Abstract
Expression of the gene for protein‐arginine deiminase 2 (PADI2) has been shown to be downregulated in colon cancer, with such downregulation being indicative of a poor prognosis in individuals with this disease. We have now examined the expression of PADI2 in matched colon cancer and normal colon tissue specimens as well as in colon cancer cell lines. We found that isoform 1 of PADI2 is the predominant isoform in colon tissue and is downregulated during colon carcinogenesis. Immunohistochemical analysis showed that PADI2 is expressed in normal colonic epithelial cells. Overexpression of PADI2 isoform 1 suppressed the proliferation of colon cancer cells in vitro in association with increased protein citrullination. Expression of a catalytically inactive mutant (C647A) of PADI2 or of PADI2 isoform 2 did not induce such effects, indicating that the protein citrullination activity of PADI2 is required for inhibition of cell growth. The growth defect induced by PADI2 was not attributable to increased apoptosis but rather was accompanied by arrest of cell cycle progression in G1 phase. Finally, we detected citrullinated proteins in normal colon tissue by immunoblot analysis. Our data thus suggest that PADI2 suppresses the proliferation of colonic epithelial cells through catalysis of protein citrullination, and that downregulation of PADI2 expression might therefore contribute to colon carcinogenesis.
Keywords: Cell proliferation, colorectal cancer, epigenomic regulation, PADI2, protein citrullination
Citrulline is an α‐amino acid that is not incorporated into proteins during their synthesis but is generated from arginine residues in protein molecules by a family of enzymes known as protein‐arginine deiminases (PADIs).1 The conversion of positively charged arginine to uncharged citrulline by these enzymes alters the tertiary structure of target proteins. Protein citrullination mediated by PADIs is thus thought to modulate the function or interactions of target proteins and thereby to regulate cellular processes.2
Five PADI isozymes have been identified in mammals.3, 4 They are Ca2+‐dependent enzymes and share ~50–60% amino acid sequence identity. Although each isozyme targets a distinct set of proteins for citrullination, there is some overlap among these sets.5 In addition, the expression patterns of the PADI isozymes differ, suggesting that regulation of their expression is of biological relevance and may shed light on the function of both PADIs and protein citrullination.1
Protein citrullination and PADIs have been implicated in the pathogenesis of various inflammatory diseases and cancers.6, 7, 8, 9 Inhibitors of these enzymes have been shown to suppress inflammatory conditions such as rheumatoid arthritis as well as neutrophil extracellular trap (NET) formation in mouse models.10, 11 The NET formation is promoted by PADI4‐mediated histone citrullination; PADI4 KO mice are unable to form NETs and therefore show an increased susceptibility to bacterial infection, suggesting that citrullination by PADI4 is an important mediator of innate immunity.12
Expression of PADI4 is regulated by the estrogen receptor.13 Given that estrogen serves as a mitogenic stimulus in female cancers, the relevance of PADI4 expression or protein citrullination to carcinogenesis has been a subject of recent interest. Expression of PADI2 was recently shown to be downregulated in colon carcinoma, with such downregulation being associated with poor prognosis, suggesting that protein citrullination might suppress carcinogenesis.14
We now show that PADI2 expression is downregulated in colon tumors as well as in colon cancer cell lines, and that PADI2 expression in such cell lines is dependent on epigenomic status. Overexpression of PADI2 halted cell cycle progression in G1 phase in association with increased protein citrullination in colon cancer cells. Furthermore, protein citrullination was suppressed in colon tumor tissue compared with normal colon tissue, likely as a result of attenuation of PADI2 expression.
Materials and Methods
Surgical specimens
We obtained paired tumor and corresponding normal tissue specimens from colon cancer patients at Tohoku University Hospital (Sendai, Japan) (Table S1). This aspect of the study was approved by the Institutional Review Board of Tohoku University Graduate School of Medicine, and the patients provided written informed consent. Total RNA was isolated from the tissue samples with the use of an SV Total RNA Isolation System (Promega, Madison, WI, USA), and the quality of the RNA was analyzed with an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) (Table S1).
Immunohistochemistry and immunofluorescence analysis
Paraffin‐embedded tissue sections were stained with antibodies to PADI2 (Proteintech, Rosemont, IL, USA), and immune complexes were detected with biotin‐labeled secondary antibodies, HRP‐labeled streptavidin, and 3,3ʹ‐diaminobenzidine (SAB‐PO Kit; Nichirei Bioscience, Tokyo, Japan). The sections were counterstained with hematoxylin. For immunofluorescence analysis, cells were stained with antibodies to FLAG and immune complexes were detected with Alexa Fluor 488‐labeled secondary antibodies (Thermo Fisher Scientific, Waltham, MA, USA). All samples were observed with a BZ‐9000 microscope (Keyence, Osaka, Japan).
