Abstract
Pancreatic ductal adenocarcinoma (PDAC) is one of the most lethal malignancies. To improve its outcome, reliable biomarkers are urgently needed. In this study, we aimed to elucidate the key molecules involved in PDAC progression using proteomics approaches. First, we undertook 2‐D electrophoresis to identify the proteins overexpressed in PDAC tissues. Following the analysis of agarose gel spots, cofilin‐1 was identified and verified as a candidate protein commonly upregulated in PDAC tissues. In immunohistochemistry, cofilin‐1 was strongly expressed in the cytoplasm of PDAC cells. Samples were divided into two groups based on the level of cofilin‐1 expression. The high expression group showed significantly higher incidence of hematogenous dissemination in relapsed patients than the low expression group (P = 0.0083). In in vitro experiments, knockdown of cofilin‐1 significantly decreased chemotaxis in PDAC cell lines. After we confirmed that cofilin‐1 was secreted from PDAC cells, we established a detection system for the immune‐complex of cofilin‐1 in sera. Using this system, we measured the IC levels of cofilin‐1 in sera and observed that the IC levels of cofilin‐1 in PDAC patients were higher than those in healthy volunteers and patients with pancreatitis (PDAC vs. healthy volunteers, P < 0.0001; PDAC vs. patients with pancreatitis, P < 0.026). Notably, the IC levels of cofilin‐1 showed a stepwise increase during PDAC progression (P = 0.0034), and high IC levels of cofilin‐1 indicated poor prognosis of patients after surgery (P = 0.039). These results suggest that the IC of cofilin‐1 in sera is a potentially attractive serum biomarker for the prognosis of PDAC.
Keywords: Cofilin‐1, immune‐complex, pancreatic cancer, prognosis, serum biomarker
Abbreviations
- 2‐DE
two‐dimensional gel electrophoresis
- BAG3
BCL2‐associated athanogene 3
- CA19‐9
Cancer antigen 19‐9
- HV
healthy volunteer
- IC
immune‐complex
- IHC
immunohistochemistry
- MS
mass spectrometry
- PDAC
pancreatic ductal adenocarcinoma
- PD‐ECGF
platelet‐derived endothelial cell growth factor
- PT
pancreatitis
- ROC
receiver operating characteristic
- SSH
slingshot
Pancreatic ductal adenocarcinoma is one of the most lethal malignancies among various solid tumors with an extremely low 5‐year survival rate of 7%.1 Advances in the field of clinical biomarker discovery are developing to not only provide useful tools for patient stratification, but also to offer potential therapeutic targets and revealing molecular mechanistic insights behind disease progression, thereby facilitating exploitation of novel therapies. However, identifying a desired biomarker still faces significant challenges.2 In line with this, some attractive biomarkers for PDAC have been discovered and tested in a clinical setting as diagnostic and prognostic indicators. Despite the identification of candidate proteins or peptides that have potentially been useful and reliable in translational research, there are still some critical limitations for clinical application.
Of the various possible human samples, serum or plasma samples are reasonable to use as they contain the richest and most detailed source of information (e.g. DNA, RNA, proteins, and peptides) about the physiological state of the body as well as being less invasive to collect. Thus, blood samples would potentially have a pivotal role in ascertaining the status of cancer progression in patients. Emerging new technology for serum proteomics using mass spectrometry, such as surface‐enhanced laser desorption/ionization–time of flight or matrix‐assisted laser desorption/ionization–time of flight, enables the identification of protein profiles composed of isolated or clustered peaks that differ according to molecular weight.3 Although these techniques have some advantages for detecting differences in protein profiling, including post‐translational modifications, it is still difficult to apply in clinical scenarios due to the complexity of optimization for standard use.
Antibody‐based proteomic approaches such as the standard ELISA method or the measurement of autoimmune antibodies are primarily carried out in the examination of target protein or peptide levels in blood samples.4 Focusing on these targets in sera, the IC has recently been highlighted as a novel biomarker in blood samples of cancer patients.5, 6, 7, 8 Recently, it has been reported that a high‐throughput screening method for native autoantigen–autoantibody complexes using antibody microarrays provides serum biomarkers as a useful tool for disease detection and characterization.9
In this study, we initially used a gel‐based proteomic approach, 2‐DE, to analyze the comprehensive protein profile and identify biomarkers for PDAC. Subsequently, we have attempted to establish a detection system for candidate proteins involved with PDAC progression by measuring the serum levels of patients. Finally, we revealed that the IC level in sera is associated with cancer progression and may potentially provide a useful prognostic serum biomarker for PDAC.
Materials and Methods
Patient samples
Excised samples of both cancer tissue and adjacent normal pancreatic tissue were collected from 10 PDAC patients (Table 1). Pathological samples were obtained from 59 PDAC patients who had undergone pancreatectomy in the Department of General Surgery, Chiba University (Chiba, Japan, between October 2006 and December 2008. All patients were diagnosed histologically with primary PDAC. Excised tumor and adjacent non‐tumor samples were obtained within 1 h of the resection. All excised tissues were immediately placed in liquid nitrogen and stored at −80°C until proteome and IHC analysis was carried out.
Table 1.
Clinical features of 10 patients with pancreatic cancer
| Sample number | Age, years | Gender | UICC stage | Histology | |
|---|---|---|---|---|---|
| 2‐DE | W.B. | ||||
| I | 74 | Male | IIB | Well differentiated | |
| II | 1 | 68 | Female | IIB | Mod. differentiated |
| III | 2 | 63 | Male | IIB | Mod. differentiated |
| 3 | 68 | Female | IIB | Mod. differentiated | |
| 4 | 61 | Female | III | Mod. differentiated | |
| 5 | 45 | Female | IIB | Mod. differentiated | |
| 6 | 75 | Female | IIB | Mod. differentiated | |
| 7 | 66 | Male | IIB | Mod. differentiated | |
| 8 | 69 | Male | IIB | Mod. differentiated | |
| 9 | 61 | Female | IIB | Mod. differentiated | |
2‐DE, 2‐D electrophoresis; Mod, moderately; UICC, Union for International Cancer Control; W.B, Western blotting.
