Abstract
Gene therapy is a potentially effective treatment for retinal degenerative diseases. Clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) system has been developed as a new genome-editing tool in ophthalmic studies. Recent advances in researches showed that CRISPR/Cas9 has been applied in generating animal models as well as gene therapy in vivo of retinitis pigmentosa (RP) and leber congenital amaurosis (LCA). It has also been shown as a potential attempt for clinic by combining with other technologies such as adeno-associated virus (AAV) and induced pluripotent stem cells (iPSCs). In this review, we highlight the main points of further prospect of using CRISPR/Cas9 in targeting retinal degeneration. We also emphasize the potential applications of this technique in treating retinal degenerative diseases.
Keywords: CRISPR/Cas9, gene therapy, genome editing, retinal degeneration, retinitis pigmentosa, leber congenital amaurosis
INTRODUCTION
Retinal degeneration is a main cause of a large number of blinding diseases, such as retinitis pigmentosa (RP), age-related macular degeneration (AMD) and leber congenital amaurosis (LCA)[1]–[3]. Gene therapy has been regarded as a novel, potential and effective therapeutic method in treating those retinal degenerative diseases[4]–[8]. Recently, the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) system has been developed as a novel genome-editing tool in lots of medical aspects including ocular diseases[9]. Unlike other gene-editing tools such as transcription activator-like effector nucleases (TALENs) and zinc-finger nucleases (ZFNs), CRISPR/Cas9 system has multiplexed gene-editing ability as well as significantly higher efficiency[10]. Thus, CRISPR/Cas9 could be considered as a novel tool by ophthalmologists in targeting retinal degenerative diseases. Herein we present an overview of research advances of CRISPR/Cas9 system as a therapeutic tool for retinal degenerative diseases. In this review, we also emphasize some important points regarding the potential applications of CRISPR/Cas9 in retinal degeneration.
OVERVIEW OF CRISPR/CAS9 SYSTEM
Originally reported in 1987 as a set of short repeats located downstream of the iap gene in E. coli[11], CRISPR was then found both in bacteria and archaea as a immune system to protect them from plasmids, invading viruses, and other foreign nucleic acids[12]–[14]. And the endonuclease Cas9 can cleave DNA according to the sequence within an RNA duplex and create site-specific double-strand breaks (DSBs)[15]. These site-specific DSBs in the target DNA make CRISPR/Cas9 system a useful tool in genome editing with the ability to either cut or introduce a new gene to the genome. In the past decades, CRISPR/Cas9 has been increasingly recognized and applied as a powerful tool for regenerative medicine and gene/cell based therapy as well as animal model establishment[16]–[20]. With a breakthrough in 2015, CRISPR/Cas9 system has attracted tremendous attention from both scientists and clinicians and publications came out in different fields using this “magic” technology, especially with those diseases that remained untreatable for a long time such as AIDS, cancers and degenerative diseases. Scientists demonstrated that CRISPR/Cas9 technologies had the capability for a promising and sustained genetic therapy for HIV[21]–[22] and might contribute to HIV-1 therapeutic resistance[23]; CRISPR/Cas9 was thought to be an useful tool for cancer therapy by inactivating tumor mutations[24] or knocking out a specific gene to decrease the malignant potential of cancer cells[25]; CRISPR/Cas9 has also been applied in neurodegenerative diseases[26] and retinal degenerative disease[27] as a choice of possible gene therapy. All these researches made CRISPR/Cas9 a super star in the scientific world and brought this new technology to an unprecedented level.
MECHANISMS OF CRISPR/CAS9
According to an updated evolutionary classification of CRISPR/Cas system[28], CRISPR/Cas system can be divided into two groups, class I and class II, depending on whether the systems possess multisubunit CRISPR RNA (crRNA)-effector complex such as CRISPR-associated complex for antiviral defense (Cascade) complex or a single protein, such as Cas9. In this classification, class I includes type I, II and the putative new type IV CRISPR/Cas systems, which all present a multisubunit crRNA-effector complex. However, class II contains the type II CRISPR/Cas system as well as a putative new classification, type V, which dramatically differs from the other three by requiring only one protein for gene editing instead of a multisubunit crRNA-effector complex. And Cas9 is the signature gene of type II. Because of its simplicity, CRISPR/Cas9 has been studied and applied the most by scientists and clinicians. The mechanism of this system is also studied and reported the most by a great many scholars.
