Abstract
It is well documented that human epidermal growth factor receptor 2 (HER2) overexpression/amplification is associated with poor survival in breast cancer patients. However, it is largely unknown whether HER2 somatic mutations are associated with survival in HER2‐negative breast cancer patients. Here, we identified HER2 somatic mutations in tumors from 1348 unselected breast cancer patients by sequencing the entire HER2 coding region. All of these mutations were tested for in corresponding blood samples to determine whether they were somatic or germline mutations. We further investigated the associations between HER2 somatic mutations and recurrence‐free survival and distant recurrence‐free survival in this cohort of patients. We found that 27 of 1348 (2.0%) of these patients carried a HER2 somatic mutation. In vitro experiments indicated that some of the novel mutations and those with unknown functions increased HER2 activity. HER2 status was available for 1306 patients, and the HER2 somatic mutation rates in HER2‐positive (n = 353) and HER2‐negative breast cancers (n = 953) were 1.4% and 2.3%, respectively. Among the HER2‐negative patients, those with a HER2 somatic mutation had a significantly worse recurrence‐free survival (unadjusted hazard ratio = 2.67; 95% confidence interval, 1.25–5.72, P = 0.002) and distant recurrence‐free survival (unadjusted hazard ratio = 2.50; 95% confidence interval, 1.10–5.68, P = 0.004) than those with wild‐type HER2. Taken together, our findings suggested that HER2 somatic mutations occur at a higher frequency in HER2‐negative breast cancer, and HER2‐negative breast cancer patients with these mutations have poor survival. Therefore, HER2‐negative patients with a HER2 somatic mutation are potentially good candidates for HER2‐targeted therapy.
Keywords: Breast cancer, HER2, somatic mutations, survival, targeted therapy
Human epidermal growth factor 2 is a major proliferative stimulator that activates downstream signaling through the phosphoinositide 3‐kinase/AKT and MAPK pathways.1, 2, 3, 4, 5 Amplification/overexpression of HER2 occurs in 20%–25% of breast cancers, and is associated with poor survival.6, 7The use of HER2‐targeted drugs, which currently include trastuzumab, pertuzumab, and lapatinib, have dramatically improved the outcomes in HER2‐positive breast cancers.8, 9, 10, 11, 12, 13
Human epidermal growth factor receptor 2 somatic mutations were initially identified in the tyrosine kinase domain of the HER2 gene in breast cancer patients in 2006.14 Recently, such mutations have been identified in HER2‐negative breast cancer patients by sequencing assays, including whole‐cancer genome sequencing.14, 15, 16, 17, 18, 19, 20, 21 Although the mutation rate of this gene is very low (<2%), in vitro and in vivo experiments have shown that some somatic mutations can activate the HER2 signaling pathway in HER2‐negative cells and that these cells are sensitive to some HER2‐targeted drugs.22, 23, 24, 25 These findings suggest that HER2 somatic mutations represent an alternative mechanism for the activation of HER2 in HER2‐negative breast cancers, raising an interesting question of whether somatic mutations in HER2‐negative breast cancer patients influence the clinical outcome. Therefore, the purposes of this study were as follows: (i) to identify HER2 somatic mutations in tumor tissues from a large cohort of 1348 patients; (ii) to investigate whether the somatic mutations with novel or unknown functions identified in this study affect HER2 function by undertaking in vitro experiments; and (iii) to investigate whether HER2 somatic mutations influence patient survival in the entire study population and, specifically, in HER2‐negative breast cancer patients.
Materials and Methods
Patients
From November 2003 to July 2012, fresh‐frozen tumor tissues were obtained by core‐needle biopsy prior to therapy or were procured during surgery from 1496 primary breast cancer patients (stages I–III) at the Breast Center, Peking University Cancer Hospital (Beijing, China). Human epidermal growth factor receptor 2 somatic mutation status was successfully determined in 1348 of these patients. Tumor stage was classified according to the TNM classification of the Union for International Cancer Control. Tumor size was defined as the maximum tumor diameter as measured by ultrasound at the time of diagnosis. Tumors were graded histologically according to the modified Bloom–Richardson grading system. This study was carried out in accordance with the ethics principles of the Declaration of Helsinki and approved by the Research and Ethics Committee of Peking University Cancer Hospital. All patients provided written informed consent.
Analysis of HER2 mutations in tumor tissue
Tumor samples were obtained by core‐needle biopsy or at the time of surgery and immediately stored at −80°C. Total RNA was extracted using TRIzol reagent (Life Technologies, Gaithersburg, MD, USA) and reverse‐transcribed to cDNA using the standard procedure. The complete HER2 coding sequence was amplified with nine sets of primers. All fragments were sequenced using a BigDye Terminator Cycle Sequencing Kit and an ABI3730 automated sequencer (Applied Biosystems, Foster City, CA, USA). All sequence variants were confirmed in duplicate.
Analysis of HER2 germline mutations
Genomic DNA was extracted from peripheral blood leukocytes using a Whole Blood Genome DNA Isolation Kit (Bioteke, Beijing, China). Patients with a HER2 mutation in their breast tumor were further investigated to determine whether the mutation was present in their blood DNA.
