Abstract
Emerging evidence has indicated that deregulation of long non‐coding RNAs (lncRNAs) can contribute to the progression and metastasis of human cancer, including hepatocellular carcinoma (HCC). However, the roles of most lncRNAs in HCC remain largely unknown. Here we found a long noncoding RNA termed SchLAH (seven chromosome locus associated with HCC; also called BC035072) was generally downregulated in HCC. Low expression of SchLAH was significantly correlated with shorter overall survival of HCC patients. In vitro and in vivo assays indicated that overexpression of SchLAH inhibited the migration and lung metastasis of HCC cells. Knockdown of SchLAH by siRNA pool promoted the migration of HCC cells. RNA pull‐down and RNA immunoprecipitation assays demonstrated SchLAH physically interacted with fused in sarcoma (FUS). PCR array analysis showed that RhoA and Rac1 were the downstream effector molecules of SchLAH during HCC metastasis. Knockdown of FUS rescued the mRNA levels of RhoA and Rac1 that were repressed by SchLAH. These results suggest that SchLAH may suppress the metastasis of HCC cells by interacting with FUS, which indicates potential of SchLAH for the prognosis and treatment of HCC.
Keywords: Fused in sarcoma, hepatocellular carcinoma, long noncoding RNA, metastasis, SchLAH
Hepatocellular carcinoma (HCC) is currently the fifth most common tumor worldwide and the second leading cause of cancer‐related death.1, 2 Although significant progress in HCC treatment has been made in recent years, the 5‐year survival rates are still low and approximately 422 100 HCC patients die each year in China.3, 4 The poor prognosis and high recurrence rate is due to the high rate of metastases.5, 6 However, the underlying molecular mechanisms that mediate the metastatic procedure remain unclear.7 Elucidation of the metastatic mechanisms may promote the development of effective diagnosis and treatment, and improve the overall prognosis of patients with HCC.
Long non‐coding RNAs (lncRNAs) represent a subgroup of noncoding RNAs that are longer than 200 nucleotides. lncRNA‐mediated biological regulation has been implicated in a wide variety of cellular processes, and in cancer lncRNAs are involved in multilevel regulation of gene expression,8, 9 often by interacting with epigenetic complexes,10, 11, 12 proteins,13 miRNAs14, 15 or mRNA.16, 17 Recently, many studies have shown that lncRNAs are frequently deregulated in HCC and play important roles in cell proliferation,18, 19 apoptosis,20, 21 and metastasis.22, 23 However, the clinical significance and roles of most deregulated lncRNAs in HCC remain largely unknown.
Fused in sarcoma/translocated in liposarcoma (FUS/TLS or FUS) is a multifunctional DNA/RNA‐binding protein associated with cancer and neurodegeneration. Some articles revealed that FUS acted as a tumor suppressor and its knockdown increased cell proliferation.24, 25 Recently, FUS has been shown to interact with a number of lncRNAs in neuron diseases.26 Wang et al.27 reported that FUS could be directed to the regulatory regions of target genes by lncRNA transcripts induced by DNA damage signals. However, relationship between FUS and lncRNAs in HCC has only been implicated in bioinformatic analysis28 and needs to further explore.
In this study, we first found a lncRNA termed SchLAH that was downregulated in HCC tumor tissues, compared to the adjacent noncancerous tissues. Low expression of SchLAH was correlated with poor prognosis of HCC patients. SchLAH could suppress the migration of HCC cells in vitro and lung metastasis in vivo. Furthermore, we demonstrated that SchLAH bound to FUS and inhibition of this binding between SchLAH and FUS could contribute to promoting migration in HCC cells.
Materials and Methods
Ethics statement
The present study was approved by the research ethics committee of Zhongshan hospital, and the experiments were undertaken with the understanding and written consent of each subject.
Patients and cell lines
Frozen samples of HCC tissues and paired adjacent noncancerous liver tissues were randomly selected from patients undergoing hepatectomy at Zhongshan hospital (Shanghai, China) between 2004 and 2005. Ethical approval was obtained from the research ethics committee of Zhongshan hospital, and informed consent was obtained from each patient. All patients were followed up until October 2010. Overall survival (OS) was defined as the interval between the dates of surgery or the last follow‐up.
Cell culture
The human HCC cell lines HepG2, Hep3B were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). HCC cell line SMMC7721 was purchased from Shanghai Institute of Cell Biology, Chinese Academy of Sciences. Huh7 was purchased from RIKEN BRC cell Bank, Tsukuba, Japan. All the cell lines were maintained in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and antibiotics (100 U/mL penicillin, 100 mg/mL streptomycin), in a 5% CO2 atmosphere at 37°C.
Quantitative real‐time PCR
Real‐time PCR analyses were performed according to the manufacturer's instructions (Takara Biotechnology, Dalian, China). The primers used are listed in Table S1. The expression levels were normalized to β‐actin, and the relative expression levels were calculated using the 2−ΔCt method.
Western blotting
Total cell lysates were prepared in 6× SDS loading buffer. Proteins were separated by sodium dodecyl sulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) and transferred onto nitrocellulose membranes. Membranes were incubated with the specific primary antibodies and then with HRP‐conjugated secondary antibodies. Protein bands were visualized using chemiluminescence detection. The following antibodies were used: FUS (1:500; Abcam, Cambridge, MA, USA), β‐actin (1:10 000; Sigma, St. Louis, MO, USA).
Race
5′‐RACE and 3′‐RACE were performed using SMART RACE cDNA Amplification Kit (Clontech, Palo Alto, CA, USA) according to the manufacturer's instructions. The gene‐specific primers used for RACE analysis were presented in Table S2.
SchLAH overexpression and RNA interference
For overexpression, SchLAH was cloned into pWPT vector. HepG2 and Hep3B cells were infected with virus containing the plasmid SchLAH‐pWPT. siRNA transfections were done with 50 nM siRNA pool (Ribobio, Guangzhou, China) and Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA).