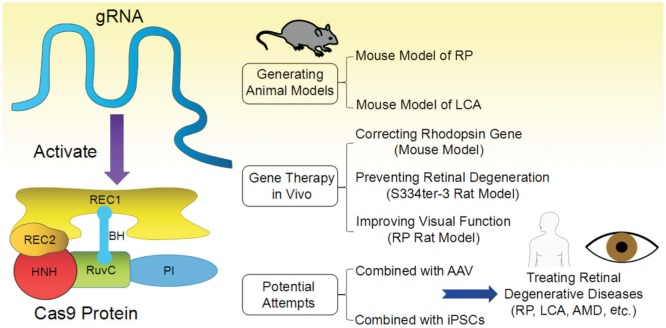
In CRISPR/Cas9 system, the Cas9 protein is responsible for locating and cleaving target DNA. The Cas9 protein has 6 domains (Figure 1), recognition (REC)1, REC2, Bridge Helix, protospacer adjacent motif (PAM) Interacting, HNH and RuvC (Nuclease domains)[29]–[30]. The Cas9 protein is activated after binding guide RNA (gRNA or sgRNA) by REC1 following a conformational change in the protein. Then, it searches for target DNA stochastically by binding with sequences that matches its PAM sequence [31] and immediately melts the bases of the PAM, paring them with the complementary region on the gRNA. By now, initiated by the PAM-interacting domain, target DNA binding is finished [29]. If the matching region and the target region are properly paired, the nuclease domains, RuvC and HNH, will cut the target DNA after the third nucleotide base upstream of the PAM. In the whole process, arginine-rich bridge helix is the key for initiating cleavage activity upon binding of target DNA[30].
Figure 1. Structure and applications of CRISPR/Cas9 in retinal degenerative diseases.
BH: Bridge helix; PI: PAM interacting. The structure of Cas9 protein of this figure is adapted from Cavanagh&Garrity, “CRISPR Mechanism”, CRISPR/Cas9, Tufts University, 2014. Available at https://sites.tufts.edu/crispr/; accessed on Dec. 23, 2016.
Gene Knockout
gRNA or sgRNA are designed to a specific genomic sequence. sgRNAs and Cas9 can be cloned into plasmids and then introduced into mammalian cells by transfection, directing Cas9 to knockout the gene[32]–[33]. For long-term expression which will result in stable knockout, Cas9 protein associated with sgRNAs can be pre-packed into lentiviral vectors, and then transduced into target cells[34]. Both the sgRNA and Cas9 are integrated stably into the genome of host cells, and have the ability to pass along to their daughter cells when the cells divide. This will provide permanent expression of shRNA and Cas9, generating long-term and stable knockout gene expression.
Gene Insertion/Knockin
CRISPR/Cas9-induced site-specific DNA DSBs can be repaired by homology-directed repair (HDR) or non-homologous end joining (NHEJ) pathways[35]. The mechanisms of these two pathways were studied by scientists and explained as follows: the HDR pathway repairs DNA damage accurately based on existing homologous DNA sequences mediated by a strand-exchange process[36], which we can intentionally replace the endogenous genome segments with plasmid sequences to insert targeted DNA into the genome and precisely induce genetic modification in living cells; the NHEJ pathway joins the broken ends through a homology-independent mechanistically flexible process to repair DNA DSBs, which often leads to random small insertions or deletions (indels)[37].
Gene Correction
After CRISPR/Cas9 located the specific target locus, deleting the mutated gene which considered to cause diseases, gene correction at the target locus can be achieved by the insertion of the corrected sequences via NHEJ-mediated insertions/deletions (indels) or HDR based on an exogenously supplied oligonucleotide[38]–[39].
Advantages of CRISPR/Cas9
As we presented, with the mechanism of CRISPR/Cas9, gene editing has become much easier than before. Most scientists and clinicians believe that the CRISPR/Cas9 system has created a new era for establishing animal models/generating cell lines and gene therapy. Compared with other tools for gene editing like ZFNs and TALENs, CRISPR/Cas9 has its own merits: a) target design simplicity: only a short guide RNA is needed[40]; b) high efficiency: when compared with other gene editing tools, it is more efficient[41]–[42]; c) multiplexed gene deletion or insertion. It can introduce in or knock out multiple genes at the same time by simply injecting them with multiple gRNAs[43]–[45].