Serum samples were collected preoperatively from 49 patients with PDAC, and 18 patients with PT, and the sera of 48 age/gender‐matched HVs were collected as controls. Blood samples were obtained from all patients histologically diagnosed with PDAC and PT in the Department of General Surgery, Chiba University Hospital. Samples were also obtained from HVs at Kashiwado Hospital (Chiba, Japan). All blood samples were processed according to a standardized protocol, and serum sample aliquots were frozen at −80°C until subsequent analysis. None of the patients underwent any therapeutic measures, such as radiation, chemotherapy, or surgery until serum samples were collected. The Ethics Committee of each institute approved this protocol and written informed consent was obtained from each patient and healthy controls.
Agarose 2‐DE and image analysis
Agarose 2‐DE and image analysis were carried out as described previously10, 11 and are described in detail in Appendix S1.
Purification and identification of candidate proteins
Proteins separated by agarose 2‐DE were identified by in‐gel tryptic digestion of the proteins followed by MS. In‐gel tryptic digestion was carried as previously described11 (for details see Appendix S1).
Western blot analysis
Protein extracts were separated on SDS‐PAGE in 12.5% gels (DRC, Tokyo, Japan) and transferred onto PVDF membranes (0.2‐μm pore size; Millipore, Billerica, MA, USA) at 10 V overnight. The membranes were blocked for 2 h at room temperature with 0.3% low‐fat milk in TBS‐T (Tris‐buffered saline, 0.1% Tween 20). The reaction time for the primary and secondary antibodies was 2 h. Antigens on the membrane were detected by a chemiluminescence imager (LPR‐400EX; TAITEC, Tokyo, Japan) with enhanced chemiluminescence detection reagents (ECL plus; GE Healthcare, Tokyo, Japan). Band intensities of Western blot images were quantified by LumiVison IMAGER imaging analysis software (TAITEC). Details and information of antibodies are described in Appendix S1.
Immunohistochemistry
Pancreatic tumor and adjacent normal tissues were cut in 4‐μm‐thick serial sections and deparaffinized. Slides were incubated in 300 mL citric acid buffer (10 mM, pH 6.0) with 0.1% Tween‐20 at 97°C for 40 min for antigen retrieval. Immunohistochemical staining was carried out using the labeled streptavidin–biotin–peroxidase method (LSAB+ System; Dako, Kyoto, Japan) according to the manufacturer's protocol. All slides were incubated with the primary antibody, anti‐cofilin‐1 mouse mAb (1:50; Abnova, Taipei, Taiwan), in a humidified chamber for 2 h at room temperature. Tissue sections were counterstained with hematoxylin for 30 s and dehydrated with 100% ethanol and xylene, and coverslips were mounted with Malinol (Mito Pure Chemicals, Tokyo, Japan). Staining intensities were categorized on the following scale: 0, no staining; 1 + , weak staining; 2 + , moderate staining; 3 + , strong staining. For statistical analysis, samples were divided into weak (0/1 + staining), and strong (2 + /3 + staining) groups. Microscopic examination was independently undertaken and evaluated by two authors.
Cell culture and gene knockdown using siRNA
Human primary and metastatic PDAC cell lines, PANC‐1 and Capan‐1 (ATCC, Manassas, VA, USA) used in this study were cultured in DMEM (Gibco BRL, Grand Island, NY, USA) supplemented with 10% heat‐inactivated FBS, incubated in a humidified atmosphere containing 5% CO2 at 37°C. Cofilin‐1 siRNA duplexes were purchased from Sigma‐Aldrich Japan (Tokyo, Japan). The target sequence for cofilin‐1 RNA interference was 5′‐CCCUCUAUGAUGCAACCUA‐3′. Luciferase (GL2) siRNA was used as a negative control and purchased from Sigma‐Aldrich Japan. Transient transfection of siRNA was carried out using Lipofectamine 2000 (Invitrogen, Tokyo, Japan) according to the manufacturer's instructions.
Cell proliferation assay
After siRNA transfection, cells were incubated for 24 h and then harvested for the cell proliferation assay. A total of 5.0 × 104 PANC‐1 and 2.5 × 104 Capan‐1 human PDAC cell lines were cultured in 6‐well plates in the appropriate medium and incubated in a humidified atmosphere containing 5% CO2 at 37°C. After washing with PBS, these cells were transfected with GL2 siRNA (20 nM final concentration) or cofilin‐1 siRNA (1, 5, and 10 nM final concentration). Both attached and floating cells were collected with trypsin. After staining with Trypan blue, the number of Trypan blue‐positive cells was counted on days 3 and 5 after transfection. All of these experiments were carried out in triplicate twice, independently. The medium was changed every 3 days.
Chemotaxis assay
After siRNA transfection, cells were incubated for 3 days and then harvested for chemotaxis assay. The chemotaxis assay was carried out using the BD Falcon Cell Culture Insert (8‐μm pore size; BD Biosciences, Sparks, MA, USA) according to the manufacturer's instructions. PANC‐1 and Capan‐1 cells were harvested, resuspended in FBS‐free appropriate medium (DMEM) for 2 h and seeded into a cell culture insert (2 × 105 cells/well). Lower chambers were filled with culture medium containing 10% FBS as a chemoattractant. After dispersed cells were cultured at 37°C for 22 h, cells on the upper side of the membrane were then removed using cotton swabs, and the filters were washed, fixed, and stained using the Diff‐Quik kit (Sysmex, Kobe, Japan). Cell counting was undertaken by photographing the four fields of a membrane through the microscope. All of these experiments were carried out in independent triplicates.
Detection for IC of cofilin‐1
Anti‐cofilin‐1 antibody (5 μg/mL; Abnova) was dispensed into a 96‐well polystyrene microtiter plate (Maxisorp; Thermo Fisher Scientific, Yokohama, Japan) at 0.5 μg/well and incubated overnight at 4°C. The plate was washed three times with PBS wash buffer containing 0.05% Tween‐20, coated with 1.5% BSA (Proliant, Ankeny, IA, USA) containing 10% sucrose at 200 μL/well overnight at 4°C. After washing the microtiter plate with PBS wash buffer, 100 μL aliquots of 100‐fold diluted serum samples were added to wells. The plates were incubated at 37°C for 1 h and then washed three times with PBS wash buffer. After washing, mouse anti‐human IgG conjugated to HRP (1:4000; Zymed Laboratories, San Francisco, CA, USA) in PBS containing 0.05% Tween‐20 (100 μL) was added to each well and the plate was incubated at 37°C for 1 h. The plate was washed three times with 200 μL PBS wash buffer and then 100 μL 3,3′,5,5′‐tetramethylbenzidine solution was added. After incubation at room temperature for 10 min, 100 μL stop solution (1 N sulfuric acid) was added and absorbance at 450 nm was measured.