Subcellular fractionation location
The separation of the nuclear and cytosolic fractions was performed using the PARIS Kit (Life Technologies, Carlsbad, CA, USA) according to the manufacturer's instructions.
Fluorescence in situ hybridization
Cells were fixed in 4% formaldehyde then incubated in 5% acetic acid for 15 min followed by washes with phosphate‐buffered saline (PBS). The fixed cells were further treated with pepsin (1% in 10 mM HCl) and subsequent dehydration through 70%, 90%, and 100% ethanol. The air‐dried cells were subjected to incubation with 40 nM FISH probe in hybridization buffer (100 mg/mL dextran sulfate, 10% formamide in 2× saline sodium citrate [SSC]) at 80°C for 2 min. The hybridization was performed at 55°C for 2 h and the slide was washed with 2× SSC at 65°C followed by dehydration through 70%, 90%, and 100% ethanol. The air‐dried slide was mounted with Prolong Gold Antifade Reagent with DAPI for detection. RNA FISH probes were designed and synthesized by Shanjing Biological Science and Technology Co., Ltd (Shanghai, China). Probe sequences are listed in Table S3.
Transwell assays
For the transwell migration assay, cells were trypsinized and resuspended in serum‐free DMEM. 5 × 104 cells (300 μL) were planted on the top chamber of each insert (BD Biosciences, Franklin Lakes, NJ, USA) with 8‐μm‐diameter pores on its membrane. To conduct migration assay of HCC cells, 800 μL DMEM supplemented with 10% FBS was injected into the lower chambers. After incubation at 37°C, cells remaining in the top chamber of the inserts were carefully removed. After fixation and staining in a dye solution containing 0.1% crystal violet, cells adhering to the lower side of the inserts were counted and imaged through an IX71 inverted microscope (Olympus, Tokyo, Japan).
Cell proliferation assays
Cell proliferation was assayed by Cell Counting Kit‐8 (Dojindo Laboratories, Kumamoto, Japan). HCC cells were plated in 96‐well plates (1–2 × 103 cells per well), and the numbers of cells per well were measured at the indicated time points.
In vivo metastasis assays
The tail vein metastasis model was established to validate the role of SchLAH on HCC metastasis ability in vivo. A total of 2 × 106 HepG2 cells suspended in 200 μL serum‐free DMEM were injected into the tail vein of nude mice. After 6 weeks, all of the mice were euthanized. The lung tissues were dissected and fixed with 4% formalin for at least 72 h. Lung tissues were analyzed by hematoxylin and eosin (HE) staining. SPF level BALB/c nude mice, male, age 6–7 week, weight 18–22 g, were provided by Shanghai Cancer Institute. The production license number is SCXK (Shanghai) 2007‐0001 and the use certificate number is SYXK (Shanghai) 2007‐0001. All mice were received human care and manipulated according to protocols approved by the Shanghai Medical Experimental Animal Care Commission.
PCR array analysis for tumor migration
The Cancer Motility RT2 Profiler PCR Arrays (Qiagen, Hilden, Germany), which is designed to represent 84 genes known to be involved in migration, was used to profile HepG2‐SchLAH, Hep3B‐SchLAH cells, and their relative control cells. PCR array was performed by Shanghai OE Biotech. Co, Ltd, Shanghai, China.
RNA pull‐down assay
RNA pull‐down and deletion mapping assays were performed as described previously.10 Briefly, biotinylated SchLAH or antisense RNA was incubated with cell protein extractions (1 mg), which were then targeted with streptavidin beads (Invitrogen) and washed. The associated proteins were resolved by gel electrophoresis. Specific bands were excised and identified by mass spectrometry. The length of synthetic biotinylated SchLAH is 2872 nt. Biotin RNA Labeling Mix (Roche diagnostics, Indianapolis, IN, USA) and DIG RNA Labeling Kit (SP6‐T7) (Roche) were used to perform RNA pull‐down experiment. The transcription buffer was from DIG RNA Labeling Kit (SP6‐T7). Oligo Anealing buffer (Beyotime, Shanghai, China), RIPA lysis buffer (Beyotime), 5× RNA Oligo Anealing buffer (Beyotime) were used in this assay.
Chromatin immunoprecipitation and RIP
RIP assays were performed using a Millipore EZ‐Magna RIP RNA‐Binding Protein Immunoprecipitation kit (Millipore, Bedford, MA, USA) according to the manufacturer's instructions. RIP PCR was performed as RT‐qPCR using total RNA as input controls. Antibodies used for RIP included rabit polyclonal IgG (Millipore, PP64) and antibodies to FUS/TLS (Abcam, ab23439). The gene‐specific primers used for detecting SchLAH are presented in Table S2.
Statistical evaluation
All statistical analyses were performed with SPSS 17.0 Software, Chicago, USA. Data were presented as mean ± standard deviation (SD) from at least three separate experiments. The significance of mean values between two groups was analyzed by Student's t‐test. Kaplan–Meier survival analysis was used to compare survival of HCC patients with high or low expression of SchLAH by log‐rank test. A P‐value < 0.05 was considered significant (*P < 0.05, **P < 0.01, and ***P < 0.001).
Results
SchLAH is downregulated in HCC tissues and asscociated with poor prognosis
By comparing HCC tissues to nontumor liver tissues using GEO database (GSE45436), we found a lncRNA (NCBI: BC035072) was lowly expressed in HCC tissues (P < 0.0001) (Fig. 1a). To confirm expression of BC035072 in HCC, we examined 132 paired samples of tumor and nontumor liver tissues. Notably, BC035072 transcript was expressed at lower levels in over 90% of tumor tissues compared to the paired adjacent liver tissues of the same patients after normalizing to β‐actin expression (P < 0.0001) (Fig. 1b–d). We next examined relationship between expression levels of BC035072 and clinicopathological characteristics of 95 HCC patients (Table S4). Kaplan–Meier analysis showed significant correlations between the low expression level of BC035072 and poor overall survival (P = 0.0124) (Fig. 1e). Therefore, we focused on this uncharacterized lncRNA and named it SchLAH (seven chromosome locus associated with HCC) (Fig. 1f). SchLAH is composed of one exon with a full length of 2872 nt determined by RACE (rapid amplification of cDNA ends) assay (Fig. S1a,b). The sequence of SchLAH we obtained was identical with that in NCBI database.