Reverse transcription–quantitative PCR, immunoblot analysis, and in vitro citrullination assay
Reverse transcription–quantitative PCR (qPCR) and immunoblot analysis were carried out as described previously.15, 16 Reverse transcription‐qPCR data were normalized by the amount of β‐glucuronidase or β2‐microglobulin mRNAs. An in vitro citrullination assay was also undertaken as described previously.17 In brief, 3 × FLAG‐tagged PADI2 was immunoprecipitated from HCT 116 cell lysates with antibodies to FLAG. The immunoprecipitates were suspended in Citrullination Assay Buffer, consisting of 100 mM Tris‐HCl (pH 7.4), 5 mM DTT, 10 mM CaCl2, and a crude histone fraction from 293T cells (10 μg/mL), and were then incubated for 1 h at 37°C. The reaction products were detected by immunoblot analysis.
Additional methods
Methods for cell culture and reagents, RNA sequencing (RNA‐seq), microarray data analysis, in vitro cell growth assays, and flow cytometry are described in Appendix S1. Primary antibodies and qPCR primers are listed in Table S2.
Statistical analysis
Data are presented as mean ± SD for biological (not technical) replicates. Statistical analysis was carried out with the R package version 3.3.0 (The R Foundation for Statistical Computing, Vienna, Austria). A P‐value of <0.05 was considered statistically significant.
Results
PADI2 is downregulated in colon cancer
Expression of PADI2 was previously shown to be downregulated in colorectal cancer.14 We determined the gene expression profiles for three sets of paired colon cancer and normal colon tissue specimens as well as for five human colon cancer cell lines by RNA‐seq analysis (Table S1). Among the five PADI family genes, we found that PADI2 was the only one to be expressed in normal colon tissue (Fig. S1a). Furthermore, the expression level of PADI2 was greatly reduced in tumor tissue and colon cancer cell lines compared with normal colon tissue, whereas that of housekeeping genes (GUSB and B2M) was similar among all the samples (Fig. 1a, Fig. S1a–c). We validated these results for PADI2 expression by RT‐qPCR analysis (Fig. 1a). Analysis of a microarray dataset in the Gene Expression Omnibus database also revealed that the expression level of PADI2 was significantly reduced in human colon polyps compared with normal colon tissue and was further reduced in primary colon tumors and metastatic lesions of colon cancer (Fig. 1b).18 These data were confirmed by analysis of different datasets with different probes (Fig. S1d,e).
Figure 1.
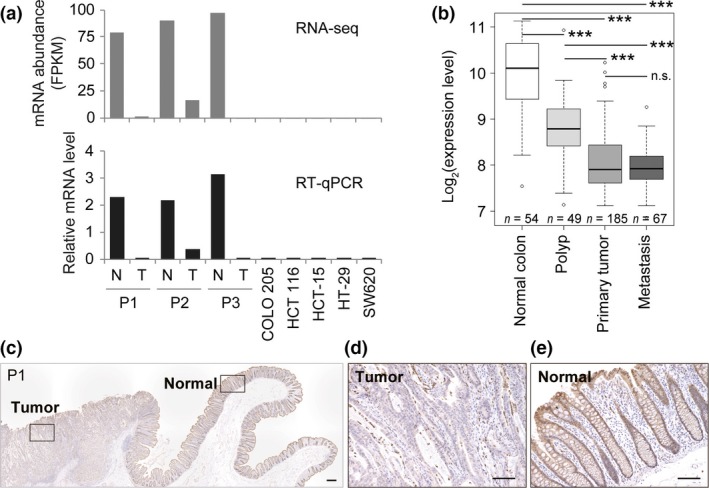
Expression of PADI2 in normal colon and colon cancer tissue. (a) RNA sequencing and quantitative RT‐PCR analysis of PADI2 expression in paired normal colon (N) and colon tumor (T) tissue from patients (P)1–3 as well as in five colorectal cancer cell lines. (b) Box plots of PADI2 expression level were constructed from the Gene Expression Omnibus dataset GSE41258. ***P < 0.001 (Student's t‐test with Welch's correction). n.s., not significant. (c–e) Immunohistochemical staining of PADI2 in a tissue section of patient 1. Boxed regions in (c) are shown at higher magnification in (d) and (e). Scale bar = 300 μm (c) or 100 μm (d,e).
We also examined expression of the PADI2 protein in paired colon tumor and normal colon specimens by immunohistochemistry. Immunoreactivity of PADI2 was detected in epithelial cells, predominantly in the cytosol but also to a lesser extent in the nucleus (Figs. 1c–e, Fig. S2). In contrast, PADI2 expression was virtually undetectable in cancerous epithelial cells, consistent with our data for PADI2 mRNA. On the basis of these results, we concluded that expression of PADI2 is prominent in normal colon tissue but is markedly down‐regulated during tumorigenesis.
