Abstract
Multiple myeloma (MM) is characterized by the accumulation of a population of malignant plasma cells within the bone marrow and its microenvironment. A hypoxic niche is located within the microenvironment, which causes myeloma cells to become quiescent, anti‐apoptotic, glycolytic, and immature. Cell heterogeneity may be related to distinct gene expression profiles under hypoxic and normoxic conditions. During hypoxia, myeloma cells acquire these phenotypes by downregulating interferon regulatory factor 4 (IRF4), an essential transcription factor in myeloma oncogenesis. To identify essential microRNAs and their targets regulated under hypoxic conditions, we undertook microRNA and cDNA microarray analyses using hypoxia‐exposed primary MM samples and myeloma cell lines. Under hypoxia, only miR‐210 was highly upregulated and was accompanied by direct downregulation of an 18S rRNA base methyltransferase, DIMT1. This inverse expression correlation was validated by quantitative RT‐PCR for primary MM samples. We further determined that DIMT1 has an oncogenic potential as its knockdown reduced tumorigenicity of myeloma cells through regulation of IRF4 expression. Notably, by analyzing gene expression omnibus datasets in the National Center for Biotechnology Information database, we found that DIMT1 expression increased gradually with MM progression. In summary, by screening for targets of hypoxia‐inducible microRNA‐210, we identified DIMT1 as a novel diagnostic marker and therapeutic target for all molecular subtypes of MM.
Keywords: DIMT1, hypoxia, IRF4, miR‐210, multiple myeloma
Multiple myeloma is characterized by the accumulation of a population of malignant plasma cells within the BM and its microenvironment.1 Two distinct microenvironmental niches in the BM, the endosteal (osteoblastic) and vascular niches, are more hypoxic than BM and thought to play a crucial role in maintaining myeloma‐initiating cells and hematopoietic stem cells.2, 3, 4, 5, 6 It is known that gene expression patterns in cancer cells are heterogeneous, even in the same individual. This heterogeneity is related to epigenetic alterations and confers plasticity of stem‐like states and drug resistance to cancer cells.7 Distinct gene expression may be related in part to cell conditions such as niches. Specifically, hypoxic oxygen stress occurs in the niches, leading to various epigenetic gene alterations. Because hypoxia‐induced gene alterations may strongly contribute to resistance to chemotherapy in non‐curable diseases, such as MM, numerous clinical or basic studies have examined the phenotypic changes that occur in hypoxia‐exposed cancer cells.
Although a variety of gene expression changes occur in cancer cells during hypoxia, the master regulator may be common to all cancer subtypes, namely, the transcription factors HIF‐1α and HIF‐2α.8, 9, 10, 11, 12, 13, 14 Under hypoxic conditions, HIFs regulate a variety of downstream target genes, including those involved in cell‐cycle arrest, angiogenesis, glycolysis, and glucose transport induction.8, 9, 10, 11, 12, 13, 14 Additionally, in MM, HIFs were reported to play crucial roles in pathogenesis.3 For instance, by activating HIF‐1α, hypoxia induces activation of the glycolytic pathway and inactivation of the tricarboxylic acid cycle in myeloma cells by regulating numerous related genes.15 Hypoxia‐inducible factors may not only regulate the transcription of coding genes, but also non‐coding RNA such as miRNA, a class of small non‐coding regulatory RNA molecules that bind to the 3′‐UTR of target messenger RNAs to repress their translation in various hematological malignancies.16, 17, 18, 19, 20 Direct regulation of HIF‐1α against miR‐210 has been reported in various solid cancers,21 and its upregulation leads to inhibition of cell growth.22, 23, 24, 25 Recent studies have provided considerable evidence that miRNAs are master regulators, orchestrating a myriad of cellular pathways to control growth during physiological and pathological conditions of MM.26, 27, 28, 29, 30, 31, 32 Among them, Umezu et al. recently reported a functional analysis of exosomes discharged from myeloma cells during hypoxia. They showed that miR‐135b is highly integrated in the exosomes of myeloma cells, which upregulate HIF‐1α by downregulating factor inhibiting HIF‐1.32 Hypoxia‐inducible factor‐1α stimulation causes significant induction of vessel formation and vascular endothelial growth factors, promoting tumor formation. Although the miRNAs present in exosomes released from myeloma cells have been determined, there have been no reports describing the expression changes of miRNAs in myeloma cells during hypoxia.
The aim of this study was to detect regulated miRNA(s) and its (their) essential target(s) affected by hypoxia in myeloma cells. We first examined the phenotypic changes in hypoxia‐exposed myeloma cells and then searched for coding and non‐coding genes (miRNAs) by whole gene/miRNA array analysis.
Materials and Methods
For additional information, including sequences of siRNA, see Appendix S1.
Primary MM samples
The study included 15 cases of primary multiple myeloma from Akita University Hospital (Akita, Japan). Samples were collected under a protocol approved by the Institutional Review Boards of Akita University (no. 1313). This study was carried out with written informed consent of the study participants and the approval of these Institutional Review Boards, according to the Declaration of Helsinki, before collection of the specimens. Information about the patient samples is summarized in Table 1. Primary MM samples were subjected to hypoxia for 48 h and then CD38++ cells were sorted; total RNA was subsequently collected.
Table 1.
Characteristics of 15 patients with primary multiple myeloma
| Patient no. | Age, years | Sex | Progression at diagnosis† | ISS | TP (g/dL) | Alb (g/dL) | β2MG | Bone lesion | Hb (g/dL) | Cre (mg/dL) | Ca (mg/dL) | Ig type | IgG (mg/dL) | IgA (mg/dL) | IgM (mg/dL) | Plasma cell (%)‡ | Karyotype | p53 del§ |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 82 | M | RR | I | 7.6 | 4.2 | 4.4 | + | 7.3 | 1.19 | 9.2 | IgGκ | 2345 | 64 | 13 | NA | 46, XY, del(20)(q11.2q13.3) [7/20] | − |
| 2 | 63 | F | ND | III | 12.3 | 2.8 | 7.9 | + | 10.7 | 0.85 | 7.6 | IgGλ | 9642 | 12 | 7 | 40 | Normal | − |
| 3 | 62 | F | ND | I | 9.5 | 4.5 | 2.0 | − | 12.9 | 0.64 | 10.0 | IgGλ | 3926 | 36 | 38 | 12 | 61,X,−X,t(1,18)(p13;q21),+5, −8, −10, −12, −13, −14,+15, −16,add(17)(p11,2), −18,+19, −210, −22[1/20] | + |
| 4 | 34 | M | ND | III | 11.7 | 3.4 | 6.4 | + | 7.2 | 1.11 | 10.7 | IgAλ | 188 | 6891 | 42 | 23 | Normal | − |
| 5 | 37 | M | ND | I | 7.7 | 4.1 | 3.3 | + | 14.7 | 0.69 | 8.8 | IgGλ | 2195 | 37 | 32 | 38 | Normal | − |
| 6 | 66 | M | ND | II | 10.1 | 2.9 | 2.6 | + | 10.4 | 0.54 | 9.4 | IgGκ | 5830 | 15 | 10 | NA | NA | − |
| 7 | 63 | F | ND | III | 4.7 | 2.7 | 15.0 | + | 8.1 | 1.19 | 7.9 | BJPλ | 264 | 25 | 10 | 70 | 52, XX, +5, +7, +9, +15, +19, +21 [1/20], 52, idem, del(1)(p?)[5/20], 52, idem, del(1)(p?)[4/20] | − |
| 8 | 84 | F | ND | I | 7.5 | 4.0 | 2.1 | + | 11.5 | 0.63 | 8.5 | IgAλ | 792 | 1369 | 59 | 14 | Normal | − |
| 9 | 67 | F | ND | I | 7.3 | 3.7 | 2.5 | + | 8.2 | 0.62 | 9.2 | IgGκ | 2026 | 14 | 33 | 25 | 46, XX, t(2,8) (p23;q22) [1/20] | + |
| 10 | 82 | F | ND | III | 13.2 | 3.2 | 11.4 | + | 7.8 | 1.13 | 10.5 | IgGλ | 5253 | 14 | 16 | 37 | Normal | − |
| 11 | 63 | M | ND | II | 11.4 | 3.4 | 5.4 | + | 8.9 | 0.66 | 9.5 | IgGλ | 6410 | 381 | 14 | 14 | Normal | − |
| 12 | 57 | M | RR | III | 6.0 | 2.7 | 10.5 | − | 8.5 | 1.29 | 7.7 | IgGλ | 1871 | 5 | 12 | 21 | Normal | − |
| 13 | 71 | M | RR | III | 10.7 | 3.1 | 11.0 | + | 8.2 | 1.87 | 12.1 | IgGκ | 6695 | 4 | 7 | 21 | Normal | − |
| 14 | 61 | M | ND | I | 8.8 | 4.1 | 2.4 | − | 9.3 | 0.63 | 9.2 | IgAλ | 301 | 3906 | 15 | 12 | Normal | − |
| 15 | 74 | M | ND | I | 6.5 | 4.5 | 2.8 | + | 8.6 | 1.06 | 9.6 | BJPλ | 437 | 15 | 12 | 22 | Normal | − |
β2MG, β2‐microgloublin; Alb, albumin; BJP, Bence‐Jones protein; Cre, creatinine; F, female; Hb, hemoglobin; ISS, International Staging System; M, male; NA, not available; TP, total protein. †Newly diagnosed (ND) or relapsed/refractory sample (RR). ‡Plasma cell % in the bone marrow. §Presence (+) or absence (−) of p53 deletion (del) (p53 deletion or 17p13 deletion) assessed by FISH.