Figure 1.
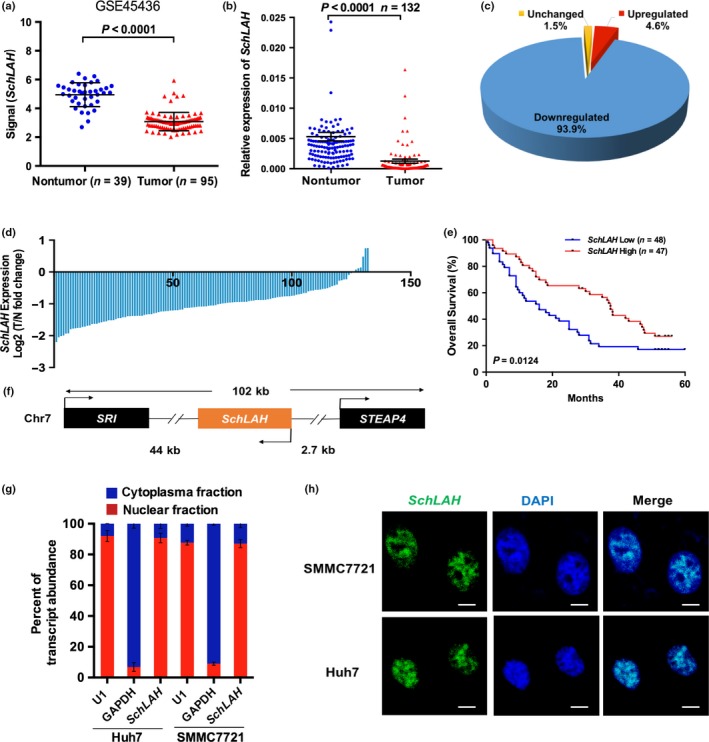
Identification of SchLAH as a HCC‐associated lncRNA. (a) SchLAH was downregulated in tumor tissues compared to nontumor liver tissues (GSE45436). (b–d) SchLAH was downregulated in tumor tissues compared to paired adjacent noncancerous liver tissues (n = 132). The expression level of SchLAH was analyzed by RT‐PCR and normalized to β‐actin. (e) Survival rates of 95 HCC patients who underwent liver surgery were compared between the SchLAH high‐expression and SchLAH low‐expression groups (log rank: P = 0.0124). The median expression level was used as the cutoff. (f) Schematic representation of SchLAH location on chromosome. (g) Fractionation of Huh7 and SMMC7721 cell lysates demonstrated nuclear expression of SchLAH. U1 RNA served as an internal control for nuclear gene expression. Values are mean ± standard deviation (n = 3). (h) Nuclear localization of SchLAH detected by RNA FISH in SMMC7721 and Huh7 cells. Scale bar, 10 μm.
Additionally, quantification of nuclear/cytoplasmic RNA in Huh7 and SMMC7721 cell extracts revealed that SchLAH transcripts were mainly enriched in the nucleus (Fig. 1g). We confirmed the localization of SchLAH in these cells using in situ hybridization assay (Fig. 1h). Open Reading Frame (ORF) finder analysis of the SchLAH sequence suggested that SchLAH had no coding potential. Consequently, SchLAH is a non‐coding RNA in nuclei of HCC cells.
Overexpression of SchLAH inhibits migration of HCC cells in vitro and in vivo
The expression levels of SchLAH in HCC cell lines were detected by qRT‐PCR (Fig. 2a). SchLAH level is relatively lower in HepG2 and Hep3B cells and higher in Huh7 and SMMC7721 cells. The hepatoma cell lines HepG2 and Hep3B were infected with the lentivirus containing the SchLAH expression vector (Fig. 2b). Notably, overexpression of SchLAH dramatically inhibited cell migration in vitro but did not affect cell proliferation and invasion (Fig. 2c,d, Fig. S2a,b). To further confirm the effects of SchLAH, we performed small antisense RNA‐mediated knockdowns in SMMC7721 and Huh7 cells to observe the impact of SchLAH depletion. Silencing of SchLAH dramatically increased the ability of these cells to migration (Fig. 2e,f).
Figure 2.
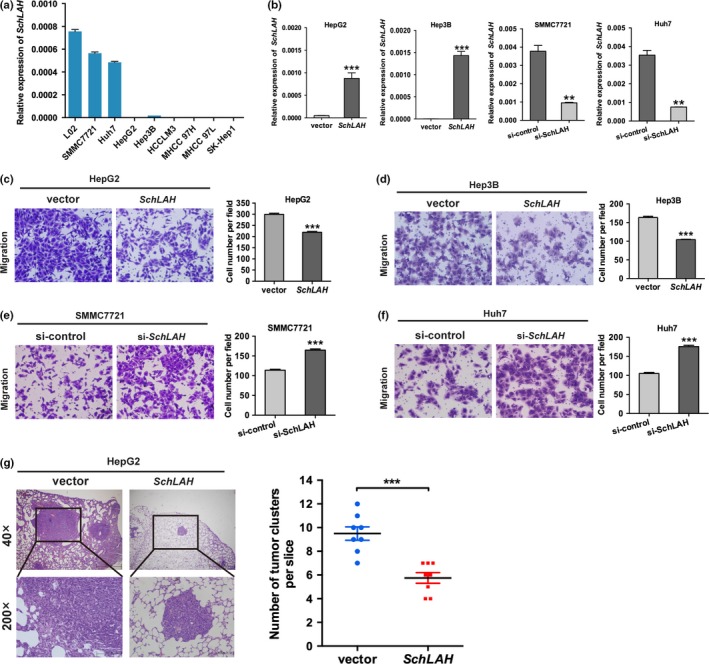
SchLAH inhibits the migration of HCC cells. (a) Relative expression of SchLAH in different HCC cell lines. (b) The expression of SchLAH in stable HepG2 cell clones and Hep3B cell clones infected with lentiviruses encoding SchLAH. Relative levels of SchLAH in the Huh7 and SMMC7721 lines after transfection with siRNA pool against SchLAH. (c, d) Migration assays showed the effects of SchLAH overexpression on migratory potentials of HepG2 and Hep3B cells. Mean ± SD are shown (n = 3). (e, f) Migration assays showed the effects of SchLAH silence on migratory potentials of SMMC7721 and Huh7 cells. (g) Histological analysis of the number of lung metastasis in each group 6 weeks after tail vein injection of vector or SchLAH overexpressed HepG2 cells (n = 8). Values are mean ± standard deviation (n = 3). **P < 0.01, ***P < 0.001.
