Abstract
Tumor stem cells with self‐renewal and multipotent capacity play critical roles in the initiation and progression of cancer. Recently, a new 3‐D culture system known as organoid culture has been developed, allowing Lgr5‐positive stem cells to form organoids that resemble the properties of original tissues. Here we established organoids derived from intestinal tumors of Apc min/+ mice and normal intestinal epithelia of C57BL/6J mice and investigated the roles of microRNA (miRNA) in intestinal tumor organoids. The results of microarray analyses revealed that expression of the cluster miRNAs, miR‐194 and miR‐215 was markedly suppressed in intestinal tumor organoids in comparison with organoids derived from normal intestinal epithelia. Enforced expression of miR‐194 resulted in inhibition of E2f3, a positive regulator of the cell cycle and growth suppression of intestinal tumor organoids. In addition, enforced expression of miR‐215 suppressed the cancer stem cell signature through downregulation of intestinal stem cell markers including Lgr5. These findings indicate that the miRNA cluster including miR‐194 and miR‐215 plays important roles in suppressing the growth and attenuating the stemness of intestinal tumor organoids.
Keywords: Intestinal tumor, microRNA, miR‐194, miR‐215, organoid
MicroRNA (miRNA) are 21–25‐nucleotide non‐coding RNA that can post‐transcriptionally downregulate the expression of various target genes. miRNA are expressed in a tissue‐specific manner and play important roles in cell proliferation, apoptosis and differentiation during mammalian development.1 Misexpression of cancer‐related miRNA leads to the initiation and progression of cancer through modulation of their target oncogenes or tumor suppressor genes.2, 3 We have reported that some important tumor suppressor miRNA are silenced by epigenetic alterations such as DNA methylation and histone modification in human cancer cells.4, 5
Accumulated evidence has clarified that cancer cells in solid cancer tissues are heterogeneous, showing a hierarchy of stemness.6 Stem cells have the ability to perpetuate themselves through self‐renewal and to generate mature cells of various tissues through differentiation. A subpopulation of cancer cells with distinct stem‐like properties is responsible for tumor initiation, invasive growth and metastasis formation, and these are defined as cancer stem cells (CSC). As CSC are considered to be resistant to conventional chemotherapies and radiation therapy, it would be desirable to develop a therapeutic strategy specifically targeting CSC.
Lgr5, a member of the Wnt signaling pathway, has been identified as a new molecular marker of stem cells in the small intestine, colon, stomach, liver and pancreas.7, 8, 9, 10, 11, 12 The newly developed technique of 3D culture allows Lgr5‐positive stem cells to form budding cyst‐like structures (organoids) that resemble the properties of original tissues.7 This type of 3D culture uses serum‐free medium that includes only predefined factors such as R‐spondin 1 (Rspo1) and epidermal growth factor (EGF). Rspo1 has been identified as a ligand for Lgr5 and is an essential factor for activation of the Wnt signaling pathway.13, 14 Thus, interaction between Lgr5 and Rspo1 plays a pivotal role in the self‐renewal of stem cells. Using this new 3D culture system, organoid models of human colon, prostate and pancreatic cancers have been established, and these have thrown light on the biology of cancer stem cells.10, 15, 16, 17
Here we established organoids derived from intestinal tumors of Apc min/+ (Min) mice and normal intestinal epithelia of C57BL/6J mice. The organoid culture system allowed expansion of Lgr5‐positive stem cells into cyst‐like structures with properties resembling the original tissues. Thus, organoids derived from tumors of Min mice are considered to be a desirable model for the initial stage of intestinal tumorigenesis according to the adenoma‐carcinoma sequence.18 In the present study, we investigated the roles of miRNA relevant to the initial stage of tumorigenesis in intestinal tumor organoids.
Materials and Methods
Organoid culture
We performed organoid culture of intestinal tumors of Min mice and normal epithelia from wild‐type C57BL/6 mice. In brief, isolated tumor cells and epithelial cells were embedded in Matrigel on ice (growth factor‐reduced, phenol red‐free; BD Biosciences, San Jose, CA, USA) and seeded in 48‐well plates. The Matrigel was polymerized for 10 min at 37°C, and overlaid with 250 μL/well basal culture medium (advanced DMEM/F12 supplemented with penicillin/streptomycin, 10 mmol/L HEPES, Glutamax, 1 × N2, 1 × B27 [all from Thermo Fisher Scientific, Waltham, MA, USA] and 1 mmol/L N‐acetylcysteine [Sigma‐Aldrich, St. Louis, MO, USA]) containing the following optimized growth factor combinations: EGF and noggin for intestinal tumors; and EGF, noggin and Rspo1 for intestinal epithelia. For drug treatment, cells were seeded 2 days before drug administration and treated with 10 nM irinotecan (Wako, Pure Chemical Industries, Osaka, Japan) for 48 h. All experiments and procedures were approved by the Keio University Animal Research Committee, and all methods were carried out in accordance with the approved guidelines.
Cell proliferation assay
Cell proliferation activity of organoids was examined by WST assay with a Cell Counting Kit‐8 (Dojindo, Kumamoto, Japan) in accordance with the manufacturer's instructions.
Lentivirus‐mediated overexpression of miR‐194 and miR‐215
Overexpression of miR‐194 and miR‐215 in tumor organoids was conducted using lentiviral vectors. A puromycin‐resistant lentiviral vector (pLVSIN‐CMV Pur) was obtained from (Takara Bio, Shiga, Japan). The sequences of miR‐194, miR‐215 and the cluster were prepared by PCR. The primers employed are listed in Table S1. To prepare the infection medium, the lentiviral vector encoding miR‐194, miR‐215, the cluster including both miR‐194 and miR‐215, and the empty control vector were transfected into 293FT cells together with ViraPower lentiviral packaging mix (Thermo Fisher Scientific). We incubated the organoids with trypsin for 10 min at 37°C, and then incubated organoid fragments with a small volume of infection medium for 2 h at 37°C. After centrifugation, we discarded the supernatant and resuspended the pellet in Matrigel. We added the culture medium to the infection medium. Twenty‐four hours after transduction, the infected organoids were selected with puromycin (1 μg/mL).
