
| Anti-lipopolysaccharide factor isoform 4 | comp20686_c1_seq1_No.1 | 126 | 47.7 | 4 | –1.86 | –2.12 |
| Physiologic proteins | ||||||
| Transketolase-like protein 2-like isoform 1 | comp18784_c0_seq2_No.1 | 3,418 | 44.3 | 21 | –2.19 | –2.2 |
| Sarcoplasmic calcium-binding protein 1 | comp40442_c0_seq1_No.1 | 83 | 16.1 | 3 | +4.78 | +8.4 |
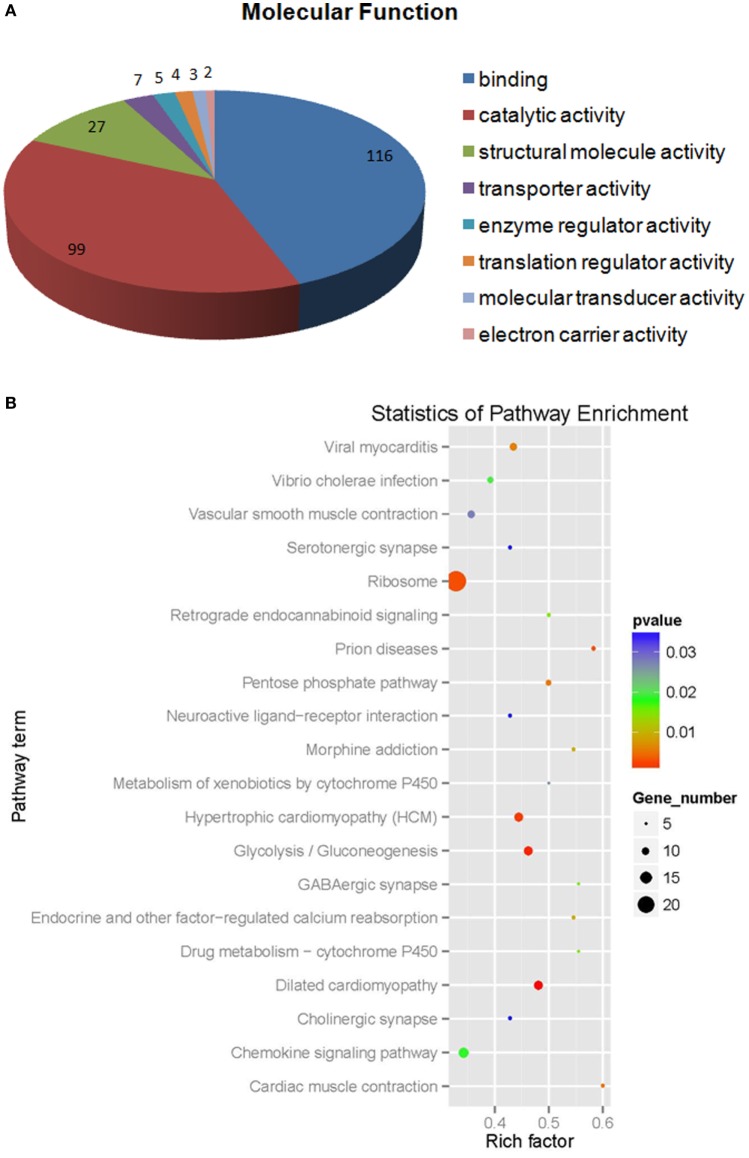
Of the differentially expressed proteins in WSSV infection, the GO enrichment scatterplot showed that the organelle and intracellular organelle proteins were the most important, and a KEGG enrichment scatterplot indicated that metabolic pathways were the most important (Figure 4; Figure S1A in Supplementary Material). The differentially expressed proteins (over twofold) and the related iTRAQ data are presented in Table 1. The upregulated proteins included copine-8 with 6.29-fold change, troponin C isoform 2b with 5.35-fold change, sarcoplasmic calcium-binding protein 1 with 4.78-fold change, beta-actin with 4.02-fold change, myosin light chain 2 with 3.58-fold change, slow muscle myosin S1 heavy chain with 2.69-fold change, clathrin light chain with 2.67-fold change, ubiquitin carboxyl-terminal esterase L3 with 2.65-fold change, 2-phospho-d-glycerate hydrolase with 2.65-fold change, membrane-associated protein with 2.64-fold change, alpha actin with 2.56-fold change, myosin-9 with 2.1-fold change, myosin heavy chain type 1with 2.04-fold change and some enzymes with above a twofold change (Table 1). The data indicate that many endocytosis-related cytoskeleton proteins of host hemocytes contribute to WSSV infection. The upregulated proteins and the related iTRAQ data are presented in Supplementary Material Table S1. The downregulated proteins included Sacsin with 8.76-fold change, pacifastin-like serine protease inhibitor with 5.45-fold change, male reproductive tract-specific Kazal-type proteinase inhibitor with 3.77-fold change, PAPI-1 with 3.1-fold change, ALF 4 with 2.59-fold change, crustin AMP with 2.47-fold change, heat shock protein 90 with twofold change and some enzymes with above a twofold change.
