Abstract
Background
TNM staging alone does not accurately predict outcome in colon cancer (CC) patients who may be eligible for adjuvant chemotherapy. It is unknown to what extent the molecular markers microsatellite instability (MSI) and mutations in BRAF or KRAS improve prognostic estimation in multivariable models that include detailed clinicopathological annotation.
Patients and methods
After imputation of missing at random data, a subset of patients accrued in phase 3 trials with adjuvant chemotherapy (n = 3016)—N0147 (NCT00079274) and PETACC3 (NCT00026273)—was aggregated to construct multivariable Cox models for 5-year overall survival that were subsequently validated internally in the remaining clinical trial samples (n = 1499), and also externally in different population cohorts of chemotherapy-treated (n = 949) or -untreated (n = 1080) CC patients, and an additional series without treatment annotation (n = 782).
Results
TNM staging, MSI and BRAFV600E mutation status remained independent prognostic factors in multivariable models across clinical trials cohorts and observational studies. Concordance indices increased from 0.61–0.68 in the TNM alone model to 0.63–0.71 in models with added molecular markers, 0.65–0.73 with clinicopathological features and 0.66–0.74 with all covariates. In validation cohorts with complete annotation, the integrated time-dependent AUC rose from 0.64 for the TNM alone model to 0.67 for models that included clinicopathological features, with or without molecular markers. In patient cohorts that received adjuvant chemotherapy, the relative proportion of variance explained (R2) by TNM, clinicopathological features and molecular markers was on an average 65%, 25% and 10%, respectively.
Conclusions
Incorporation of MSI, BRAFV600E and KRAS mutation status to overall survival models with TNM staging improves the ability to precisely prognosticate in stage II and III CC patients, but only modestly increases prediction accuracy in multivariable models that include clinicopathological features, particularly in chemotherapy-treated patients.
Keywords: colon cancer, BRAF mutation, KRAS mutation, microsatellite instability, prognosis
Introduction
Colon cancer (CC) is a leading cause of cancer mortality worldwide [1, 2]. Initial patient management is defined by TNM stage at diagnosis, based on depth of tumor wall invasion, lymph node involvement and distant metastasis. This simple staging system, now at its seventh edition, is widely used, clinically useful and is highly associated with 5-year overall survival (OS), ranging from 92% in stage I to 11% in stage IV [3]. However, in patients with stage II and III CC, TNM staging less clearly distinguishes groups of patients with different prognosis, particularly in those who receive adjuvant chemotherapy, with 5-year OS between 50% and 90% [3]. Additional patient or disease characteristics known to affect survival in CC, including age, sex, primary site location, tumor grade, number of positive lymph nodes (LNs), number of examined LNs, lymphovascular and perineural invasion, bowel obstruction or perforation, and adjuvant treatment (fluoropyrimidine single agent or in combination with oxaliplatin) [4–6], are currently not directly included in American Joint Commission on Cancer (AJCC)-based risk determinations.
The AJCC has increasingly recognized the growing need for more accurate and probabilistically based outcome prediction for precision medicine by incorporating additional anatomic and nonanatomic prognostic factors. This could be achieved through the use of accurate risk models and web-based calculators. The AJCC Precision Medicine Core has recently established inclusion and exclusion criteria necessary for a risk model to potentially be endorsed by the AJCC, with the emphasis centered on performance metrics, implementation clarity, and clinical relevance [7]. Recommended performance measures in survival analysis include concordance index (c-index) and time-dependent area under the curve (tAUC). The c-index represents the probability that a model will correctly predict which of two randomly selected patients will experience an event before the other. The tAUC involves computing sensitivity and specificity at multiple time points, which provides more comprehensive information about the model predictive power than the c-index. The integrated (iAUC) is a weighted time-averaged summary of tAUC.
Previously, to estimate the value of clinicopathological covariates in prognosis prediction in stage III CC, data from close to 16 000 patients accrued to phase III clinical trials were aggregated (ACCENT database) and used to construct multivariable Cox models for OS [4]. Authors have shown that both patient and disease characteristics provide clinically significant information for OS prediction and developed calculators that better discriminate patient risk than does TNM staging alone [8]. Of note, the c-index rose from 0.58 for the TNM system model to 0.66 for the model that included TNM and detailed clinicopathological annotation [4]. These numbers confirm that there is room for improvement of prognosis prediction in intermediate-risk CC. Indeed, biomarker analysis of multiple studies strongly supports the feasibility of refining risk stratification by factoring in molecular characteristics with tumor staging [9]. In stage II disease, e.g. microsatellite instability (MSI) defines a population with low recurrence rates and very good outcome without adjuvant treatment [10, 11]. BRAFV600E or KRAS mutations confer worse survival in patients with stage II and III disease, and their value as additional risk factors is greater when MSI status and primary tumor location are taken into account [12–14].