APPLICATION OF CRISPR/CAS9 IN RETINAL DEGENERATION
With all these advantages we listed above, CRISPR/Cas9 technology has now gotten increasingly high attention and has been widely used in creating knock out animal models and cell lines[16]–[17],[46]–[47] to mimic diseases. At the same time, it has been broadly used for studying gene therapy for a great number of diseases[26],[48]–[49], including retinal diseases[50]. For decades, retinal degenerative diseases in ocular have been challenging lots of ophthalmologists and researchers. With the advent of this magic CRISPR/Cas9 system, many animal models in the eye can be created and studied and several diseases that could not be treated in the eye now might have a promising way to cure. In this review, we are going to present the advances of CRISPR/Cas9 technology applied in retinal degenerative diseases.
Research Progress in Generating Animal Models
RP is the most frequent form of inherited retinal degeneration that mainly caused by gene mutations and can gradually lead to irreversible blindness[51]. This disease mainly affects rod photoreceptors and after rods die, cone photoreceptors die secondarily. And it can be passed from parents to offspring through one of three genetic inheritance patterns: autosomal dominant (AD), autosomal recessive (AR) and X-linked (XL) recessive traits[52]. In order to treat this disease, scientist has created numbers of animal models such as rd mice, N-methyl-N-nitrosourea (MNU)-induced mice and zebrafish model for XL RP, etc. All these models are trying to mimic the degeneration of the photoreceptors. However, as we already know, RP is a genetic disease resulted by gene mutations such as RP2 mutation, retinitis pigmentosa GTPase regulator (RPGR) mutation[53], mutations in pre-mRNA processing factor 31 (PRPF31)[54], and c-mer proto-oncogene tyrosine kinase (MERTK) mutations[55] etc, the best way to generate animal models to represent this disease is gene editing. Thus, the appearance of CRISPR/Cas9 has provided a powerful tool to generate RP animal models for researchers and doctors to study.
Up to now, there are already some studies using this CRISPR/Cas9 technology to create animal models for RP. Arno et al[56] reported a mouse model using gene editing mediated by CRISPR/Cas9 technology. They produced knock-in mice with the p.Leu135Pro RP-associated variant identified in one RP-affected mouse. By mimicking the clinical phenotypes of RP, including progressive photoreceptor degeneration and dysfunction of the rod photoreceptors, these homozygous knock-in mice can provide an better animal model for scientist to study[56]. In another study of the causes of rd1 mice, they performed gene editing via the CRISPR/Cas9 system and illustrated that the Y347X mutation is the contributing variant of the disease[50].
LCA is another challenging congenital retinal dystrophy for ophthalmologists and scholars since patients with LCA usually end up with significant vision loss at an early age[57]. CRISPR/Cas9 was demonstrated to be a useful tool to generate a LCA mouse model to mimic human KCNJ13-related LCA disease[58].
Additionally, not only animal models of retinal degenerative diseases such as RP and LCA can be created by CRISPR/Cas9 technology, animal models for other ocular diseases using this technology also have being studied widely. By injecting multiplex CRISPR/Cas9 gRNAs, a highly penetrant and rapid retinoblastoma (Rb) animal model was generated the first time and it will be a good model facilitating rapid identification of targets that allow therapeutic intervention[59].
Gene Therapies
Gene therapy for ocular diseases is currently a hot stone worldwide. Scientists are working extremely hard to find good ways to treat inherited eye diseases. As CRSPR/Cas9 technology showed up with its ability to edit target genome specifically and efficiently as well as the capacity of targeting multiple genes at the same time, it soon has become a valuable and powerful tool for gene editing, which is perfect for gene therapeutic intentions in retinal degenerative diseases and gives ophthalmologists the hope for permanent treatment of ocular genetic diseases.
Latella et al[60] reported a successful in vivo editing of the human mutant Rhodopsin gene, which is a common cause of RP, by application of CRISPR/Cas9 system. Thus, the genome editing by CRISPR/Cas9 system might be a great method to generate genomic deletions and targeted frameshifts in the retina, which provided us a new therapeutic tool for treating retinal degenerative diseases such as RP.