RNA extraction and microarray analysis
Total RNA were extracted from cancer cell lines using the mirVana miRNA Isolation Kit (Applied Biosystems, Foster City, CA, USA). Extracted total RNA was checked with a Bioanalyzer (Agilent Technologies, CA, USA) and labeled with a 3D‐Gene miRNA labeling kit (Toray Industries, Kamakura, Japan). Labeled RNA were hybridized onto 3D‐Gene Mouse miRNA Oligo chips (Toray, Kamakura, Japan). The annotation and oligonucleotide sequences of the probes conformed to the miRBase Release 19 database (www.mirbase.org). After stringent washes, the fluorescent signals were scanned with a 3D‐Gene Scanner (Toray Industries) and analyzed using the 3D‐Gene Extraction software (Toray Industries). The raw data for each spot was normalized by subtraction with the mean intensity of the background signal determined from the signal intensities of all blank spots with 95% confidence intervals. Measurements for spots with signal intensities greater than two standard deviations (SD) of the background signal intensity were considered to be valid. The relative expression level of a given miRNA was calculated by comparing the signal intensities of the valid spots throughout the microarray experiments. The normalized data were globally normalized per array, so that the median of the signal intensity was adjusted to 25. All data were submitted to the GEO database, under the accession number GSE75482.
Quantitative RT–PCR
Expression levels of genes were analyzed by TaqMan quantitative RT–PCR assay for miR‐215, miR‐194, miR‐192 and Lgr5 (Applied Biosystems) in accordance with the manufacturer's instructions. Expression levels for Mdm2, E2f3, Ctnnbip1 and Ereg were analyzed by Universal SYBR Select Master in accordance with the manufacturer's instructions. The primers are listed in Table S2. U6 was used as an internal control for miR‐215, miR‐194 and miR‐192. Gapdh was used as an internal control for Lgr5, Mdm2, E2f3, Ctnnbip1 and Ereg. All experiments were carried out in triplicate.
Western blotting
Western blotting was performed with antibodies against p53 (FL‐393) and Lgr5 (Anti‐GPCR GPR49 antibody) purchased from Santa Cruz Biotechnology and Abcam, respectively. HRP‐conjugated β‐actin (Santa Cruz Biotechnology, Santa Cruz, CA, USA) was used as an internal control. Quantitative analysis was performed using the Image‐J.
Apoptosis assay
Apoptosis assay was performed using a GFP‐Certified Apoptosis/Necrosis Detection Kit for microscopy and flow cytometry (Enzo Life Sciences, Farmingdale, NY, USA).
Cell cycle assay
Cells were harvested by trypsinization, washed with PBS and fixed in 70% ice‐cold ethanol overnight at 4°C. They were then washed with PBS and treated with RNAe A (1 mg/mL) at 37°C for 60 min and incubated with propidium iodide (50 mg/mL) for 30 min at room temperature. After incubation, flow cytometry analysis was performed.
Patients and tissue specimens
Paired samples of primary colorectal cancer and corresponding non‐tumor colorectal mucosa were obtained from surgically resected specimens of five patients treated at the National Cancer Center Hospital, Tokyo, Japan. These fresh frozen tissues were subjected to extraction of total RNA. The clinicopathological features of these patients, including age, sex, tumor location, tumor size, histologic type and TNM stage, are described in Table S3. All experiments and procedures were approved by the Ethics Committees of the National Cancer Center, and all methods were carried out in accordance with the approved guidelines. Written informed consent was obtained from all of the patients.
Luciferase assay
In brief, the 3′ untranslated region (UTR) including miRNA target sites of the Mdm2, E2f3, Ctnnbip1 and Ereg genes were amplified by PCR from mouse genomic DNA in accordance with previous reports19, 20, 21 and TargetScan (http://www.targetscan.org). The luciferase constructs were made by ligating the 3′ UTR of the Mdm2, E2f3, Ctnnbip1 and Ereg genes into the Xba I site of the pGL3‐control vector (Promega, Madison, WI, USA). NIH3T3 cells were cultured in DMEM medium supplemented with 10% FBS. The cells were then transfected with the firefly luciferase reporter vector containing the 3′ UTR of the Mdm2, E2f3, Ctnnbip1 and Ereg genes and the control vector containing Renilla luciferase pRL‐CMV (Promega) using lipofectamine 3000 (Thermo Fisher Scientific) in six‐well plates. The luciferase assays were performed 24 h after transfection using the Dual Luciferase Reporter Assay System (Promega). The activity of firefly luciferase was normalized to that of Renilla luciferase.
Flow cytometry
Flow cytometry analysis was performed with the PE‐conjugated mouse anti‐Lgr5 monoclonal antibody (1:50, clone 2A2; Origene, Rockville, MD, USA) using a BD LSR‐II flow cytometer (BD Bioscience). PE‐conjugated mouse IgG (R&D Systems, Minneapolis, MN, USA) was used as an isotype control.
Statistical analysis
All statistical analyses were performed using R studio. Dunnett's test and Tukey's test were used (*P < 0.05, **P < 0.01, ***P < 0.001. All error bars represent SD).
Results
miR‐194 and miR‐215 are significantly downregulated in intestinal tumor organoids
Here we established cultured organoids from normal intestinal epithelia of wild‐type mice and intestinal tumors of Min mice. Because mutation of the Apc gene is an early event in the multi‐step carcinogenesis according to the adenoma‐carcinoma sequence theory,18 Apc‐deficient tumor organoids may be a desirable model for investigating the molecular mechanism underlying colorectal carcinogenesis.