PADI2 isoforms and mechanism of PADI2 repression
Two transcripts of the PADI2 gene, isoforms 1 and 2, were annotated by GENCODE version 24 (Wellcome Trust Sanger Institute, Cambridge, UK). Both transcripts share exons 1–11, but isoform 2 does not undergo splicing out of intron 11 and so has a longer exon 11 that includes a stop codon and 3′‐UTR (Fig. 2a). We found that the abundance of isoform 2 was much lower than that of isoform 1 and did not differ between paired normal colon and colon tumor tissues (Fig. 2b).
Figure 2.
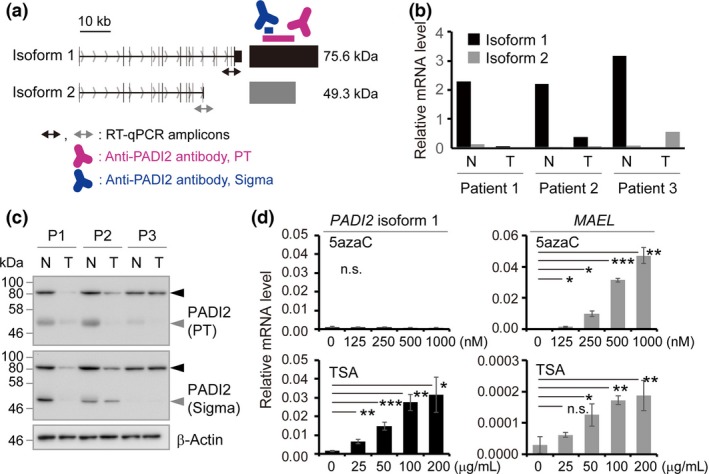
PADI2 isoform expression in normal colon and colon cancer tissue. (a) Schematic representation of two PADI2 isoforms. Bidirectional arrows indicate quantitative PCR (qPCR) amplicons specific to isoform 1 (black) or isoform 2 (gray). Antibodies to PADI2 used in this study are shown in red (Proteintech [PT]) or blue (Sigma‐Aldrich [Sigma]). (b) RT‐qPCR analysis of the expression of PADI2 isoforms 1 and 2 in normal colon (N) and colon tumor (T) tissue from patients 1–3. (c) Immunoblot analysis of normal and tumor tissue from patients 1–3. Black and gray arrowheads indicate PADI2 isoform 1 and an ~50‐kDa protein, respectively. (d) RT‐qPCR analysis of HCT 116 cells exposed to 5‐azacytidine (5azaC) or trichostatin A (TSA). Data are mean ± SD (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001 (one‐way anova [upper left panel] or Student's t‐test with Welch's correction [other three panels]).
Immunoblot analysis revealed downregulation of the protein encoded by transcript isoform 1 in colon tumor tissue compared with the paired normal colon tissue for all of the nine patients examined with the exception of patient 3 (Fig. 2c, Fig. S3a). For patient 3, the extent of PADI2 expression at the mRNA or protein level was not consistent among RT‐qPCR (Fig. 2b), immunohistochemical (Fig. S2d–f), and immunoblot (Fig. 2c) analyses, possibly because different tissue blocks were used for these analyses and normal cells might have been included in the tumor tissue block used for immunoblot analysis. We also observed an ~50‐kDa immunoreactive protein band consistent with the expected size of the protein encoded by transcript isoform 2 in normal colon tissue specimens (Fig. 2c, Fig. S3b). However, we were not able to confirm whether this signal was due to PADI2 isoform 2 or to a non‐specific protein.
To investigate whether a change in epigenomic status might contribute to the downregulation of PADI2 expression associated with colon tumorigenesis, we examined the effects of a DNA methyltransferase inhibitor (5‐azacytidine) and a histone deacetylase inhibitor (trichostatin A) on PADI2 isoform 1 expression. Trichostatin A, but not 5‐azacytidine, increased PADI2 expression in a concentration‐dependent manner in both HCT 116 (Fig. 2d) and COLO 205 (Fig. S3c) cells. The expression of MAEL analyzed as a positive control was found to be induced by both 5‐azacytidine and trichostatin A. These results thus suggested that histone deacetylation contributes to the repression of PADI2 expression during colon carcinogenesis.
PADI2 induces protein citrullination and suppresses cell proliferation
To examine PADI2 function in tumorigenesis, we overexpressed PADI2 in the colon cancer cell line HCT 116, which expresses the endogenous gene at only a low level (Fig. 1a). Protein‐arginine deiminase 2 is a deiminase that converts proteinaceous arginine to citrulline. Cysteine‐647 of PADI2 is thought to be localized in the active site, and mutation of this residue to alanine (C647A) results in a loss of catalytic activity.19 Given that it lacks amino acids 438–665 of isoform 1 (Fig. 3a), which are required for enzyme activity, isoform 2 of PADI2 likely does not catalyze protein citrullination.19
Figure 3.