The attempt of CRISPR/Cas9 system for treating retinal degeneration has also been done in 2015 in the respective S334ter-3 rat model[61]. A single subretinal injection of gRNA/Cas9 plasmid in combination with electroporation, and the generation of allele-specific disruption of the murine S334ter allele were achieved, resulting in retinal degeneration prevention and visual function improvement. In another study, Suzuki et al[27] devised a homology-independent targeted integration (HITI) strategy through CRISPR/Cas9 technology, demonstrating that the efficacy of this strategy improved visual function in rat model of RP, which proved the therapeutic potential of this technology.
As a kind of small non-pathogenic dependovirus, adeno-associated virus (AAV) has been recognized to show great potential for safe and long-term genetic pay-load in treating retinal diseases[62]. Da Costa et al[63] showed a novel method that combined vitreous aspiration and intravitreal injection of AAV2/8 could result in a widespread transduction of the retina (including photoreceptors and RPE cells), which also provided a new approach for CRISPR/Cas9 application in treating retinal diseases. Cereso et al[64] performed gene therapy mediated by AAV2/5 to induced pluripotent stem cells (iPSCs)-derived RPE cells from a choroideremia patient. As delivery of combined use of CRISPR/Cas9 and AAV has been reported successfully in mouse hepatocytes[65], Zheng et al[66] suggested to use AAV-CRISPR to deliver directly and locally for treating retinal diseases. CRISPR/Cas9 genome editing was also used for precise correction of a pathogenic RP mutation in patient-derived iPSCs[67]. Thus, CRISPR/Cas9 can also be considered as a tool to be combined with iPSCs for clinical applications in gene therapy. Further investigations are needed for combined use of these novel technologies for gene and cell therapy.
CONCLUSION
In this review, we summarized recent studies on CRISPR/Cas9 in retinal degenerative diseases, and we believe that this new technology can be used not only in basic research such as creating animal models of retinal degenerative diseases like RP, but also in clinical applications such as developing therapeutic methods for those diseases in the future. We also believe that it is also promising in gene therapy to perform the combined utility with other technologies such as AAVs and iPSCs. Thus, CRISPR/Cas9 should be a useful and powerful tool for genome editing in targeting retinal degenerative diseases.
Acknowledgments
Conflicts of Interest: Peng YQ, None; Tang LS, None; Yoshida S, None; Zhou YD, None.
REFERENCES
- 1.Sundaramurthy S, Swaminathan M, Sen P, Arokiasamy T, Deshpande S, John N, Gadkari RA, Mannan AU, Soumittra N. Homozygosity mapping guided next generation sequencing to identify the causative genetic variation in inherited retinal degenerative diseases. J Hum Genet. 2016;61(11):951–958. doi: 10.1038/jhg.2016.83. [DOI] [PubMed] [Google Scholar]
- 2.Jones BW, Pfeiffer RL, Ferrell WD, Watt CB, Marmor M, Marc RE. Retinal remodeling in human retinitis pigmentosa. Exp Eye Res. 2016;150:149–165. doi: 10.1016/j.exer.2016.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nowak JZ. AMD-the retinal disease with an unprecised etiopathogenesis: in search of effective therapeutics. Acta Pol Pharm. 2014;71(6):900–916. [PubMed] [Google Scholar]
- 4.Scholl HP, Strauss RW, Singh MS, Dalkara D, Roska B, Picaud S, Sahel JA. Emerging therapies for inherited retinal degeneration. Sci Transl Med. 2016;8(368):368rv6. doi: 10.1126/scitranslmed.aaf2838. [DOI] [PubMed] [Google Scholar]