In this article, we assess whether the addition of cancer cell molecular markers (MSI, BRAFV600E mutations and KRAS mutations) can improve prediction of OS over traditional clinicopathological covariates in patients with nonmetastatic CC. To meet the high levels of evidence required by the AJCC for potential endorsement, the project involved multiple institutions capable of contributing expertise and large cohorts, both clinical trial data and observational studies. Here we present the results of this effort, first with a description of how molecular markers of interest associate with selected clinicopathological features, then with a detailed performance analysis of models containing TNM staging, clinicopathological and/or molecular markers in multiple validation cohorts.
Methods—patients
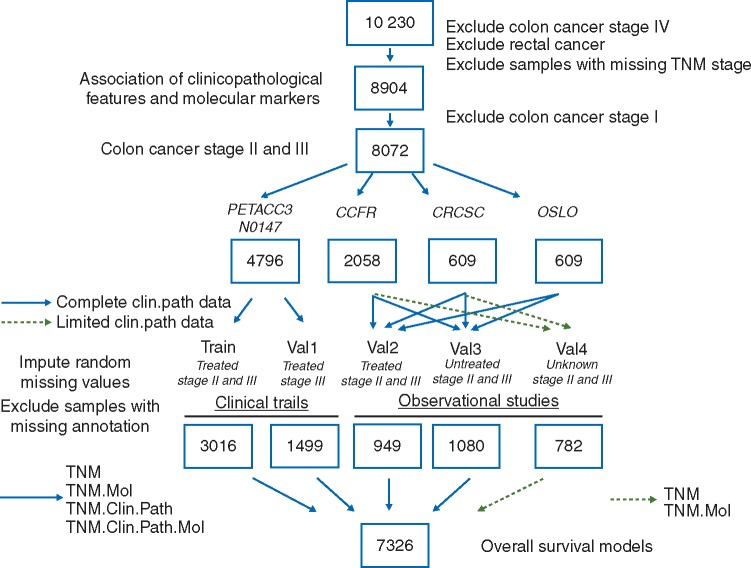
Our study population included 8904 patients diagnosed with stage I–III CC, treated or untreated with adjuvant chemotherapy, with clinicopathological and molecular annotation for variables of interest. Patients diagnosed with rectal cancer or stage IV CC were not eligible, and those with stage I CC (n = 832) were included only in the descriptive correlative analysis and excluded from survival models (Figure 1). The endpoint of interest was 5-year OS, measured from time of cancer diagnosis until death from any cause, censored at last follow-up or at 5-year post-diagnosis. The covariates considered for inclusion in the models were as follows:
Figure 1.
Study workflow, with schematic representation of population used for initial correlative analysis, followed by data splits in training and validation cohorts, data imputation, and survival models.
TNM stage AJCC version 7: pathological tumor stage (pT-stage; pT1, pT2, pT3, pT4) and pathological nodal stage (pN-stage: pN0, pN1, pN2);
Clinicopathological features: age (continuous), sex, primary site location (right [caecum to transverse colon] versus left [splenic flexure to sigmoid]), number of positive LNs (continuous), number of examined LNs (continuous), tumor grade (low or medium versus high), type of adjuvant chemotherapy type (oral/infusional/bolus 5FU variations versus 5FU and oxaliplatin, 5FU and oxaliplatin plus cetuximab, 5FU and irinotecan, 5FU and irinotecan plus cetuximab).
Molecular markers: MSI status (MSI-high versus microsatellite stable [MSS] or MSI-low), mutations in BRAFV600E or KRAS codons 12/13.
Datasets splits, summarized in Figure 1 and Table 1, were as follows:
Table 1.