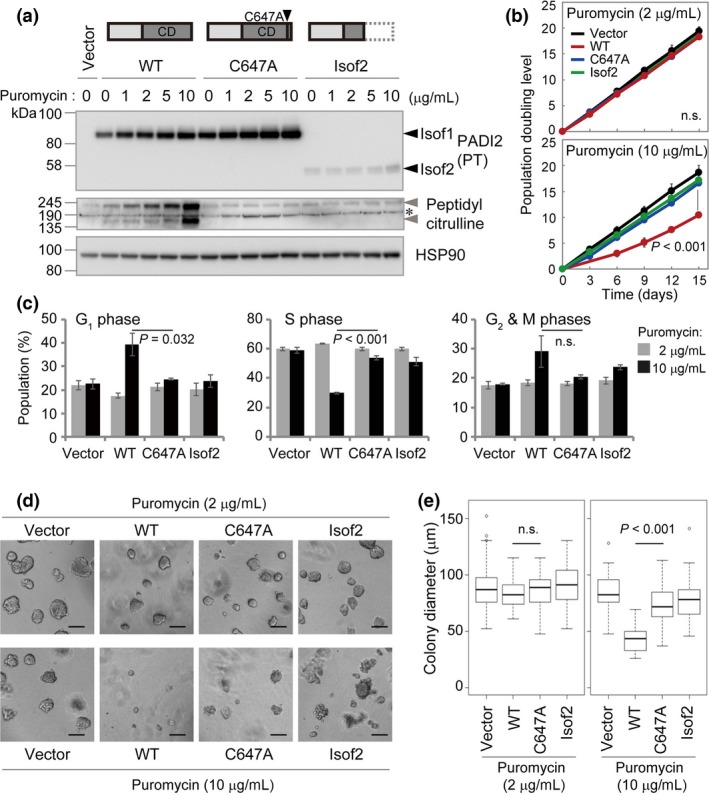
Effect of PADI2 overexpression on the proliferation of HCT 116 colon cancer cells. (a) Immunoblot analysis of HCT 116 cells expressing FLAG‐tagged WT or C647A mutant forms of PADI2 isoform 1 (Isof1) or PADI2 isoform 2 (Isof2). Structures of the exogenous PADI2 proteins are depicted above the blot. Black arrowheads indicate PADI2 isoforms 1 and 2, gray arrowheads indicate specific citrullinated protein bands, and the asterisk indicates non‐specific bands. CD, catalytic domain; HSP90, heat shock protein 90. (b) Growth curves for cells transfected as in (a). Data are mean ± SD (n = 3). P‐values: one‐way anova (upper panel) or Student's t‐test (WT vs. C647A) for day 15. (c) Cell cycle analysis for cells transfected as in (a). Data are mean ± SD (n = 3). P‐values: Student's t‐test with Welch's correction (WT vs. C647A at a puromycin concentration of 10 μg/mL). (d,e) Matrigel growth assay for cells transfected as in (a). Colony diameter (n = 50) in (e) was determined from phase‐contrast images. Scale bar = 100 μm. P‐values: Student's t‐test with Welch's correction (WT vs. C647A).
We transfected HCT 116 cells with expression vectors for FLAG epitope‐tagged PADI2 that also contained a puromycin resistance gene and then maintained the cells in puromycin‐containing medium. Immunofluorescence analysis revealed that WT or C647A mutant forms of PADI2 isoform 1 as well as PADI2 isoform 2 localized to the cytosol and nucleus of the cells (Fig. S4a), with the expression pattern being reminiscent of that apparent for PADI2 in immunohistochemical analysis of normal colon tissue (Fig. 1e). The cells expressing WT PADI2 isoform 1 (PADI2(WT)) manifested protein citrullination activity both in vivo in the presence of the Ca2+ ionophore A23187 (Fig. S4b) as well as in vitro in the presence of Ca2+ (Fig. S4c), whereas those expressing the C647A mutant or isoform 2 showed no such activity, suggesting that PADI2(C647A) and isoform 2 do not possess enzymatic activity.
We then cultured the cells in medium containing various concentrations of puromycin, with puromycin increasing the expression of PADI2 in a concentration‐dependent manner (Fig. 3a). The increase in the abundance of PADI2(WT) was accompanied by an increase in the extent of protein citrullination (Fig. 3a). Such citrullination was detected in the absence of A23187, suggesting that PADI2 was active under normal cell culture conditions without any stimulation. Again, we did not detect protein citrullination in cells expressing the C647A mutant or isoform 2 of PADI2 (Fig. 3a).