Statistical analyses
All numerical data are presented as mean ± SD. Statistical significance of observed differences was assessed using Welch's t‐test. The ROC curves were estimated by a logistic regression model and values of the area under the ROC curves were calculated by R Ver. 3.3.2 (http://cran.r-project.org/). Overall survival time was calculated according to the Kaplan–Meier method and compared by the log–rank test. Prognostic data were evaluated using univariate and multivariate Cox proportional regression analyses. Differences were considered statistically significant at P < 0.05.
Results
Identification and validation of candidate proteins by proteomic analyses
To identify useful biomarkers for PDAC, as the first step, we used the agarose 2‐DE method to explore proteins overexpressed in PDAC tissues compared to their expression in adjacent normal pancreas tissues. Although proteome analyses can be both gel‐ and MS‐based, the gel‐based method was selected for use in this study because the concentration of proteins or peptides in sera of patients was most likely to be dependent on the expression in cancer tissues. A total of 36 spots were identified as overexpressing proteins in PDAC tissues by 2‐DE analysis for three patients (Fig. 1a, Table 1). The minimum criterion of the image analysis was set at a 2‐fold increase in tumor tissues compared to expression in normal tissues. These spots were analyzed by LC‐MS/MS, and 11 proteins were identified as candidate proteins. To confirm differences in protein expression between normal and tumor tissues, a validation study was undertaken using Western blotting (Table 1). Five of 11 proteins were confirmed to be more highly expressed in tumor tissues than in normal tissues (Table S1). Among the five identified proteins, vimentin, annexin A1, endophilin B2, and PD‐ECGF were overexpressed in most tumor tissue samples (Fig. 1b,c), and cofilin‐1 was markedly overexpressed in all nine tumor tissue samples compared to its expression in normal tissues (Fig. 1d). Therefore, we selected cofilin‐1 as a candidate biomarker of PDAC for further analyses.
Figure 1.
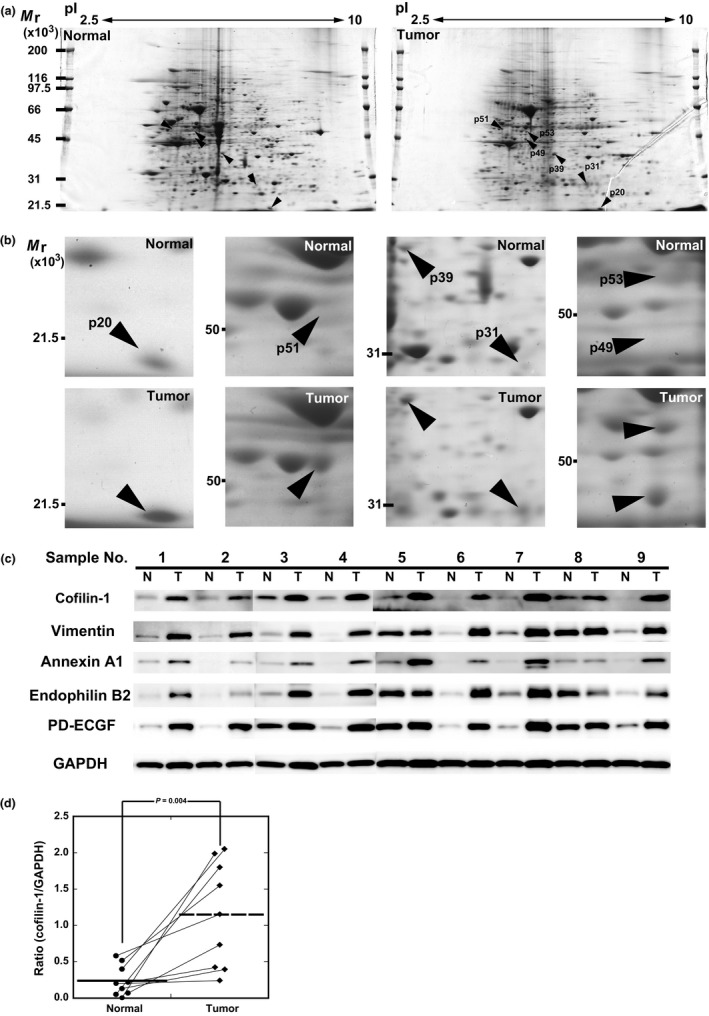
Comparison of protein profiling in 2‐D electrophoresis (2‐DE) patterns and Western blot analysis. (a) Comparison of 2‐DE patterns of normal and tumor pancreatic tissues. The 2‐DE gel was stained with Coomassie brilliant blue. (b) Expanded views of six overexpressed protein spots (p20, p51, p39, p31, p53, and p49) in tumor tissues. Protein spots are named according to their apparent molecular mass. Basic end of the gel is to the right. (c) Western blot analysis of overexpressed proteins in tumor tissues. Proteins were identified by mass spectrometry. Total protein lysates were prepared from nine paired samples of normal (N) and tumor tissue (T). Anti‐GAPDH antibody was used as a loading control. PD‐ECGF, platelet‐derived endothelial cell growth factor. (d) Image analysis of Western blot data of cofilin‐1. The intensity and area of each band was measured, and these protein levels between normal and tumor tissue, normalized with GAPDH, were calculated. The expression of cofilin‐1 was increased in all tumor tissues.
