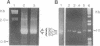Abstract
The sequence of the HA1 region of the hemagglutinin gene of an influenza virus has been determined without growing the virus in eggs or in cultured cells. The virus used was an H1 strain of influenza A from a clinical specimen taken from a patient in 1987. RNA was extracted directly from virus that had been sedimented out of the transport medium in which the sample had been stored. DNA copies of the hemagglutinin gene, obtained by reverse transcription, were then amplified by the polymerase chain reaction and were sequenced by the dideoxy termination method. The deduced amino acid sequence is highly similar to that of other H1 viruses that had been isolated at about the same time and cultured for a limited number of passages in eggs. Furthermore, the HA1 sequence of progeny virus from this isolate obtained after one passage in chicken embryos is identical to that of the virus obtained directly from the nasopharynx. The results suggest that H1 isolates that have been grown for a limited number of passages in embryonated eggs have HA1 subunits that faithfully represent the virus population in the clinical samples from which they were derived.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bosch F. X., Garten W., Klenk H. D., Rott R. Proteolytic cleavage of influenza virus hemagglutinins: primary structure of the connecting peptide between HA1 and HA2 determines proteolytic cleavability and pathogenicity of Avian influenza viruses. Virology. 1981 Sep;113(2):725–735. doi: 10.1016/0042-6822(81)90201-4. [DOI] [PubMed] [Google Scholar]
- Both G. W., Shi C. H., Kilbourne E. D. Hemagglutinin of swine influenza virus: a single amino acid change pleiotropically affects viral antigenicity and replication. Proc Natl Acad Sci U S A. 1983 Nov;80(22):6996–7000. doi: 10.1073/pnas.80.22.6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caton A. J., Brownlee G. G., Yewdell J. W., Gerhard W. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype). Cell. 1982 Dec;31(2 Pt 1):417–427. doi: 10.1016/0092-8674(82)90135-0. [DOI] [PubMed] [Google Scholar]
- Concannon P., Cummings I. W., Salser W. A. Nucleotide sequence of the influenza virus A/USSR/90/77 hemagglutinin gene. J Virol. 1984 Jan;49(1):276–278. doi: 10.1128/jvi.49.1.276-278.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox N. J., Black R. A., Kendal A. P. Pathways of evolution of influenza A (H1N1) viruses from 1977 to 1986 as determined by oligonucleotide mapping and sequencing studies. J Gen Virol. 1989 Feb;70(Pt 2):299–313. doi: 10.1099/0022-1317-70-2-299. [DOI] [PubMed] [Google Scholar]
- Crecelius D. M., Deom C. M., Schulze I. T. Biological properties of a hemagglutinin mutant of influenza virus selected by host cells. Virology. 1984 Nov;139(1):164–177. doi: 10.1016/0042-6822(84)90337-4. [DOI] [PubMed] [Google Scholar]
- Daniels P. S., Jeffries S., Yates P., Schild G. C., Rogers G. N., Paulson J. C., Wharton S. A., Douglas A. R., Skehel J. J., Wiley D. C. The receptor-binding and membrane-fusion properties of influenza virus variants selected using anti-haemagglutinin monoclonal antibodies. EMBO J. 1987 May;6(5):1459–1465. doi: 10.1002/j.1460-2075.1987.tb02387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deom C. M., Caton A. J., Schulze I. T. Host cell-mediated selection of a mutant influenza A virus that has lost a complex oligosaccharide from the tip of the hemagglutinin. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3771–3775. doi: 10.1073/pnas.83.11.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desselberger U., Racaniello V. R., Zazra J. J., Palese P. The 3' and 5'-terminal sequences of influenza A, B and C virus RNA segments are highly conserved and show partial inverted complementarity. Gene. 1980 Feb;8(3):315–328. doi: 10.1016/0378-1119(80)90007-4. [DOI] [PubMed] [Google Scholar]
- Hiti A. L., Davis A. R., Nayak D. P. Complete sequence analysis shows that the hemagglutinins of the H0 and H2 subtypes of human influenza virus are closely related. Virology. 1981 May;111(1):113–124. doi: 10.1016/0042-6822(81)90658-9. [DOI] [PubMed] [Google Scholar]
- Katz J. M., Naeve C. W., Webster R. G. Host cell-mediated variation in H3N2 influenza viruses. Virology. 1987 Feb;156(2):386–395. doi: 10.1016/0042-6822(87)90418-1. [DOI] [PubMed] [Google Scholar]
- Katz J. M., Webster R. G. Antigenic and structural characterization of multiple subpopulations of H3N2 influenza virus from an individual. Virology. 1988 Aug;165(2):446–456. doi: 10.1016/0042-6822(88)90588-0. [DOI] [PubMed] [Google Scholar]