- 5.Dalkara D, Goureau O, Marazova K, Sahel JA. Let There Be Light: Gene and Cell Therapy for Blindness. Hum Gene Ther. 2016;27(2):134–147. doi: 10.1089/hum.2015.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim J, Mirando AC, Popel AS, Green JJ. Gene delivery nanoparticles to modulate angiogenesis. Adv Drug Deliv Rev. 2016;pii doi: 10.1016/j.addr.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Veleri S, Lazar CH, Chang B, Sieving PA, Banin E, Swaroop A. Biology and therapy of inherited retinal degenerative disease: insights from mouse models. Dis Model Mech. 2015;8(2):109–129. doi: 10.1242/dmm.017913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rakoczy EP, Lai CM, Magno AL, Wikstrom ME, French MA, Piercce CM, Schwartz SD, Blumenkranz MS, Chalberg TW, Degli-Esposti MA, Constable IJ. Gene therapy with recombinant adeno-associated vectors for neovascular age-related macular degeneration: 1 year follow-up of a phase 1 randomised clinical trial. Lancet. 2015;386(10011):2395–2403. doi: 10.1016/S0140-6736(15)00345-1. [DOI] [PubMed] [Google Scholar]
- 9.Hung SS, McCaughey T, Swann O, Pébay A, Hewitt AW. Genome engineering in ophthalmology: application of CRISPR/Cas to the treatment of eye disease. Prog Retin Eye Res. 2016;53:1–20. doi: 10.1016/j.preteyeres.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 10.Pennisi E. The CRISPR craze. Science. 2013;341(6148):833–836. doi: 10.1126/science.341.6148.833. [DOI] [PubMed] [Google Scholar]
- 11.Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol. 1987;(12):5429–5433. doi: 10.1128/jb.169.12.5429-5433.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fineran PC, Charpentier E. Memory of viral infections by CRISPR-Cas adaptive immune systems: acquisition of new information. Virology. 2012;434(2):202–209. doi: 10.1016/j.virol.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 13.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315(5819):1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 14.Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327(5962):167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 15.Nafissi N, Foldvari M. Neuroprotective therapies in glaucoma: II. Genetic nanotechnology tools. Front Neurosci. 2015;9:355. doi: 10.3389/fnins.2015.00355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tu Z, Yang W, Yan S, Guo X, Li XJ. CRISPR/Cas9: a powerful genetic engineering tool for establishing large animal models of neurodegenerative diseases. Mol Neurodegener. 2015;10:35. doi: 10.1186/s13024-015-0031-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jinwei Z, Qipin X, Jing Y, Shumin Y, Suizhong C. CRISPR/Cas9 genome editing technique and its application in site-directed genome modification of animals. Yi Chuan. 2015;37(10):1011–1020. doi: 10.16288/j.yczz.15-066. [DOI] [PubMed] [Google Scholar]
- 18.Yao S, He Z, Chen C. CRISPR/Cas9-mediated genome editing of epigenetic factors for cancer therapy. Hum Gene Ther. 2015;26(7):463–471. doi: 10.1089/hum.2015.067. [DOI] [PubMed] [Google Scholar]
- 19.Lee PC, Truong B, Vega-Crespo A, Gilmore WB, Hermann K, Angarita SA, Tang JK, Chang KM, Wininger AE, Lam AK, Schoenberg BE, Cederbaum SD, Pyle AD, Byrne JA, Lipshutz GS. Restoring ureagenesis in hepatocytes by CRISPR/Cas9-mediated genomic addition to arginase-deficient induced pluripotent stem cells. Mol Ther Nucleic Acids. 2016;5(11):394. doi: 10.1038/mtna.2016.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang WC, Liang PP, Wong CW, Ng TB, Huang JJ, Zhang JF, Waye MM, Fu WM. CRISPR/Cas9 technology targeting fas gene protects mice from concanavalin-A induced fulminant hepatic failure. J Cell Biochem. 2017;118(3):530–536. doi: 10.1002/jcb.25722. [DOI] [PubMed] [Google Scholar]