Myeloma cell lines
We used 12 well‐known MM cell lines with various molecular subtypes,33 RPMI‐8226 (8226), KMS‐12‐BM (KMS12BM), KMS‐11 (KMS11), U266, JJN3, Amo1, MM.1S, NCI‐H929 (H929), KMS‐20 (KMS20), KMS‐21‐PE (KMS21PE), SKMM1, and KMS‐28 (KMS28).
Gene or miRNA array
We analyzed gene expression using an Agilent DNA microarray and an Agilent miRNA microarray (Agilent, Santa Clara, CA, USA). The array scanner was an Agilent G2600A SureScan Microarray Scanner System. Experiments protocols were according to Agilent Protocol version 6.7 (for gene expression), or Agilent Protocol version 2.4 (for miRNA expression). Data were analyzed by GeneSpring (Agilent), computed normalized signal intensity with 75 percentile (for gene) or 90 percentile (for miRNA) normalization. Data were uploaded at GSE80769 (cell line hypoxia miRNA array), GSE80770 (patient samples hypoxia miRNA array), GSE80140 (cell line hypoxia DNA array), and GSE80545 (patient samples hypoxia DNA array).
Quantitative RT‐PCR analysis
Quantitative RT‐PCR was carried out using the TaqMan method (Applied Biosystems, Foster City, CA, USA). TaqMan probes of GAPDH (Hs02758991_g1), DIMT1 (Hs00917508_m1), CIAPIN1 (Hs00938899_m1), TTC13 (Hs00225960_m1), NOL12 (Hs04194561_s1), ARMC1 (Hs00982339_m1), U47 (001223), and miR‐210 (000512) were purchased from Applied Biosystems. Expression levels were separately normalized with GAPDH (for gene) or U47 (for miRNA), and the relative expression level of specific mRNA or miRNA was presented by or Quantitative RT‐PCR was carried out using LightCycler Nano (Roche, Basel, Switzerland) with the TaqMan method. Total RNA was extracted using TRIzol (Life Technologies, Palo Alto, CA, USA). Reverse transcription was carried out using a Transcriptor First Strand cDNA Synthesis Kit (Roche).
Western blot analysis
Western blot analysis was undertaken according to the manufacturer's protocol. Antibodies of HIF‐1α (#3716S), IRF4 (#4964), c‐Myc (#5605S), and Bcl‐2 (#2870P) were purchased from Cell Signaling Technology (Danvers, MA, USA). Antibody of DIMT1 (sc135130) was from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Tubulin (MS‐581‐P0) was from NeoMarkers (Fremont, CA, USA).
Xenograft mouse model
MM.1S cells (2 × 106 each) were s.c. injected into the right or left side of the body of 6–8‐week‐old female NOG mice (Central Institute for Experimental Animals, Kawasaki, Japan). The protocols for animal experimentation described in this paper were previously approved by the Animal Committee, Akita University (approval no. b‐1‐2301).
Cell cycle analysis
The cells were suspended in a mixture containing 0.2 mL of 0.9% NaCl and 3.0 mL of 70% EtOH, after which the nuclei were stained with propidium iodide (Sigma‐Aldrich, St. Louis, MO, USA). The cellular DNA content was then measured using a FACS Canto flow cytometer running the FACS Diva program (BD Biosciences, San Jose, CA, USA).
Apoptosis analysis
An annexin V–FITC apoptosis detection kit (Sigma‐Aldrich) was used to assess the incidence of apoptosis over 72 h (for nucleotransfection) or 48 h (for drugs). The assays were carried out according to manufacturer's protocol.
Cell viability assay
Cell viability assays were carried out using an In Vitro Toxicology Assay Kit, XTT based, according to manufacturers’ protocol (Sigma‐Aldrich).
Chemicals
PX‐478 was purchased from MedKoo Biosciences (Chapel Hill, NC, USA). Bortezomib was purchased from LC Laboratories (Woburn, MA, USA).
Statistical analysis
Data was analyzed by either Student's t‐test or paired t‐test. Bars represent mean ± 95% confidence interval of three independent experiments. Asterisks (*) indicate statistical significance: *P < 0.05; **P < 0.01; ***P < 0.001.
Results
Downregulation of IRF4 oncogenic axis and upregulation of miR‐210 during chronic hypoxia in myeloma cells
Oxygen pressure in endosteal or vascular niches is thought to be <10 mmHg.3 In our in vitro experiments, because 1% O2 equates to 7.6 mmHg, these conditions mimic the microenvironment. To validate the phenotypic changes in MM, we first examined myeloma cell lines under conditions of chronic hypoxia (1% O2 for 24, 48, and 72 h) and observed changes in cell phenotypes. We confirmed that hypoxia could induce following phenotypical changes in myeloma cells in vitro such as cell growth inhibition (Fig. S1a) accompanied by an increased proportion of G1‐arrested cells (Fig. S1b), acidification of the medium by accelerated glycolysis (Fig. S1c), increased drug resistance (Fig. S1d), and decreased expression of CD138 (Fig. S1e). These alterations are identical to those reported in previous studies.15, 34 However, we could not detect significant changes between the percentage of sub‐G1 cells in normoxia and hypoxia, suggesting that hypoxia maintains anti‐apoptotic phenotypes in myeloma cells. These results also suggested that myeloma cells could survive under conditions of hypoxia by epigenetically affecting regulated oncogenic factors. The most promising candidate oncogenic factor was IRF4, which controls a myeloma‐specific gene expression program that fuses the IRF4 regulatory programs from activated B cells and CD138‐expressing plasma cells.35, 36 We undertook qRT‐PCR and Western blot analyses to examine the effect of hypoxia on the expression of IRF4/IRF4 and its target MYC/MYC, and noted reduced expression of these genes/products during hypoxia in all myeloma cell lines examined (Fig. S1f,g). This might be partly responsible for cell growth inhibition (by MYC) and reduction of CD138 expression (by IRF4) in myeloma cells, although it was unclear why hypoxia could not affect cell survival despite promoting IRF4 downregulation. Our results suggested that MM might demonstrate an anti‐apoptotic phenotype in both hypoxia and normoxia through different gene initiation mechanisms.