To test SchLAH in vivo, we established tail vein injected models and compared the rates of lung colonization as measured by hematoxylin and eosin (HE) staining. The number of micrometastatic lesions of each mouse was detected and counted under microscope. Compared to mice injected with HepG2 cells expressing a control vector, mice injected with HepG2 cells stably overexpressing SchLAH had fewer metastatic tumor clusters and smaller metastatic tumor size (Fig. 2g). These results indicate functional significance of SchLAH in HCC metastasis.
The interaction of SchLAH with FUS
Recently, several studies have found that many lncRNAs are involved in multiple regulation pathways through their interaction with proteins. SchLAH might affect cellular function in a similar manner. To test this hypothesis, we sought to identify proteins that are associated with SchLAH by an RNA pull‐down experiment. We resolved the RNA‐associated proteins on a SDS‐PAGE gel, cut out the bands specific to SchLAH, and subjected them to mass spectrometry (Fig. 3a, Table S1). Among all of the proteins identified by mass spectrometry, only FUS was detected by western blotting from three independent RNA pull‐down assays (Fig. 3b). To further validate the interaction between SchLAH and FUS, we performed RNA immunoprecipitation (RIP) with an antibody against FUS using cell extracts from SMMC7721 HCC cell lines. We observed an enrichment of SchLAH (but not other unrelated RNAs) with FUS antibody as compared to the nonspecific antibody (IgG control) (Fig. 3c). We further performed deletion‐mapping experiments to determine whether FUS interacts with a specific region of SchLAH. We carried out RNA pull‐down experiments with truncated versions of SchLAH followed by western blot detection of bound FUS (Fig. 3d). These analyses identified a region between 800 and 1800 nt of SchLAH containing GGUG or GUGGU motif might be required for the interaction with FUS. As is reported, our results also indicate that FUS preferentially recognizes the UG‐rich motif on RNA sequence.29
Figure 3.
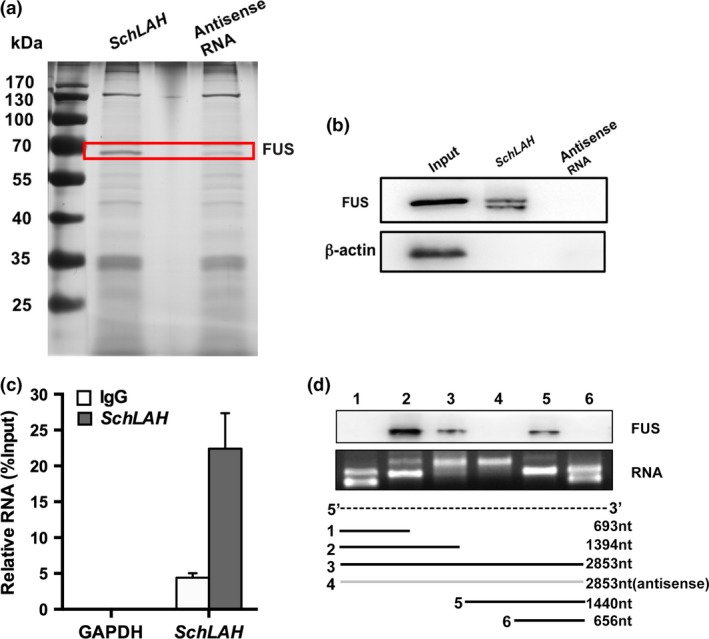
The Interaction of SchLAH with FUS. (a) SchLAH and FUS specifically interacted in vitro. SDS‐PAGE gel of proteins bound to SchLAH (left lane) or antisense RNA (right lane). The highlighted region was submitted for mass spectrometry identifying FUS as the band unique to SchLAH. (b) Western blot analysis of the specific association of FUS with SchLAH. A nonspecific protein (β‐actin) is shown as a control (n = 3). (c) RNA immunoprecipitation (RIP) experiments were performed using the FUS antibody, and specific primers were used to detect SchLAH or GAPDH. Values are mean ± standard deviation (n = 3). (d) SchLAH bound to FUS/TLS through a region between 800 and 1800 nt which contains “GU” rich motif.
SchLAH inhibits migration of HCC cells through RhoA and Rac1
To further investigate downstream molecular targets that were regulated by SchLAH in HCC migration, we analyzed migration‐related genes for HepG2‐SchLAH cells, Hep3B‐SchLAH cells, and their relative control cells using a Cancer Motility RT2 Profiler PCR Arrays (Fig. 4a). This analysis revealed a total of 11 downregulated migration‐related genes in mRNA levels in HepG2‐SchLAH cells and 12 in Hep3B‐SchLAH cells, which had a more than 1.5‐fold change, compared with their controlled cells, respectively (Fig. 4b). Among these genes, six genes were downregulated in both cell lines that overexpressing SchLAH. Subsequently, these six candidates were validated by qRT‐PCR assay (Fig. 4c–f). When SchLAH was overexpressed, RhoA and Rac1 were notably downregulated, which was verified with the upregulation of RhoA and Rac1 when SchLAH was knocked down. These results indicate that RhoA and Rac1 may be the downstream targets of SchLAH during the metastasis of HCC.