Cofilin‐1 is associated with hematogenous dissemination in PDAC
To examine the localization of cofilin‐1 and the correlation between its expression and clinicopathological features in PDAC patients, we undertook IHC staining for cofilin‐1 in PDAC tissues. Cofilin‐1 was weakly expressed in normal pancreatic ductal cells (Fig. 2a) and acinar cells (Fig. 2b). Contrary to this, positive cofilin‐1 expression was observed in invasive cancer cells in 57 of 59 samples (96.6%: Fig. 2c–f). Cofilin‐1 was mainly expressed in the cytoplasm of tumor cells and was not found in the stromal cells surrounding the tumor cells.
Figure 2.
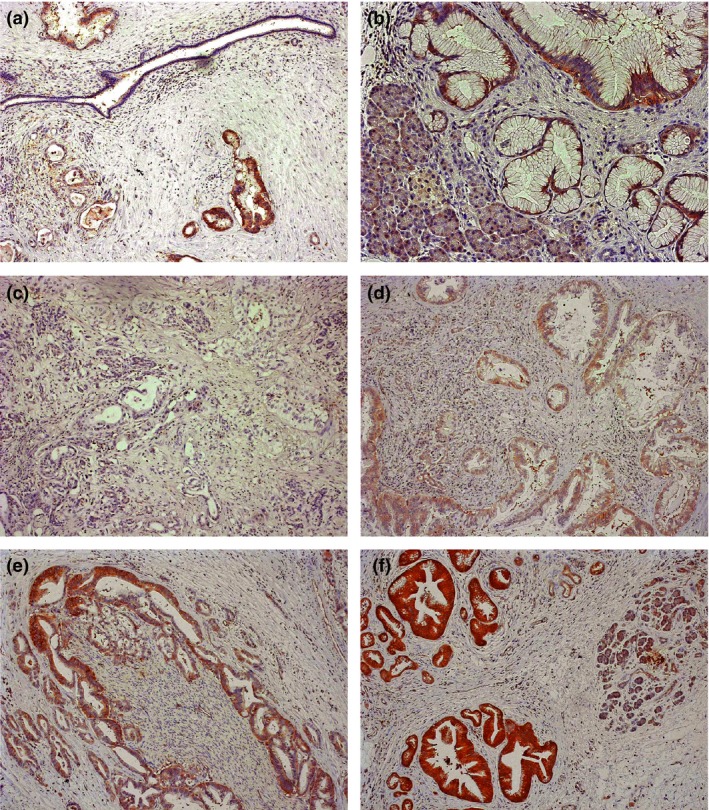
Immunohistochemistry for cofilin‐1 in pancreatic ductal adenocarcinoma tissues. (a) Cofilin‐1 is weakly expressed in normal pancreatic ductal cells (×100). (b) Cofilin‐1 is weakly expressed in acinar cells (×200), and strongly overexpressed in the cytoplasm of tumor cells. (c–f) Immunohistochemical staining patterns of cofilin‐1 in resected pancreatic cancer tissues. Representative staining of low expression (c,d) and high expression (e,f) are indicated (×100).
Next, we investigated whether the expression levels of cofilin‐1 in PDAC cells correlated with the clinical outcomes of PDAC patients. Fifty‐nine PDAC patients, in whom cofilin‐1 expression levels were analyzed by IHC, were divided into two groups based on the immunostaining score (Fig. 2c–f). The intensity of staining of islet cells was used as an internal positive control to evaluate the staining score. Fifteen cases (25.4%) were classified in the weak expression group (Fig. 2c,d), and 44 cases (74.6%) were classified in the strong expression group (Fig. 2e,f). Between these two groups, there were no significant differences in terms of sex, age, histological grade, nodal status, or resection status (data not shown). Interestingly, all six (100%) patients with stage IV cancer who had distant metastasis were categorized in the strong expression group. Furthermore, we examined the correlation with recurrent forms (local, lymph node, peritoneum, and distant metastasis [liver/lung]) of the PDAC patients after surgery. Notably, in 50 of the 59 patients (84.7%) in which a recurrence of disease occurred, the cofilin‐1 strong expression group (20 of 36 patients [55.6%]) showed significantly higher frequency of liver and/or lung metastases as the first recurrent site than did the weak expression group (2 of 14 patients [14.3%]) (P = 0.0083, χ2‐test) (Table 2). These results suggested that cofilin‐1 was associated with the incidence of hematogenous metastasis in PDAC patients.
Table 2.
Recurrence form of patients with pancreatic cancer after surgery in cofilin‐1 immunohistochemical (IHC) analysis
| Cofilin‐1 IHC staining | |||
|---|---|---|---|
| Recurrence form | Total | Low expression | High expression |
| 50 | 14 | 36 | |
| Local recurrence | 15 | 7 | 8 |
| Lymph node | 4 | 1 | 3 |
| Peritoneum | 9 | 4 | 5 |
| Distant metastasis (liver/lung) | 22 | 2 | 20 |
Knockdown of cofilin‐1 decreases migration, not proliferation, in PDAC cells
We next analyzed the biological functions of cofilin‐1 in PDAC cells. To investigate this, we examined whether cofilin‐1 contributes to cell proliferation and migration in PDAC cells. After siRNA transfection, cells were incubated for 72 h and then harvested for Western blotting. Cofilin‐1 expression levels were successfully reduced by cofilin‐1‐specific siRNA in a concentration‐dependent manner (Fig. 3a). The knockdown of cofilin‐1 by siRNA did not affect cell proliferation in either PANC‐1 or Capan‐1 cells (Fig. 3b). To examine whether cofilin‐1 is involved in the migration capacity of PDAC cells, chemotaxis assay was carried out using a Boyden chamber. Chemotaxis was significantly decreased by siRNA in a concentration‐dependent manner in both PANC‐1 and Capan‐1 cell lines (Fig. 3c). These results suggest that cofilin‐1 is functionally involved in the cell motility of PDAC cells.
Figure 3.