Estrogen receptor, PR, and HER2 status
Estrogen receptor, PR, and HER2 status were determined using breast cancer tissue obtained from the core‐needle biopsy or tumor tissues after surgery. Estrogen receptor or PR immunostaining was considered positive when ≥1% of tumor cells showed positive nuclear staining. The HER2 staining was scored according to the standard method. Scores of 0 and 1+ were considered negative, and a score of 3+ was considered positive. If a score was 2+, we further evaluated the HER2 status using FISH (Vysis, Downers Grove, IL, USA) of core biopsies according to the manufacturer's instructions.
Functional characterization of somatic HER2 mutations in vitro
MCF‐7 (human breast cancer cells) and HEK293T (human embryonic kidney cells) cells were recultured in DMEM (HyClone, Logan, UT, USA) with 10% FBS (HyClone). The HER2 WT‐pXJ40‐myc plasmid was kindly provided by Dr. Qinong Ye (Academy of Military Medical Sciences, Beijing, China). Fifteen missense mutations (L12R, E139G, E139D, A466V, C515R, T526A, L755S, G776R, S783P, T862R, L869R, P885S, R897G, F1030C, and P1074S) were introduced by site‐directed mutagenesis and confirmed by sequencing. V777L, a well‐characterized activating mutation, served as a positive control. Transient transfection of the plasmids into the MCF‐7 and HEK293T cells was carried out using VigoFect (Vigorous Biotechnology, Beijing, China), according to the manufacturer's protocol. After 24 h, the cells were washed twice with 1 × PBS and starved in serum‐free medium for another 12 h.
The following antibodies were used: AKT (C67E7), phospho‐AKT (Ser473) (D9E), ERK1/2 (137F5), and phospho‐ERK1/2 (Thr202/Tyr204) (D13.14.4E) (Cell Signaling Technology, Boston, MA, USA). Phospho‐HER2 (pY1248, 06‐229) was from Millipore (CA, USA); HER2 (C‐18) and GAPDH (FL‐335) were from Santa Cruz Biotechnology (CA, USA). GAPDH was used as a protein loading control. The relative intensities of individual protein bands were quantified by analysis of digitized images using ImageJ software (https://imagej.nih.gov/ij/download.html). For the phospho‐specific detection of proteins, the acquired density was compared with the corresponding total antibody signal. All data shown are representative of at least three experiments.
Statistical analysis
Differences in clinicopathological characteristics between patients with a HER2 somatic mutation and those with wild‐type HER2 were determined using Pearson's chi‐square‐test or Fisher's exact test. Survival curves were generated and compared using the Kaplan–Meier method and Breslow tests. Recurrence‐free survival was calculated from the time of diagnosis to the first recurrence (local or distant) or death from breast cancer (for patients without a recorded relapse) or to the date of the last follow‐up. Distant recurrence‐free survival was calculated from the time of diagnosis to the first distant metastasis or death from breast cancer (for patients without a recorded relapse) or to the date of the last follow‐up. The Cox proportional hazards model was used to determine the association of HER2 somatic mutation status with the risk of local or distant recurrence after adjustments for patient and tumor characteristics. Two‐sided P‐values less than 0.05 were considered statistically significant. All analyses were carried out using spss 20.0 software (Chicago, IL, USA).
Results
Somatic mutations in the HER2 gene
A total of 33 patients carried HER2 mutations in their tumor tissues in this cohort of 1348 breast cancer patients (Table S1). Four patients had two HER2 mutations, resulting in a total of 37 mutations in 33 patients. The mutations in seven patients (one of the seven patients also had a somatic HER2 mutation) were also present in the corresponding blood DNA samples, indicating that they were germline (Table S1). The remaining 30 mutations (27 patients) were absent from matched blood samples, indicating that they were somatic (Table S1). Thus, we finally confirmed that 27 of 1348 patients (2.0%) carried a HER2 somatic mutation in this cohort (Table 1).
Table 1.