To identify hypoxia‐induced miRNA, we carried out miRNA screening using myeloma cell lines and four primary samples that were subjected to chronic hypoxic conditions. Because our experiment showed a significant decrease in cell numbers on exposure to 1% O2 for 24 h, and other reports defined a duration of more than 24 h as chronic hypoxia, we also defined chronic hypoxia as exposure of cells to 1% O2 for 24 h or more.3, 14, 37 From the 2006 array probes that were used for expression analysis, 238 probes could be detected among all four examined cell lines and 198 probes among primary samples. Among these miRNAs with altered expression, we found upregulation of miR‐210 in all examined cell lines (Figs 1a,S2), and this expression was also confirmed by qRT‐PCR (Fig. 1b) and northern blot analysis (Fig. 1c). MicroRNA microarray analysis revealed that all four primary cases for which CD38++ cells were purified (Fig. S3) also showed miR‐210 upregulation during chronic hypoxia (Figs 1a,S2). These data strongly suggest that miR‐210 might be upregulated by hypoxia in MM. We validated expression of miR‐210 by qRT‐PCR analysis in 15 primary samples, and found significant upregulation of miR‐210 in all examined primary samples (Fig. 1d). Upregulation of miR‐210 might be a common occurrence in various molecular subtypes of MM. MicroRNA‐210 expression has been well documented in various solid tumors and is a target of HIF‐1α.21, 22, 23, 24, 38) It is well documented that HIF‐1α upregulates miR‐210 by direct interaction with the miR‐210 promoter region during hypoxia in various solid tumors.21 We confirmed that HIF‐1α was upregulated 3 h after hypoxia induction in myeloma cells, as described previously.14 Using an inhibitor of HIF‐1α, it was shown that PX‐478,39 at 25 μM, inhibited accumulation of HIF‐1α during acute hypoxia (3 h) (Fig. S4a). It was also determined that PX‐478 could inhibit upregulation of miR‐210 during hypoxia in a time‐dependent manner (Fig. S4b). Although we did not examine its direct interaction with miR‐210, this result strongly suggests that hypoxia can upregulate miR‐210 through upregulation of HIF‐1α in MM. For the next part of our study, we examined essential oncogenic targets of miR‐210 in MM.
Figure 1.
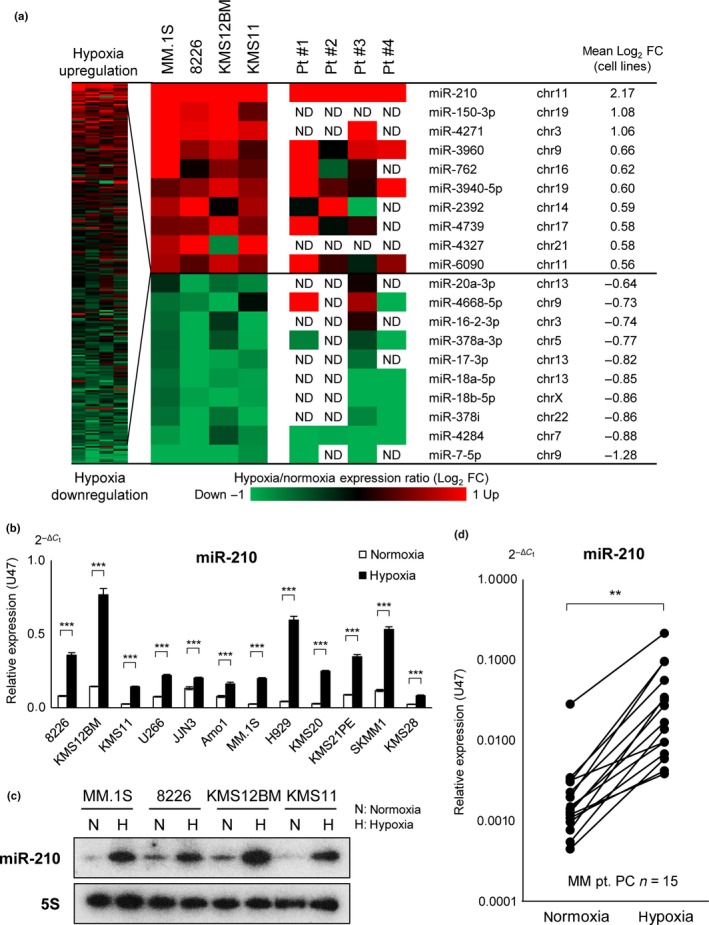
Chronic hypoxia induces microRNA (miR)‐210 upregulation in multiple myeloma (MM). (a) MicroRNA expression profiling for four myeloma cell lines and four primary samples under 1% O2 over 48 h. chr, chromosome; FC, fold change; ND, not detected; Pt., patient. (b) Quantitative (q)RT‐PCR of miR‐210 for indicated myeloma cell lines under 1% O2 over 48 h. Bars represent mean ± 95% confidence interval of three independent experiments. ***P < 0.001. Student's t‐test was used to test for significance. (c) Northern blot analysis of miR‐210 in myeloma cell lines under chronic hypoxia. (d) qRT‐PCR analysis of patient samples (n = 15) under 1% O2 over 48 h. Samples were sorted by CD38++ cells and examined by qRT‐PCR analysis. **P < 0.01. Paired t‐test was used to test for significance. PC, plasma cell.
MicroRNA‐210 directly downregulates DIMT1 in MM
It is well documented that miRNA translationally represses mRNA protein production by binding to the seed sequence in the 3′‐UTR of the target RNA in various hematological malignancies.16, 17, 18, 19, 20 It is also known that miRNA occasionally represses expression of mRNA itself.40, 41 We therefore undertook cDNA array analyses using the same samples subjected to miRNA analysis. Among genes that are downregulated by hypoxia, we detected five commonly downregulated genes that possessed a seed sequence of miR‐210 with a high rate of context score by using the TargetScan program (Fig. 2a).40 These genes were DIMT1, CIAPIN1, TTC13, ARMC1, and NOL12 (Fig. 2a,b). We then transiently transduced miR‐210 into MM.1S, H929, and KMS11 cells (Fig. S5), and examined these genes through qRT‐PCR (Fig. 2c). We found that DIMT1, TTC13, and ARMC1 were significantly reduced in all examined cells compared to controls (Scrambled). Among them, because DIMT1 was the most significantly downregulated gene, it was thought to be the best candidate for hypoxia‐induced miR‐210 regulation in MM.
Figure 2.
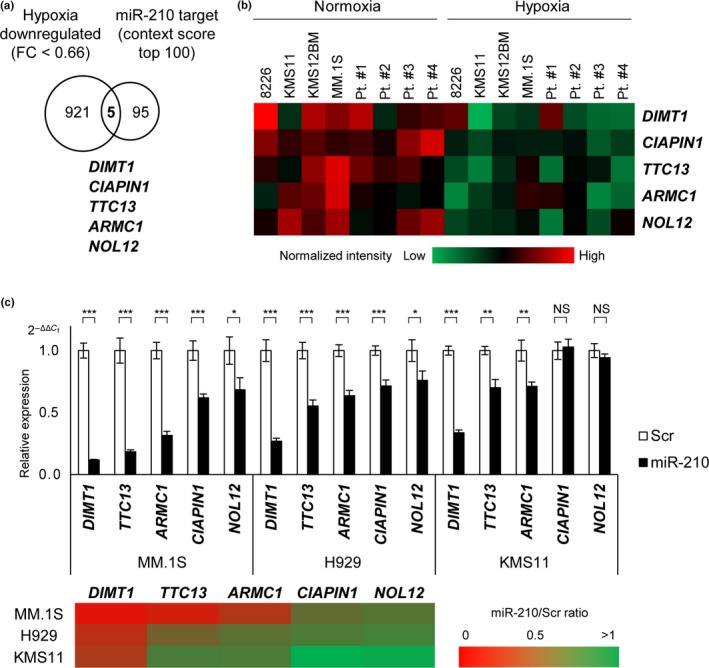
DIMT1 is the most likely target of microRNA (miR)‐210 in multiple myeloma. (a) Diagram of predicted high scoring candidate genes of miR‐210 showing fold change (FC) <0.66 in four myeloma cell lines during hypoxia. (b) Heat map of candidate gene expression for myeloma cell lines (n = 4) and primary samples (n = 4). (c) Quantitative expression of the candidate targets for miR‐210 transduced myeloma cells. Upper panel: quantitative RT‐PCR for candidate targets of miR‐210 (24 h after miR‐210 transduction in indicated myeloma cell lines). Lower panel: Heat map. Bars represent mean ± 95% confidence interval of three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001. Student's t‐test was used to test for significance. NS, not significant; Scr, scrambled siRNA.