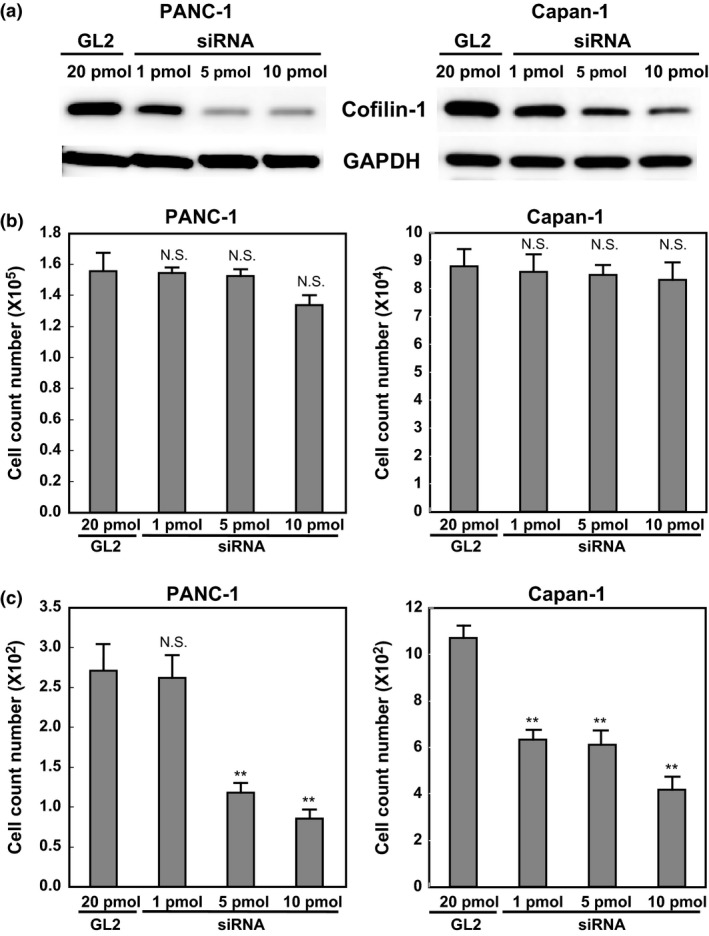
Knockdown of cofilin‐1 decreases the chemotaxis in PANC‐1 and Capan‐1 pancreatic ductal adenocarcinoma cells. (a) Expression of cofilin‐1 in PANC‐1 and Capan‐1 cells using control siRNA (GL2) or cofilin‐1 siRNA, as determined by Western blot analysis. GAPDH was used as a control for internal protein loading. Cofilin‐1 siRNA dose‐dependently inhibits cofilin‐1 protein expression in PANC‐1 and Capan‐1 cells. (b) Proliferation assay of PANC‐1 and Capan‐1 cells using GL2 or cofilin‐1 siRNA at 3 days after siRNA transfection. (c) Chemotaxis assay of two cell lines using a Boyden chamber. Four randomly selected fields were photographed and the number of migrated cells on the back of the filter was counted. *P < 0.05, **P < 0.01, control siRNA versus cofilin‐1 siRNA. N.S., not significant.
Cofilin‐1 is secreted from PDAC cells
We sought to develop a role for cofilin‐1 in clinical use. To investigate the expression of biomarkers, obtaining blood samples from patients is one of the easiest approaches with minimal invasiveness. Thus, we first investigated whether cofilin‐1 is secreted from PDAC cells. Western blot analysis showed that cofilin‐1 protein with a 21‐kDa band was increased in the medium with cultured PANC‐1 and Capan‐1 cells (Fig. 4a). This result indicated that cofilin‐1 was secreted from PANC‐1 and Capan‐1 cells.
Figure 4.
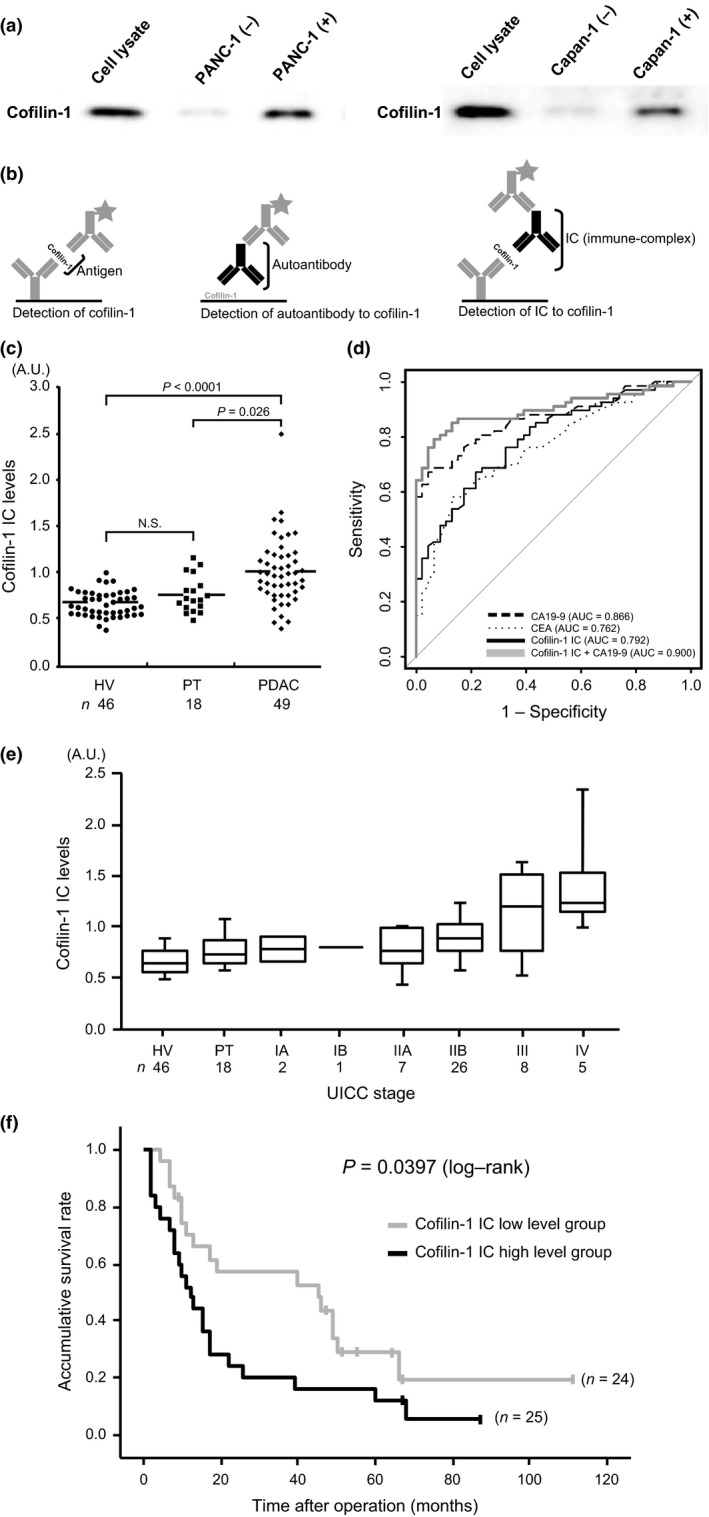
Detection and clinical utility of the cofilin‐1 immune‐complex in sera of patients with pancreatic ductal adenocarcinoma (PDAC). (a) Left lane, cell lysates were applied as a loading control. Middle lane, supernatant of the medium (containing 10% FBS) in which no cells were cultured was used as a negative control. Cofilin‐1 was detected very weakly. Right lane, supernatant of the medium in which PANC‐1 and Capan‐1 cells were cultured for 24 h. Cultured supernatant was incubated in new medium for 24 h. Intense expression of cofilin‐1 was detected. (b) Schema of three detection systems of sandwich ELISA (left, cofilin‐1 antigen; middle, cofilin‐1 autoantibody; right: cofilin‐1 immune‐complex [IC]). (c) Measurement of IC to cofilin‐1 in sera of healthy volunteers (HV), patients with pancreatitis (PT), and PDAC patients. (d) Comparison of receiver operating characteristic curves for classification of cancer (PDAC) and non‐cancer (HV and PT) patients. Dashed line, cancer antigen 19‐9 (CA19‐9); dotted line, carcinoembryonic antigen (CEA); black line, cofilin‐1 IC; bold gray line, combination score of cofilin‐1 IC and CA19‐9 estimated by the logistic regression model. Thin gray line represents the random classification. (e) IC levels in HVs, PT, and each stage of PDAC (according to Union for International Cancer Control criteria). (f) Gray and black lines indicate cofilin‐1 IC low and high level groups, respectively. Kaplan–Meier survival curves show unfavorable prognosis in the cofilin‐1 IC high level group (P = 0.0397).
Establishment of a detection system for IC to cofilin‐1 in sera
We next considered multiple approaches to detect and measure the expression of cofilin‐1 in sera by ELISA. Among the experiments for the detection of cofilin‐1 in sera, including the detection of cofilin‐1 antigen and autoantibody to cofilin‐1, we successfully established a detection system of the IC of cofilin‐1 in sera (Fig. 4b). Using this novel system, we attempted to measure the levels of IC to cofilin‐1 in the sera of PDAC patients. To assess the discriminatory power of serum expression levels of IC to cofilin‐1, we compared the serum IC level among 49 PDAC patients and age‐matched benign controls, including 46 HVs and 18 patients with PT. As shown in Figure 4(c), the serum IC levels in PDAC patients were significantly greater than in the HV and PT patients (PDAC vs. HV, P < 0.0001; PDAC vs. PT, P < 0.026, Mann–Whitney U‐test). Importantly, there was no significant difference in the levels of IC to cofilin‐1 between HV and PT patients (Fig. 4c).