Figure 4.
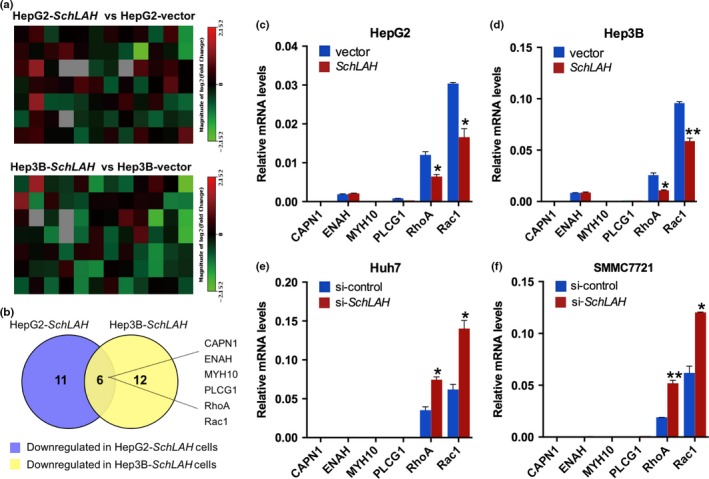
SchLAH inhibits migratory potential of HCC cells through RhoA and Rac1. (a, b) Changes of gene expression were analyzed by a Cancer Motility RT 2 Profiler PCR array. The downegulated genes that fold change >1.5 in HepG2‐SchLAH and Hep3B‐SchLAH were intersected to generate an overlapping gene signature. (c–f) Overlapping genes in (b) were confirmed by qRT‐PCR in HepG2, Hep3B, Huh7, and SMMC7721 cells. β‐actin serves as a control. Values are mean ± standard deviation (n = 3). *P < 0.05, **P < 0.01.
SchLAH functions through interaction with FUS
FUS is a RNA‐binding protein that is often associated with oncogenesis.30, 31 FUS serves as a key molecule in transcriptional regulation and RNA processing including processes such as pre‐messenger RNA (pre‐mRNA) splicing and polyadenylation.32, 33 Therefore, we sought to validate whether FUS is required for SchLAH to regulate the mRNA levels of RhoA and Rac1 indirectly. We performed siRNA against FUS in SchLAH overexpressed cells (Fig. 5a,b). Notably, depletion of FUS reversed the ability of SchLAH to suppress HCC cell migration (Fig. 5c,d). Quantitative RT‐PCR confirmed that SchLAH‐repressed target genes, such as RhoA and Rac1, were transcriptionally derepressed upon FUS depletion (Fig. 5e,f). These results suggest that FUS is required for SchLAH to suppress the migration of HCC cells and affects the expression of RhoA and Rac1 in an indirect way.
Figure 5.
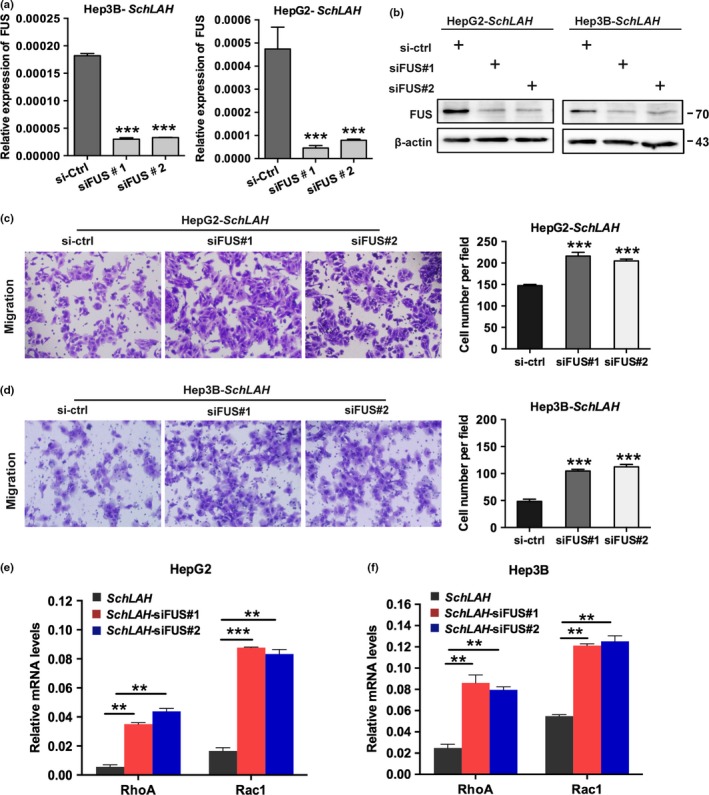
SchLAH function through its binding to FUS. (a, b) The detection of knockdown of FUS in HepG2‐SchLAH and Hep3B‐SchLAH cells by qRT‐PCR and western blotting. (c, d) Migration assay in vector or SchLAH cells transfected by the indicated siRNAs against FUS. (e, f) qRT‐PCR detection of RhoA and Rac1 expression in vector or SchLAH cells with siRNAs targeting FUS Values are mean ± standard deviation (n = 3). *P < 0.05, **P < 0.01, and ***P < 0.001.