Figure 4.
The molecular function (A) statistics of pathway enrichment (B) of differential expressed proteins (more than 1.2-fold) from white spot syndrome virus group vs control group.
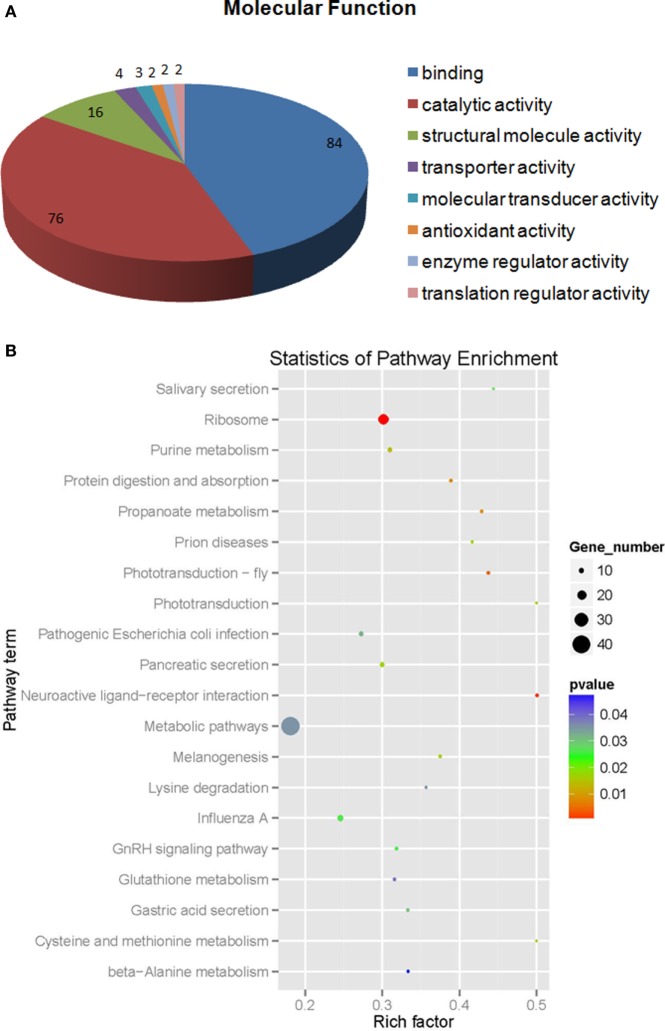
Of the differentially expressed proteins in V. alginolyticus infection, the GO enrichment scatterplot showed that the organelle and intracellular organelle are the most important, and a KEGG enrichment scatterplot indicated that metabolic pathways were the most important (Figure 5; Figure S1B in Supplementary Material). The differentially expressed proteins (over twofold) and the related iTRAQ data are presented in Table 2. The upregulated proteins included sarcoplasmic calcium-binding protein 1 with 8.4-fold change, troponin C isoform 2b with 2.78-fold change, clip domain serine proteinase 3 with 2.4-fold change, putative secreted salivary gland peptide with 2.23-fold change, copine-8 with 2.17-fold change, slow muscle myosin S1 heavy chain with 2.13-fold change, chloride intracellular channel isoform 1 with 2.11-fold change, and myophilin with 2.03-fold change (Table 2). The downregulated proteins included Sacsin with 31.39-fold change, phospholipase D1 with 6.97-fold change, low-density lipoprotein receptor with 5.77-fold change, PAPI I with 3.02-fold change, pacifastin-like serine protease inhibitor with 2.99-fold change, ALF 4 with 2.82-fold change and ALF iso4 with 2.12-fold change. It was showed that intracellular non-membrane-bounded organelle is the most important in statistics of GO enrichment of differentially expressed proteins (more than 1.2-fold) from WSSV vs V. alginolyticus, and phagosome is the most important in statistics of GO enrichment (Figure S2 in Supplementary Material).
Figure 5.
The molecular function (A) statistics for pathway enrichment (B) of differentially expressed proteins (more than 1.2-fold) from Vibrio alginolyticus vs control.