Clinical information of breast cancer patients with HER2 somatic mutations (n = 27)
| Patient ID | Protein change | Impact | Tumor type | Grade | ER | PR | HER2 | Lymph nodes status |
|---|---|---|---|---|---|---|---|---|
| 469 | p.S310F | Activating(26) | IDC | 1 | − | − | + | − |
| 3044 | p.S310F | Activating(26) | IDC | 2 | + | − | − | − |
| 3456 | p.S310F | Activating(26) | IDC | 2 | + | + | − | − |
| 5547 | p.S310F | Activating(26) | ILC | – | + | + | − | − |
| 9603 | p.D769H | Activating(22) | IDC | 2 | − | − | + | − |
| 4393 | p.A775‐G776insYVMA | Activating(27) | IDC | 2 | + | + | − | − |
| 2619 | p.A775‐G776insYVMA | Activating(27) | IDC | 2 | + | + | − | + |
| 2373 | p.V777L | Activating(22) | ILC | – | − | − | − | + |
| 3624 | p.V777L | Activating(22) | IDC | 2 | + | − | − | − |
| 4223 | p.V777L | Activating(22) | IDC | 1 | + | + | − | − |
| 5669 | p.V777L | Activating(22) | IDC | 2 | − | − | + | + |
| 6795 | p.V777L | Activating(22) | IDC | 2 | + | − | + | − |
| 3943 | p.V777L | Activating(22) | IDC | 2 | − | − | − | − |
| 3943 | p.T862A | Unknowna | IDC | 2 | − | − | − | − |
| 1510 | p.L12R | Novel | IDC | 2 | − | − | − | + |
| 327 | p.E139D | Novel | IDC | 3 | + | − | − | − |
| 930 | p.E139G | Novel | IDC | 2 | + | + | − | − |
| 3860 | p.A466V | Novel | IDC | 3 | + | − | − | + |
| 146 | p.C515R | Novel | IDC | 2 | − | − | − | − |
| 407 | p.T526A | Novel | ACC | – | − | − | − | − |
| 10001 | p.G776R | Novel | IDC | 2 | + | + | − | + |
| 1028 | p.L869R | Unknowna | IDC | 2 | + | + | + | − |
| 3733 | p.L869R | Unknowna | IDC | 2 | − | − | − | + |
| 3733 | p.R897G | Novel | IDC | 2 | − | − | − | + |
| 4137 | p.P885S | Novel | IDC | 2 | − | − | − | − |
| 4892 | p.F1030C | Novel | IDC | 3 | + | + | − | − |
| 4663 | p.P1074S | Novel | IDC | 2 | + | + | − | − |
| 2476 | p.L755S | Unknown(22) | IDC | 2 | + | − | − | + |
| 3896 | p.L755S | Unknown(22) | IDC | 3 | + | + | − | + |
Reported in COSMIC (http://cancer.sanger.ac.uk/cancergenome/projects/cosmic/).
ACC, adenoid cystic carcinoma; ER, estrogen receptor; HER2, human epidermal growth factor receptor 2; IDC, invasive ductal carcinoma; ILC, invasive lobular carcinoma; PR, progesterone receptor.
Information regarding HER2 status was available for 1306 of the 1348 patients in this study. Of these, 353 (27.0%) were HER2‐positive, and 953 (73.0%) were HER2‐negative (Table 2). Among the 27 patients with a HER2 somatic mutation, the frequencies of these mutations in those with HER2‐negative and HER2‐positive breast cancers were 2.3% (22/953) and 1.4% (5/353), respectively.
Table 2.
Association of patient/tumor characteristics with HER2 somatic mutation in the full cohort of breast cancer patients (n = 1348)
| Characteristic | No. of patients | Non‐carriers (n = 1321) | Carriers (n = 27) | a P‐value | ||
|---|---|---|---|---|---|---|
| No. | % | No. | % | |||
| Age, years | ||||||
| ≤50 | 596 | 589 | 44.6 | 7 | 25.9 | <0.001 |
| >50 | 752 | 732 | 55.4 | 20 | 74.1 | |
| Tumor size, cm | ||||||
| ≤2 | 515 | 504 | 38.2 | 11 | 40.7 | 0.789 |
| >2 | 831 | 815 | 61.8 | 16 | 59.3 | |
| Unknown | 2 | 2 | 0 | |||
| Tumor grade | ||||||
| 1 | 146 | 144 | 12.0 | 2 | 8.0 | 0.790 |
| 2 | 907 | 888 | 74.1 | 19 | 76.0 | |
| 3 | 170 | 166 | 13.9 | 4 | 16.0 | |
| Unknown | 125 | 123 | 2 | |||
| Tumor stage | ||||||
| I | 316 | 308 | 24.1 | 8 | 29.6 | 0.793 |
| II | 764 | 749 | 58.7 | 15 | 55.6 | |
| III | 224 | 220 | 17.2 | 4 | 14.8 | |
| Unknown | 44 | 44 | 0 | |||
| Lymph node status | ||||||
| Negative | 742 | 724 | 56.7 | 18 | 66.7 | 0.298 |
| Positive | 563 | 554 | 43.3 | 9 | 33.3 | |
| Unknown | 43 | 43 | 0 | |||
| ER status | ||||||
| Negative | 388 | 378 | 28.7 | 10 | 37.0 | 0.347 |
| Positive | 954 | 937 | 71.3 | 17 | 63.0 | |
| Unknown | 6 | 6 | 0 | |||
| PR status | ||||||
| Negative | 530 | 514 | 39.4 | 16 | 59.3 | 0.037 |
| Positive | 802 | 791 | 60.6 | 11 | 40.7 | |
| Unknown | 16 | 16 | 0 | |||
| HER2 status | ||||||
| Negative | 953 | 931 | 72.8 | 22 | 81.5 | 0.314 |
| Positive | 353 | 348 | 27.2 | 5 | 18.5 | |
| Unknown | 42 | 42 | 0 | |||
| Triple‐negative status | ||||||
| No | 1144 | 1124 | 85.5 | 20 | 74.1 | 0.098 |
| Yes | 198 | 191 | 14.5 | 7 | 25.9 | |
| Unknown | 6 | 6 | 0 | |||
| Surgery type | ||||||
| BCS | 478 | 472 | 36.7 | 6 | 22.2 | 0.122 |
| Mastectomy | 835 | 814 | 63.3 | 21 | 77.8 | |
| Unknown | 35 | 35 | 0 | |||
| Trastuzumab use | ||||||
| No | 1286 | 1260 | 95.4 | 26 | 96.3 | 1.000 |
| Yes | 62 | 61 | 4.6 | 1 | 3.7 | |
| Adjuvant therapy | ||||||
| C | 374 | 362 | 28.6 | 12 | 44.4 | 0.060 |
| E | 246 | 239 | 18.9 | 7 | 25.9 | |
| C plus E | 672 | 664 | 52.5 | 8 | 29.6 | |
| None | 56 | 56 | 0 | |||
Patients with HER2 somatic mutations versus patients with wild‐type.