- Kawaoka Y., Webster R. G. Interplay between carbohydrate in the stalk and the length of the connecting peptide determines the cleavability of influenza virus hemagglutinin. J Virol. 1989 Aug;63(8):3296–3300. doi: 10.1128/jvi.63.8.3296-3300.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilbourne E. D. Genetic dimorphism in influenza viruses: characterization of stably associated hemagglutinin mutants differing in antigenicity and biological properties. Proc Natl Acad Sci U S A. 1978 Dec;75(12):6258–6262. doi: 10.1073/pnas.75.12.6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noronha-Blob L., Schulze I. T. Viral interference-mediated selection of a plaque-type variant of influenza virus. Virology. 1976 Jan;69(1):314–322. doi: 10.1016/0042-6822(76)90218-x. [DOI] [PubMed] [Google Scholar]
- Raymond F. L., Caton A. J., Cox N. J., Kendal A. P., Brownlee G. G. The antigenicity and evolution of influenza H1 haemagglutinin, from 1950-1957 and 1977-1983: two pathways from one gene. Virology. 1986 Jan 30;148(2):275–287. doi: 10.1016/0042-6822(86)90325-9. [DOI] [PubMed] [Google Scholar]
- Robertson J. S. 5' and 3' terminal nucleotide sequences of the RNA genome segments of influenza virus. Nucleic Acids Res. 1979 Aug 24;6(12):3745–3757. doi: 10.1093/nar/6.12.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson J. S., Bootman J. S., Newman R., Oxford J. S., Daniels R. S., Webster R. G., Schild G. C. Structural changes in the haemagglutinin which accompany egg adaptation of an influenza A(H1N1) virus. Virology. 1987 Sep;160(1):31–37. doi: 10.1016/0042-6822(87)90040-7. [DOI] [PubMed] [Google Scholar]
- Robertson J. S., Naeve C. W., Webster R. G., Bootman J. S., Newman R., Schild G. C. Alterations in the hemagglutinin associated with adaptation of influenza B virus to growth in eggs. Virology. 1985 May;143(1):166–174. doi: 10.1016/0042-6822(85)90105-9. [DOI] [PubMed] [Google Scholar]
- Robertson J. S. Sequence analysis of the haemagglutinin of A/Taiwan/1/86, a new variant of human influenza A(H1N1) virus. J Gen Virol. 1987 Apr;68(Pt 4):1205–1208. doi: 10.1099/0022-1317-68-4-1205. [DOI] [PubMed] [Google Scholar]
- Rogers G. N., Paulson J. C., Daniels R. S., Skehel J. J., Wilson I. A., Wiley D. C. Single amino acid substitutions in influenza haemagglutinin change receptor binding specificity. Nature. 1983 Jul 7;304(5921):76–78. doi: 10.1038/304076a0. [DOI] [PubMed] [Google Scholar]
- Rogers G. N., Paulson J. C. Receptor determinants of human and animal influenza virus isolates: differences in receptor specificity of the H3 hemagglutinin based on species of origin. Virology. 1983 Jun;127(2):361–373. doi: 10.1016/0042-6822(83)90150-2. [DOI] [PubMed] [Google Scholar]
- Rott R., Orlich M., Klenk H. D., Wang M. L., Skehel J. J., Wiley D. C. Studies on the adaptation of influenza viruses to MDCK cells. EMBO J. 1984 Dec 20;3(13):3329–3332. doi: 10.1002/j.1460-2075.1984.tb02299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schild G. C., Oxford J. S., de Jong J. C., Webster R. G. Evidence for host-cell selection of influenza virus antigenic variants. Nature. 1983 Jun 23;303(5919):706–709. doi: 10.1038/303706a0. [DOI] [PubMed] [Google Scholar]
- Skehel J. J., Hay A. J. Nucleotide sequences at the 5' termini of influenza virus RNAs and their transcripts. Nucleic Acids Res. 1978 Apr;5(4):1207–1219. doi: 10.1093/nar/5.4.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skehel J. J., Stevens D. J., Daniels R. S., Douglas A. R., Knossow M., Wilson I. A., Wiley D. C. A carbohydrate side chain on hemagglutinins of Hong Kong influenza viruses inhibits recognition by a monoclonal antibody. Proc Natl Acad Sci U S A. 1984 Mar;81(6):1779–1783. doi: 10.1073/pnas.81.6.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y., Kato H., Naeve C. W., Webster R. G. Single-amino-acid substitution in an antigenic site of influenza virus hemagglutinin can alter the specificity of binding to cell membrane-associated gangliosides. J Virol. 1989 Oct;63(10):4298–4302. doi: 10.1128/jvi.63.10.4298-4302.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster R. G., Laver W. G., Air G. M., Schild G. C. Molecular mechanisms of variation in influenza viruses. Nature. 1982 Mar 11;296(5853):115–121. doi: 10.1038/296115a0. [DOI] [PubMed] [Google Scholar]
- Weis W., Brown J. H., Cusack S., Paulson J. C., Skehel J. J., Wiley D. C. Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature. 1988 Jun 2;333(6172):426–431. doi: 10.1038/333426a0. [DOI] [PubMed] [Google Scholar]
- Wiley D. C., Skehel J. J. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu Rev Biochem. 1987;56:365–394. doi: 10.1146/annurev.bi.56.070187.002053. [DOI] [PubMed] [Google Scholar]
- Wilson I. A., Skehel J. J., Wiley D. C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature. 1981 Jan 29;289(5796):366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- Winter G., Fields S., Brownlee G. G. Nucleotide sequence of the haemagglutinin gene of a human influenza virus H1 subtype. Nature. 1981 Jul 2;292(5818):72–75. doi: 10.1038/292072a0. [DOI] [PubMed] [Google Scholar]
- Winter G., Fields S., Gait M. J., Brownlee G. G. The use of synthetic oligodeoxynucleotide primers in cloning and sequencing segment of 8 influenza virus (A/PR/8/34). Nucleic Acids Res. 1981 Jan 24;9(2):237–245. doi: 10.1093/nar/9.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]