We also found that PADI2(WT) suppressed cell proliferation in a manner dependent on its expression level (Fig. S5a), whereas the C647A mutant or isoform 2 of PADI2 had no such effect. This effect of PADI2(WT) was thus apparent at a puromycin concentration of 10 μg/mL but not at 2 μg/mL (Fig. 3b). These results thus suggested that PADI2(WT) attenuated cell proliferation in a manner dependent on protein citrullination. To examine the mechanism of the growth inhibition induced by PADI2(WT), we determined the cell cycle distribution by measurement of BrdU incorporation. We found that overexpression of PADI2(WT) significantly increased the fraction of cells in G1 phase and reduced that of those in S phase for cells cultured in medium containing puromycin at 10 μg/mL (Fig. 3c, Fig. S5b).
To assess the effect of PADI2 on cell proliferation under more physiological conditions, we carried out a Matrigel growth assay20 (Fig. 3d,e). Culture of cells for 3 days revealed that overexpression of PADI2(WT) in the presence of puromycin at 10 μg/mL markedly limited colony growth, whereas PADI2(WT) at a puromycin concentration of 2 μg/mL or the C647A mutant or isoform 2 of PADI2 at either puromycin concentration had no such effect.
Examination of apoptosis by staining with annexin V and propidium iodide revealed that the proportion of cells in the late stage of apoptosis (those positive for staining with both annexin V and propidium iodide) was low for cells under all the conditions examined and was actually decreased by overexpression of PADI2(WT) in medium containing puromycin at either 2 or 10 μg/mL (Fig. S6a–c). Consistent with these data, we did not detect the cleaved form of caspase‐3 in any of the cells examined (Fig. S6d). Together, our findings thus suggested that protein citrullination by PADI2 halted cell cycle progression in G1 phase but did not induce apoptosis in colon cancer cells.
Knockdown of PADI2 in HCT 116 colon cancer cells does not affect cell proliferation
All five colon cancer cell lines examined in the present study expressed PADI2 at a low (compared with normal colon tissue) or undetectable level (Fig. 1a). Among these cell lines, HCT 116 showed the highest level of PADI2 expression (Fig. S7a). To investigate whether this low level of PADI2 expression affects cell proliferation, we generated HCT 116 cells that stably express shRNAs targeted to either PADI2 or luciferase mRNAs. The cells expressing PADI2 shRNAs manifested depletion of PADI2 isoform 1 mRNA to levels as low as ~20% of that in the parental cells (Fig. S7b), although we were not able to detect PADI2 protein in these cells by immunoblot analysis, presumably as a result of its low abundance (Fig. S7c). Knockdown of PADI2 did not affect cell proliferation under normal culture conditions (Fig. S7d), and we did not detect a correlation between PADI2 expression level and cell proliferation in the manipulated cells (Fig. S7e). These results, together with the previous observation that PADI2 KO mice survive at least until the preweaning stage,21 suggest that PADI2 is not essential for cell cycle progression.
Protein citrullination is suppressed in colon cancer
We next examined citrullination of endogenous proteins in normal colon and colon tumor tissue by immunoblot analysis (Fig. 4, Fig. S8). A prominent band corresponding to a citrullinated protein (or proteins) was apparent at a molecular size of ~80 kDa, and the intensity of this band was decreased in tumor tissue compared with normal tissue from all nine patients examined. These data suggested that the ~80‐kDa citrullinated protein is a substrate of PADI2 and that its citrullination is downregulated in tumor tissue. However, a similar ~80‐kDa band was not detected in HCT 116 or HCT‐15 cells overexpressing PADI2. Two low‐intensity bands were also detected at ~240 kDa and ~30 kDa in colon tissue of some patients. A similar ~240‐kDa band, but not ~30‐kDa band, was detected in HCT 116 and HCT‐15 cells overexpressing PADI2. Although the citrullinated proteins in colon tissue appeared to differ from those in PADI2‐overexpressing cell lines, these data suggested that PADI2 is active and citrullinates proteins in normal colon tissue and that the extent of protein citrullination is reduced in tumors, probably as a result of the downregulation of PADI2 expression.
Figure 4.