Histone deacetylation is involved in the downregulation of SchLAH
We next explored the reason for SchLAH downregulation in HCC. Recent study showed lncRNAs could be regulated by inhibitors of histone deacetylation in HCC cell lines.34 We determined that SchLAH was upregulated by the histione deacetylase inhibitor trichostatin A (TSA) in HepG2 (Fig. 6a) and Huh7 cells (Fig. 6b). To validate this result in vivo, we measured total histone H3 and H4 acetylation levels across the SchLAH promoter in HepG2 and Huh7 cells. Chromatin immunoprecipitation (ChIP) assay showed the enrichment of total histone H3 acetylation but not total H4 acetylation across the SchLAH promoter region (Fig. 6c). In addition, the histone H3 acetylation levels across SchLAH in HepG2 cells was lower than that in Huh7 cells which has higher levels of this lncRNA. These results suggest that downregulation of SchLAH may be caused by the H3 deacetylation.
Figure 6.
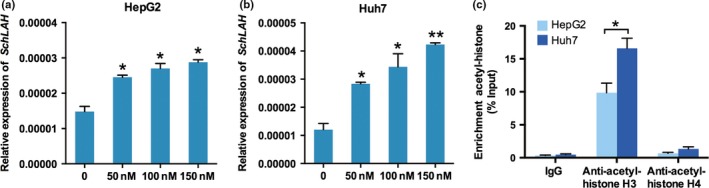
Histone deacetylation contributes to SchLAH downregulation. (a, b) HepG2 and Huh7 cells were stimulated with different concentrations of the histone deacetylase inhibitor trichostatin A (TSA) for 24 h. The expression of SchLAH was detected using qRT‐PCR and normalized to β‐actin. (c) ChIP analyses were conducted on the SchLAH promoter regions using anti‐acetyl‐histone H3 and anti‐acetyl‐histone H4. Enrichment was determined relative to input controls. Values are mean ± standard deviation (n = 3). *P < 0.05, **P < 0.01.
Discussion
Metastasis and recurrence are the leading causes of poor survival of HCC patients. Although numerous genes involved in tumor metastasis were identified, metastatic mechanism of HCC remains poorly understood. Recently, increasing evidence has revealed functional roles of lncRNAs in cellular transformation and metastasis of HCC. In this study, we reported that SchLAH is frequently downregulated in 132 HCC tissues. With a cohort of 95 randomly selected HCC samples, we determined that low levels of SchLAH were associated with poor overall survival. This result indicates that SchLAH may act as a tumor suppressor. We further observed that overexpression of SchLAH inhibited HCC cell migration in vitro and lung metastasis in vivo, while inhibition of SchLAH promoted HCC cell migration.
Many lncRNAs regulate gene transcription by interfacing with correspondent RNA binding proteins (RBPs) in a specific manner.35 Computational analysis predicted that FUS is the most implicated RBPs in interaction with HCC related lncRNAs.28 In this study, we identified a specific interaction between SchLAH and FUS. Wang et al.27 have shown that FUS preferentially interact with GGUG RNA oligonucleotides. The latest cross linking‐immunoprecipitation and high‐throughput sequencing (CLIP‐seq) data using mouse and human brain revealed the enrichment of GUGGU motif in FUS clusters.36 Interestingly, SchLAH has several binding sites containing the FUS motif GGUG or GUGGU and our analyses indicate that SchLAH may bind to FUS through these motifs.
Fused in sarcoma is known to regulate transcription events through interaction with complexes such as transcription machinery.37 The physical interaction between SchLAH and FUS is likely for SchLAH‐mediated gene repression, as loss of FUS results in derepression of genes (such as RhoA and Rac1) that are repressed by SchLAH. Honda et al.38 revealed that “small GTPase‐mediated signal transduction” was enriched in the list of the top 20 GO terms for genes with FUS‐regulated expression in FUS‐silenced neuron. It has been shown that RhoA and Rac1 were frequently upregulated in HCC tissue and is associated with poor prognosis.39, 40, 41 Activation of Rho GTPases could stimulate cell motility and metastasis and inhibition of RhoA and Rac1 could suppress metastatic potential of HCC cells.42, 43, 44 Our study showed that RhoA and Rac1 might be the downstream effector molecules of SchLAH and overexpression of SchLAH suppressed migration of HCC cells through downregulating the mRNA level of RhoA and Rac1. However, it is not clear how SchLAH inhibits the transcription of RhoA and Rac1 by its binding to FUS. Our results also showed that SchLAH did not change the mRNA and protein level of FUS (data not shown). A previous study indicated that FUS was recruited by lncRNAs to the genomic locus encoding cyclin D1 and inhibited CBP/p300 HAT function, resulting in increasing apoptosis tolerance.27 We suggested that FUS may acted as a repressor of RhoA and Rac1 at promoter region with interaction of CBP/p300 or HDACs.45 Further study needs to be investigated. Additionally, FUS knockdown in Huh7 and HepG2 also enhanced migration (Fig. S3). These results indicated that FUS played a tumor‐suppressive role in HCC.
Lastly, our results indicated that SchLAH downregulation in HCC could be the result of histone H3 deacetylation. Epigenetic regulatory factors, such as histone acetylation and methylation or DNA methylation can manipulate the expression of lncRNA.34, 46, 47 Yang48 showed that lncRNA‐LET was repressed by hypoxia‐induced histone deacetylase 3 (HDAC3). Our experiments also showed that the level of RhoA and Rac1 increased by use of TSA, while FUS slightly increased (Fig. S4). It is possible that RhoA and Rac1 were also regulated by histone deacetylase.49, 50, 51 In this case, it is hard to prove the overall hypothetical mechanism that overexpression of SchLAH could downregulate RhoA and Rac1 by TSA treatment. However, mechanisms of SchLAH downregulation in HCC needs to be further investigated.
Taken together, our results indicate that SchLAH, which is frequently downregulated in HCC, could promote HCC metastasis through interacting with FUS. These findings suggest that SchLAH may serve as an indicator for HCC prognosis and could be a potential therapeutic target.
Disclosure Statement
The authors have no conflict of interest.
Supporting information
Fig. S1. Cloning the full‐length of human SchLAH gene.
Fig. S2. Effects of SchLAH overexpression on cell invasion and proliferation.