- 21.Saayman S, Ali SA, Morris KV, Weinberg MS. The therapeutic application of CRISPR/Cas9 technologies for HIV. Expert Opin Biol Ther. 2015;15(6):819–830. doi: 10.1517/14712598.2015.1036736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaminski R, Chen Y, Fischer T, Tedaldi E, Napoli A, Zhang Y, Karn J, Hu W, Khalili K. Elimination of HIV-1 Genomes from human T-lymphoid cells by CRISPR/Cas9 gene editing. Sci Rep. 2016;6:22555. doi: 10.1038/srep22555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoder KE, Bundschuh R. Host double strand break repair generates HIV-1 strains resistant to CRISPR/Cas9. Sci Rep. 2016;6:29530. doi: 10.1038/srep29530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gebler C, Lohoff T, Paszkowski-Rogacz M, Mircetic J, Chakraborty D, Camgoz A, Hamann MV, Theis M, Thiede C, Buchholz F. Inactivation of cancer mutations utilizing CRISPR/Cas9. J Natl Cancer Inst. 2017;109(1):pii:djw183. doi: 10.1093/jnci/djw183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawamura N, Nimura K, Nagano H, Yamaguchi S, Nonomura N, Kaneda Y. CRISPR/Cas9-mediated gene knockout of NANOG and NANOGP8 decreases the malignant potential of prostate cancer cells. Oncotarget. 2015;6(26):22361–22374. doi: 10.18632/oncotarget.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang W, Tu Z, Sun Q, Li XJ. CRISPR/Cas9: implications for modeling and therapy of neurodegenerative diseases. Front Mol Neurosci. 2016;9:30. doi: 10.3389/fnmol.2016.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suzuki K, Tsunekawa Y, Hernandez-Benitez R, et al. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature. 2016;540(7631):144–149. doi: 10.1038/nature20565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Makarova KS, Wolf YI, Alkhnbashi OS, et al. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol. 2015;13(11):722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anders C, Niewoehner O, Duerst A, Jinek M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. 2014;513(7519):569–573. doi: 10.1038/nature13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, Ishitani R, Zhang F, Nureki O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156(5):935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507(7490):62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ni W, Qiao J, Hu S, Zhao X, Regouski M, Yang M, Polejaeva IA, Chen C. Efficient gene knockout in goats using CRISPR/Cas9 system. PLoS One. 2014;9(9):e106718. doi: 10.1371/journal.pone.0106718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang J, Song X, Guo C, Wang Y, Yin Y. Establishment of MAGEC2-knockout cells and functional investigation of MAGEC2 in tumor cells. Cancer Sci. 2016;107(12):1888–1897. doi: 10.1111/cas.13082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choi JG, Dang Y, Abraham S, Ma H, Zhang J, Guo H, Cai Y, Mikkelsen JG, Wu H, Shankar P, Manjunath N. Lentivirus pre-packed with Cas9 protein for safer gene editing. Gene Ther. 2016;23(7):627–633. doi: 10.1038/gt.2016.27. [DOI] [PubMed] [Google Scholar]
- 35.He X, Tan C, Wang F, Wang Y, Zhou R, Cui D, You W, Zhao H, Ren J, Feng B. Knock-in of large reporter genes in human cells via CRISPR/Cas9-induced homology-dependent and independent DNA repair. Nucleic Acids Res. 2016;44(9):e85. doi: 10.1093/nar/gkw064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heyer WD, Ehmsen KT, Liu J. Regulation of homologous recombination in eukaryotes. Annu Rev Genet. 2010;44:113–139. doi: 10.1146/annurev-genet-051710-150955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu Y, Zhou H, Fan X, Zhang Y, Zhang M, Wang Y, Xie Z, Bai M, Yin Q, Liang D, Tang W, Liao J, Zhou C, Liu W, Zhu P, Guo H, Pan H, Wu C, Shi H, Wu L, Tang F, Li J. Correction of a genetic disease by CRISPR-Cas9-mediated gene editing in mouse spermatogonial stem cells. Cell Res. 2015;25(1):67–79. doi: 10.1038/cr.2014.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Finotti A, Borgatti M, Gambari R. Ground state naive pluripotent stem cells and CRISPR/Cas9 gene correction for beta-thalassemia. Stem Cell Investig. 