As described previously, organoids derived from Apc‐deficient tumors formed cystic organoid structures without the budding that is observed in organoids derived from normal intestinal epithelia. Because Apc loss constitutively activates the Wnt pathway, Rspo1 was dispensable for the culture of Apc‐deficient tumor organoids (Fig. 1a). We dissected three Min mice, and established tumor organoids from two to three tumors of each mouse. We successfully established tumor organoids from all of the tumors examined, and confirmed that the organoids showed no morphological or biological changes.
Figure 1.
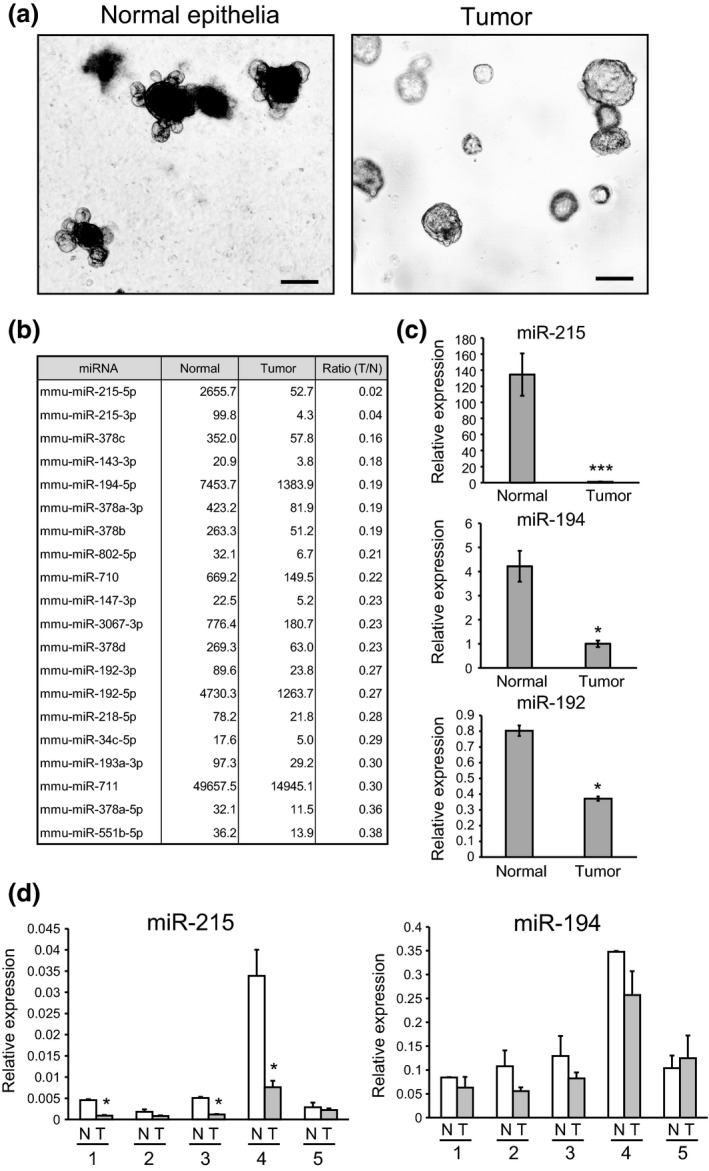
Difference in morphology and miRNA expression profiles between normal intestinal organoids and intestinal tumor organoids. (a) Left: Organoid derived from normal intestinal epithelia of C57BL/6J mice (normal epithelia). Right: Organoid derived from intestinal tumors of Min mice. Scale bars: 1000 μm. (b) MiRNA that were downregulated in intestinal tumor organoids identified by microarray analysis. (c) Relative expression levels of miR‐215, miR‐194 and miR‐192 in normal intestinal organoids and intestinal tumor organoids. U6 was used as an internal control. (d) Relative expression levels of miR‐194 and miR‐215 in human colon cancer tissues (T) as well as non‐tumor epithelial tissues (N) obtained from colon cancer patients. U6 was used as an internal control. The clinicopathological features of colorectal cancer patients, including age, sex, tumor location, tumor size, histologic type and TNM stage, are described in Table S3. *P < 0.05, ***P < 0.001.
We comprehensively examined the expression levels of 1265 miRNA transcripts by microarray analyses. This revealed that 55 miRNA were significantly upregulated and 35 miRNA were significantly downregulated in intestinal tumor organoids in comparison with normal epithelial organoids. miR‐215 was the main miRNA downregulated in intestinal tumor organoids, in comparison with organoids derived from normal epithelia (Fig. 1b). miR‐215 is located in the miRNA cluster with miR‐194‐1 on chromosome 1. miR‐194 is also downregulated in intestinal tumor organoids, and the expression of both miRNA is considered to alternate in a coordinated manner. To validate the microarray data, we performed quantitative RT‐PCR of miR‐215 and miR‐194. The relative expression levels of miR‐215 and miR‐194 were significantly decreased in tumor organoids relative to normal organoids (Fig. 1c). Expression of miR‐194 may be influenced by miR‐194‐2, which is located in the other miRNA cluster with miR‐192 on chromosome 19. We examined the expression levels of miR‐192 by quantitative RT‐PCR. As shown in Figure 1(c), the expression level of miR‐192 was also downregulated in tumor organoids relative to normal organoids, but the ratio (Tumor/Normal) of miR‐192 expression was higher than those of miR‐215 and miR‐194. Therefore, we focused our study on miR‐215 and miR‐194.