Table 2.
The differential expressed proteins in Scylla paramamosain hemocytes with above two-fold change post injection with V. anguillarum.
| Protein name | Accession | Score | Coverage | Peptide | Fold change |
|---|---|---|---|---|---|
| Upregulated proteins | |||||
| Sarcoplasmic calcium-binding protein 1 | comp40442_c0_seq1_No.1 | 83 | 16.1 | 3 | 8.40 |
| Troponin C isoform 2b | comp22592_c0_seq1_No.1 | 69 | 26.3 | 3 | 2.78 |
| Clip domain serine proteinase 3 | comp19522_c0_seq1_No.1 | 520 | 33.8 | 9 | 2.40 |
| Copine-8 | comp20718_c0_seq2_No.1 | 841 | 30.9 | 14 | 2.19 |
| Slow muscle myosin S1 heavy chain | comp20546_c0_seq16_No.1 | 2,058 | 43.7 | 26 | 2.13 |
| Putative secreted salivary gland peptide | comp12272_c1_seq1_No.1 | 83 | 22 | 3 | 2.23 |
| Chloride intracellular Channel isoform 1 | comp20820_c0_seq1_No.1 | 100 | 25.7 | 5 | 2.11 |
| Myophilin | comp18304_c1_seq2_No.1 | 1,662 | 62.4 | 8 | 2.03 |
| Spermine synthase | comp18763_c0_seq1_No.1 | 200 | 16.1 | 4 | 2.02 |
| Putative phosphoglycerate kinase | comp12041_c0_seq1_No.1 | 897 | 33.5 | 13 | 2.02 |
| Downregulated proteins | |||||
| Sacsin | comp19443_c0_seq1_No.1 | 42 | 0.2 | 1 | 31.39 |
| Phospholipase D1 | comp1157_c0_seq1_No.1 | 211 | 17.4 | 5 | 6.97 |
| Low-density lipoprotein receptor | comp20609_c0_seq1_No.1 | 283 | 10.2 | 6 | 5.77 |
| Pseudomonas aeruginosa pathogenicity island I | comp20408_c0_seq1_No.1 | 54 | 7.6 | 1 | 3.02 |
| pacifastin-like serine protease inhibitor | comp3991_c0_seq1_No.1 | 82 | 15.1 | 2 | 2.99 |
| Anti-lipopolysaccharide factor 4 | comp12264_c0_seq1_No.1 | 179 | 27.6 | 2 | 2.82 |
| Anti-lipopolysaccharide factor isoform 4 | comp20686_c1_seq1_No.1 | 126 | 47.7 | 4 | 2.12 |
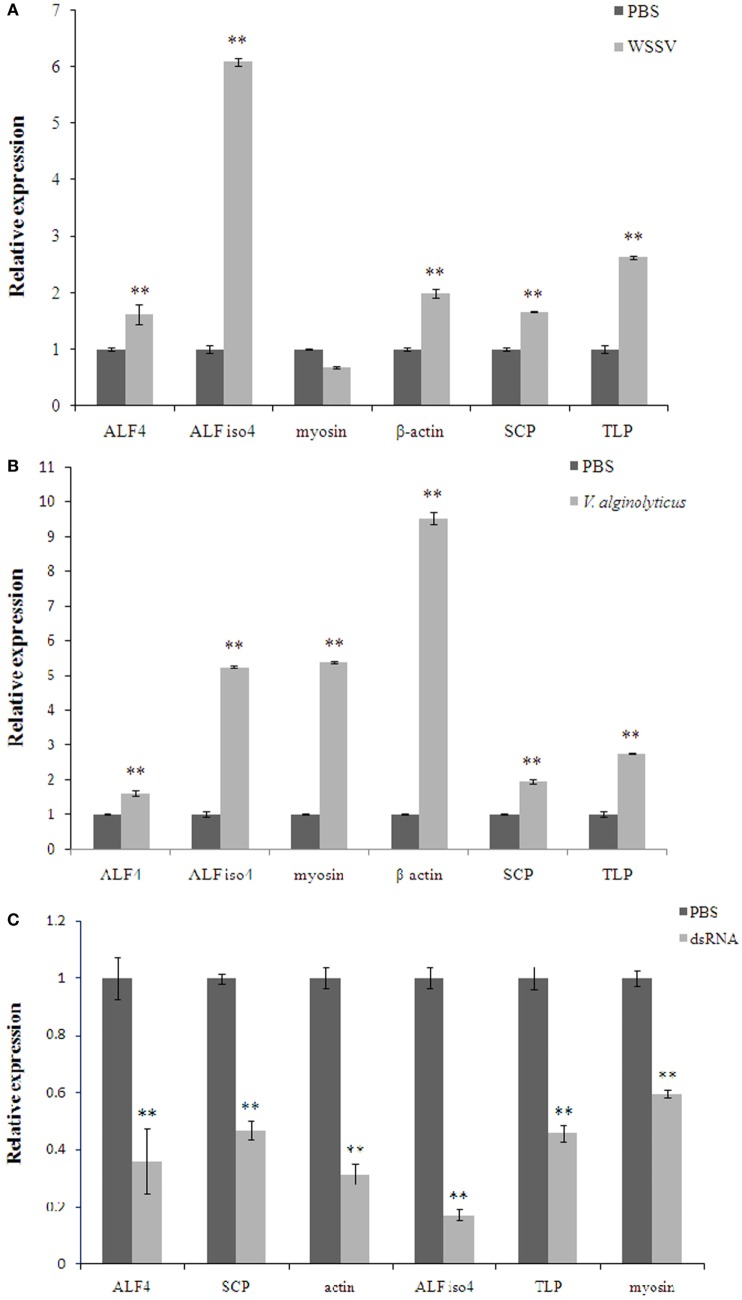
Interestingly, some proteins were changed significantly (P < 0.01) in WSSV and V. alginolyticus infection (Table S2 in Supplementary Material; Table 3). Six differentially expressed genes, β-actin, myosin-9, ALF iso4, ALF 4, transketolase-like protein 2-like isoform 1, and sarcoplasmic calcium-binding protein 1 were characterized by real-time PCR assay (Figures 6A,B). The expression of these