Demographics and disease characteristics of patients included in overall survival models postimputation
| Train | Val1 | Val2 | Val3 | Val4 | P value | |
|---|---|---|---|---|---|---|
| Dataset | ||||||
| N0147 | 2318 | 943 | 0 | 0 | 0 | |
| PETACC-3 | 698 | 556 | 0 | 0 | 0 | |
| CRCSC | 0 | 0 | 368 | 189 | 52 | |
| CCFR | 0 | 0 | 473 | 401 | 730 | |
| Oslo | 0 | 0 | 108 | 490 | 0 | |
| Total | 3016 | 1499 | 949 | 1080 | 782 | |
| TNM staging | <0.001 | |||||
| IIA | 318 (10%) | 0 | 336 (35%) | 756 (70%) | 320 (40%) | |
| IIB/IIC | 77 (3%) | 0 | 61 (7%) | 78 (7%) | 22 (3%) | |
| IIIA | 292 (10%) | 170 (11%) | 50 (5%) | 11 (1%) | 59 (8%) | |
| IIIB | 1795 (59%) | 1034 (69%) | 359 (38%) | 147 (14%) | 342 (44%) | |
| IIIC | 534 (18%) | 295 (20%) | 143 (15%) | 88 (8%) | 39 (5%) | |
| Clinicopathological | ||||||
| Age in years | <0.001 | |||||
| Median (range) | 59 (19–85) | 59 (21–86) | 62 (22–96) | 70 (24–94) | NA | |
| Sex | 0.004 | |||||
| Female | 1374 (46%) | 694 (46%) | 465 (46%) | 551 (51%) | NA | |
| Male | 1642 (54%) | 805 (54%) | 484 (54%) | 529 (49%) | NA | |
| Lymph nodes assessed | <0.001 | |||||
| Median (IQR) | 17 (12–23) | 16 (11–22) | 11 (6–19) | 10 (6–21) | NA | |
| Tumor grade | <0.001 | |||||
| Low/medium | 2382 (79%) | 1212 (81%) | 811 (84%) | 981 (91%) | NA | |
| High | 634 (21%) | 287 (19%) | 148 (16%) | 99 (9%) | NA | |
| Primary tumor site | <0.001 | |||||
| Right colon | 1464 (49%) | 688 (46%) | 482 (51%) | 647 (60%) | NA | |
| Left colon | 1552 (51%) | 811 (54%) | 467 (49%) | 433 (40%) | NA | |
| Adjuvant chemotherapy | <0.001 | |||||
| None | 0 | 0 | 0 | 1080 (100%) | NA | |
| 5FU/capecitabine | 359 (12%) | 274 (18%) | 852 (90%) | 0 | NA | |
| FOLFIRIa | 415 (14%) | 313 (21%) | 4 (1%) | 0 | NA | |
| FOLFIRIa/cetuximab | 32 (1%) | 9 (1%) | 0 | 0 | NA | |
| FOLFOXb | 1228 (41%) | 550 (37%) | 93 (9%) | 0 | NA | |
| FOLFOXb/cetuximab | 982 (32%) | 353 (23%) | 0 | 0 | NA | |
| Molecular | <0.001 | |||||
| MSI status | ||||||
| MSI-high | 383 (13%) | 183 (12%) | 182 (19%) | 276 (26%) | 135 (17%) | |
| MSS/MSI-low | 2633 (87%) | 1316 (88%) | 767 (81%) | 804 (74%) | 647 (83%) | |
| BRAF V600E status | <0.001 | |||||
| Mutated | 345 (11%) | 162 (11%) | 124 (13%) | 205 (19%) | 102 (13%) | |
| Wild-type | 2671 (89%) | 1337 (89%) | 825 (87%) | 875 (81%) | 680 (87%) | |
| KRAS codons 12/13 status | <0.001 | |||||
| Mutated | 1078 (36%) | 566 (38%) | 349 (37%) | 331 (31%) | 239 (31%) | |
| Wild-type | 1938 (64%) | 933 (62%) | 600 (63%) | 749 (69%) | 543 (69%) | |
Infusional 5FU with irinotecan.
Infusional 5FU with oxaliplatin.
NA, not applicable.
-
Training (train) and internal validation (val1): Clinical trial cohorts
N0147 (NCT00079274) (n=3392) [13]: Treated with adjuvant infusional 5FU and oxaliplatin (FOLFOX) or irinotecan (FOLFIRI) with or without cetuximab in stage III CC patients;
PETACC-3 (NCT00026273) (n=1404) [15]: Treated with adjuvant infusional 5FU with or without irinotecan (FOLFIRI) in high-risk stage II or stage III CC patients.
-
External validation cohorts (val2-val4): Observational studies
CCFR (Colon Cancer Family Registry; n=2058) [16]: Four datasets of stage II or III CC patients (treated, untreated, unknown adjuvant therapy status);
CRCSC (Colorectal Cancer Subtyping Consortium; n=609) [17]: Four datasets of stage II or III CC patients (treated, untreated, unknown adjuvant therapy status);
OSLO (Oslo University Hospital; n=609) [18]: Consecutive, population-based series of stage II or III CC (treated or untreated).
All patients with stage II tumors from clinical trial cohorts (PETACC-3 only) were allocated to the train data, and those with stage III tumors were randomly distributed in train (2/3 of samples) and val1 (1/3 of samples). Data split in the external validation cohorts was based on availability of complete clinicopathological annotation and adjuvant therapy: val2 (chemotherapy-treated with complete annotation); val3 (untreated with complete annotation); val4 (unknown adjuvant therapy status or missing clinicopathological annotation).
Methods—statistical considerations
In total, 2821 samples had missing values for one or more covariates. Patterns of co-missingness among multiple covariates were generally attributable to the specific data collection patterns of individual studies. As a result, missing data was not missing at random (MAR), resulting in a subset of samples (n = 1910) with missing parameters that could not be imputed. For the remaining samples (n = 991), multiple imputation was carried out via the “mice” R package [19], separately for each cohort, with all variables except adjuvant therapy. Samples from validation cohorts that could not be imputed were aggregated in val4. A subset of these samples (n = 782) had annotation on TNM staging plus molecular markers and was used to validate models that did not include clinicopathological features or had unknown adjuvant treatment information.