To examine whether miR‐210 could directly regulate DIMT1 by binding to its seed sequence of the 3′‐UTR, we used a luciferase reporter assay for KMS11 cells transiently transduced with the 3′‐UTR of wild‐type or mutated DIMT1 (Fig. S6a). For this analysis, we established KMS11 transfectants stably expressing miR‐210 (KMS11‐miR‐210‐puro) and its associated control (KMS11‐scrambled‐puro). Luciferase activity of the wild type showed remarkable repression in KMS11‐miR‐210‐puro during normoxia, indicating that miR‐210 could directly bind to the 3′‐UTR of DIMT1 (Fig. S6b). An interesting finding was that, although there was no significant difference in miR‐210 expression between wild‐type and mutated DIMT1 during hypoxia, a significant decrease was detected during normoxia (Fig. S6c). This must be attributable to the fact that, in hypoxia, there were no significant differences in expression of miR‐210 between KMS11‐miR‐210‐puro and KMS11‐scrambled‐puro.
Hypoxia downregulates DIMT1, and hypoxia‐induced miR‐210 suppresses DIMT1 and IRF4 in MM
To validate whether chronic hypoxia could induce reduction in DIMT1 expression, we carried out qRT‐PCR on 12 MM cell lines, and confirmed that all examined cells showed significant reduction of DIMT1 (Fig. 3a) and its product during chronic hypoxia (Fig. 3b). Significant downregulation of DIMT1 by hypoxia was also detected in all primary myeloma samples examined (n = 15) (Fig. 3c). Reduction in DIMT1 in primary samples was also confirmed by immunohistochemical analysis of three primary cases (Figs 3d,S7). Notably, this analysis revealed that DIMT1 was strongly localized to the cytoplasm of primary myeloma cells during normoxia but not in other BM cells, and also that CD138 and DIMT1 expression was reduced during hypoxia. Moreover, qRT‐PCR analysis revealed that DIMT1 in myeloma cell lines (n = 12) was expressed at a significantly higher level than that of B‐cell lymphoma (n = 28) (Fig. S8). These results suggested that DIMT1 expression is myeloma cell‐specific. We further carried out transient transduction of miR‐210 in myeloma cell lines to examine its effect on DIMT1 expression, and found reduced expression of DIMT1 in a time‐dependent manner (Fig. 3e). Notably, IRF4 was also downregulated in miR‐210 transiently transduced cells. Together, these results strongly suggest that hypoxia‐induced miR‐210 expression leads to direct reduction in DIMT1/DIMT1 in myeloma cells, resulting in downregulation of IRF4 under hypoxia.
Figure 3.
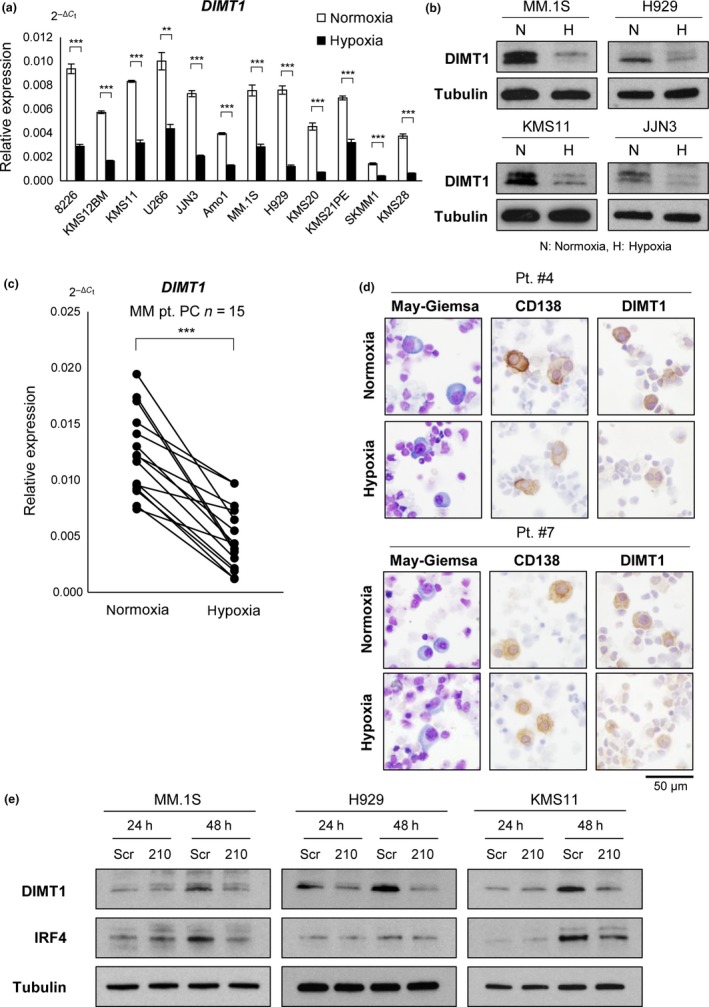
Hypoxia downregulates dimethyladenosine transferase 1 homolog 1 (DIMT1), and hypoxia‐inducible microRNA‐210 suppresses DIMT1 and interferon regulatory factor 4 (IRF4) production in multiple myeloma. (a) Quantitative RT‐PCR of DIMT1 for indicated cell lines under hypoxia (48 h). Bars represent mean ± 95% confidence interval of three independent experiments. ***P < 0.001. Student's t‐test was used to test for significance. (b) Western blot analysis of DIMT1 in indicated cell lines under normoxia (N) and hypoxia (H). (c) Quantitative RT‐PCR for primary myeloma samples (n = 15) treated with 1% O2 over 48 h. ***P < 0.001. Paired t‐test was used to test for significance. PC, plasma cell. (d) Immunostaining of DIMT1 and CD138 in two primary samples (pt. #4 and pt. #7) with 1% O2 over 48 h. May–Giemsa staining is also shown. (e) Western blot of DIMT1 and IRF4 for indicated microRNA‐210 transiently transduced myeloma cells (MM.1S, H929, and KMS11). Scr, scrambled siRNA.
Oncogenic potential of DIMT1 in MM
It has been reported that 18S rRNA base methyltransferases, including DIMT1 and WBSCR22, are required for distinct pre‐rRNA processing reactions leading to synthesis of ribosome.42, 43 A study using siRNA screening identified an association between WBSCR22 and myeloma cell survival;44 however, the association between DIMT1 and myeloma oncogenesis is largely unknown. To examine its function in myeloma oncogenesis, we undertook a knockdown study of DIMT1 in myeloma cell lines (Fig. 4a). Because both hypoxia and miR‐210 transduction could downregulate IRF4, we reasoned that it might be a downstream target of DIMT1. Thus, we undertook transient knockdown of DIMT1 and investigated the relationship between DIMT1 and IRF4 expression. Transient transduction of siDIMT1 (siDIMT1#1 and siDIMT1#2) repressed IRF4 protein expression (Fig. 4b), although siDIMT1 did not affect IRF4 gene expression (data not shown). In addition, MYC/MYC and BCL2/Bcl‐2 did not show any expression changes. This result suggested that DIMT1 might have target selectivity and that it could regulate IRF4‐IRF4 translation.
Figure 4.