six genes showed a trend of upregulation at 24 h post WSSV or V. alginolyticus challenge except for that of myosin-9 in response to WSSV. Only β-actin and sarcoplasmic calcium-binding protein 1 showed the same trend in the proteomic results. dsRNA silencing results showed that when the dsRNA was present expression of these six genes was significantly decreased (P < 0.01) (Figure 6C).
Figure 6.
The expression of the selected genes in Scylla paramamosain hemocytes in response to white spot syndrome virus (WSSV) (A), Vibrio alginolyticus (B), and specific dsRNA (C). Data are shown as means ± SD of three separate individuals in the tissues. Capital letters indicate expression of crustin-like different adult tissues. Double asterisks indicate a significant difference (P < 0.01) between two samples. ALF 4, anti-lipopolysaccharide factor 4; ALF iso4, anti-lipopolysaccharide factor isoform 4; myosin-9, myosin; TLP, transketolase-like protein 2-like isoform 1; SCP, sarcoplasmic calcium-binding protein 1.
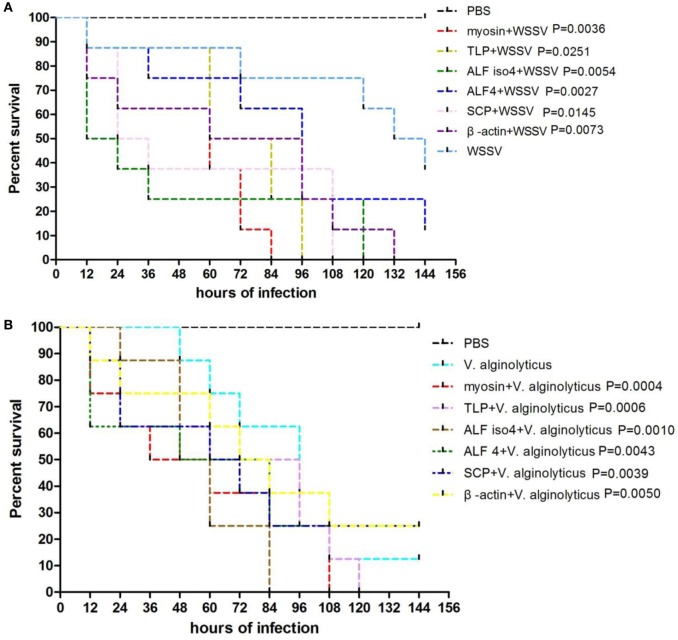
To confirm the protective effects of these six genes, S. paramamosain were injected intramuscularly with specific dsRNAs before WSSV or V. alginolyticus challenge. There were 16 crabs in each group and experiments were conducted for 144 hours. The control group was injected with PBS solution followed by WSSV challenge. Each point represents the means of a triplicate assays within the SD. Then we tested the mortality rates of crabs when they were infected by WSSV or V. alginolyticus. The data showed that the knockdown of these six genes led to a faster morbidity rate in the WSSV-infected crabs and significantly (P < 0.01) increased the mortality of crabs compared with that of the control WSSV group (Figure 7A). In the first 84 h, knockdown of these six genes also led to a faster morbidity rate in the V. alginolyticus-infected crabs but did not change the mortality rate significantly when compared with that of the V. alginolyticus control group (Figure 7B). However, four genes did show a higher mortality rate than that of the control after they were knocked down.