We investigated the expression levels of miR‐215 and miR‐194 in human colon cancer tissues as well as non‐tumor epithelial tissues obtained from colon cancer patients. As shown in Figure 1(d), the expression levels of miR‐215 and miR‐194 were lower in colon cancer tissues than in non‐tumor epithelial tissues, consistent with the findings obtained using tumor and normal organoids derived from Min and wild‐type mice.
Enforced expression of miR‐194 suppresses the growth of intestinal tumor organoids
To investigate the function of miR‐194 and miR‐215, we overexpressed them in tumor organoids, in which these miRNA are markedly suppressed. We performed lentivirus‐mediated overexpression of miR‐194, miR‐215 and the cluster containing both miR‐194 and miR‐215 in tumor organoids. As shown in Figure 2(a), significant overexpression of miR‐194 and miR‐215 was confirmed in tumor organoids, in which each miRNA was transfected. To examine the roles of miR‐194 and miR‐215 on cell growth, WST cell proliferation assays were performed using the miRNA‐transfected tumor organoids. Significant inhibition of cell proliferation was observed in both miR‐194 and cluster‐transfected organoids, whereas overexpression of miR‐215 did not significantly suppress cell growth (Fig. 2b). The inhibitory effect of cell proliferation was more profound in organoids overexpressing both miR‐194 and miR‐215 by induction of cluster than in organoids transfected with only miR‐194 (Fig. 2b). The proliferative potential of organoids was analyzed in terms of their size (Fig. 2c,d). Figure 2(f) shows the size distribution of organoids normalized to the total number of colonies for each treatment. The tumor organoids transfected with miR‐194 and cluster were smaller than those transfected with the empty vector (control) and miR‐215 (Fig. 2c,d,f).
Figure 2.
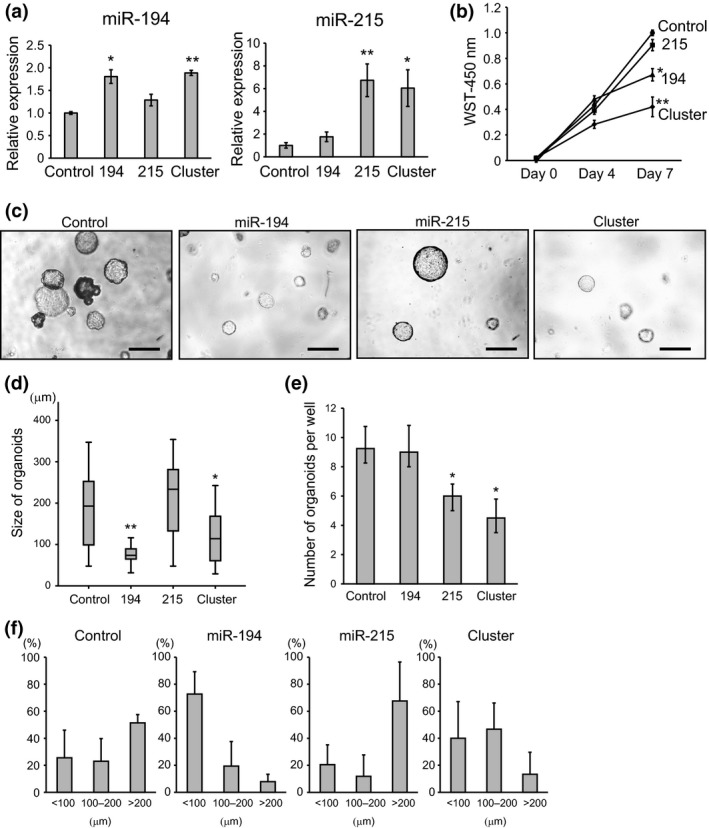
Growth and morphological change in intestinal tumor organoids transfected with miR‐194 and miR‐215. (a) Relative expression levels of miR‐194 and miR‐215 in miRNA‐transfected intestinal tumor organoids. U6 was used as an internal control. (b) WST cell proliferation assay of intestinal tumor organoids transfected with miRNA. (c) Representative images of intestinal tumor organoids transfected with miR‐194, miR‐215 and cluster. Scale bars: 1000 μm. (d) Size of intestinal tumor organoids transfected with miR‐194, miR‐215 and cluster. (e) Number of intestinal tumor organoids transfected with miR‐194, miR‐215 and cluster. (f) Size distribution of organoids normalized to the total number of organoids for each treatment. *P < 0.05, **P < 0.01. Control, 194, 215 and cluster represent tumor organoids transfected with empty vector, miR‐194, miR‐215 and cluster, including miR‐194 and miR‐215, respectively.
To clarify the molecular mechanism underlying cell growth suppression by miR‐194, we examined the expression of Mdm2, which is a direct target of miR‐194, and miR‐215, a negative regulator of p53.22 As expected, we found that enforced expression of both miR‐194 and miR‐215 significantly repressed Mdm2 expression, resulting in upregulation of p53 (Fig. 3a). In addition, we investigated the relative expression levels of other target genes of miR‐194 and miR‐215. E2f3 and Ctnnbip1 are reported to be potential targets of miR‐194 and miR‐215, respectively.20, 23 As shown in Figure 3(b), the expression levels of E2f3 and Ctnnbip1 were significantly decreased in tumor organoids transfected with miR‐194 and miR‐215, respectively. The luciferase assay to confirm the direct relationship between miRNA and their target genes showed that the relative luciferase activities of the constructs containing the target sites of miR‐194 and miR‐215 in the 3′ UTR of the Mdm2, E2f3 and Ctnnbip1 genes were significantly lower than those of the constructs lacking these regions (*P < 0.05, Fig. 3a–c). Thus, enforced expression of miR‐194 may lead to the growth arrest of tumor organoids via overexpression of p53 and an increase of G0/G1 phase cells. Because E2f3 and Ctnnbip1 play important roles in cell cycle and cell proliferation, we performed cell cycle and apoptosis assays using miRNA‐transfected organoids. The cell cycle assay demonstrated that suppression of E2f3 by transfection of miR‐194 and the cluster increased the G0/G1 phase, indicating cell cycle arrest (Fig. 3d). Flow cytometric analyses revealed that annexin V‐positive cells were increased in cluster‐transfected organoids, indicating that induction of both miR‐194 and miR‐215 caused cell apoptosis (Fig. 3d).