Following imputation, multivariable Cox proportional hazards models were formulated using factors that demonstrated statistical significance for OS in univariate models (with P< 0.10 according to a log-rank test). Age was modeled using restricted cubic splines. The proportional hazards assumption was verified (P=0.22 across different cohorts; P>0.05 across different datasets within each cohort) and all Cox models were stratified by dataset.
We constructed models with TNM staging alone (TNM), TNM plus molecular markers (TNM.Mol), TNM plus clinicopathological features (TNM.Clin.Path) and all covariates (TNM.Clin.Path.Mol). Final models were validated both internally (train and val1) and externally (val2–val4) to obtain optimism-corrected discrimination for survival data via c-indices, tAUCs and iAUC (1–5 years) using “timeROC” R package [20]. Stage-proportional 5-fold data splitting was performed for internal validation in the train set. We selected a Bayesian method for model comparison—in validation cohorts with complete annotation on markers of interest (val1–val3), the corresponding iAUCs were cross-compared using Bayes factor estimation [21] and bootstrapping procedure. We calculated the relative proportion of explained variance (Cox & Snell pseudo R2) in OS that is accounted for by different categories of predictor covariates using “survival” R package. Calibration plots of predicted probability versus observed proportion were generated using the “rms” R package. All analyses were performed using R statistical software version 3.2.5 [22].
Results
Association among molecular markers and clinocopathological features
We explored the association of molecular markers and TNM stage and primary site location in the complete patient cohort of stage I, II and III CC (supplementary Figure S1, available at Annals of Oncology online). There was a stepwise decrease in prevalence of KRAS or BRAFV600E mutations (mutually exclusive in our cohort) moving from the right to left colon (supplementary Figure S1A, available at Annals of Oncology online). KRAS or BRAFV600E mutations were ∼2 times more likely to be found in the caecum as compared to sigmoid colon (supplementary Figure S1A, available at Annals of Oncology online). The prevalence of BRAFV600E mutations and MSI-high status paralleled each other and exhibited a bell-shaped distribution, with an increase from the caecum to hepatic flexure, then gradual decrease through sigmoid colon (supplementary Figure S1A, available at Annals of Oncology online). MSI-high status was ∼4 times more prevalent in tumors of the right colon as compared to the left, and 2 times higher in stage II versus stage III tumors (supplementary Figure S1B, available at Annals of Oncology online). KRAS and BRAFV600E mutations were equally distributed across stages (supplementary Figure S1C and D, available at Annals of Oncology online). KRAS mutations were 2 times more frequent in MSS/MSI-low compared to MSI-high tumors (supplementary Figure S1C, available at Annals of Oncology online). On the contrary, BRAFV600E mutations were 8 times more prevalent in MSI-high as compared to MSS/MSI-low tumors (supplementary Figure S1D, available at Annals of Oncology online). Finally, we observed a higher proportion of high-grade tumors with increasing pT (pT1–pT4) and pN (pN0–pN2) stage, with ∼2 times higher prevalence of poorly differentiated carcinomas in MSI-high tumors as compared to MSS/MSI-low (supplementary Figure S1E, available at Annals of Oncology online).
Prognostic associations
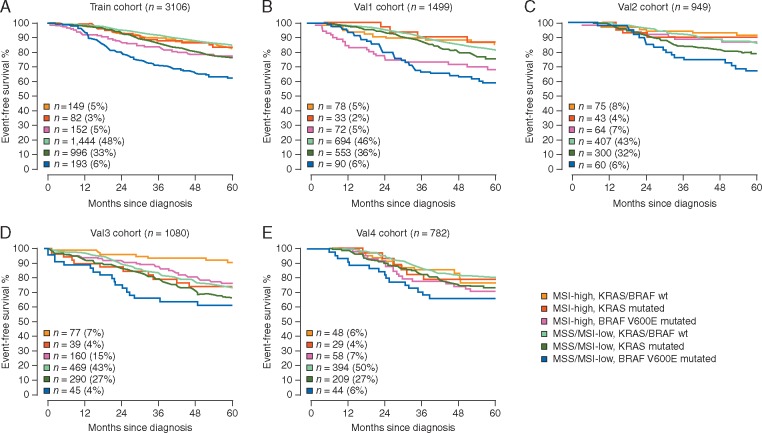
Demographics, tumor-related characteristics and molecular markers of the different cohorts of patients with stage II or III CC included in survival models are described in Table 1. As a result of our data split approach, the training and external validation cohorts are heterogeneous with respect to adjuvant chemotherapy treatment and additional covariates. In univariate Cox models, all clinicopathological features and molecular markers demonstrated a statistically significant association with OS in both the training and all validation cohorts (data not presented). Figure 2 illustrates Kaplan–Meier OS curves in clinical trial cohorts and observational studies, with stratification by molecular subgroup. Overall, hazard ratios for the association of BRAFV600E or KRAS mutations with worse OS were repeatedly significant in MSS/MSI-low tumors (supplementary Table S1, available at Annals of Oncology online). Patients whose tumors were MSS/MSI-low and BRAFV600E mutated had the highest risk of death across all cohorts. Five-year OS rates in stage III CC patients treated with oxaliplatin-based adjuvant chemotherapy (train) ranged from 62% (95% CI 56–70%) if the tumors were MSS/MSI-low BRAFV600E mutated to 83% (95% CI 77–90%) if the tumors were MSI-high KRAS/BRAFV600E wild-type. The corresponding numbers in stage II untreated CC patients (val3) were 66% (95% CI 52–85%) and 90% (95% CI 80–99%), respectively.