Figure 7.
Comparison of protective effects between six selected genes. The percentage survival of challenged crabs treated with dsRNA. Juvenile Scylla paramamosain were injected intramuscularly with dsRNAs 24 h before white spot syndrome virus (WSSV) challenge (A) or Vibrio alginolyticus challenge (B). Control group was injected with phosphate-buffered saline (PBS) solution, and WSSV or V. alginolyticus group was challenged by WSSV or V. alginolyticus. The treatment is shown on the right. The P-value is the data of experimental groups compared to that of WSSV or V. alginolyticus group.
Discussion
In this study, we identified 1,799 proteins through the iTRAQ analysis of the S. paramamosain hemocytes proteome and showed that proteins with a mass between 10–60 kDa occupied 59% of all proteins in these animals. The major pathways included metabolism, 226 proteins, translation, 212 proteins, and transport and catabolism, 160 proteins. The important biological processes included cellular and metabolic processes, and the major molecular functions were binding and catalytic activity. No matter for WSSV or V. alginolyticus infection, the organelle and intracellular organelle are the most important, and metabolic pathways were the key pathways. WSSV was found to induce metabolic changes (17), and endocytosis is vital for its replication in crustacean (18). V. alginolyticus infection would damage the mitochondria of hemocytes and then cause hemocyte death in crabs (19). Interestingly, we found that WSSV infection significantly affected more proteins’ expression in host hemocytes than V. alginolyticus infection. Many cytoskeleton proteins contribute to WSSV infection indicating that phagocytosis is the most important cellular process during infection. This finding is accordance with previous reports (11, 20). Interestingly, we found that 106 proteins were differentially expressed simultaneously in WSSV infection and V. alginolyticus infection. These proteins can be categorized into some important pathways including carbon metabolism, ribosome, MAPK signaling pathway, Ras signaling pathway, phagosome, and Hippo signaling pathway. The major molecular functions of these proteins were binding, catalytic activity, and organelle composition.
Then, we chose six differentially expressed genes to investigate their expression and the protective effects against WSSV infection or V. alginolyticus infection. We found that these six genes showed a trend of upregulation at 24 h post-WSSV or V. alginolyticus challenge except for that of myosin-9 in response to WSSV, and only β-actin and sarcoplasmic calcium-binding protein 1 showed the same trend in the proteomic results. We found that the knockdown of these six genes led to an increase in morbidity and mortality in the WSSV-infected crabs when compared with that of WSSV control group. Similarly, knockdown of these six genes also led to an increased morbidity rate in the V. alginolyticus-infected crabs in the first 84 h and four genes showed an increase in the mortality rate when compared to the V. alginolyticus control group. β-Actin and myosin are known to regulate phagocytosis in shrimp and participate in the response to WSSV (11, 20, 21). Phagocytosis is also the key immune response to bacterial challenge in invertebrates (22). In this study, β-actin and myosin-9 were upregulated significantly (P < 0.01) in WSSV infection, but β-actin was downregulated and myosin-9 upregulated in V. alginolyticus infection. When β-actin or myosin-9 was knocked down during WSSV or V. alginolyticus infection, there was an increase in the morbidity and mortality rates. The data indicated that β-actin and myosin-9 play an important role in the immune response to WSSV or V. alginolyticus infection.