Figure 3.
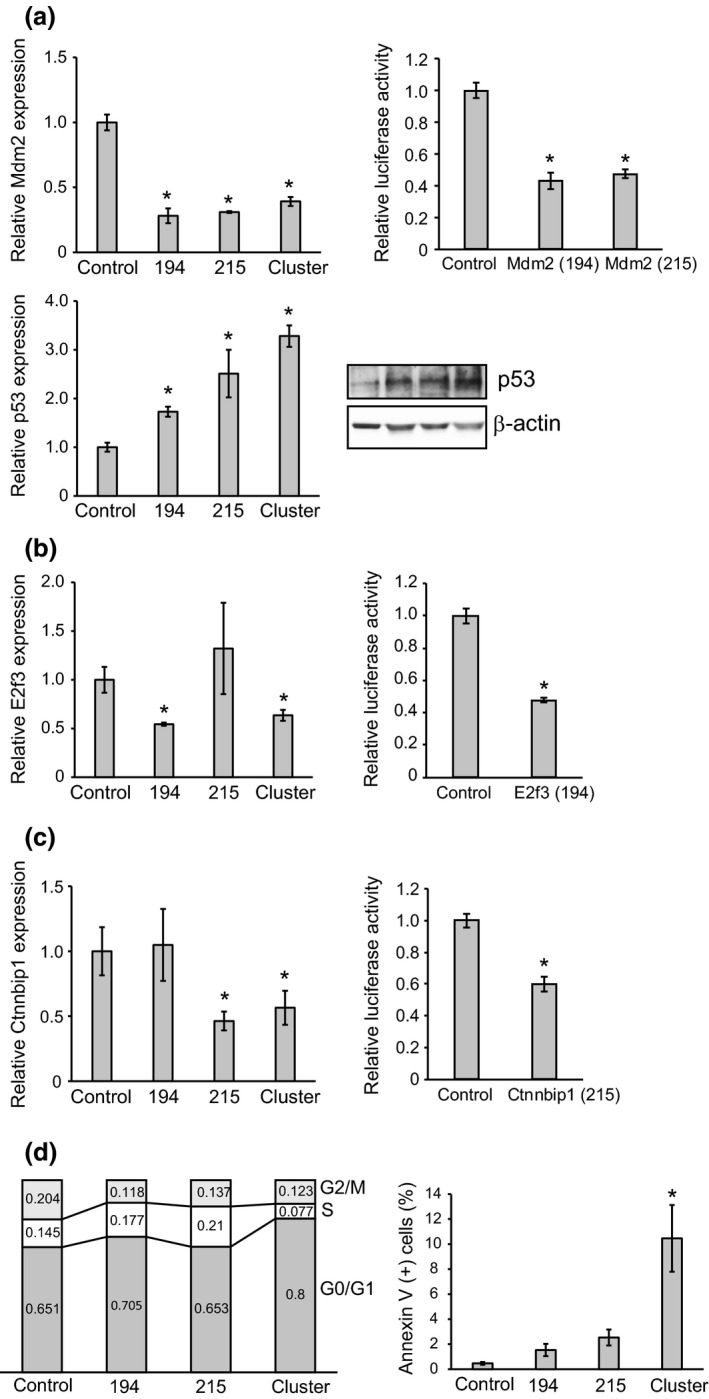
Relative expression levels of target genes of miR‐194 and miR‐215 in intestinal tumor organoids. (a) Quantitative RT‐PCR analysis of Mdm2 normalized to Gapdh and relative luciferase activities of the constructs containing the target sites of miR‐194 and miR‐215 in the 3′ UTR of the Mdm2 gene and the constructs lacking these regions (upper). Quantitative analysis for western blotting of p53 normalized to β‐actin in intestinal tumor organoids transfected with miR‐194, miR‐215 and the cluster (lower). (b) Quantitative RT‐PCR analysis of E2f3 normalized to Gapdh in intestinal tumor organoids transfected with miR‐194, miR‐215 and the cluster (left). Relative luciferase activities of the construct containing the target site of miR‐194 in the 3′ UTR of the E2f3 gene and the construct lacking this region (right). (c) Quantitative RT‐PCR analysis of Ctnnbip1 normalized to Gapdh in intestinal tumor organoids transfected with miR‐194, miR‐215 and the cluster (left). Relative luciferase activities of the construct containing the target site of miR‐215 in the 3′ UTR of the Ctnnbip1 gene and the construct lacking this region (right). (d) Cell cycle assay and apoptosis assay of intestinal tumor organoids transfected with miR‐194, miR‐215 and the cluster. *P < 0.05.