Figure 2.
Overall survival Kaplan–Meier estimates across clinical trial cohorts of chemotherapy-treated patients (A and B), and multiple validation cohorts of chemotherapy-treated (C), -untreated (D) or unknown adjuvant therapy status (E). Univariate Cox models are detailed in supplementary Table S1, available at Annals of Oncology online.
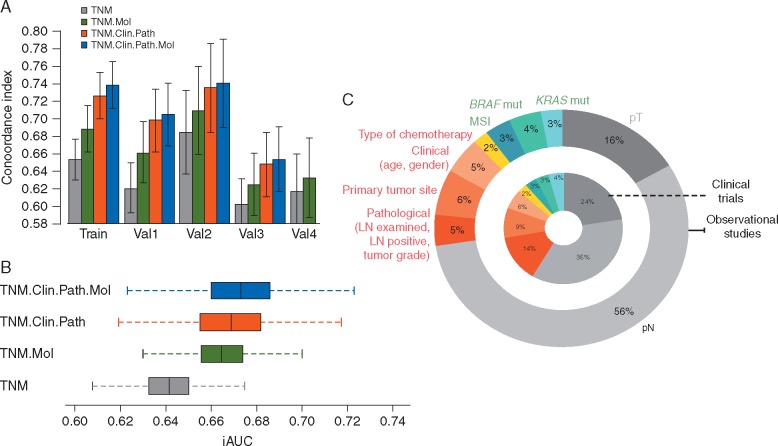
In multivariable analyses, detailed in Table 2, all examined factors were statistically significant for OS prediction in the train and val1 cohorts (clinical trial data). In val2 and val3 cohorts (complete clinicopathological annotation), MSI status and BRAFV600E mutations remained independent prognostic factors, together with pT and pN staging. We found no statistically significant interaction between molecular markers and clinicopathological features with impact on patient outcomes. The TNM model for OS demonstrated a c-index of 0.66 and iAUC of 0.68 in the training set, while the full TNM.Clin.Path.Mol model had c-index of 0.73 and iAUC of 0.76. The calibration of observed versus predicted 5-year OS rates in the training cohort was strong across the spectrum of ordered risk groups (supplementary Figure S2, available at Annals of Oncology online). Model’s c-indices in the training and validation cohorts are shown in Figure 3A. Across different validation cohorts, c-indices increased from 0.61–0.68 in the TNM alone model to 0.63–0.71 in models with added molecular markers, 0.65–0.73 with clinicopathological features and 0.66–0.74 with all covariates (supplementary Table S2, available at Annals of Oncology online). The iAUC increased from 0.644 for the TNM system model to 0.667 for the TNM.Mol model, 0.671 for the TNM.Clin.Path model and 0.675 for the TNM.Clin.Path.Mol model. Inspection of K-values from Bayes factor estimation in different validation cohorts with complete annotation (supplementary Table S3, available at Annals of Oncology online) and corresponding bootstrapped iAUCs (Figure 3B and supplementary Table S4, available at Annals of Oncology online) confirmed that there is strong evidence that all models with clinicopathological features and/or molecular markers provide significantly improved OS prediction when compared to TNM-only models. On the other hand, when comparing the TNM.Clin.Path.Mol model with gold standard TNM.Clin.Path model in patient cohorts patients treated with adjuvant chemotherapy (val1 and val2), we did not find evidence for superiority, with K-values less than 2 (supplementary Table S4, available at Annals of Oncology online). Detailed tAUCs of models are shown in supplementary Figure S3, available at Annals of Oncology online.
Table 2.