Fig. S3. Effects of FUS knockdown on HepG2 and Huh7 migration.
Fig. S4. The expression of FUS, RhoA and Rac1 by TSA treatment.
Table S1. Proteins in the band specific to SchLAH identified by mass spectrometry.
Table S2. PCR primer pairs for qRT‐PCR.
Table S3. Probes for RNA FISH.
Table S4. Clinical characteristics of 95 HCC patients according to SchLAH expression levels using β‐actin as internal control.
Acknowledgments
This work was supported by National Key Basic Research Program of China (973 Program: 2015CB553905), National Natural Science Foundation of China (81201626, 81301759, 81402278, 81421001), Project of State Key Laboratory of Oncogenes and Related Genes (91‐1411, 91‐1502).
Cancer Sci 108 (2017) 653–662
Funding Information
National Key Basic Research Program of China (2015CB553905) Project of State Key Laboratory of Oncogenes and Related Genes (91‐1411, 91‐1502); National Natural Science Foundation of China (81201626, 81301759, 81402278, 81421001).
Contributor Information
Zhouhong Ge, Email: gezhouhong37@hotmail.com.
Jia Fan, Email: jiafan99@yahoo.com.
References
- 1. El‐Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology 2007; 132: 2557–76. [DOI] [PubMed] [Google Scholar]
- 2. Maluccio M, Covey A. Recent progress in understanding, diagnosing, and treating hepatocellular carcinoma. CA Cancer J Clin 2012; 62: 394–9. [DOI] [PubMed] [Google Scholar]
- 3. Takayama T. Surgical treatment for hepatocellular carcinoma. Jpn J Clin Oncol 2011; 41: 447–54. [DOI] [PubMed] [Google Scholar]
- 4. Chen W, Zheng R, Baade PD et al Cancer statistics in China, 2015. CA Cancer J Clin 2016; 66: 115–32. [DOI] [PubMed] [Google Scholar]
- 5. Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet 2012; 379: 1245–55. [DOI] [PubMed] [Google Scholar]
- 6. Giannelli G, Koudelkova P, Dituri F, Mikulits W. Role of epithelial to mesenchymal transition in hepatocellular carcinoma. J Hepatol 2016; 65: 798–808. [DOI] [PubMed] [Google Scholar]
- 7. Aravalli RN, Cressman EN, Steer CJ. Cellular and molecular mechanisms of hepatocellular carcinoma: an update. Arch Toxicol 2013; 87: 227–47. [DOI] [PubMed] [Google Scholar]
- 8. Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov 2011; 1: 391–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell 2016; 29: 452–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rinn JL, Kertesz M, Wang JK et al Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007; 129: 1311–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tsai MC, Manor O, Wan Y et al Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010; 329: 689–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gupta RA, Shah N, Wang KC et al Long non‐coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010; 464: 1071–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xing Z, Lin A, Li C et al lncRNA directs cooperative epigenetic regulation downstream of chemokine signals. Cell 2014; 159: 1110–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Karreth FA, Tay Y, Perna D et al In vivo identification of tumor‐ suppressive PTEN ceRNAs in an oncogenic BRAF‐induced mouse model of melanoma. Cell 2011; 147: 382–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gong C, Maquat LE. lncRNAs transactivate STAU1‐mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature 2011; 470: 284–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang P, Xue Y, Han Y et al The STAT3‐binding long noncoding RNA lnc‐DC controls human dendritic cell differentiation. Science 2014; 344: 310–3. [DOI] [PubMed] [Google Scholar]
- 17. Yuan JH, Yang F, Wang F et al A long noncoding RNA activated by TGF‐beta promotes the invasion‐metastasis cascade in hepatocellular carcinoma. Cancer Cell 2014; 25: 666–81. [DOI] [PubMed] [Google Scholar]
- 18. Yang F, Zhang L, Huo XS et al Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology 2011; 54: 1679–89. [DOI] [PubMed] [Google Scholar]
- 19. Xu D, Yang F, Yuan JH et al Long noncoding RNAs associated with liver regeneration 1 accelerates hepatocyte proliferation during liver regeneration by activating Wnt/beta‐catenin signaling. Hepatology 2013; 58: 739–51. [DOI] [PubMed] [Google Scholar]
- 20. Cao C, Sun J, Zhang D et al The long intergenic noncoding RNA UFC1, a target of MicroRNA 34a, interacts with the mRNA stabilizing protein HuR to increase levels of beta‐catenin in HCC cells. Gastroenterology 2015; 148: 415–26 e18. [DOI] [PubMed] [Google Scholar]
- 21. Zhou CC, Yang F, Yuan SX et al Systemic genome screening identifies the outcome associated focal loss of long noncoding RNA PRAL in hepatocellular carcinoma. Hepatology 2016; 63: 850–63. [DOI] [PubMed] [Google Scholar]
- 22. Wang J, Wang H, Zhang Y et al Mutual inhibition between YAP and SRSF1 maintains long non‐coding RNA, Malat1‐induced tumourigenesis in liver cancer. Cell Signal 2014; 26: 1048–59. [DOI] [PubMed] [Google Scholar]
- 23. Quagliata L, Matter MS, Piscuoglio S et al Long noncoding RNA HOTTIP/HOXA13 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. Hepatology 2014; 59: 911–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brooke GN, Culley RL, Dart DA et al FUS/TLS is a novel mediator of androgen‐dependent cell‐cycle progression and prostate cancer growth. Can Res 2011; 71: 914–24. [DOI] [PubMed] [Google Scholar]
- 25. Ward CL, Boggio KJ, Johnson BN et al A loss of FUS/TLS function leads to impaired cellular proliferation. Cell Death Dis 2014; 5: e1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lourenco GF, Janitz M, Huang Y, Halliday GM. Long noncoding RNAs in TDP‐43 and FUS/TLS‐related frontotemporal lobar degeneration (FTLD). Neurobiol Dis 2015; 82: 445–54. [DOI] [PubMed] [Google Scholar]