In addition, we estimated the diagnostic ability to distinguish between cancer (PDAC) and non‐cancer (HVs and PT) patients among the levels of cofilin‐1 IC, two representative serum markers for PDAC, CA19‐9, carcinoembryonic antigen, and the combination of cofilin‐1 IC and CA19‐9 in ROC analyses. Although there was no significant difference between the combination model and CA19‐9 alone (P = 0.130, DeLong's test), the area under the ROC curve value of the combination model (0.900) was higher than that of CA19‐9 (0.866) (Fig. 4d).
Level of cofilin‐1 IC in sera is associated with PDAC progression and poor prognosis
Finally, to evaluate the clinical significance of the expression levels of IC to cofilin‐1 in the sera of PDAC patients, we analyzed the correlation between IC levels and clinical outcomes. Cofilin‐1 IC levels showed a stepwise increase and were significantly correlated with Union for International Cancer Control stage during PDAC progression (P = 0.0034, χ2‐test) (Fig. 4e). Dividing all 49 patients into two groups by the median levels of IC to cofilin‐1, the Kaplan–Meier analysis indicated that the high level group predicted poor prognosis of patients with PDAC after surgery (P = 0.039, log–rank test) (Fig. 4f). Moreover, cofilin‐1 IC level was significantly correlated with overall survival of patients with PDAC on univariate analysis (P = 0.042, Cox's proportional hazards model), but was not an independent prognostic factor on multivariate analysis (Table S2). These results suggested that the level of serum cofilin‐1 IC is positively correlated with the degree of PDAC progression and poor prognosis of patients.
Discussion
In this study, we have shown that the serum level of IC to cofilin‐1 identified by a gel‐based proteomic approach is associated with PDAC progression and poor prognosis. We then compared protein expression in PDAC and normal pancreatic tissues. A total of five overexpressed proteins in PDAC, namely vimentin, annexin A1, endophilin B2, PD‐ECGF, and cofilin‐1 were identified and validated.
Vimentin is well known as a predominant intermediate filament protein in mesenchymal cells (a mesenchymal marker) and Nakajima et al.12 reported that vimentin was observed in a few cancer cells of pancreatic primary tumors, but was substantially expressed in liver metastasis. Annexin A1 is known as a member of the family of calcium/phospholipid‐binding proteins, and reports of its expression levels in various cancer tissues are contradictory.13, 14, 15, 16 In PDAC, Bai et al.17 showed that annexin A1 was highly expressed by IHC, and de Graauw et al.18 also suggested that annexin A1 promotes metastasis formation by enhancing transforming growth factor‐β/Smad signaling and actin reorganization, which facilitates an epithelial–mesenchymal transition‐like switch, thereby allowing efficient cell migration and invasion of metastatic breast cancer cells. The relationship between endophilin B2 and PDAC has not been reported. In the present study we have shown, for the first time, the high expression of endophilin B2 in PDAC. The relationship between endophilin B2 and metastasis is reported in prostate cancer, although the molecular mechanism has yet to be elucidated. Takao et al.19 investigated the correlation between PD‐ECGF expression and clinicopathological factors and clinical outcome by IHC in PDAC. They suggested that PD‐ECGF expression in PDAC enhances tumor invasion and/or metastasis through its angiogenic properties.