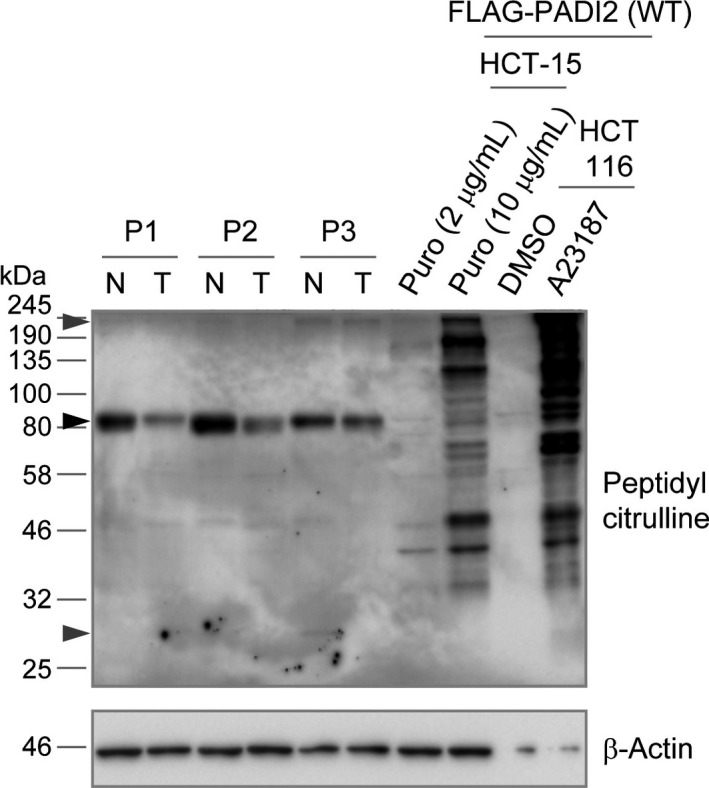
Protein citrullination in colon tissue and colon cancer cell lines. Normal colon (N) and colon tumor (T) tissue from patients 1–3 as well as HCT‐15 and HCT 116 cells overexpressing FLAG‐tagged PADI2(WT) were subjected to immunoblot analysis. The HCT‐15 cells were maintained in medium containing puromycin (Puro) at 2 or 10 μg/mL, whereas the HCT 116 cells were treated with A23187 or DMSO vehicle. The black arrowhead indicates a prominent ~80‐kDa band, and gray arrowheads indicate lower‐intensity ~240‐kDa and ~30‐kDa bands. The ~240‐kDa band was detected in normal and tumor tissue from patient 3; the ~30‐kDa band was detected in normal tissue from patient 3.
Discussion
Suppression of cell cycle progression by PADI2
Inhibition of PADI4 with Cl‐amidine in HCT 116 cells was previously shown to induce cell cycle arrest in a p53‐dependent manner,22 suggesting that PADI4 promotes cell cycle progression. We have now shown that overexpression of PADI2 suppressed cell cycle progression in HCT 116 cells. PADI4 catalyzes the citrullination of histone H4 at the R3 residue, and citrullinated H4 regulates the transcription of genes related to cell cycle control.23 Given that we did not detect citrullinated proteins with a molecular size similar to that of histones in cells overexpressing PADI2 or in normal colon tissue, the mechanism of cell cycle regulation by PADI2 may differ from that mediated by PADI4.
The molecular mechanism by which PADI2 inhibits cell cycle progression remains to be determined. Our preliminary data have revealed the accumulation of p53 and the cyclin‐dependent kinase inhibitor p21 in HCT 116 cells overexpressing PADI2. Further studies are now warranted to investigate the role of these and other signaling molecules in the phenotypes of PADI2‐overexpressing cells.
Two groups of researchers have generated PADI2 KO mice.21, 24 The International Mouse Phenotyping Consortium reports that PADI2 KO mice manifest preweaning mortality with incomplete penetrance, although the role of PADI2 in colon carcinogenesis has not been addressed (http://www.mousephenotype.org). Analysis of such KO mouse models in future studies might prove informative with regard to our hypothesis that downregulation of PADI2 expression contributes to tumor progression in colon cancer.
Regulation of PADI2 expression
Our analysis of public microarray datasets revealed that PADI2 expression was significantly downregulated in precancerous polyps compared with normal colon tissue, consistent with the previous finding that downregulation of PADI2 expression is an early event in colonic tumorigenesis.14 Signaling pathways such as that mediated by Wnt that act at early stages of colon carcinogenesis might possibly be responsible for such suppression of PADI2 expression.
Advances in genomics technology have indicated that epigenetic changes also occur early in colon cancer and that they manifest more frequently than genetic alterations.25 We found that inhibition of histone deacetylation, but not that of DNA methylation, increased PADI2 expression in both HCT 116 and COLO 205 cells. These results are consistent with the previous finding of no correlation between DNA methylation and PADI2 expression in colon cancer tissue.14 Histone acetyltransferases or deacetylases and their associated transcription factors may thus regulate PADI2 expression during colon carcinogenesis.