BCS, breast‐conserving surgery; C, chemotherapy; E, endocrine therapy; ER, estrogen receptor; HER2, human epidermal growth factor receptor 2; PR, progesterone receptor.
HER2 somatic mutations were not significantly associated with tumor size, tumor grade, lymph node status, ER, PR, or HER2 status. However, patients with a HER2 somatic mutation were older than those with wild‐type HER2 (Table 2). Trastuzumab use, adjuvant chemotherapy, and breast‐conserving therapy did not significantly differ between the patients with a HER2 somatic mutation and those with wild‐type HER2 (Table 2).
The majority of patients with a HER2 mutation had missense mutations (92.6%, 25/27), and only two showed the same insertion mutation (A775_G776insYVMA). Examination of the locations of the somatic mutations in the HER2 domains revealed that they were clustered into two major areas: 37.0% of the patients (10/27) had an ECD mutation and 55.6% (15/27) had a KD mutation (Fig. 1). Five recurrent mutations, V777L, S310F, L755S, A775_G776insYVMA, and L869R, were found in this cohort (Table 1). Sixteen of the 27 patients (59.3%) carried a recurrent mutation (Table 1). All five recurrent mutations have been reported previously, and all were located in the KD, with the exception of S310F (Fig. 1).
Figure 1.
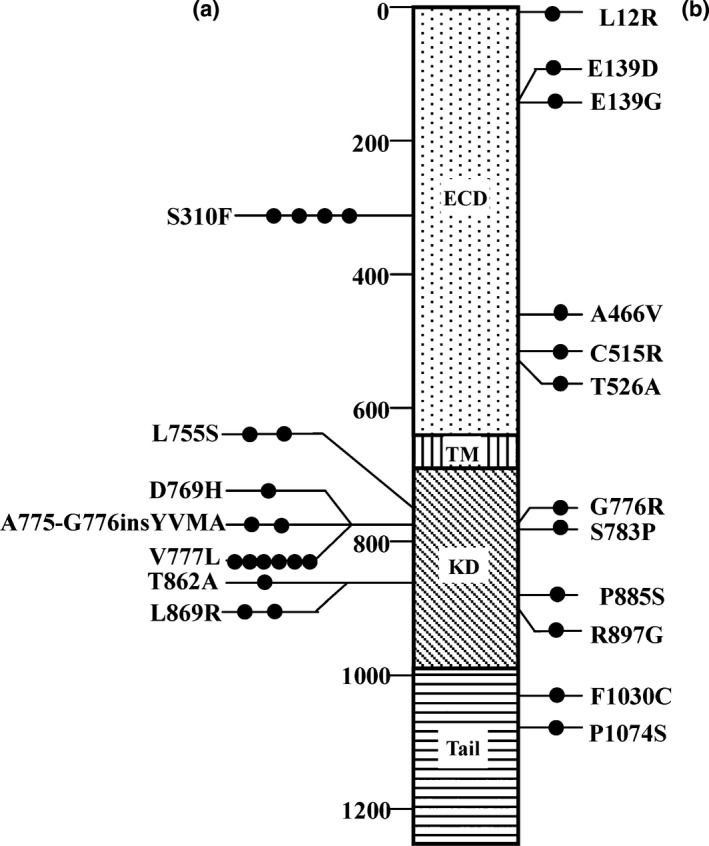
HER2 somatic mutations observed in 27 patients with primary breast cancer. (a) Mutations described previously. (b) Novel mutations. The black circles represent each case of the indicated mutation. Three patients had two HER2 somatic mutations each, resulting in a total of 30 mutations in 27 patients. ECD, extracellular domain; KD, kinase domain; TM, transmembrane region.
Functional effects of HER2 somatic mutations
In this cohort, 27 patients carried 30 somatic mutations (Table 1, Fig. 1). Of these, 18 have been reported previously22, 26, 27 and 12 are novel (Fig. 1). Based on published reports,22, 26, 27 we estimated that at least 13 of the 27 patients carried an activating HER2 mutation that was likely to be a driver event in breast cancer (Table 1). The functional effects of the remaining 15 mutations were not determined or were not fully elucidated. We therefore further characterized these mutations by undertaking in vitro experiments.