Sarcoplasmic calcium-binding protein 1 is believed to function as the invertebrate equivalent of vertebrate parvalbumin, namely, to “buffer” cytosolic Ca2+, and its expression is downregulated in pre- and postmolt stages compared with intermolt in crayfish (23). Sarcoplasmic calcium-binding protein is also an important allergen in the muscle and shell of Fenneropenaeus merguiensis (24). WSSV infection in Penaeus monodon is facilitated by housekeeping molecules like sarcoplasmic calcium-binding protein 1 (25). When sarcoplasmic calcium-binding protein 1 was knocked down, WSSV infection would cause the animals to die faster and more often, but in V. alginolyticus infection this would only reduce the lifespan of infected crabs but decreased the overall mortality rates. These results revealed that sarcoplasmic calcium-binding protein 1 plays an important role in the immune response to WSSV and plays a different role in the immune response to V. alginolyticus infection. The anti-lipopolysaccharide factors (ALFs) are well known for being the most common effectors affecting the invading microorganisms in crustaceans and its antimicrobial activity in killing Gram-positive and -negative bacteria and viruses (26–31). In this study, ALF 4 and ALF iso4 were upregulated significantly in WSSV and V. alginolyticus infection. When ALF iso4 was knocked down, in either WSSV or V. alginolyticus infection it resulted in a reduced lifespan an increase in the mortality rate. However, the knockdown of ALF 4 leads to increased morbidity and mortality in WSSV-infected crabs and increased morbidity but decreased mortality in V. alginolyticus-infected crabs. The transketolase is required for cancer growth because of its ability to affect the production of NAPDH to counteract oxidative stress (32). The transketolase-like protein 2-like isoform 1 was downregulated significantly in WSSV infection and V. alginolyticus infection. The knockdown of transketolase-like protein 2-like isoform 1 caused an increase in the morbidity and mortality rates following infection with either pathogen.
In this investigation, we reported on the proteome of the mud crab S. paramamosain in response to WSSV or V. alginolyticus infection. We investigated the molecular mechanisms underlying the immune response to WSSV infection in crustaceans. These findings will contribute to our understanding of the molecular mechanisms of immune response to WSSV or V. alginolyticus infection in crab hemocytes.
Ethics Statement
The animal subjects used in the present study are crab, which are invertebrates and are exempt from this requirement.
Author Contributions
FZ and BS designed experiments, analyzed experimental results, and wrote the manuscript; BS, ZhW, ZiW, and XM carried out experiments.
Conflict of Interest Statement
There are no patents, products in development, or marketed products to declare. This does not alter our adherence to all the gene policies on sharing data and materials.
Footnotes
Funding
This work was financially supported by National Natural Science Foundation of China (31370050 and 31430089) and the Major State Basic Research Development Program (2012CB114403).
Supplementary Material
The Supplementary Material for this article can be found online at http://journal.frontiersin.org/article/10.3389/fimmu.2017.00468/full#supplementary-material.
(A) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway classification of differential expressed proteins (more than 1.2-fold) from white spot syndrome virus group vs control group. (B) KEGG pathway classification of differentially expressed proteins (more than 1.2-fold) from Vibrio alginolyticus vs control.
The statistics of gene ontology (GO) enrichment (A) and statistics of pathway enrichment (B) of differentially expressed proteins (more than 1.2-fold) from white spot syndrome virus vs Vibrio alginolyticus.
Abbreviations
iTRAQ, isobaric tags for relative and absolute quantitation; WSSV, white spot syndrome virus; KEGG, Kyoto Encyclopedia of Genes and Genomes; COG, Cluster of Orthologous Groups of proteins; PCR, polymerase chain reaction; GO, gene ontology; MAPK, mitogen-activated protein kinase; PAPI, Pseudomonas aeruginosa pathogenicity island; TEAB, tetraethyl ammonium bromide; ACN, acetonitrile; FA, formaldehyde.