Enforced expression of miR‐215 suppresses the cancer stem cell signature
A recent study has shown that CDX1 directly activates miR‐215 expression, and that miR‐215 then represses the expression of stem cell genes and promotes cell differentiation via inhibition of stemness 21. Because organoids are formed from Lgr5‐positive stem cells, the number of organoids is considered to be correlated with stemness. We investigated the effect of miR‐215 on stemness by counting the number of tumor organoids transfected with each miRNA per well (Fig. 2c,e). As shown in Figure 2(e), the number of tumor organoids was significantly decreased by transfection of miR‐215, whereas miR‐194 had no effect on the number of tumor organoids. To investigate the molecular mechanism underlying inhibition of stemness by transfection of miR‐215, we examined the expression levels of Lgr5, a newly identified stem cell marker, by quantitative RT‐PCR and western blotting. The expression of Lgr5 mRNA and protein was significantly suppressed in tumor organoids transfected with miR‐215 and the cluster, in comparison to control and tumor organoids transfected with miR‐194 (Fig. 4a). In addition, we performed flow cytometry to determine the number of Lgr5‐positive cells. As shown in Figure S1(a), we confirmed that Lgr5‐positive cells were decreased in tumor organoids transfected with miR‐215 and the cluster, in comparison to control and tumor organoids transfected with miR‐194. These results are consistent with the findings of Western blotting and quantitative RT‐PCR. We also examined the expression level of Ereg, which has been identified as a direct target of miR‐215 and an intestinal cancer stem cell marker.21, 24 We confirmed that Ereg was downregulated in tumor organoids transfected by miR‐215 (Fig. 4a). The relative luciferase activity of the construct containing the target site of miR‐215 in the 3′ UTR of the Ereg gene was significantly lower than that of the construct lacking this region (*P < 0.05, Fig. 4a).
Figure 4.
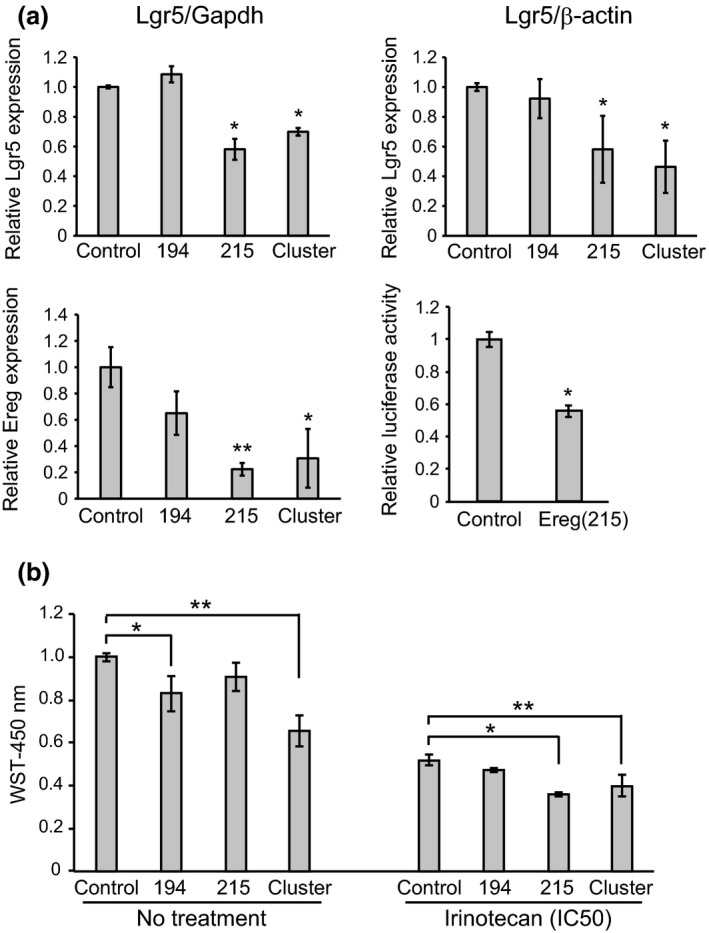
miR‐215 inhibits stemness of intestinal tumor organoids. (a) Quantitative RT‐PCR analysis of Lgr5 normalized to Gapdh and quantitative analysis of western blotting of Lgr5 normalized to β‐actin (upper). Quantitative RT‐PCR analysis of Ereg normalized to Gapdh, and relative luciferase activities of the construct containing the target site of miR‐215 in the 3′ UTR of the Ereg gene and the construct lacking this region (lower). (b) WST cell proliferation assay of miRNA‐transfected intestinal tumor organoids with or without irinotecan treatment (10 nM: IC50). *P < 0.05, **P < 0.01.
To evaluate the effect of miR‐215 on chemosensitivity, we treated miRNA‐transfected organoids with irinotecan. The results of the cell proliferation assay revealed that transfection with miR‐194 and the cluster caused significant growth inhibition in the tumor organoids, whereas after treatment with irinotecan (IC50), growth inhibition compared to the control was abrogated. In contrast, organoids transfected with miR‐215 and the cluster showed considerably increased sensitivity to irinotecan, suggesting that miR‐215 inhibits stemness and increases sensitivity to irinotecan (Fig. 4b).
Discussion
Decreased expression of miR‐194 has been reported in various types of malignancy, including liver, lung, stomach and colorectal cancers, as well as multiple myeloma.19, 25, 26, 27, 28, 29 Overexpression of miR‐194 in gastrointestinal cancer cells suppresses cell migration, invasion and metastasis.26 These reports suggest that miR‐194 functions as a tumor suppressor in various human malignancies. It has been reported that downregulation of miR‐215 may be a prognostic marker in patients with colon cancer.28, 30
Here we report that the cluster miRNAs miR‐194 and miR‐215 are downregulated in intestinal tumor organoids, as well as in human colon cancer tissues. In particular, the expression level of miR‐215 was very low in tumor organoids relative to normal epithelial organoids, whereas the observed differences between human colon cancer tissues and non‐tumor tissues were not large. Tumor tissues contain many non‐epithelial cells, such as stromal cells including fibroblasts, vascular endothelial cells and macrophages. Because the organoid culture system allows expansion of Lgr5‐positive stem cells and other epithelial cells derived from stem cells, it is possible to observe the biological signatures of the tumor cells alone, to the exclusion of non‐epithelial cells. This may explain the discrepancy in gene expression between tumor organoids and tumor tissues.