Functional analyses of novel and unknown mutations in vitro
The level of P‐HER2 was markedly higher in MCF‐7 cells with C515R, T526A, G776R, L755S, A466V, T862R, and P1074S mutations than in cells with wild‐type HER2 (Fig. 2a,Fig. S1a). No significant differences were observed in the level of P‐HER2 between cells with the remaining mutations and wild‐type HER2 (Fig. 2a,Fig. S1a). In HEK293T cells, the C515R, A466V, T862R, L869R, and R897G mutations strongly increased the level of P‐HER2 (Fig. 2b,Fig. S1b). The C515R mutation also increased the levels of P‐ERK and P‐AKT (Fig. 2b). In addition, the L12R, T526A, and G776R mutations increased the level of P‐ERK but had little effect on P‐HER2 and P‐AKT levels. The L755S mutation increased the phosphorylation of ERK and AKT but decreased the phosphorylation of HER2 (Fig. 2b). These results showed that the C515R, A466V, and T862R mutations increased HER2 activity in both cell lines, suggesting that they are activating mutations. The T526A, G776R, L755S, L869R, R897G, and P1074S mutations increased HER2 activity in either MCF‐7 or HEK293T cells, suggesting that they are also activating mutations. The other mutations (L12R, E139G, E139D, S783P, P885S, and F1030C) seemed to be neutral. Thus, nine of the 15 mutations are activating mutations.
Figure 2.
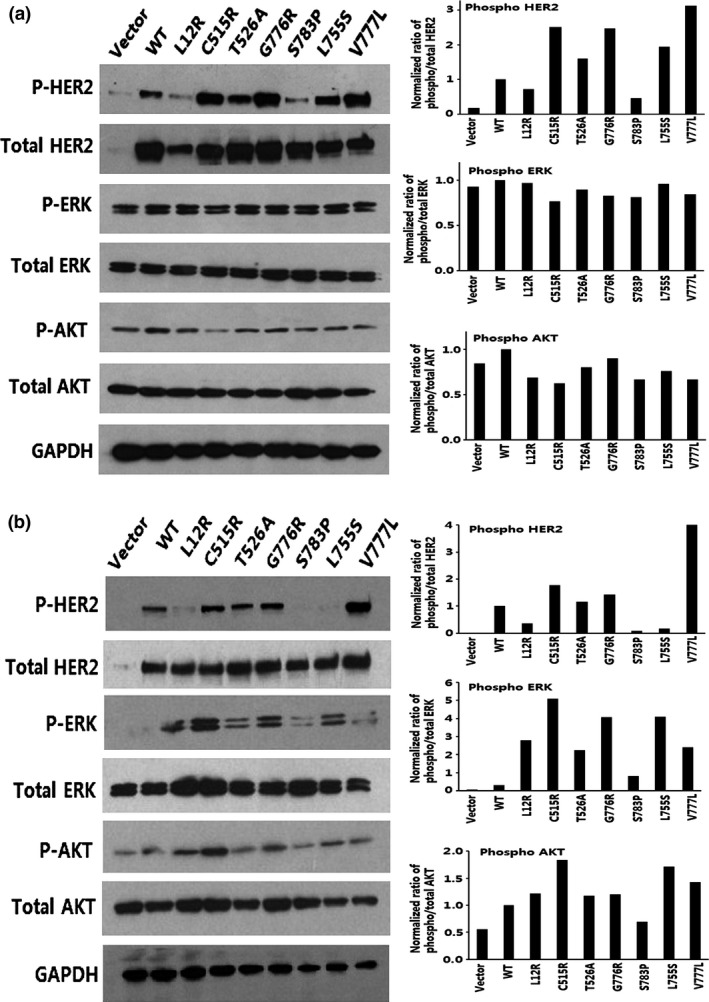
MCF‐7 (a) and HEK293T (b) cells were transfected with wild‐type HER2 or L12R, C515R, T526A, G776R, S783P, L755S, and V777L mutants, and lysates were probed with the indicated antibodies. The bar graphs show the quantifications of Western blot bands. AKT, protein kinase B; HER2, human epidermal growth factor receptor 2; P‐, phosphorylated.
HER2 somatic mutations and survival in the entire study cohort
To investigate the association between survival and HER2 somatic mutations, a total of 1348 breast cancer patients were evaluated. The median length of follow‐up was 60 months (range, 1–116 months). Two hundred and sixteen patients experienced recurrence (local or distant) or died of breast cancer in this cohort of patients during the follow‐up period.
Patients with HER2 mutations had a significantly worse RFS (unadjusted HR = 1.91; 95% CI, 0.90–4.05, P = 0.025; Fig. S2a) and DRFS (unadjusted HR = 1.91; 95% CI, 0.85–4.32, P = 0.033; Fig. S2b) than those with wild‐type HER2 in the entire study cohort.
HER2 somatic mutations and survival in HER2‐negative breast cancer patients
HER2 somatic mutations were detected at a higher frequency in HER2‐negative breast cancer patients. HER2‐negative patients with a HER2 somatic mutation (n = 22) had a significantly worse RFS (unadjusted HR = 2.67; 95% CI, 1.25–5.72, P = 0.002; Fig. 3a) and DRFS (unadjusted HR = 2.50; 95% CI, 1.10–5.68, P = 0.004; Fig. 3b) than those with wild‐type HER2 (n = 953). Multivariate analysis revealed that HER2 somatic mutation was a borderline unfavorable factor for RFS (HR = 2.47; 95% CI, 0.99‐6.16, P = 0.051) (Table S2), and it was also a non‐significantly unfavorable factor for DRFS (HR = 2.29; 95% CI, 0.92–5.70, P = 0.075) (Table S2) after adjusting for age, lymph node, tumor size, tumor grade, ER, PR, and HER2 status.