Cofilin belongs to a family of related proteins with similar biochemical activities called the actin depolymerizing factor/cofilin family. Cofilin‐1 is the most abundant isoform in the actin depolymerizing factor/cofilin family. The cofilin pathway has emerged as having a central role in the generation of free actin filament ends resulting in actin filament remodeling, which is essential in the chemotaxis, cell migration, and invasion of tumor cells.20 The functional roles of cofilin‐1 are different and contradictory among various types of tumor cells. Many of these reports describe the positive correlation between cofilin expression and invasion and metastases of tumor cells because cofilin activation is essential for the formation of stable invadopods, which are used during the migration of invasive tumor cells, linking cofilin to tumor cell invasion.21
In line with previous findings, we have shown that cofilin‐1 expression is associated with high incidence of hematogenous metastases of PDAC patients after curative surgery. Furthermore, in our in vitro experiments, cofilin‐1 has a biological role in the chemotaxis and motility of PDAC cells. Previous reports indicated that cofilin is overexpressed in C6 rat glioblastoma cell lines22 and A549 human lung cancer cells,23 as well as human pancreatic cancer cells,24 which show high invasive capacity. The cofilin pathway is composed of a group of kinases, such as LIM domain kinase 1, and phosphatases, such as SSH, that regulate cofilin and initiate actin polymerization and cell motility. Therefore, the balanced contribution of cofilin and other key molecules, including LIM domain kinase 1 and SSH, is required for chemotaxis and motility in tumor cells.20, 25 Indeed, Wang et al. recently described that SSHL1, a cofilin‐phosphatase, plays a role in actin dynamics and cell migration by reactivating cofilin‐1. Loss of its expression was associated with an increase in the phosphorylation of cofilin‐1 at serine‐3 and further inhibited cell migration in pancreatic cancer cells.26 These results suggest that the molecules related to the cofilin pathway orchestrate invasion and metastasis in PDAC progression.
We next sought to detect three different targets, cofilin‐1 antigen, autoantibody, or putative autoantigen–antibody (IC), as serum samples are readily available and very useful for the investigation of cancer biomarkers. We found that we could only measure and establish a detection system for the IC of cofilin‐1 in sera, but not for the antigen or autoantibody. The reason why the antigen or autoantibody of cofilin‐1 could not be detected is unknown; nevertheless, we have shown that cofilin‐1 is secreted from both primary and metastatic PDAC cell lines (PANC‐1 and Capan‐1). It is speculated that the levels of cofilin‐1 antigen itself produced by tumor cells is insufficient to be measured in the circulatory system. Actually, the immune system reacts to developing tumors and generates autoantibodies against tumor‐associated antigens. The production of autoantibodies can be amplified by the host immune response; therefore, low levels of cofilin‐1 antigen originating from PDAC can still lead to a robust signal.27 Moreover, there is a possibility that the binding affinity between cofilin‐1 antigen and the autoantibody is high and the IC of cofilin‐1 is also effectively enhanced by humoral immune responses against the PDAC tumor.9 Recently, Falco et al.28 have described that BAG3/anti‐BAG3 complexes in sera from PDAC patients are detected at higher levels than in sera from HVs and patients with chronic PT. Taken together, the IC in sera of PDAC patients is a potentially good candidate for detection of prognostic serum biomarkers. Further analysis will be needed to elucidate the mechanism of this phenomenon.
In this study, we first showed that the IC of cofilin‐1 is detected at higher levels in sera of PDAC patients, especially patients with advanced PDAC such as those with hematogenous metastasis, compared to sera from healthy controls or patients with chronic PT. Considering this biomarker for clinical use, it is possible that the IC of cofilin‐1 could be a potential serum biomarker for selecting PDAC patients who have occult metastasis. It would be very useful for those patients to avoid the unnecessary surgery. One of the critical limitations is that this study has been analyzed retrospectively. Clearly, validation of these results in a larger cohort of patients is required to determine the clinical applications of this serum biomarker.
Disclosure Statement
The authors have no conflict of interest.
Supporting information
Table S1. Mass spectrometric identification of pancreatic proteins.
Table S2. Prognostic factors of patients with pancreatic ductal adenocarcinoma in Cox's proportional hazards model.
Appendix S1. Supplementary materials and methods.
Acknowledgments
This work was supported by Grants‐in‐Aid for Scientific Research KAKENHI (Young Scientists B: 22700904 to M.S.), KIBAN C (15K08610 to M.S.), KIBAN B (15H04925 to S.T., H.Y., and M.M.), and Challenge Exploratory Research (16K15607 to S.T., M.S., H.Y., and M.M.), KIBAN C (16K08979 to K.S., M.S., and F.N.), and KIBAN B (26293299 to M.M., S.T., and H.Y.) from the Japan Society for the Promotion of Science.
Cancer Sci 108 (2017) 795–803
Funding Information
This study was supported by grants from the Japanese Ministry of Education, Culture, Sports Science and Technology (15H04925, 15K08610, 16K08979, 16K15607, 22700904, 26293299).