2016;3:66. doi: 10.21037/sci.2016.09.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lim Y, Bak SY, Sung K, Jeong E, Lee SH, Kim JS, Bae S, Kim SK. Structural roles of guide RNAs in the nuclease activity of Cas9 endonuclease. Nat Commun. 2016;7:13350. doi: 10.1038/ncomms13350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Auer TO, Duroure K, De Cian A, Concordet JP, Del Bene F. Highly efficient CRISPR/Cas9-mediated knock-in in zebrafish by homology-independent DNA repair. Genome Res. 2014;24(1):142–153. doi: 10.1101/gr.161638.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Petersen B, Frenzel A, Lucas-Hahn A, Herrmann D, Hassel P, Klein S, Ziegler M, Hadeler KG, Niemann H. Efficient production of biallelic GGTA1 knockout pigs by cytoplasmic microinjection of CRISPR/Cas9 into zygotes. Xenotransplantation. 2016;23(5):338–346. doi: 10.1111/xen.12258. [DOI] [PubMed] [Google Scholar]
- 43.Minkenberg B, Xie K, Yang Y. Discovery of rice essential genes by characterizing CRISPR-edited mutation of closely related rice MAP kinase genes. Plant J. 2017;89(3):636–648. doi: 10.1111/tpj.13399. [DOI] [PubMed] [Google Scholar]
- 44.Lopez-Obando M, Hoffmann B, Géry C, Guyon-Debast A, Téoulé E, Rameau C, Bonhomme S, Nogué F. Simple and efficient targeting of multiple genes through CRISPR-Cas9 in physcomitrella patens. G3 (Bethesda) 2016;pii:g3.116.033266. doi: 10.1534/g3.116.033266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zetsche B, Heidenreich M, Mohanraju P, Fedorova I, Kneppers J, DeGennaro EM, Winblad N, Choudhury SR, Abudayyeh OO, Gootenberg JS, Wu WY, Scott DA, Severinov K, van der Oost J, Zhang F. Multiplex gene editing by CRISPR-Cpf1 using a single crRNA array. Nat Biotechnol. 2017;35(1):31–34. doi: 10.1038/nbt.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kang JT, Ryu J, Cho B, Lee EJ, Yun YJ, Ahn S, Lee J, Ji DY, Lee K, Park KW. Generation of RUNX3 knockout pigs using CRISPR/Cas9-mediated gene targeting. Reprod Domest Anim. 2016;51(6):970–978. doi: 10.1111/rda.12775. [DOI] [PubMed] [Google Scholar]
- 47.Roy A, Goodman JH, Begum G, Donnelly BF, Pittman G, Weinman EJ, Sun D, Subramanya AR. Generation of WNK1 knockout cell lines by CRISPR/Cas-mediated genome editing. Am J Physiol Renal Physiol. 2015;308(4):366–376. doi: 10.1152/ajprenal.00612.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Janssen MJ, Arcolino FO, Schoor P, Kok RJ, Mastrobattista E. Gene based therapies for kidney regeneration. Eur J Pharmacol. 2016;790:99–108. doi: 10.1016/j.ejphar.2016.07.037. [DOI] [PubMed] [Google Scholar]
- 49.Savić N, Schwank G. Advances in therapeutic CRISPR/Cas9 genome editing. Transl Res. 2016;168:15–21. doi: 10.1016/j.trsl.2015.09.008. [DOI] [PubMed] [Google Scholar]
- 50.Wu WH, Tsai YT, Justus S, Lee TT, Zhang L, Lin CS, Bassuk AG, Mahajan VB, Tsang SH. CRISPR repair reveals causative mutation in a preclinical model of retinitis pigmentosa. Mol Ther. 2016;24(8):1388–1394. doi: 10.1038/mt.2016.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsubura A, Yoshizawa K, Kuwata M, Uehara N. Animal models for retinitis pigmentosa induced by MNU; disease progression, mechanisms and therapeutic trials. Histol Histopathol. 2010;25(7):933–944. doi: 10.14670/HH-25.933. [DOI] [PubMed] [Google Scholar]
- 52.Rivas MA, Vecino E. Animal models and different therapies for treatment of retinitis pigmentosa. Histol Histopathol. 2009;24(10):1295–1322. doi: 10.14670/HH-24.1295. [DOI] [PubMed] [Google Scholar]
- 53.Lyraki R, Megaw R, Hurd T. Disease mechanisms of X-linked retinitis pigmentosa due to RP2 and RPGR mutations. Biochem Soc Trans. 2016;44(5):1235–1244. doi: 10.1042/BST20160148. [DOI] [PubMed] [Google Scholar]