Final multivariable overall survival Cox models across clinical trial cohorts and different observational studies
| Cohorts with adjuvant chemotherapy exposure | Train (n=3016; 550 events) |
Val1 (n=1499; 307 events) |
Val2 (n = 949; 151 events) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | P value | HR | 95% CI | P value | HR | 95% CI | P value | ||||
| pT3 versus pT2/pT1 | 2.43 | 1.59 | 3.73 | <0.001 | 1.67 | 1.05 | 2.66 | <0.001 | 1.45 | 0.66 | 3.17 | 0.35 |
| pT4 versus pT2/pT1 | 4.83 | 3.08 | 7.59 | <0.001 | 2.81 | 1.69 | 4.66 | <0.001 | 3.39 | 1.47 | 7.84 | 0.004 |
| pN1 versus pN0 | 1.79 | 1.20 | 2.66 | 0.004 | NA | NA | NA | NA | 2.69 | 1.66 | 4.37 | <0.001 |
| pN2 versus pN0a | 3.09 | 2.04 | 4.68 | <0.001 | 1.47 | 1.09 | 1.98 | 0.01 | 3.27 | 1.86 | 5.72 | <0.001 |
| Age in years (continuous) | 1.01 | 1.00 | 1.02 | 0.01 | 1.01 | 1.00 | 1.02 | 0.02 | 1.01 | 1.00 | 1.03 | 0.07 |
| Male versus female | 1.36 | 1.15 | 1.62 | <0.001 | 1.48 | 1.17 | 1.88 | 0.001 | 1.28 | 0.92 | 1.78 | 0.14 |
| LN assessed ≥12 versus <12 | 0.74 | 0.60 | 0.91 | 0.004 | 0.63 | 0.48 | 0.83 | <0.001 | 1.17 | 0.84 | 1.62 | 0.35 |
| LN positive (continuous) | 1.06 | 1.04 | 1.08 | <0.001 | 1.11 | 1.07 | 1.14 | <0.001 | 1.08 | 1.04 | 1.12 | <0.001 |
| High grade versus low/medium | 1.35 | 1.11 | 1.64 | 0.003 | 1.35 | 1.11 | 1.64 | 0.003 | 1.38 | 0.93 | 2.05 | 0.11 |
| Right colon versus left colon | 1.46 | 1.21 | 1.77 | <0.001 | 1.65 | 1.28 | 2.12 | <0.001 | 1.31 | 0.92 | 1.85 | 0.13 |
| FOLFIRIb versus 5FU/LV | 1.15 | 0.81 | 1.62 | 0.44 | 0.86 | 0.61 | 1.20 | 0.37 | NA | NA | NA | NA |
| FOLFIRIb/cetuximab versus 5FU/LV | 0.54 | 0.19 | 1.50 | 0.24 | 0.36 | 0.05 | 2.62 | 0.31 | NA | NA | NA | NA |
| FOLFOXc versus 5FU/LV | 0.76 | 0.55 | 1.05 | 0.09 | 0.65 | 0.47 | 0.90 | 0.01 | 0.77 | 0.47 | 1.26 | 0.3 |
| FOLFOXc/cetuximab versus 5FU/LV | 0.97 | 0.70 | 1.35 | 0.88 | 0.83 | 0.59 | 1.17 | 0.29 | NA | NA | NA | NA |
| MSI-high versus MSS/MSI-low | 0.75 | 0.57 | 0.99 | 0.043 | 0.72 | 0.49 | 1.05 | 0.08 | 0.44 | 0.25 | 0.77 | 0.004 |
| BRAF V600E mutation versus wt | 1.63 | 1.25 | 2.12 | <0.001 | 2.53 | 1.78 | 3.60 | <0.001 | 1.94 | 1.19 | 3.16 | 0.007 |
| KRAS codons 12/13 mutation versus Wt | 1.49 | 1.24 | 1.81 | <0.001 | 1.42 | 1.10 | 1.84 | 0.007 | 1.35 | 0.95 | 1.93 | 0.09 |
| Cohorts without adjuvant chemotherapy exposure |
Val3 (n=1080; 286 events) |
Val4 (n=782; 179 events) |
||||||||||
| HR | 95% CI | P value | HR | 95% CI | P value | |||||||
| pT3 versus pT2/pT1 | 1.77 | 0.65 | 4.83 | 0.26 | 4.46 | 1.64 | 12.15 | 0.004 | ||||
| pT4 versus pT2/pT1 | 3.29 | 1.16 | 9.35 | 0.02 | 11.18 | 3.64 | 34.39 | <0.001 | ||||
| pN1 versus pN0 | 1.45 | 1.04 | 2.01 | 0.03 | 1.82 | 1.31 | 2.53 | <0.001 | ||||
| pN2 versus pN0 | 1.72 | 1.11 | 2.64 | 0.01 | 1.53 | 0.86 | 2.74 | 0.14 | ||||
| Age in years (continuous) | 1.05 | 1.04 | 1.07 | <0.001 | NA | NA | NA | NA | ||||
| Male versus female | 1.08 | 0.85 | 1.38 | 0.52 | NA | NA | NA | NA | ||||
| LN assessed ≥12 versus <12 | 1.22 | 0.97 | 1.55 | 0.09 | NA | NA | NA | NA | ||||
| LN positive (continuous) | 1.04 | 1.00 | 1.07 | 0.03 | NA | NA | NA | NA | ||||
| High grade versus low/medium | 0.99 | 0.63 | 1.55 | 0.97 | NA | NA | NA | NA | ||||
| Right colon versus left colon | 0.70 | 0.54 | 0.91 | 0.007 | NA | NA | NA | NA | ||||
| MSI-high versus MSS/MSI-low | 0.49 | 0.32 | 0.77 | 0.002 | 0.97 | 0.64 | 1.47 | 0.88 | ||||
| BRAF V600E mutation versus wt | 1.64 | 1.03 | 2.62 | 0.04 | 1.72 | 1.10 | 2.69 | 0.02 | ||||
| KRAS codons 12/13 mutation versus wt | 1.13 | 0.87 | 1.48 | 0.36 | 1.28 | 0.92 | 1.78 | 0.13 | ||||
In Val2, pN2 versus pN1.