Examination of the effect of C1‐amidine on colitis, a form of inflammatory bowel disease, and colitis‐associated colorectal cancer revealed that this PADI inhibitor had a beneficial action in mice when administered either prophylactically or after disease onset.26, 27 Although this previous study found that PADI2 and PADI4 levels were increased in mouse and human colitis, a more recent study showed that PADI2 is downregulated in human ulcerative colitis.14 Given that infiltrating neutrophils express PADI2 at a high level,14 further studies are required to examine PADI2 expression in mucosal epithelial cells of colitis.
We have shown here that PADI2 suppresses cell cycle progression in colon cancer cells. Whether the downregulation of PADI2 expression in colon cancer actually contributes to tumorigenesis remains to be determined definitively. Further studies are required to identify protein substrates of PADI2 that control cell cycle progression and to elucidate how citrullination of such substrates influences the cell cycle.
Disclosure Statement
The authors have no conflict of interest.
Supporting information
Fig. S1. Expression of PADI genes in colon tissue and colon cancer cell lines.
Fig. S2. Immunohistochemical staining of PADI2 in colon cancer and normal colon tissue.
Fig. S3. PADI2 isoform expression in colon tissue and COLO 205 cells.
Fig. S4. Characterization of HCT 116 cells overexpressing PADI2.
Fig. S5. Effect of PADI2 overexpression in HCT 116 cells on cell proliferation.
Fig. S6. Effect of PADI2 overexpression in HCT 116 cells on apoptosis.
Fig. S7. Effect of PADI2 knockdown on the proliferation of HCT 116 cells.
Fig. S8. Protein citrullination in colon tissue and colon cancer cell lines.
Table S1. Characteristics of colon cancer patients from whom paired tumor and corresponding normal tissue specimens were obtained
Table S2. Oligonucleotide sequences and primary antibodies used in this study
Appendix S1. Supplementary methods.
Acknowledgments
We thank H. Miyoshi for providing the CSII lentiviral vector, Y. Nagasawa, K. Kuroda, M. Tsuda, M. Kikuchi, and M. Nakagawa for technical assistance, T. Konishi for help in preparation of the manuscript, other laboratory members for discussions, and the Biomedical Research Core of Tohoku University Graduate School of Medicine for technical support. This work was supported by the Japan Society for the Promotion of Science (Kakenhi Grant Nos. JP16K07107, JP16K14664, and JP26293059).
Cancer Sci 108 (2017) 713–718
Funding Information
Japan Society for the Promotion of Science
References
- 1. Vossenaar ER, Zendman AJ, van Venrooij WJ, Pruijn GJ. PAD, a growing family of citrullinating enzymes: genes, features and involvement in disease. Bioessays 2003; 25: 1106–18. [DOI] [PubMed] [Google Scholar]
- 2. Gudmann NS, Hansen NU, Jensen AC, Karsdal MA, Siebuhr AS. Biological relevance of citrullinations: diagnostic, prognostic and therapeutic options. Autoimmunity 2015; 48: 73–9. [DOI] [PubMed] [Google Scholar]
- 3. Chavanas S, Mechin MC, Takahara H et al Comparative analysis of the mouse and human peptidylarginine deiminase gene clusters reveals highly conserved non‐coding segments and a new human gene, PADI6. Gene 2004; 330: 19–27. [DOI] [PubMed] [Google Scholar]
- 4. Witalison EE, Thompson PR, Hofseth LJ. Protein arginine Deiminases and associated citrullination: physiological functions and diseases associated with dysregulation. Curr Drug Targets 2015; 16: 700–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Knuckley B, Causey CP, Jones JE et al Substrate specificity and kinetic studies of PADs 1, 3, and 4 identify potent and selective inhibitors of protein arginine deiminase 3. Biochemistry 2010; 49: 4852–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schellekens GA, de Jong BA, van den Hoogen FH, van de Putte LB, van Venrooij WJ. Citrulline is an essential constituent of antigenic determinants recognized by rheumatoid arthritis‐specific autoantibodies. J Clin Invest 1998; 101: 273–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Opdenakker G, Proost P, Van Damme J. Microbiomic and posttranslational modifications as preludes to autoimmune diseases. Trends Mol Med 2016; 22: 746–57. [DOI] [PubMed] [Google Scholar]