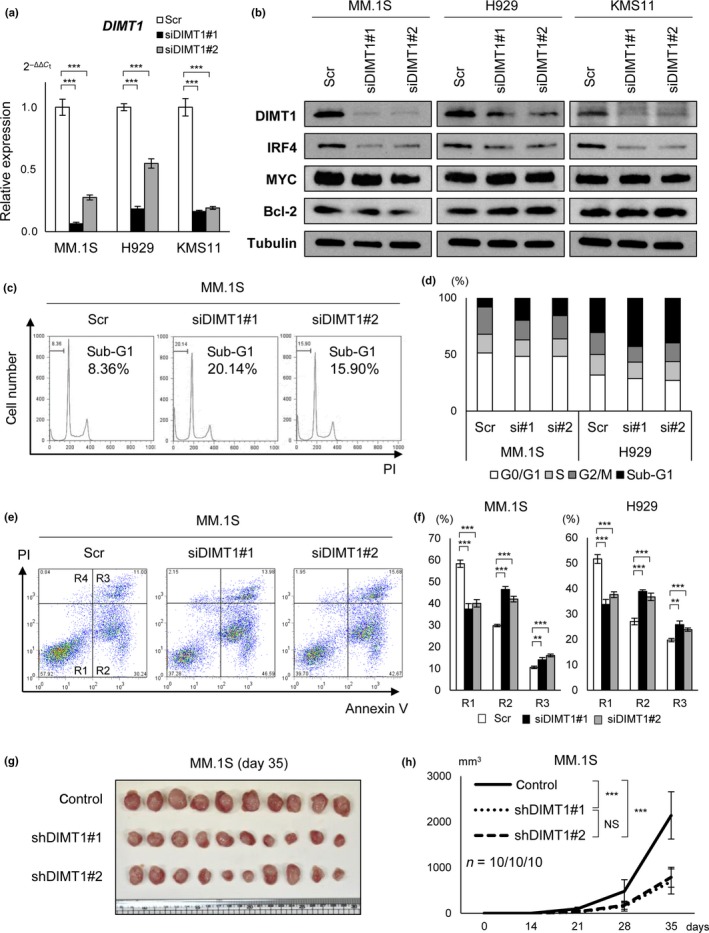
Dimethyladenosine transferase 1 homolog 1 (DIMT1) regulates myeloma cell survival. (a) Quantitative RT‐PCR for DIMT1 in MM.1S, H929, and KMS11 cells transiently transduced with siDIMT1#1, siDIMT1#2, and control scrambled siRNA (Scr) for 24 h. Bars are mean ± 95% confidence interval of three independent experiments. ***P < 0.001. Student's t‐test was used to test for significance. (b) Western blot analysis of DIMT1, interferon regulatory factor 4 (IRF4), MYC, and Bcl‐2 in MM.1S, H929, and KMS11 cells transiently transduced with siDIMT1#1, siDIMT1#2, and control siRNA (Scr) for 48 h. (c,d) Cell cycle and percentage (%) of cells in sub‐G1, G0/1, S, and G2/M, examined in siDIMT1 transduced multiple myeloma (MM) cells. (e,f) Apoptosis assay in MM cells transiently transduced with siDIMT1#1, siDIMT1#2, and control siRNA (Scr). (e) Annexin V assay. R1, viable (no‐apoptotic) cells; R2, early apoptosis; R3, late apoptosis; R4, necrosis. x‐axis: annexin V; y‐axis: propidium iodide (PI). (f) Percentage (%) of viable cells (R1) and apoptotic cells (R2 and R3). Bars represent mean ± 95% confidence interval of three independent experiments. **P < 0.01; ***P < 0.001. Student's t‐test was used to test for significance. (g,h). In vivo transplantation of shDIMT1 (shDIMT1#1‐GFP, shDIMT1#2‐GFP) or Control‐GFP stably transduced MM.1S cells into NOG mice (n = 10 each). (g) Photograph of tumors. (h) Tumor growth curve. x‐axis, time after transplantation (days); y‐axis, tumor volume (mm3, major × minor2/2). ***P < 0.001. Student's t‐test was used to test for significance. NS, not significant.
To determine its association with myeloma oncogenesis, we carried out apoptosis and cell cycle assays and found an increase in the number of apoptotic cells and sub‐G1 cells in siDIMT1‐transduced MM.1S and H929 cells (Fig. 4c–f). However, the knockdown did not affect cell cycle phases (Fig. 4c,d). Moreover, we established shDIMT1#1 and shDIMT1#2 stable transfectants in MM.1S cells and carried out in vivo transplantation in NOG mice (n = 10, each group). When these transfectants were injected s.c. into NOG mice, we observed significantly reduced tumor volume compared to that of control mice (control‐GFP, n = 10) (Fig. 4g,h). These results indicated that DIMT1 could contribute to cell survival and that aberrantly activated DIMT1 plays a role as a proto‐oncogene in MM. Moreover, these results also suggested that upregulation of DIMT1/DIMT1 in symptomatic MM might be associated with cell survival through the regulation of IRF4/IFR4 translation.
Upregulation of DIMT1 during disease progression of MM
During myeloma development, MGUS and SMM are preliminary stages of symptomatic MM.1 To examine DIMT1 expression in MM progression, we compared its expression among MGUS, SMM, symptomatic MM, and plasma cell leukemia cases using gene expression datasets previously uploaded to the NCBI GEO database (http://www.ncbi.nim.nih.gov). We used two sets of uploaded GEO datasets, GSE211345 (Fig. 5a) and GSE647746 (Fig. 5b) and found that DIMT1 was upregulated during MM progression. Notably, these data indicated that DIMT1 showed positive correlation with IRF4 (Fig. 5, bottom graphs) or MYC (Fig. S9), which were also upregulated during disease progression.36, 47 These findings strongly suggested that DIMT1 could be a diagnostic marker for MM progression. In addition, although we examined expression of other candidates such as ARMC1, TTC13, CIAPIN1, and NOL12, we could not detect gradual upregulation of these genes, as detected with DIMT1 (Fig. S10a). We also examined the relationship between DIMT1 and other representative oncogenes such as CCND1‐3, MAF, and FGFR3. Although we could not identify any significant relationships between DIMT1 and MAF or FGFR3, we found that DIMT1 expression was significantly decreased in the group of CCND1 overexpression when compared with other cyclin overexpression groups (Fig. S10b). Because overexpression of CCND1 is known to be an early event index of MM progression,48, 49, 50, 51 a reverse correlation between DIMT1 and CCND1 also supports the value of DIMT1 in predicting disease progression and as a marker of poor prognosis.
Figure 5.
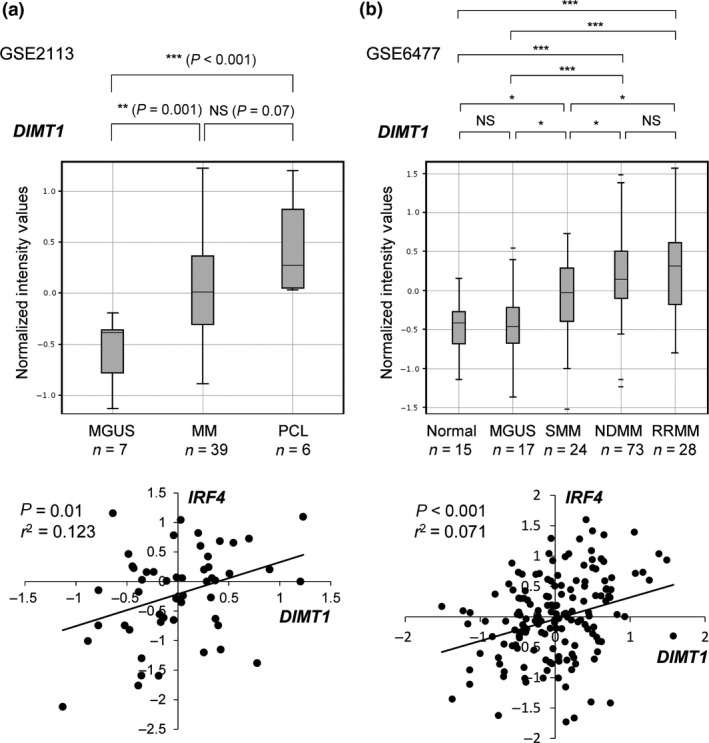
Upregulation of DIMT1 during disease progression of multiple myeloma (MM) and its correlation with IRF4 expression. (a) Comparison of DIMT1 expression from published dataset GSE2113 including primary samples of monoclonal gammopathy of undetermined significance (MGUS; n = 7), MM (n = 39), and plasma cell leukemia (PCL; n = 6). Correlation of IRF4 with DIMT1 is shown in the bottom graph. (b) Comparison of DIMT1 expression from the published dataset GSE6477 including primary samples of normal plasma cells (n = 15), MGUS (n = 17), smoldering multiple myeloma (SMM; n = 24), newly diagnosed MM (NDMM; n = 73), and relapsed/refractory MM (RRMM; n = 28). Correlation of IRF4 with DIMT1 is also shown in bottom graph. All plots show the 25th–75th percentiles (boxes) and 5th–95th percentiles (whiskers). The line in each box represents the median. *P < 0.05; **P < 0.01; ***P < 0.001; NS, not significant by Student's t‐test; r 2, correlation coefficient.