Infusional 5FU with irinotecan.
Infusional 5FU with oxaliplatin.
Figure 3.
Risk discrimination and performance of overall survival models, with (A) C-index (error bars represent 95% confidence intervals) across training and validation cohorts, and (B) boxplot of distributions of bootstrapped iAUCs for Bayes factor estimation across all validation cohorts combined (except val4 because of missing clinicopathological annotation). (C) Relative proportion of explained variance in overall survival of the full model (in patient cohorts treated with adjuvant chemotherapy) that is accounted for by different categories of predictor covariates.
Risk discrimination was consistently better in patient cohorts that received adjuvant chemotherapy, mirroring the training data. We then focused on this population (train, val1 and val2) to illustrate the relative contribution of different factors for prognosis prediction (Figure 3C). In multivariable models, the relative proportion of variance explained (R2) by TNM staging, clinicopathological features and molecular markers was 59%, 31% and 10%, respectively, in clinical trial cohorts. The corresponding numbers in observational studies were 72%, 18% and 10%.
Discussion
In an era of precision medicine and increased public awareness and access to health information, movement toward individualized prognostic estimation is critical for both treatment planning and patient–physician communication. In this study we focused on well-defined cancer cell molecular markers in intermediate-risk CC. First, we confirmed that the prevalence of KRAS and BRAFV600E mutations and MSI-high varies significantly throughout the colon [13]. Other studies have shown that the biological differences between right- and left-sided carcinomas also extend to the gene expression level, which translate into different prognosis and response to anti-EGFR therapy [23, 24]. Indeed, we found that primary tumor site was one of the strongest clinicopathological determinants of survival, with worse outcomes for those diagnosed with right-sided tumors.
Next, we investigated to what extent these markers impact the prediction of 5-year OS in multivariable models that include patient and disease characteristics. The selection of OS as the endpoint of interest is based on its proven association with the AJCC-TNM staging system, unambiguous clinical relevance and unbiased interpretation. We demonstrated that there is a statistically significant increase in the performance of models when KRAS/BRAF mutation and MSI status are added to TNM models. Therefore, from an individual patient perspective, these molecular markers significantly improve the ability to discriminate risk of death over TNM staging alone. Interestingly, the molecular markers tested in our series have high prognostic impact in small subgroups of patients, particularly those whose tumors are MSS/MSI-low and BRAFV600E mutated. On the contrary, from a population or modeling perspective, there is marginal (albeit not statistically significant) impact of adding this set of molecular markers to clinicopathological models currently available to make critical prognostic estimations. The existing data do not support routine KRAS/BRAF mutation plus MSI testing, with its added cost, for improved prognostic stratification of nonmetastatic CC patients eligible to adjuvant chemotherapy as per TNM staging and clinicopathological features. Therefore, our results do not justify a revision of existing web-calculators for widespread adoption by the medical community [4]. However, it is important to emphasize that full models described here achieved c-indices ranging from 0.70 to 0.74 in validation cohorts treated with adjuvant chemotherapy (Figure 3A), numbers exceeding those that have been reported with models lacking molecular annotation (c-index of 0.66) [4, 5]. This may be in part related to more accurate TNM staging and clinicopathological annotation in the contemporary cohorts included in our study.
Major strengths of our work include compliance with AJCC recommendations for potential model endorsement [7], and the significant clinicopathological and molecular heterogeneity in the validation datasets, reflecting major differences in clinical trial cohorts and observational studies—which increases generalizability of our findings. We recognize that clinicopathological annotation available for modeling in our series is not complete, and some covariates with well-established prognostic impact were not examined, including lymphovascular and perineural invasion, and tumor presentation with obstruction or perforation [9]. Another limitation of our study is the lack of annotation on exposure to these targeted agents in the metastatic setting, which is known to interact with KRAS/BRAF mutations. Nevertheless, the differences in OS according to molecular subgroups depicted in Figure 2 are likely related not only to patient outcome in the metastatic setting but also to the risk of disease relapse. Indeed, previous reports from clinical trials included in our study have shown that both disease-free survival and survival after relapse are affected by MSI status and KRAS/BRAFV600E mutations [12, 13].