Figure 3.
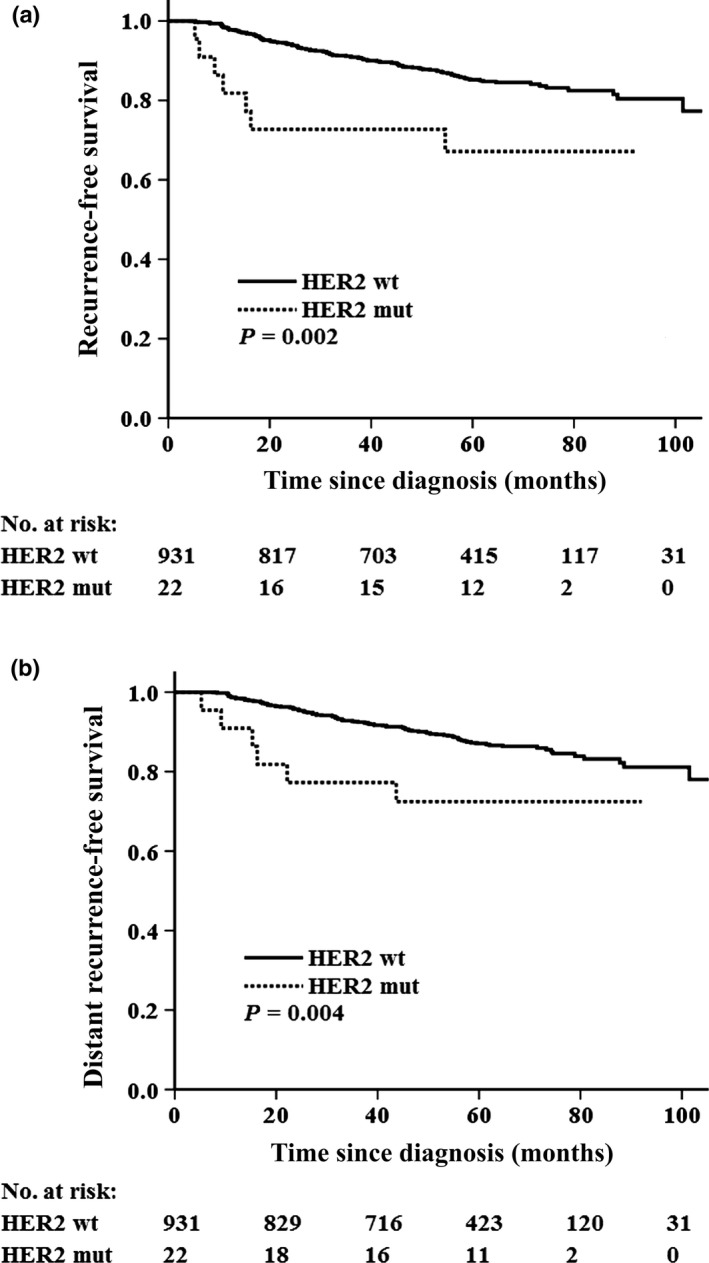
Kaplan−Meier curves for survival based on HER2 somatic mutations in HER2‐negative patients with primary breast cancer (n = 953). (a) Recurrence‐free survival. (b) Distant recurrence‐free survival. HER2, human epidermal growth factor receptor 2; mut, mutant; wt, wild‐type.
Only five patients with a HER2 somatic mutation were HER2‐positive, and none of them received trastuzumab treatment. Given the small sample size, we did not undertake survival analysis in this subgroup.
Discussion
In this study, by sequencing tumor cDNA, we found that 27 patients (2.0%) carried a HER2 somatic mutation in a cohort of 1348 operable primary breast cancer patients. These mutations were more common in HER2‐negative breast cancer patients (2.3%) than in those who were HER2‐positive (1.4%). HER2‐negative patients with a HER2 somatic mutation had a significantly worse survival than those with wild‐type HER2.
All of the recurrent mutations identified in this study have been previously reported, and of these, V777L, S310F, and A775_G776insYVMA were determined to be activating mutations.22, 26, 27 Four of the five recurrent mutations were located in the KD, and S310F was localized to the ECD. The five recurrent mutations and other mutations in the KD accounted for 74.1% (20/27) of the mutations in this study. Therefore, screening for recurrent mutations and mutations in the KD is a fast and effective assay to quickly detect HER2 somatic mutations in breast cancer patients.
We also identified 12 novel mutations, which were located throughout the entire gene. Functional analysis of the novel mutations and those with unknown function (a total of 15 mutations) showed that 9 of the 15 mutations activated the HER2 signaling pathway. Our findings, and those of previous reports,22, 26, 27 suggest that the majority of HER2 somatic mutations are disease‐associated.