References
- 1.Johnson PT. Diseases caused by viruses, rickettsia, bacteria and fungi. In: Provenzano AJ, Jr, editor. The Biology of the Crustacea Pathobiology. (Vol. 6), New York, NY: Academic Press; (1983). p. 2–78. [Google Scholar]
- 2.Xia XA, Wu QY, Li YY, Wang SQ, You CH, Lin YS. Isolation and identification of two bacterial pathogens from mixed infection mud crab Scylla serrata and drug therapy. J Trop Oceanogr (2010) 29(5):103–10. 10.11978/j.issn.1009-5470.2010.05.103 [DOI] [Google Scholar]
- 3.Bonami JR, Zhang S. Viral diseases in commercially exploited crabs: a review. J Invertebr Pathol (2011) 106(1):6–17. 10.1016/j.jip.2010.09.009 [DOI] [PubMed] [Google Scholar]
- 4.Chen DW, Zhang M, Shrestha S. Compositional characteristics and nutritional quality of Chinese mitten crab (Eriocheir sinensis). Food Chem (2007) 103(4):1343–9. 10.1016/j.foodchem.2006.10.047 [DOI] [Google Scholar]
- 5.Escobedo-Bonilla CM, Alday Sanz V, Wille M, Sorgeloos P, Pensaert MB, Nauwynck HJ. A review on the morphology, molecular characterization, morphogenesis and pathogenesis of white spot syndrome virus. J Fish Dis (2008) 31(1):1–18. 10.1111/j.1365-2761.2007.00877 [DOI] [PubMed] [Google Scholar]
- 6.Vaseeharan B, Jayakumar R, Ramasamy P. PCR-based detection of white spot syndrome virus in cultured and captured crustaceans in India. Lett Appl Microbiol (2003) 37(6):443–7. 10.1046/j.1472-765X.2003.01428 [DOI] [PubMed] [Google Scholar]
- 7.Hameed AS, Balasubramanian G, Musthaq SS, Yoganandhan K. Experimental infection of twenty species of Indian marine crabs with white spot syndrome virus (WSSV). Dis Aquat Organ (2003) 57(1–2):157–61. 10.3354/dao057157 [DOI] [PubMed] [Google Scholar]
- 8.Chen LL, Lo CF, Chiu YL, Chang CF, Kou GH. Natural and experimental infection of white spot syndrome virus (WSSV) in benthic larvae of mud crab Scylla serrata. Dis Aquat Organ (2000) 40(2):157–61. 10.3354/dao040157 [DOI] [PubMed] [Google Scholar]
- 9.Vazquez L, Alpuche J, Maldonado G, Agundis C, Pereyra-Morales A, Zenteno E. Review: immunity mechanisms in crustaceans. Innate Immun (2009) 15(3):179–88. 10.1177/1753425909102876 [DOI] [PubMed] [Google Scholar]
- 10.Haug T, Kjuul AK, Stensvag K, Sandsdalen E, Styrvold OB. Antibacterial activity in four marine crustacean decapods. Fish Shellfish Immunol (2002) 12(5):371–85. 10.1006/fsim.2001.0378 [DOI] [PubMed] [Google Scholar]
- 11.Li S, Li F, Sun Z, Zhang X, Xiang J. Differentially proteomic analysis of the Chinese shrimp at WSSV latent and acute infection stages by iTRAQ approach. Fish Shellfish Immunol (2016) 54:629–38. 10.1016/j.fsi.2016.05.016 [DOI] [PubMed] [Google Scholar]
- 12.Jeswin J, Xie XL, Ji QL, Wang KJ, Liu HP. Proteomic analysis by iTRAQ in red claw crayfish, Cherax quadricarinatus, hematopoietic tissue cells post white spot syndrome virus infection. Fish Shellfish Immunol (2016) 50:288–96. 10.1016/j.fsi.2016.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics (2004) 3(12):1154–69. 10.1074/mcp.M400129 [DOI] [PubMed] [Google Scholar]
- 14.Meng Q, Hou L, Zhao Y, Huang X, Huang Y, Xia S, et al. iTRAQ-based proteomic study of the effects of Spiroplasma eriocheiris on Chinese mitten crab Eriocheir sinensis hemocytes. Fish Shellfish Immunol (2014) 40(1):182–9. 10.1016/j.fsi.2014.06.029 [DOI] [PubMed] [Google Scholar]
- 15.Zhu F, Zhang XB. The Wnt signaling pathway is involved in the regulation of phagocytosis of virus in Drosophila. Sci Rep (2013) 3:2069. 10.1038/srep02069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang M, Liu Y, Xie C, Wang WN. LvDJ-1 plays an important role in resistance against Vibrio alginolyticus in Litopenaeus vannamei. Fish Shellfish Immunol (2015) 44(1):180–6. 10.1016/j.fsi.2015.02.022 [DOI] [PubMed] [Google Scholar]
- 17.Chen IT, Aoki T, Huang YT, Hirono I, Chen TC, Huang JY, et al. White spot syndrome virus induces metabolic changes resembling the Warburg effect in shrimp hemocytes in the early stage of infection. J Virol (2011) 85(24):12919–28. 10.1128/JVI.05385-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verbruggen B, Bickley LK, van-Aerle R, Bateman KS, Stentiford GD, Santos EM, et al. Molecular mechanisms of white spot syndrome virus infection and perspectives on treatments. Viruses (2016) 8(1):23. 10.3390/v8010023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu SL, Qiu CG, Zhou W, Wang DL, Jia CY, Wang CL. Pathological analysis of hemolymphs of Charybdis japonica infected with Vibrio alginolyticus. Fish Shellfish Immunol (2013) 35(5):1577–84. 10.1016/j.fsi.2013.08.025 [DOI] [PubMed] [Google Scholar]