Our results demonstrated that induction of miR‐194 suppressed the growth of tumor organoids by repressing expression of Mdm2 and E2f3. In contrast, induction of miR‐215 did not suppress the growth of tumor organoids. Ctnnbip1 is a negative regulator of the Wnt/β‐catenin signaling pathway. Induction of miR‐215 suppressed Ctnnbip1 as its target gene, suggesting that it may promote the proliferation of tumor organoids via activation of the Wnt/β‐catenin signaling pathway. Stem cell markers such as Lgr5 and Ereg were suppressed in tumor organoids after induction of miR‐215, although Lgr5 is not a direct target of miR‐215. We speculate that in tumor organoids transfected with miR‐215, Lgr5 expression is suppressed due to loss of stemness.
Induction of miR‐215 reduced the number of organoids, whereas the size of the organoids was increased. The finding that miR‐215 decreased the number of tumor organoids recapitulates a recent study showing that the number of colonies formed by colon cancer cell lines overexpressing miR‐215 was decreased.21 In addition, miR‐215‐transfected tumor organoids showed significantly increased sensitivity to irinotecan, suggesting that miR‐215 inhibits the stemness of tumor organoids. These results are consistent with the report described above in which miR‐215 inhibited the stemness of colon cancer cells in 3D culture.21 Thus, miR‐215 is considered to suppress the stemness of cancer cells and tumor organoids. However, as shown in Figure 2(f), miR‐215 induced the formation of larger tumor organoids, suggesting that the cells in large organoids transfected with miR‐215 are more differentiated than those in small organoids.
To examine the expression levels of miRNA in stem cells, we performed flow cytometry to sort Lgr5‐positive stem cells in tumor organoids, as shown in Figure S1(b). We then examined the expression levels of miR‐192, miR‐194 and miR‐215 in both Lgr5‐positive stem cells and Lgr5‐negative cells by quantitative RT‐PCR. As shown in Figure S1(b), no expression of miR‐194 and miR‐215 was detected in Lgr5‐positive stem cells, suggesting that miR‐194 and miR‐215 were expressed in differentiated cells, but not in Lgr5‐positive stem cells. These findings are consistent with our observation that miR‐215 suppressed the stem cell signatures, as shown in Figure 4.
To confirm the tumorigenesis of organoids derived from tumors of Apc min/+ mice, we implanted tumor organoids (1.0 × 106 cells) subcutaneously into the backs of SCID mice. However, at approximately 2 months after implantation, no tumors had formed (data not shown). Because tumors of Apc min/+ mice are adenomas, and, thus, at an early stage of tumorigenesis, they do not have the ability to form tumors in SCID mice. In order to produce in vivo models of tumorigenesis, additional driver gene mutations such as Kras, Tp53 and Smad4 in the tumor organoids might be necessary. Further experiments such as introduction of driver gene mutations in tumor organoids using the CRISPR/Cas9 system will be necessary.
Taken together, these results indicate that miR‐194 is relevant to growth suppression, whereas miR‐215 inhibits the stemness of tumor organoids. miR‐215 alone did not affect the growth of organoids, but miR‐194 and 215 showed synergistic effect on apoptosis and growth suppression of tumor organoids. In the future, replacement of miR‐194 and miR‐215 may lead to the prevention and treatment of colorectal cancer. The concept of this therapy is to restore expression of miR‐194 and miR‐215 in colorectal cancers to a level comparable to that in surrounding non‐cancer tissues. Further studies to investigate tissue‐specific delivery of miRNA and potential off‐target side effects will be necessary to develop an effective miRNA‐based cancer therapy.
Disclosure Statement
The authors have no conflicts of interest to declare.
Supporting information
Fig. S1. Flow cytometry analysis of Lgr5 in intestinal tumor organoids. (a) Flow cytometry analysis of Lgr5 expression in intestinal tumor organoids transfected with miR‐194, miR‐215 and the cluster. (b) Flow cytometry analysis for sorting Lgr5‐positive stem cells in intestinal tumor organoids (upper). Quantitative RT‐PCR analysis of miR‐192, 194 and 215 normalized to U6 in Lgr5‐positive and negative cells (lower).
Table S1. Primer sequences used for constructing vectors encoding miRNA
Table S2. Primer sequences used for quantitative RT‐PCR
Table S3. Clinicopathological features of the colorectal cancer patients, including age, sex, tumor location, tumor size, histologic type and TNM stage.
Acknowledgments
This work was supported by a Keio University Special Grant‐in‐Aid for Innovative Collaborative Research Projects (to Y. Saito), a Grant‐in‐Aid for Scientific Research B (26290049) from the Japan Society for Promotion of Science (to Y. Saito) and the Takeda Science Foundation (to Y. Saito).