References
- 1. Miller KD, Siegel RL, Lin CC et al Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin 2016; 66: 271–89. [DOI] [PubMed] [Google Scholar]
- 2. de Gramont A, Watson S, Ellis LM et al Pragmatic issues in biomarker evaluation for targeted therapies in cancer. Nat Rev Clin Oncol 2015; 12: 197–212. [DOI] [PubMed] [Google Scholar]
- 3. Takano S, Yoshitomi H, Togawa A et al Apolipoprotein C‐1 maintains cell survival by preventing from apoptosis in pancreatic cancer cells. Oncogene 2008; 27: 2810–22. [DOI] [PubMed] [Google Scholar]
- 4. Capello M, Cappello P, Linty FC et al Autoantibodies to Ezrin are an early sign of pancreatic cancer in humans and in genetically engineered mouse models. J Hematol Oncol 2013; 6: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Beneduce L, Prayer‐Galetti T, Giustinian AM et al Detection of prostate‐specific antigen coupled to immunoglobulin M in prostate cancer patients. Cancer Detect Prev 2007; 31: 402–7. [DOI] [PubMed] [Google Scholar]
- 6. Makrantonakis P, Pectasides D, Aggouridakis C, Visvikis A, Daniilidis J, Fountzilas G. Squamous cell carcinoma antigen, circulating immune complexes, and immunoglobulins in monitoring squamous cell carcinoma of head and neck: a study of the hellenic co‐operative oncology group (HeCOG). Am J Clin Oncol 1999; 22: 542–9. [DOI] [PubMed] [Google Scholar]
- 7. Fuchs C, Krapf F, Kern P, Hoferichter S, Jäger W, Kalden JR. CEA‐containing immune complexes in sera of patients with colorectal and breast cancer–analysis of complexed immunoglobulin classes. Cancer Immunol Immunother 1988; 26: 180–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cramer DW, O'Rourke DJ, Vitonis AF et al CA125 immune complexes in ovarian cancer patients with low CA125 concentrations. Clin Chem 2010; 56: 1889–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rho JH, Lampe PD. High‐throughput screening for native autoantigen‐autoantibody complexes using antibody microarrays. J Proteome Res 2013; 12: 2311–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Takano S, Togawa A, Yoshitomi H et al Annexin II overexpression predicts rapid recurrence after surgery in pancreatic cancer patients undergoing gemcitabine‐adjuvant chemotherapy. Ann Surg Oncol 2008; 15: 3157–68. [DOI] [PubMed] [Google Scholar]
- 11. Satoh M, Haruta‐Satoh E, Omori A et al Effect of thyroxine on abnormal pancreatic proteomes of the hypothyroid rdw rat. Proteomics 2005; 5: 1113–24. [DOI] [PubMed] [Google Scholar]
- 12. Nakajima S, Doi R, Toyoda E et al N‐cadherin expression and epithelial‐mesenchymal transition in pancreatic carcinoma. Clin Cancer Res 2004; 10: 4125–33. [DOI] [PubMed] [Google Scholar]
- 13. Ahn SH, Sawada H, Ro JY et al Differential expression of annexin I in human mammary ductal epithelial cells in normal and benign and malignant breast tissues. Clin Exp Metastasis 1997; 15: 151–6. [DOI] [PubMed] [Google Scholar]
- 14. Paweletz CP, Ornstein DK, Roth MJ et al Loss of annexin 1 correlates with early onset of tumorigenesis in esophageal and prostate carcinoma. Cancer Res 2000; 60: 6293–7. [PubMed] [Google Scholar]
- 15. Garcia Pedrero JM, Fernandez MP, Morgan RO et al Annexin A1 down‐regulation in head and neck cancer is associated with epithelial differentiation status. Am J Pathol 2004; 164: 73–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Duncan R, Carpenter B, Main LC, Telfer C, Murray GI. Characterisation and protein expression profiling of annexins in colorectal cancer. Br J Cancer 2008; 98: 426–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bai XF, Ni XG, Zhao P et al Overexpression of annexin 1 in pancreatic cancer and its clinical significance. World J Gastroenterol 2004; 10: 1466–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. de Graauw M, van Miltenburg MH, Schmidt MK et al Annexin A1 regulates TGF‐beta signaling and promotes metastasis formation of basal‐like breast cancer cells. Proc Natl Acad Sci U S A 2010; 107: 6340–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Takao S, Takebayashi Y, Che X et al Expression of thymidine phosphorylase is associated with a poor prognosis in patients with ductal adenocarcinoma of the pancreas. Clin Cancer Res 1998; 4: 1619–24. [PubMed] [Google Scholar]
- 20. Wang W, Eddy R, Condeelis J. The cofilin pathway in breast cancer invasion and metastasis. Nat Rev Cancer 2007; 7: 429–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yamaguchi H, Wyckoff J, Condeelis J. Cell migration in tumors. Curr Opin Cell Biol 2005; 17: 559–64. [DOI] [PubMed] [Google Scholar]
- 22. Gunnersen JM, Spirkoska V, Smith PE, Danks RA, Tan SS. Growth and migration markers of rat C6 glioma cells identified by serial analysis of gene expression. Glia 2000; 32: 146–54. [PubMed] [Google Scholar]
- 23. Keshamouni VG, Michailidis G, Grasso CS et al Differential protein expression profiling by iTRAQ‐2DLC‐MS/MS of lung cancer cells undergoing epithelial‐mesenchymal transition reveals a migratory/invasive phenotype. J Proteome Res 2006; 5: 1143–54. [DOI] [PubMed] [Google Scholar]
- 24. Sinha P, Hütter G, Köttgen E, Dietel M, Schadendorf D, Lage H. Increased expression of epidermal fatty acid binding protein, cofilin, and 14–3‐3‐σ (stratifin) detected by two‐dimensional gel electrophoresis, mass spectrometry and microsequencing of drug‐resistant human adenocarcinoma of the pancreas. Electrophoresis 1999; 20: 2952–60. [DOI] [PubMed] [Google Scholar]
- 25. Mizuno K. Signaling mechanisms and functional roles of Cofilin phosphorylation and dephosphorylation. Cell Signal 2013; 25: 457–69. [DOI] [PubMed] [Google Scholar]
- 26. Wang Y, Kuramitsu Y, Kitagawa T et al Cofilin‐phosphatase slingshot‐1L (SSH1L) is over‐expressed in pancreatic cancer (PC) and contributes to tumor cell migration. Cancer Lett 2015; 360: 171–6. [DOI] [PubMed] [Google Scholar]
- 27. Dumstrei K, Chen H, Brenner H. A systematic review of serum autoantibodies as biomarkers for pancreatic cancer detection. Oncotarget 2016; 7: 11151–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Falco A, Rosati A, Festa M et al BAG3 is a novel serum biomarker for pancreatic adenocarcinomas. Am J Gastroenterol 2013; 108: 1178–80. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Mass spectrometric identification of pancreatic proteins.
Table S2. Prognostic factors of patients with pancreatic ductal adenocarcinoma in Cox's proportional hazards model.
Appendix S1. Supplementary materials and methods.