- 8. Wang S, Wang Y. Peptidylarginine deiminases in citrullination, gene regulation, health and pathogenesis. Biochim Biophys Acta 2013; 1829: 1126–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mohanan S, Cherrington BD, Horibata S, McElwee JL, Thompson PR, Coonrod SA. Potential role of peptidylarginine deiminase enzymes and protein citrullination in cancer pathogenesis. Biochem Res Int 2012; 2012: 895343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Willis VC, Gizinski AM, Banda NK et al N‐alpha‐benzoyl‐N5‐(2‐chloro‐1‐iminoethyl)‐L‐ornithine amide, a protein arginine deiminase inhibitor, reduces the severity of murine collagen‐induced arthritis. J Immunol 2011; 186: 4396–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Subramanian V, Knight JS, Parelkar S et al Design, synthesis, and biological evaluation of tetrazole analogs of Cl‐amidine as protein arginine deiminase inhibitors. J Med Chem 2015; 58: 1337–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li P, Li M, Lindberg MR, Kennett MJ, Xiong N, Wang Y. PAD4 is essential for antibacterial innate immunity mediated by neutrophil extracellular traps. J Exp Med 2010; 207: 1853–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dong S, Zhang Z, Takahara H. Estrogen‐enhanced peptidylarginine deiminase type IV gene (PADI4) expression in MCF‐7 cells is mediated by estrogen receptor‐alpha‐promoted transfactors activator protein‐1, nuclear factor‐Y, and Sp1. Mol Endocrinol 2007; 21: 1617–29. [DOI] [PubMed] [Google Scholar]
- 14. Cantarino N, Musulen E, Valero V et al Downregulation of the deiminase PADI2 is an early event in colorectal carcinogenesis and indicates poor prognosis. Mol Cancer Res 2016; 14: 841–8. [DOI] [PubMed] [Google Scholar]
- 15. Hosogane M, Funayama R, Nishida Y, Nagashima T, Nakayama K. Ras‐induced changes in H3K27me3 occur after those in transcriptional activity. PLoS Genet 2013; 9: e1003698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kobayashi M, Funayama R, Ohnuma S, Unno M, Nakayama K. Wnt‐beta‐catenin signaling regulates ABCC3 (MRP3) transporter expression in colorectal cancer. Cancer Sci 2016; 107: 1776–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cuthbert GL, Daujat S, Snowden AW et al Histone deimination antagonizes arginine methylation. Cell 2004; 118: 545–53. [DOI] [PubMed] [Google Scholar]
- 18. Sheffer M, Bacolod MD, Zuk O et al Association of survival and disease progression with chromosomal instability: a genomic exploration of colorectal cancer. Proc Natl Acad Sci U S A 2009; 106: 7131–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Slade DJ, Fang P, Dreyton CJ et al Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design. ACS Chem Biol 2015; 10: 1043–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Luca AC, Mersch S, Deenen R et al Impact of the 3D microenvironment on phenotype, gene expression, and EGFR inhibition of colorectal cancer cell lines. PLoS One 2013; 8: e59689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dickinson ME, Flenniken AM, Ji X et al High‐throughput discovery of novel developmental phenotypes. Nature 2016; 537: 508–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cui X, Witalison EE, Chumanevich AP et al The induction of microRNA‐16 in colon cancer cells by protein arginine deiminase inhibition causes a p53‐dependent cell cycle arrest. PLoS One 2013; 8: e53791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li P, Yao H, Zhang Z et al Regulation of p53 target gene expression by peptidylarginine deiminase 4. Mol Cell Biol 2008; 28: 4745–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Raijmakers R, Vogelzangs J, Raats J et al Experimental autoimmune encephalomyelitis induction in peptidylarginine deiminase 2 knockout mice. J Comp Neurol 2006; 498: 217–26. [DOI] [PubMed] [Google Scholar]
- 25. Okugawa Y, Grady WM, Goel A. Epigenetic alterations in colorectal cancer: emerging biomarkers. Gastroenterology 2015; 149: 1204–25.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chumanevich AA, Causey CP, Knuckley BA et al Suppression of colitis in mice by Cl‐amidine: a novel peptidylarginine deiminase inhibitor. Am J Physiol Gastrointest Liver Physiol 2011; 300: G929–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Witalison EE, Cui X, Causey CP, Thompson PR, Hofseth LJ. Molecular targeting of protein arginine deiminases to suppress colitis and prevent colon cancer. Oncotarget 2015; 6: 36053–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Expression of PADI genes in colon tissue and colon cancer cell lines.
Fig. S2. Immunohistochemical staining of PADI2 in colon cancer and normal colon tissue.
Fig. S3. PADI2 isoform expression in colon tissue and COLO 205 cells.
Fig. S4. Characterization of HCT 116 cells overexpressing PADI2.
Fig. S5. Effect of PADI2 overexpression in HCT 116 cells on cell proliferation.
Fig. S6. Effect of PADI2 overexpression in HCT 116 cells on apoptosis.
Fig. S7. Effect of PADI2 knockdown on the proliferation of HCT 116 cells.
Fig. S8. Protein citrullination in colon tissue and colon cancer cell lines.
Table S1. Characteristics of colon cancer patients from whom paired tumor and corresponding normal tissue specimens were obtained
Table S2. Oligonucleotide sequences and primary antibodies used in this study
Appendix S1. Supplementary methods.