In this study, we also found six HER2 germline mutations (R157W, A466V, K681N, R849W, R1146W, and E1195G) in seven breast cancer patients. Among them, R157W was previously reported as a somatic mutation,28 which was found in micropapillary urothelial carcinoma located in the ECD and was predicted to be pathogenic by FATHMM prediction (Cosmic database). The A466V mutation was also determined as somatic in this cohort and increased HER2 activity in MCF‐7 and HEK293T cell lines. The remaining germline mutations (K681N, R849W, R1146W, and E1195G) were novel and need further functional studies.
Clinically, HER2‐negative breast cancers are not typically treated with HER2‐targeted therapy. Recent studies have suggested that the HER2‐targeted drugs trastuzumab and lapatinib inhibit the growth of cells with HER2 somatic mutations in vitro and in vivo.22, 23, 24 In addition, neratinib, a novel HER2‐targeted drug, has shown a strong inhibitory effect in patients with a HER2 somatic mutation.22, 29, 30 Therefore, given the poor survival of HER2‐negative breast cancer patients who carry a HER2 somatic mutation, they are good candidates for receiving HER2‐targeted therapy or for recruitment into ongoing clinical trials. In addition, five HER2‐positive patients carried an activating HER2 somatic mutation. Two of them had contralateral breast cancer and one died from suicide. Given the small sample size, we did not undertake survival analysis in the HER2‐positive subgroup.
The 2.3% somatic mutation rate in HER2‐negative breast cancer patients was remarkable, considering that 70%–80% of all patients had HER2‐negative breast cancer. Therefore, it will not be difficult to gather enough patients for clinical trials. HER2‐negative patients had a generally favorable survival compared with the HER2‐positive patients, and when the HER2‐negative patients were grouped according to the presence/absence of a HER2 somatic mutation, those without a somatic mutation had better survival.
In summary, we found that HER2 somatic mutations occurred more frequently in the HER2‐negative breast cancer patients compared with those with HER2‐positive breast cancer, and that approximately 2.3% of these HER2‐negative patients harbored a HER2 somatic mutation. HER2‐negative patients carrying a HER2 somatic mutation were determined to have an unfavorable outcome. Therefore, HER2‐negative patients with this type of mutation are potentially good candidates for HER2‐targeted therapy.
Disclosure Statement
The authors have no conflict of interest.
Abbreviations
- AKT
protein kinase B
- CI
confidence interval
- DRFS
distant recurrence‐free survival
- ECD
extracellular domain
- ER
estrogen receptor
- HER2
human epidermal growth factor receptor 2
- HR
hazard ratio
- KD
kinase domain
- PR
progesterone receptor
- RFS
recurrence‐free survival
Supporting information
Fig. S1. MCF‐7 (a) and HEK293T (b) cells were transfected with wild‐type HER2 or mutants.
Fig. S2. Kaplan–Meier curves for survival.
Table S1. HER2 somatic and germline mutations in the entire cohort of primary breast cancer
Table S2. Multivariate analyses of recurrence‐free survival (RFS) and distant recurrence‐free survival (DRFS) in HER2‐negative subgroup.
Acknowledgments
This research was funded by the National Key Technology Research and Development Program of the Ministry of Science and Technology of China (Grant No. 2014BAI09B08), the 973 Project (Grant No. 2013CB911004), and the National Natural Science Foundation of China (Grant Nos. 30973436 and 81071629).
Cancer Sci 108 (2017) 671–677
Funding Information
Ministry of Science and Technology, China; National Natural Science Foundation of China.
References
- 1. Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol 2001; 2: 127–37. [DOI] [PubMed] [Google Scholar]
- 2. Olayioye MA, Neve RM, Lane HA, Hynes NE. The ErbB signaling network: receptor heterodimerization in development and cancer. EMBO J 2000; 19: 3159–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Deb TB, Su L, Wong L et al Epidermal growth factor (EGF) receptor kinase‐independent signaling by EGF. J Biol Chem 2001; 276: 15554–60. [DOI] [PubMed] [Google Scholar]
- 4. Yarden Y, Pines G. The ERBB network: at last, cancer therapy meets systems biology. Nat Rev Cancer 2012; 12: 553–63. [DOI] [PubMed] [Google Scholar]
- 5. Alimandi M, Romano A, Curia MC et al Cooperative signaling of ErbB3 and ErbB2 in neoplastic transformation and human mammary carcinomas. Oncogene 1995; 10: 1813–21. [PubMed] [Google Scholar]
- 6. Slamon DJ, Clark GM, Wong SG et al Human breast cancer: correlation of relapse and survival with amplification of the HER‐2/neu oncogene. Science 1987; 235: 177–82. [DOI] [PubMed] [Google Scholar]
- 7. Slamon DJ, Godolphin W, Jones LA et al Studies of the HER‐2/neu proto‐oncogene in human breast and ovarian cancer. Science 1989; 244: 707–12. [DOI] [PubMed] [Google Scholar]