- 27. Wang X, Arai S, Song X et al Induced ncRNAs allosterically modify RNA‐binding proteins in cis to inhibit transcription. Nature 2008; 454: 126–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mohamadkhani A. Long noncoding RNAs in interaction with RNA binding proteins in hepatocellular carcinoma. Hepat Mon 2014; 14: e18794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lerga A, Hallier M, Delva L et al Identification of an RNA binding specificity for the potential splicing factor TLS. J Biol Chem 2001; 276: 6807–16. [DOI] [PubMed] [Google Scholar]
- 30. Tan AY, Manley JL. The TET family of proteins: functions and roles in disease. J Mol Cell Biol 2009; 1: 82–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell 2009; 136: 777–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ishigaki S, Masuda A, Fujioka Y et al Position‐dependent FUS‐RNA interactions regulate alternative splicing events and transcriptions. Sci Rep 2012; 2: 529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sama RR, Ward CL, Bosco DA. Functions of FUS/TLS from DNA repair to stress response: implications for ALS. ASN Neuro 2014; 6: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yang H, Zhong Y, Xie H et al Induction of the liver cancer‐down‐regulated long noncoding RNA uc002mbe.2 mediates trichostatin‐induced apoptosis of liver cancer cells. Biochem Pharmacol 2013; 85: 1761–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chu C, Zhang QC, da Rocha ST et al Systematic discovery of Xist RNA binding proteins. Cell 2015; 161: 404–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lagier‐Tourenne C, Polymenidou M, Hutt KR et al Divergent roles of ALS‐linked proteins FUS/TLS and TDP‐43 intersect in processing long pre‐mRNAs. Nat Neurosci 2012; 15: 1488–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Masuda A, Takeda J, Ohno K. FUS‐mediated regulation of alternative RNA processing in neurons: insights from global transcriptome analysis. Wiley Interdiscip Rev RNA 2016; 7: 330–40. [DOI] [PubMed] [Google Scholar]
- 38. Honda D, Ishigaki S, Iguchi Y et al The ALS/FTLD‐related RNA‐binding proteins TDP‐43 and FUS have common downstream RNA targets in cortical neurons. FEBS Open Bio 2013; 4: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gou L, Wang W, Tong A et al Proteomic identification of RhoA as a potential biomarker for proliferation and metastasis in hepatocellular carcinoma. J Mol Med (Berl) 2011; 89: 817–27. [DOI] [PubMed] [Google Scholar]
- 40. Li XR, Ji F, Ouyang J, Wu W, Qian LY, Yang KY. Overexpression of RhoA is associated with poor prognosis in hepatocellular carcinoma. Eur J Surg Oncol 2006; 32: 1130–4. [DOI] [PubMed] [Google Scholar]
- 41. Yang W, Lv S, Liu X et al Up‐regulation of Tiam1 and Rac1 correlates with poor prognosis in hepatocellular carcinoma. Jpn J Clin Oncol 2010; 40: 1053–9. [DOI] [PubMed] [Google Scholar]
- 42. Chen J, Xia H, Zhang X et al ECT2 regulates the Rho/ERK signalling axis to promote early recurrence in human hepatocellular carcinoma. J Hepatol 2015; 62: 1287–95. [DOI] [PubMed] [Google Scholar]
- 43. Lee TK, Man K, Ho JW et al Significance of the Rac signaling pathway in HCC cell motility: implications for a new therapeutic target. Carcinogenesis 2005; 26: 681–7. [DOI] [PubMed] [Google Scholar]
- 44. Wong CM, Wei L, Au SL et al MiR‐200b/200c/429 subfamily negatively regulates Rho/ROCK signaling pathway to suppress hepatocellular carcinoma metastasis. Oncotarget 2015; 6: 13658–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang WY, Pan L, Su SC et al Interaction of FUS and HDAC1 regulates DNA damage response and repair in neurons. Nat Neurosci 2013; 16: 1383–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang J, Liu X, Wu H et al CREB up‐regulates long non‐coding RNA, HULC expression through interaction with microRNA‐372 in liver cancer. Nucleic Acids Res 2010; 38: 5366–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wu M, An J, Zheng Q et al Double mutant P53 (N340Q/L344R) promotes hepatocarcinogenesis through upregulation of Pim1 mediated by PKM2 and LncRNA CUDR. Oncotarget 2016; 7: 66525–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yang F, Huo XS, Yuan SX et al Repression of the long noncoding RNA‐LET by histone deacetylase 3 contributes to hypoxia‐mediated metastasis. Mol Cell 2013; 49: 1083–96. [DOI] [PubMed] [Google Scholar]
- 49. Diaz‐Nunez M, Diez‐Torre A, De Wever O et al Histone deacetylase inhibitors induce invasion of human melanoma cells in vitro via differential regulation of N‐cadherin expression and RhoA activity. BMC Cancer 2016; 16: 667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gao YS, Hubbert CC, Lu J, Lee YS, Lee JY, Yao TP. Histone deacetylase 6 regulates growth factor‐induced actin remodeling and endocytosis. Mol Cell Biol 2007; 27: 8637–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Golden SA, Christoffel DJ, Heshmati M et al Epigenetic regulation of RAC1 induces synaptic remodeling in stress disorders and depression. Nat Med 2013; 19: 337–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Cloning the full‐length of human SchLAH gene.
Fig. S2. Effects of SchLAH overexpression on cell invasion and proliferation.
Fig. S3. Effects of FUS knockdown on HepG2 and Huh7 migration.
Fig. S4. The expression of FUS, RhoA and Rac1 by TSA treatment.
Table S1. Proteins in the band specific to SchLAH identified by mass spectrometry.
Table S2. PCR primer pairs for qRT‐PCR.
Table S3. Probes for RNA FISH.
Table S4. Clinical characteristics of 95 HCC patients according to SchLAH expression levels using β‐actin as internal control.