- 20.Wang Z, Zhu F. Minichromosome maintenance protein 7 regulates phagocytosis in kuruma shrimp Marsupenaeus japonicas against white spot syndrome virus. Fish Shellfish Immunol (2016) 55:293–303. 10.1016/j.fsi.2016.06.002 [DOI] [PubMed] [Google Scholar]
- 21.Han F, Wang Z, Wang X. Characterization of myosin light chain in shrimp hemocytic phagocytosis. Fish Shellfish Immunol (2010) 29(5):875–83. 10.1016/j.fsi.2010.07.030 [DOI] [PubMed] [Google Scholar]
- 22.Lynda M, Lan-Ezekowitz SR. Phagocytosis and comparative innate immunity: learning on the fly. Nat Rev Immunol (2008) 8(2):131–41. 10.1038/nri2240 [DOI] [PubMed] [Google Scholar]
- 23.Gao Y, Gillen CM, Wheatly MG. Molecular characterization of the sarcoplasmic calcium-binding protein (SCP) from crayfish Procambarus clarkii. Comp Biochem Physiol B Biochem Mol Biol (2006) 144(4):478–87. 10.1016/j.cbpb.2006.04.007 [DOI] [PubMed] [Google Scholar]
- 24.Khanaruksombat S, Srisomsap C, Chokchaichamnankit D, Punyarit P, Phiriyangkul P. Identification of a novel allergen from muscle and various organs in banana shrimp (Fenneropenaeus merguiensis). Ann Allergy Asthma Immunol (2014) 113(3):301–6. 10.1016/j.anai.2014.06.002 [DOI] [PubMed] [Google Scholar]
- 25.Biradar V, Narwade S, Paingankar M, Deobagkar D. White spot syndrome virus infection in Penaeus monodon is facilitated by housekeeping molecules. J Biosci (2013) 38(5):917–24. 10.1007/s12038-013-9386-8 [DOI] [PubMed] [Google Scholar]
- 26.de la Vega E, O’Leary NA, Shockey JE, Robalino J, Payne C, Browdy CL, et al. Anti-lipopolysaccharide factor in Litopenaeus vannamei (LvALF): a broad spectrum antimicrobial peptide essential for shrimp immunity against bacterial and fungal infection. Mol Immunol (2008) 45(7):1916–25. 10.1016/j.molimm.2007.10.039 [DOI] [PubMed] [Google Scholar]
- 27.Zhu L, Lan JF, Huang YQ, Zhang C, Zhou JF, Fang WH, et al. SpALF4: a newly identified anti-lipopolysaccharide factor from the mud crab Scylla paramamosain with broad spectrum antimicrobial activity. Fish Shellfish Immunol (2014) 36(1):172–80. 10.1016/j.fsi.2013.10.023 [DOI] [PubMed] [Google Scholar]
- 28.Sun W, Wan W, Zhu S, Wang S, Wang S, Wen X, et al. Characterization of a novel anti-lipopolysaccharide factor isoform (SpALF5) in mud crab, Scylla paramamosain. Mol Immunol (2015) 64(2):262–75. 10.1016/j.molimm.2014.12.006 [DOI] [PubMed] [Google Scholar]
- 29.Suraprasit S, Methatham T, Jaree P, Phiwsaiya K, Senapin S, Hirono I, et al. Anti-lipopolysaccharide factor isoform 3 from Penaeus monodon (ALFPm3) exhibits antiviral activity by interacting with WSSV structural proteins. Antiviral Res (2014) 110:142–50. 10.1016/j.antiviral.2014.08.005 [DOI] [PubMed] [Google Scholar]
- 30.Methatham T, Boonchuen P, Jaree P, Tassanakajon A, Somboonwiwat K. Antiviral action of the antimicrobial peptide ALFPm3 from Penaeus monodon against white spot syndrome virus. Dev Comp Immunol (2016) 69:23–32. 10.1016/j.dci.2016.11.023 [DOI] [PubMed] [Google Scholar]
- 31.Li S, Guo S, Li F, Xiang J. Functional diversity of anti-lipopolysaccharide factor isoforms in shrimp and their characters related to antiviral activity. Mar Drugs (2015) 13(5):2602–16. 10.3390/md13052602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu IM, Lai RK, Lin SH, Tse AP, Chiu DK, Koh HY, et al. Transketolase counteracts oxidative stress to drive cancer development. Proc Natl Acad Sci U S A (2016) 113(6):E725–34. 10.1073/pnas.1508779113 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway classification of differential expressed proteins (more than 1.2-fold) from white spot syndrome virus group vs control group. (B) KEGG pathway classification of differentially expressed proteins (more than 1.2-fold) from Vibrio alginolyticus vs control.
The statistics of gene ontology (GO) enrichment (A) and statistics of pathway enrichment (B) of differentially expressed proteins (more than 1.2-fold) from white spot syndrome virus vs Vibrio alginolyticus.