- 8. Joensuu H, Kellokumpu‐Lehtinen PL, Bono P et al Adjuvant docetaxel or vinorelbine with or without trastuzumab for breast cancer. N Engl J Med 2006; 354: 809–20. [DOI] [PubMed] [Google Scholar]
- 9. Romond EH, Perez EA, Bryant J et al Trastuzumab plus adjuvant chemotherapy for operable HER2‐positive breast cancer. N Engl J Med 2005; 353: 1673–84. [DOI] [PubMed] [Google Scholar]
- 10. Piccart‐Gebhart MJ, Procter M, Leyland‐Jones B et al Trastuzumab after adjuvant chemotherapy in HER2‐positive breast cancer. N Engl J Med 2005; 353: 1659–72. [DOI] [PubMed] [Google Scholar]
- 11. Slamon DJ, Leyland‐Jones B, Shak S et al Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2 . N Engl J Med 2001; 344: 783–92. [DOI] [PubMed] [Google Scholar]
- 12. Geyer CE, Forster J, Lindquist D et al Lapatinib plus capecitabine for HER2‐positive advanced breast cancer. N Engl J Med 2006; 355: 2733–43. [DOI] [PubMed] [Google Scholar]
- 13. Baselga J, Cortes J, Kim SB et al Pertuzumab plus trastuzumab plus docetaxel for metastatic breast cancer. N Engl J Med 2012; 366: 109–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee JW. Somatic mutations of ERBB2 kinase domain in gastric, colorectal, and breast carcinomas. Clin Cancer Res 2006; 12: 57–61. [DOI] [PubMed] [Google Scholar]
- 15. Ellis MJ, Ding L, Shen D et al Whole‐genome analysis informs breast cancer response to aromatase inhibition. Nature 2012; 486: 353–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shah SP, Morin RD, Khattra J et al Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature 2009; 461: 809–13. [DOI] [PubMed] [Google Scholar]
- 17. Kan Z, Jaiswal BS, Stinson J et al Diverse somatic mutation patterns and pathway alterations in human cancers. Nature 2010; 466: 869–73. [DOI] [PubMed] [Google Scholar]
- 18. Shah SP, Roth A, Goya R et al The clonal and mutational evolution spectrum of primary triple‐negative breast cancers. Nature 2012; 486: 395–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stephens PJ, Tarpey PS, Davies H et al The landscape of cancer genes and mutational processes in breast cancer. Nature 2012; 486: 400–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Banerji S, Cibulskis K, Rangel‐Escareno C et al Sequence analysis of mutations and translocations across breast cancer subtypes. Nature 2012; 486: 405–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Endo Y, Dong Y, Yoshimoto N et al HER2 mutation status in Japanese HER2‐negative breast cancer patients. Jpn J Clin Oncol 2014; 44: 619–23. [DOI] [PubMed] [Google Scholar]
- 22. Bose R, Kavuri SM, Searleman AC et al Activating HER2 mutations in HER2 gene amplification negative breast cancer. Cancer Discov 2012; 3: 224–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Serra V, Vivancos A, Puente XS et al Clinical response to a lapatinib‐based therapy for a Li‐Fraumeni syndrome patient with a novel HER2V659E mutation. Cancer Discov 2013; 3: 1238–44. [DOI] [PubMed] [Google Scholar]
- 24. Rexer BN, Ghosh R, Narasanna A et al Human breast cancer cells harboring a gatekeeper T798M mutation in HER2 overexpress EGFR ligands and are sensitive to dual inhibition of EGFR and HER2 . Clin Cancer Res 2013; 19: 5390–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kancha RK, von Bubnoff N, Bartosch N et al Differential sensitivity of ERBB2 kinase domain mutations towards lapatinib. PLoS ONE 2011; 6: e26760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Greulich H, Kaplan B, Mertins P et al Functional analysis of receptor tyrosine kinase mutations in lung cancer identifies oncogenic extracellular domain mutations of ERBB2. Proc Natl Acad Sci USA 2012; 109: 14476–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang SE, Narasanna A, Perez‐Torres M et al HER2 kinase domain mutation results in constitutive phosphorylation and activation of HER2 and EGFR and resistance to EGFR tyrosine kinase inhibitors. Cancer Cell 2006; 10: 25–38. [DOI] [PubMed] [Google Scholar]
- 28. Ross JS, Wang K, Gay LM et al A high frequency of activating extracellular domain ERBB2 (HER2) mutation in micropapillary urothelial carcinoma. Clin Cancer Res 2014; 20: 68–75. [DOI] [PubMed] [Google Scholar]
- 29. Awada A, Dirix L, Manso Sanchez L et al Safety and efficacy of neratinib (HKI‐272) plus vinorelbine in the treatment of patients with ErbB2‐positive metastatic breast cancer pretreated with anti‐HER2 therapy. Ann Oncol 2013; 24: 109–16. [DOI] [PubMed] [Google Scholar]
- 30. Burstein HJ, Sun Y, Dirix LY et al Neratinib, an irreversible ErbB receptor tyrosine kinase inhibitor, in patients with advanced ErbB2‐positive breast cancer. J Clin Oncol 2010; 28: 1301–7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. MCF‐7 (a) and HEK293T (b) cells were transfected with wild‐type HER2 or mutants.
Fig. S2. Kaplan–Meier curves for survival.
Table S1. HER2 somatic and germline mutations in the entire cohort of primary breast cancer
Table S2. Multivariate analyses of recurrence‐free survival (RFS) and distant recurrence‐free survival (DRFS) in HER2‐negative subgroup.