Cancer Sci 108 (2017) 678–684
Funding Information
Keio University Special Grant‐in‐Aid for Innovative Collaborative Research Projects; Japan Society for Promotion of Science (26290049); Takeda Science Foundation
References
- 1. He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 2004; 5: 522–31. [DOI] [PubMed] [Google Scholar]
- 2. Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet 2009; 10: 704–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer 2006; 6: 857–66. [DOI] [PubMed] [Google Scholar]
- 4. Saito Y, Liang G, Egger G et al Specific activation of microRNA‐127 with downregulation of the proto‐oncogene BCL6 by chromatin‐modifying drugs in human cancer cells. Cancer Cell 2006; 9: 435–43. [DOI] [PubMed] [Google Scholar]
- 5. Saito Y, Jones PA. Epigenetic activation of tumor suppressor microRNAs in human cancer cells. Cell Cycle 2006; 5: 2220–2. [DOI] [PubMed] [Google Scholar]
- 6. Clevers H. The cancer stem cell: premises, promises and challenges. Nat Med 2011; 17: 313–9. [DOI] [PubMed] [Google Scholar]
- 7. Sato T, Vries RG, Snippert HJ et al Single Lgr5 stem cells build crypt‐villus structures in vitro without a mesenchymal niche. Nature 2009; 459: 262–5. [DOI] [PubMed] [Google Scholar]
- 8. Barker N, Huch M, Kujala P et al Lgr5(+ve) stem cells drive self‐renewal in the stomach and build long‐lived gastric units in vitro. Cell Stem Cell 2010; 6: 25–36. [DOI] [PubMed] [Google Scholar]
- 9. Sato T, van Es JH, Snippert HJ et al Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 2011; 469: 415–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sato T, Stange DE, Ferrante M et al Long‐term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology 2011; 141: 1762–72. [DOI] [PubMed] [Google Scholar]
- 11. Huch M, Dorrell C, Boj SF et al In vitro expansion of single Lgr5+ liver stem cells induced by Wnt‐driven regeneration. Nature 2013; 494: 247–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huch M, Bonfanti P, Boj SF et al Unlimited in vitro expansion of adult bi‐potent pancreas progenitors through the Lgr5/R‐spondin axis. EMBO J 2013; 32: 2708–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Carmon KS, Gong X, Lin Q, Thomas A, Liu Q. R‐spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/beta‐catenin signaling. Proc Natl Acad Sci USA 2011; 108: 11452–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. de Lau W, Barker N, Low TY et al Lgr5 homologues associate with Wnt receptors and mediate R‐spondin signalling. Nature 2011; 476: 293–7. [DOI] [PubMed] [Google Scholar]
- 15. Gao D, Vela I, Sboner A et al Organoid cultures derived from patients with advanced prostate cancer. Cell 2014; 159: 176–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Boj SF, Hwang CI, Baker LA et al Organoid models of human and mouse ductal pancreatic cancer. Cell 2015; 160: 324–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Matano M, Date S, Shimokawa M et al Modeling colorectal cancer using CRISPR‐Cas9‐mediated engineering of human intestinal organoids. Nat Med 2015; 21: 256–62. [DOI] [PubMed] [Google Scholar]
- 18. Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell 1990; 61: 759–67. [DOI] [PubMed] [Google Scholar]
- 19. Pichiorri F, Suh SS, Rocci A et al Downregulation of p53‐inducible microRNAs 192, 194, and 215 impairs the p53/MDM2 autoregulatory loop in multiple myeloma development. Cancer Cell 2010; 18: 367–81. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20. Mu J, Pang Q, Guo YH et al Functional implications of microRNA‐215 in TGF‐beta1‐induced phenotypic transition of mesangial cells by targeting CTNNBIP1. PLoS ONE 2013; 8: e58622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jones MF, Hara T, Francis P et al The CDX1‐microRNA‐215 axis regulates colorectal cancer stem cell differentiation. Proc Natl Acad Sci USA 2015; 112: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shmueli A, Oren M. Regulation of p53 by Mdm2: fate is in the numbers. Mol Cell 2004; 13: 4–5. [DOI] [PubMed] [Google Scholar]
- 23. Khella HW, Bakhet M, Allo G et al miR‐192, miR‐194 and miR‐215: a convergent microRNA network suppressing tumor progression in renal cell carcinoma. Carcinogenesis 2013; 34: 2231–9. [DOI] [PubMed] [Google Scholar]
- 24. Kobayashi S, Yamada‐Okabe H, Suzuki M et al LGR5‐positive colon cancer stem cells interconvert with drug‐resistant LGR5‐negative cells and are capable of tumor reconstitution. Stem Cells 2012; 30: 2631–44. [DOI] [PubMed] [Google Scholar]
- 25. Braun CJ, Zhang X, Savelyeva I et al p53‐Responsive micrornas 192 and 215 are capable of inducing cell cycle arrest. Cancer Res 2008; 68: 10094–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Meng Z, Fu X, Chen X et al miR‐194 is a marker of hepatic epithelial cells and suppresses metastasis of liver cancer cells in mice. Hepatology 2010; 52: 2148–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Song Y, Zhao F, Wang Z et al Inverse association between miR‐194 expression and tumor invasion in gastric cancer. Ann Surg Oncol 2012; 19(Suppl 3): S509–17. [DOI] [PubMed] [Google Scholar]
- 28. Chiang Y, Song Y, Wang Z et al microRNA‐192, ‐194 and ‐215 are frequently downregulated in colorectal cancer. Exp Ther Med 2012; 3: 560–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wu X, Liu T, Fang O, Leach LJ, Hu X, Luo Z. miR‐194 suppresses metastasis of non‐small cell lung cancer through regulating expression of BMP1 and p27(kip1). Oncogene 2014; 33: 1506–14. [DOI] [PubMed] [Google Scholar]
- 30. Karaayvaz M, Pal T, Song B et al Prognostic significance of miR‐215 in colon cancer. Clin Colorectal Cancer 2011; 10: 340–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Flow cytometry analysis of Lgr5 in intestinal tumor organoids. (a) Flow cytometry analysis of Lgr5 expression in intestinal tumor organoids transfected with miR‐194, miR‐215 and the cluster. (b) Flow cytometry analysis for sorting Lgr5‐positive stem cells in intestinal tumor organoids (upper). Quantitative RT‐PCR analysis of miR‐192, 194 and 215 normalized to U6 in Lgr5‐positive and negative cells (lower).
Table S1. Primer sequences used for constructing vectors encoding miRNA
Table S2. Primer sequences used for quantitative RT‐PCR
Table S3. Clinicopathological features of the colorectal cancer patients, including age, sex, tumor location, tumor size, histologic type and TNM stage.