Discussion
In previous genomic analyses of MM, numerous DNA alteration/mutations were identified.1, 51 Despite the variety of genomic alterations, MM is a single tumor entity derived from plasma cells. Thus, disease‐specific genetic alterations, which may explain myeloma oncogenesis in all MM subtypes, may be epigenetically regulated. An important finding was that of IRF4 addiction.36 This oncogene was identified as a target of recurrent translocation of t(6,14)(p25;q32) in <5% of MM cases.35 However, epigenetic addiction in the remaining cases was found to be crucially involved in all MM molecular subtypes.36 Based on experiments carried out under normoxic conditions, Shaffer et al.36 showed that IRF4 addiction essentially contributes to myeloma oncogenesis by conferring an anti‐apoptotic phenotype. We found that hypoxic myeloma cells still possessed an anti‐apoptotic phenotype, despite downregulation of IRF4, suggesting that hypoxia‐adapted myeloma cells are not addicted to IRF4. A similar phenomenon is also observed in stem cells of chronic myeloid leukemia, which shows BCR‐ABL oncogene addiction. Corbin et al.52 reported that imatinib, which targets BCR‐ABL, was not effective against chronic myeloid leukemia stem cells because survival of these cells in the BM microenvironment is independent of BCR‐ABL addiction. Based on our results and those of previous studies, attention should be given to distinct targets of both BM and its microenvironment for therapy against myeloma and other hematological malignancies.
Although many studies have examined aberrant miRNAs in MM,26, 27, 28, 29, 30, 31, 32 there have been no reports on miR‐210. This may be because miR‐210 is not expressed during normoxia, and nearly all studies were carried out under normoxic conditions. This is the first study examining the role of miR‐210 in MM pathogenesis. Moreover, our findings suggest that miR‐210 is ectopically expressed in hypoxic environments such as the endosteal or vascular niches of the BM. Interestingly, miR‐210 shows both oncogenic and tumor suppressive functions.21, 22, 23, 24, 38 As an oncogenic factor, miR‐210 promotes angiogenesis and cell migration in various cancer cells.21 As a tumor suppressive factor, miR‐210 reduces mitosis through G2/M or G1 arrest.22, 23, 24, 25 Our study further showed that miR‐210 acts as a tumor suppressor in hypoxic conditions by downregulating the DIMT1–IRF4 axis, suggesting that downregulation of this axis enhances the “sensitivity” of stimulation of apoptotic stresses in the BM microenvironment. To explain why myeloma cells show anti‐apoptotic capability even in hypoxic conditions, we propose the following (Fig. 6). In normoxia, because HIF‐1α–miR‐210 is not activated, the DIMT1–IRF4 axis is highly expressed/activated in myeloma cells, which may convert myeloma cells to the anti‐apoptotic phenotype. However, in hypoxia, the HIF‐1α itself may convert myeloma cells to an anti‐apoptotic phenotype by transcriptionally regulating other anti‐apoptotic factors, one of which is the HIF‐1α glycolysis pathway,15 the activation of which may be partly responsible for the anti‐apoptotic phenotype in MM.
Figure 6.
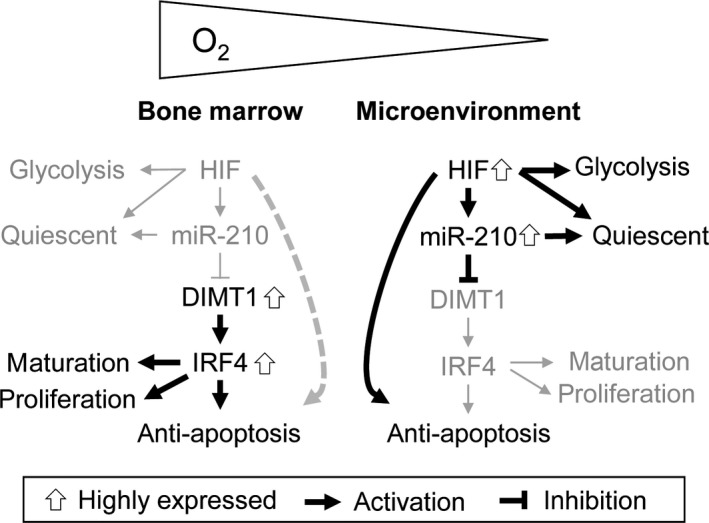
Model of phenotypical changes of myeloma cells in the bone marrow and its microenvironment. During normoxia, without the expression of hypoxia‐inducible factor‐1α (HIF‐1α)–microRNA‐210 (miR‐210), the dimethyladenosine transferase 1 homolog 1 (DIMT1)–interferon regulatory factor 4 (IRF4) axis might contribute to myeloma cell survival. During hypoxia, despite IRF4 reduction, myeloma cells did not show increased apoptosis. This might be because HIF‐1α could also have the potential to inhibit apoptosis by activating another anti‐apoptotic factor in MM. Increased expression of HIF‐1α (anti‐apoptotic) and decreased expression of IRF4 (pro‐apoptotic) might be mutually competitive during hypoxia. Bold lines/arrows, functional; gray lines/arrows, non‐functional.
As an 18S rRNA base methyltransferase, DIMT1 was previously found to regulate ribosomal proteins.42, 43 Our results strongly suggest that it is an upstream regulator of IRF4/IRF4 translation. Interestingly, DIMT1 knockdown repressed IRF4 but no other oncoproteins such as MYC and Bcl‐2, suggesting that DIMT1 translationally regulates selective oncogenetic targets. Our study also suggests that DIMT1 has oncogenic potential in all subtypes of MM, as DIMT1 is upregulated continuously during disease progression. Thus, DIMT1/DIMT1 may not only be a novel diagnostic marker/prognosis factor but also a therapeutic target of MM. Although we could not determine the exact mechanism of DIMT1 in myeloma pathogenesis, regulators of ribosomal proteins have been found to play crucial roles in carcinogenesis. For instance, WBSCR22, another 18S rRNA base methyltransferase, was found to be associated with metastasis/invasion in solid tumors and promoting survival in myeloma cells.43, 44, 53 In addition, alteration of ribosomal proteins, such as RPS14 and RPL22, have been detected in cases of hematological malignancies.54, 55 These reports support that deregulation of particular rRNAs is linked to selective translational regulation of oncogene or tumor suppressor genes. Although ribosomal proteins and their regulators should be examined in detail to determine their relationship with carcinogenesis, more detailed functional analyses of these proteins, particularly DIMT1, are necessary to elucidate their contribution to MM pathogenesis.
Disclosure Statement
The authors have no conflict of interest.