- 54.Hafler BP, Comander J, Weigel DiFranco C, Place EM, Pierce EA. Course of ocular function in PRPF31 retinitis pigmentosa. Semin Ophthalmol. 2016;31(1-2):49–52. doi: 10.3109/08820538.2015.1114856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parinot C, Nandrot EF. A comprehensive review of mutations in the MERTK proto-oncogene. Adv Exp Med Biol. 2016;854:259–265. doi: 10.1007/978-3-319-17121-0_35. [DOI] [PubMed] [Google Scholar]
- 56.Arno G, Agrawal SA, Eblimit A, et al. Mutations in REEP6 cause autosomal-recessive retinitis pigmentosa. Am J Hum Genet. 2016;99(6):1305–1315. doi: 10.1016/j.ajhg.2016.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hufnagel RB, Ahmed ZM, Corrêa ZM, Sisk RA. Gene therapy for Leber congenital amaurosis: advances and future directions. Graefes Arch Clin Exp Ophthalmol. 2012;250(8):1117–1128. doi: 10.1007/s00417-012-2028-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhong H, Chen Y, Li Y, Chen R, Mardon G. CRISPR-engineered mosaicism rapidly reveals that loss of Kcnj13 function in mice mimics human disease phenotypes. Sci Rep. 2015;5:8366. doi: 10.1038/srep08366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Naert T, Colpaert R, Van Nieuwenhuysen T, Dimitrakopoulou D, Leoen J, Haustraete J, Boel A, Steyaert W, Lepez T, Deforce D, Willaert A, Creytens D, Vleminckx K. CRISPR/Cas9 mediated knockout of rb1 and rbl1 leads to rapid and penetrant retinoblastoma development in Xenopus tropicalis. Sci Rep. 2016;6:35264. doi: 10.1038/srep35264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Latella MC, Di Salvo MT, Cocchiarella F, Benati D, Grisendi G, Comitato A, Marigo V, Recchia A. In vivo editing of the human mutant rhodopsin gene by electroporation of plasmid-based CRISPR/Cas9 in the mouse retina. Mol Ther Nucleic Acids. 2016;5(11):e389. doi: 10.1038/mtna.2016.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bakondi B, Lv W, Lu B, Jones MK, Tsai Y, Kim KJ, Levy R, Akhtar AA, Breunig JJ, Svendsen CN, Wang S. In vivo CRISPR/Cas9 gene editing corrects retinal dystrophy in the S334ter-3 rat model of autosomal dominant retinitis pigmentosa. Mol Ther. 2016;24(3):556–563. doi: 10.1038/mt.2015.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Day TP, Byrne LC, Schaffer DV, Flannery JG. Advances in AAV vector development for gene therapy in the retina. Adv Exp Med Biol. 2014;801:687–693. doi: 10.1007/978-1-4614-3209-8_86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Da Costa R, Röger C, Segelken J, Barben M, Grimm C, Neidhardt J. A novel method combining vitreous aspiration and intravitreal AAV2/8 injection results in retina-wide transduction in adult mice. Invest Ophthalmol Vis Sci. 2016;57(13):5326–5334. doi: 10.1167/iovs.16-19701. [DOI] [PubMed] [Google Scholar]
- 64.Cereso N, Pequignot MO, Robert L, Becker F, De Luca V, Nabholz N, Rigau V, De Vos J, Hamel CP6, Kalatzis V. Proof of concept for AAV2/5-mediated gene therapy in iPSC-derived retinal pigment epithelium of a choroideremia patient. Mol Ther Methods Clin Dev. 2014;1:14011. doi: 10.1038/mtm.2014.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Senís E, Fatouros C, Große S, Wiedtke E, Niopek D, Mueller AK, Börner K, Grimm D. CRISPR/Cas9-mediated genome engineering: an adeno-associated viral (AAV) vector toolbox. Biotechnol J. 2014;9(11):1402–1412. doi: 10.1002/biot.201400046. [DOI] [PubMed] [Google Scholar]
- 66.Zheng A, Li Y, Tsang SH. Personalized therapeutic strategies for patients with retinitis pigmentosa. Expert Opin Biol Ther. 2015;15(3):391–402. doi: 10.1517/14712598.2015.1006192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bassuk AG, Zheng A, Li Y, Tsang SH, Mahajan VB. Precision medicine: genetic repair of retinitis pigmentosa in patient-derived stem cells. Sci Rep. 2016;6:19969. doi: 10.1038/srep19969. [DOI] [PMC free article] [PubMed] [Google Scholar]