Abbreviations
- ARMC1
armadillo repeat containing 1
- BM
bone marrow
- CIAPIN1
cytokine‐induced apoptosis inhibitor 1
- DIMT1
DIM1 dimethyladenosine transferase 1 homolog
- HIF
hypoxia‐inducible factor
- IRF4
interferon regulatory factor 4
- MGUS
monoclonal gammopathy of undetermined significance
- miRNA
microRNA
- MM
multiple myeloma
- NOG
NOD/Shi‐scid IL‐2γnul
- NOL12
nucleolar protein 12
- qRT‐PCR
quantitative RT‐PCR
- SMM
smoldering multiple myeloma
- TTC13
tetratricopeptide repeat domain 13
- WBSCR22
Williams–Beuren syndrome chromosome region 22
Supporting information
Fig. S1. Cell cycle inhibition, acceleration of glycolysis, and drug resistance in myeloma cells with downregulation of interferon regulatory factor 4 (IRF4) during chronic hypoxia.
Fig. S2. Scatter plot of microRNA array for myeloma cell lines and primary samples affected by hypoxia.
Fig. S3. Sorting of CD38++ cells of a primary multiple myeloma sample under normoxia and hypoxia.
Fig. S4. Expression of hypoxia‐inducible factor‐1α (HIF‐1α) and quantitative RT‐PCR for U266 treated with PX‐478.
Fig. S5. Quantitative RT‐PCR of microRNA‐210 (miR‐210) for myeloma cells (MM.1S, H929, and KMS11) transiently transduced with miR‐210.
Fig. S6. Luciferase assay for KMS11 transiently transfected with wild‐type or mutated seed sequence of dimethyladenosine transferase 1 homolog (DIMT1) 3′‐UTR.
Fig. S7. Photographs of CD138 and dimethyladenosine transferase 1 homolog (DIMT1) staining for patient #11 treated with hypoxia and normoxia.
Fig. S8. Comparison of DIMT1 expression in various B‐cell lymphoma cell lines with myeloma cell lines.
Fig. S10. Gene expression data by using the dataset GSE6477.
Appendix S1. Supplementary materials and methods. Northern blot analysis, transient siRNA or miRNA transfection, siDIMT1 constructs and lentivirus virus infection, premicro‐RNA constructs and retrovirus infection, luciferase reporter assay, and pH measurement.
Acknowledgments
We thank Ms. Etsuko Kobayashi, Hiromi Kataho, Yuko Chiba, and Yukiko Abe for their outstanding technical assistance. This work was supported by the Japan Society for the Promotion of Science (KAKENHI Grant‐in‐Aid for Scientific Research to H.T.).
Cancer Sci 108 (2017) 641–652
Funding Information
Japan Society for the Promotion of Science.
References
- 1. Röllig C, Knop S, Bornhäuser M. Multiple myeloma. Lancet 2015; 385: 2197–208. [DOI] [PubMed] [Google Scholar]
- 2. Takubo K, Goda N, Yamada W et al Regulation of the HIF‐1alpha level is essential for hematopoietic stem cells. Cell Stem Cell 2010; 7: 391–402. [DOI] [PubMed] [Google Scholar]
- 3. Martin SK, Diamond P, Gronthos S, Peet DJ, Zannettino AC. The emerging role of hypoxia, HIF‐1 and HIF‐2 in multiple myeloma. Leukemia 2011; 25: 1533–42. [DOI] [PubMed] [Google Scholar]
- 4. Iriuchishima H, Takubo K, Miyakawa Y et al Neovascular niche for human myeloma cells in immunodeficient mouse bone. PLoS One 2012; 7: e30557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Silberstein LE, Lin CP. A new image of the hematopoietic stem cell vascular niche. Cell Stem Cell 2013; 13: 514–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Morrison SJ, Scadden DT. The bone marrow niche for haematopoietic stem cells. Nature 2014; 505: 327–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Easwaran H, Tsai HC, Baylin SB. Cancer epigenetics: tumor heterogeneity, plasticity of stem‐like states, and drug resistance. Mol Cell 2014; 54: 716–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Azab AK, Hu J, Quang P et al Hypoxia promotes dissemination of multiple myeloma through acquisition of epithelial to mesenchymal transition‐like features. Blood 2012; 119: 5782–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Storti P, Bolzoni M, Donofrio G et al Hypoxia‐inducible factor (HIF)‐1α suppression in myeloma cells blocks tumoral growth in vivo inhibiting angiogenesis and bone destruction. Leukemia 2013; 27: 1697–706. [DOI] [PubMed] [Google Scholar]
- 10. Carmeliet P, Dor Y, Herbert JM et al Role of HIF‐1alpha in hypoxia‐mediated apoptosis, cell proliferation and tumour angiogenesis. Nature 1998; 394: 485–90. [DOI] [PubMed] [Google Scholar]
- 11. Gardner LB, Li Q, Park MS, Flanagan WM, Semenza GL, Dang CV. Hypoxia inhibits G1/S transition through regulation of p27 expression. J Biol Chem 2001; 276: 7919–26. [DOI] [PubMed] [Google Scholar]
- 12. Goda N, Ryan HE, Khadivi B, McNulty W, Rickert RC, Johnson RS. Hypoxia‐inducible factor 1alpha is essential for cell cycle arrest during hypoxia. Mol Cell Biol 2003; 23: 359–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kim JY, Ahn HJ, Ryu JH, Suk K, Park JH. BH3‐only protein Noxa is a mediator of hypoxic cell death induced by hypoxia‐inducible factor 1alpha. J Exp Med 2004; 199: 113–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Koh MY, Powis G. Passing the baton: the HIF switch. Trends Biochem Sci 2012; 37: 364–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Maiso P, Huynh D, Moschetta M et al Metabolic signature identifies novel targets for drug resistance in multiple myeloma. Cancer Res 2015; 75: 2071–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ikeda S, Tagawa H. Dysregulation of microRNAs and their association in the pathogenesis of T‐cell lymphoma/leukemias. Int J Hematol 2014; 99: 542–52. [DOI] [PubMed] [Google Scholar]
- 17. Tagawa H, Ikeda S, Sawada K. Role of microRNA in the pathogenesis of malignant lymphoma. Cancer Sci 2013; 104: 801–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kitadate A, Ikeda S, Teshima K et al MicroRNA‐16 mediates the regulation of a senescence‐apoptosis switch in cutaneous T‐cell and other non‐Hodgkin lymphomas. Oncogene 2016; 35: 3692–704. [DOI] [PubMed] [Google Scholar]
- 19. Ito M, Teshima K, Ikeda S et al MicroRNA‐150 inhibits tumor invasion and metastasis by targeting the chemokine receptor CCR6, in advanced cutaneous T‐cell lymphoma. Blood 2014; 123: 1499–511. [DOI] [PubMed] [Google Scholar]
- 20. Teshima K, Nara M, Watanabe A et al Dysregulation of BMI1 and microRNA‐16 collaborate to enhance an anti‐apoptotic potential in the side population of refractory mantle cell lymphoma. Oncogene 2014; 33: 2191–203. [DOI] [PubMed] [Google Scholar]
- 21. Huang X, Ding L, Bennewith KL et al Hypoxia‐inducible mir‐210 regulates normoxic gene expression involved in tumor initiation. Mol Cell 2009; 35: 856–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang Z, Sun H, Dai H et al MicroRNA miR‐210 modulates cellular response to hypoxia through the MYC antagonist MNT. Cell Cycle 2009; 8: 2756–68. [DOI] [PubMed] [Google Scholar]
- 23. He J, Wu J, Xu N et al MiR‐210 disturbs mitotic progression through regulating a group of mitosis‐related genes. Nucleic Acids Res 2013; 41: 498–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tsuchiya S, Fujiwara T, Sato F et al MicroRNA‐210 regulates cancer cell proliferation through targeting fibroblast growth factor receptor‐like 1 (FGFRL1). J Biol Chem 2011; 286: 420–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kiga K, Mimuro H, Suzuki M et al Epigenetic silencing of miR‐210 increases the proliferation of gastric epithelium during chronic Helicobacter pylori infection. Nat Commun 2014; 5: 4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pichiorri F, Suh SS, Ladetto M et al MicroRNAs regulate critical genes associated with multiple myeloma pathogenesis. Proc Natl Acad Sci USA 2008; 105: 12885–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Roccaro AM, Sacco A, Thompson B et al MicroRNAs 15a and 16 regulate tumor proliferation in multiple myeloma. Blood 2009; 113: 6669–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lionetti M, Biasiolo M, Agnelli L et al Identification of microRNA expression patterns and definition of a microRNA/mRNA regulatory network in distinct molecular groups of multiple myeloma. Blood 2009; 114: e20–6. [DOI] [PubMed] [Google Scholar]
- 29. Gutiérrez NC, Sarasquete ME, Misiewicz‐Krzeminska I et al Deregulation of microRNA expression in the different genetic subtypes of multiple myeloma and correlation with gene expression profiling. Leukemia 2010; 24: 629–37. [DOI] [PubMed] [Google Scholar]
- 30. Pichiorri F, Suh SS, Rocci A et al Downregulation of p53‐inducible microRNAs 192, 194, and 215 impairs the p53/MDM2 autoregulatory loop in multiple myeloma development. Cancer Cell 2010; 18: 367–81. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31. Morelli E, Leone E, Cantafio ME et al Selective targeting of IRF4 by synthetic microRNA‐125b‐5p mimics induces anti‐multiple myeloma activity in vitro and in vivo . Leukemia 2015; 29: 2173–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Umezu T, Tadokoro H, Azuma K, Yoshizawa S, Ohyashiki K, Ohyashiki JH. Exosomal miR‐135b shed from hypoxic multiple myeloma cells enhances angiogenesis by targeting factor‐inhibiting HIF‐1. Blood 2014; 124: 3748–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nara M, Teshima K, Watanabe A et al Bortezomib reduces the tumorigenicity of multiple myeloma via downregulation of upregulated targets in clonogenic side population cells. PLoS One 2013; 8: e56954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kawano Y, Kikukawa Y, Fujiwara S et al Hypoxia reduces CD138 expression and induces an immature and stem cell‐like transcriptional program in myeloma cells. Int J Oncol 2013; 43: 1809–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Iida S, Rao PH, Butler M et al Deregulation of MUM1/IRF4 by chromosomal translocation in multiple myeloma. Nat Genet 1997; 17: 226–30. [DOI] [PubMed] [Google Scholar]
- 36. Shaffer AL, Emre NC, Lamy L et al IRF4 addiction in multiple myeloma. Nature 2008; 454: 226–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bayer C, Vaupel P. Acute versus chronic hypoxia in tumors: controversial data concerning time frames and biological consequences. Strahlenther Onkol 2012; 188: 616–27. [DOI] [PubMed] [Google Scholar]
- 38. Chan SY, Loscalzo J. MicroRNA‐210: a unique and pleiotropic hypoxamir. Cell Cycle 2010; 9: 1072–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Welsh S, Williams R, Kirkpatrick L, Paine‐Murrieta G, Powis G. Antitumor activity and pharmacodynamic properties of PX‐478, an inhibitor of hypoxia‐inducible factor‐1alpha. Mol Cancer Ther 2004; 3: 233–44. [PubMed] [Google Scholar]
- 40. Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015; 12: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lim LP, Lau NC, Garrett‐Engele P et al Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 2005; 433: 769–73. [DOI] [PubMed] [Google Scholar]
- 42. Lafontaine D, Delcour J, Glasser AL, Desgrès J, Vandenhaute J. The DIM1 gene responsible for the conserved m6(2)Am 6(2)A dimethylation in the 3’‐terminal loop of 18 S rRNA is essential in yeast. J Mol Biol 1994; 241: 492–7. [DOI] [PubMed] [Google Scholar]
- 43. Zorbas C, Nicolas E, Wacheul L, Huvelle E, Heurgué‐Hamard V, Lafontaine DL. The human 18S rRNA base methyltransferases DIMT1L and WBSCR22‐TRMT112 but not rRNA modification are required for ribosome biogenesis. Mol Biol Cell 2015; 26: 2080–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tiedemann RE, Zhu YX, Schmidt J et al Identification of molecular vulnerabilities in human multiple myeloma cells by RNA interference lethality screening of the druggable genome. Cancer Res 2012; 72: 757–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mattioli M, Agnelli L, Fabris S et al Gene expression profiling of plasma cell dyscrasias reveals molecular patterns associated with distinct IGH translocations in multiple myeloma. Oncogene 2005; 24: 2461–73. [DOI] [PubMed] [Google Scholar]
- 46. Chng WJ, Kumar S, Vanwier S et al Molecular dissection of hyperdiploid multiple myeloma by gene expression profiling. Cancer Res 2007; 67: 2982–9. [DOI] [PubMed] [Google Scholar]
- 47. Uranishi M, Iida S, Sanda T et al Multiple myeloma oncogene 1 (MUM1)/interferon regulatory factor 4 (IRF4) upregulates monokine induced by interferon‐gamma (MIG) gene expression in B‐cell malignancy. Leukemia 2005; 19: 1471–8. [DOI] [PubMed] [Google Scholar]
- 48. Agnelli L, Bicciato S, Mattioli M et al Molecular classification of multiple myeloma: a distinct transcriptional profile characterizes patients expressing CCND1 and negative for 14q32 translocations. J Clin Oncol 2005; 23: 7296–306. [DOI] [PubMed] [Google Scholar]
- 49. Bergsagel PL, Kuehl WM, Zhan F, Sawyer J, Barlogie B, Shaughnessy J Jr. Cyclin D dysregulation: an early and unifying pathogenic event in multiple myeloma. Blood 2005; 106: 296–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhan F, Huang Y, Colla S et al The molecular classification of multiple myeloma. Blood 2006; 108: 2020–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chapman MA, Lawrence MS, Keats JJ et al Initial genome sequencing and analysis of multiple myeloma. Nature 2011; 471: 467–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Corbin AS, Agarwal A, Loriaux M et al Human chronic myeloid leukemia stem cells are insensitive to imatinib despite inhibition of BCR‐ABL activity. J Clin Invest 2011; 121: 396–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nakazawa Y, Arai H, Fujita N. The novel metastasis promoter Merm1/Wbscr22 enhances tumor cell survival in the vasculature by suppressing Zac1/p53‐dependent apoptosis. Cancer Res 2011; 71: 1146–55. [DOI] [PubMed] [Google Scholar]
- 54. Ebert BL, Pretz J, Bosco J et al Identification of RPS14 as a 5q‐ syndrome gene by RNA interference screen. Nature 2008; 451: 335–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rao S, Lee SY, Gutierrez A et al Inactivation of ribosomal protein L22 promotes transformation by induction of the stemness factor, Lin28B. Blood 2012; 120: 3764–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Cell cycle inhibition, acceleration of glycolysis, and drug resistance in myeloma cells with downregulation of interferon regulatory factor 4 (IRF4) during chronic hypoxia.
Fig. S2. Scatter plot of microRNA array for myeloma cell lines and primary samples affected by hypoxia.
Fig. S3. Sorting of CD38++ cells of a primary multiple myeloma sample under normoxia and hypoxia.
Fig. S4. Expression of hypoxia‐inducible factor‐1α (HIF‐1α) and quantitative RT‐PCR for U266 treated with PX‐478.
Fig. S5. Quantitative RT‐PCR of microRNA‐210 (miR‐210) for myeloma cells (MM.1S, H929, and KMS11) transiently transduced with miR‐210.
Fig. S6. Luciferase assay for KMS11 transiently transfected with wild‐type or mutated seed sequence of dimethyladenosine transferase 1 homolog (DIMT1) 3′‐UTR.
Fig. S7. Photographs of CD138 and dimethyladenosine transferase 1 homolog (DIMT1) staining for patient #11 treated with hypoxia and normoxia.
Fig. S8. Comparison of DIMT1 expression in various B‐cell lymphoma cell lines with myeloma cell lines.
Fig. S10. Gene expression data by using the dataset GSE6477.
Appendix S1. Supplementary materials and methods. Northern blot analysis, transient siRNA or miRNA transfection, siDIMT1 constructs and lentivirus virus infection, premicro‐RNA constructs and retrovirus infection, luciferase reporter assay, and pH measurement.