Lastly, in an era of next-generation sequencing in clinical labs, other patient subgroups with specific molecular changes that have strong impact for precision medicine may be identified, such as the recently described subpopulation with POLE mutations [25]. Additionally, nongenetic markers linked to microenvironment components have demonstrated independent prognostic value in stage I to III CC. These include immune infiltration patterns—tumor infiltrating lymphocytes [26] and immunoscore [27], e.g.—and gene expression signatures that reflect stromal infiltration with epithelial–mesenchymal transition [17] or activated cancer-associated fibroblasts [28, 29]. Interestingly, these signatures may correlate with an immunosuppressive microenvironment [30] and resistance to standard adjuvant chemotherapy [31, 32], supporting the idea that an activated immune microenvironment in early stage CC is a strong determinant of the risk of distant dissemination. Future studies evaluating predictive performance of survival models in CC should incorporate larger mutation panels and signatures that reflect immune and stromal activation status, and take into account therapies received after relapse, particularly targeted agents and immunotherapies that potentially interact with molecular markers.
To conclude, we believe that our work represents a foundation for similar efforts trying to optimize prognostic stratification of CC by integrating molecular markers with clinicopathological features. Such an individualized and comprehensive approach to prognostication, with model-based staging, will aid in patient–physician communication, development of rational follow-up schedules and hopefully risk-adaptive therapies.
Supplementary Material
Acknowledgements
The authors wish to thank the following CCFR Principal Investigators for the contributing data used in this work: Noralane M. Lindor, Mark A. Jenkins, John L. Hopper, and Steve Gallinger. Thanks to “Fondation Medic, Lausanne”.
Funding
This work was partially supported by grant UM1 CA167551 from the National Cancer Institute (NCI), National Institutes of Health (NIH), and through cooperative agreements with members of the Colon Cancer Family Registry (CCFR) and Principal Investigators. Centers contributing to this analysis include the Seattle Colorectal Cancer Family Registry (U01/U24CA074794), Australasian Colorectal Cancer Family Registry (U01 CA074778 and U01/U24 CA097735), Mayo Clinic Cooperative Family Registry for Colon Cancer Studies (U01/U24 CA074800), and Ontario Familial Colorectal Cancer Registry (U01/U24 CA074783). This work was also supported by NCI/NIH grant K07CA172298 and K05CA152715 (to AIP). The content of this article does not necessarily reflect the views or policies of the NIH or any of the collaborating centers in the CCFR, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government. This study was partly supported by the “Norwegian Cancer Society” and the “Research Council of Norway” (no grants apply).
Disclosure
The authors have declared no conflicts of interest.
References
- 1. Siegel R, Desantis C, Jemal A.. Colorectal cancer statistics, 2014. CA Cancer J Clin 2014; 64: 104–117. [DOI] [PubMed] [Google Scholar]
- 2. Ferlay J, Steliarova-Foucher E, Lortet-Tieulent J. et al. Cancer incidence and mortality patterns in Europe: estimates for 40 countries in 2012. Eur J Cancer 2013; 49: 1374–1403. [DOI] [PubMed] [Google Scholar]
- 3. National Cancer Institute’s SEER database. http://seer.cancer.gov/ (26 August 2016, date last accessed).
- 4. Renfro LA, Grothey A, Xue Y. et al. ACCENT-based web calculators to predict recurrence and overall survival in stage III colon cancer. J Natl Cancer Inst 2014; 106: pii: dju333.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Weiser MR, Gonen M, Chou JF. et al. Predicting survival after curative colectomy for cancer: individualizing colon cancer staging. J Clin Oncol 2011; 29: 4796–4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sjo OH, Merok MA, Svindland A. et al. Prognostic impact of lymph node harvest and lymph node ratio in patients with colon cancer. Dis Colon Rectum 2012; 55: 307–315. [DOI] [PubMed] [Google Scholar]
- 7. Kattan MW, Hess KR, Amin MB. et al. American Joint Committee on Cancer acceptance criteria for inclusion of risk models for individualized prognosis in the practice of precision medicine. CA Cancer J Clin 2016; 66: 370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. ACCENT stage III survival calculator. http://www.mayoclinic.org/medical professionals/cancer-prediction-tools/colon-cancer (26 August 2016, date last accessed).
- 9. Dienstmann R, Salazar R, Tabernero J.. Personalizing colon cancer adjuvant therapy: selecting optimal treatments for individual patients. J Clin Oncol 2015; 33: 1787–1796. [DOI] [PubMed] [Google Scholar]
- 10. Hutchins G, Southward K, Handley K. et al. Value of mismatch repair, KRAS, and BRAF mutations in predicting recurrence and benefits from chemotherapy in colorectal cancer. J Clin Oncol 2011; 29: 1261–1270. [DOI] [PubMed] [Google Scholar]
- 11. Sargent D, Shi Q, Yothers G. et al. Prognostic impact of deficient mismatch repair (dMMR) in 7,803 stage II/III colon cancer (CC) patients (pts): a pooled individual pt data analysis of 17 adjuvant trials in the ACCENT database. J Clin Oncol 2014; 32: 5s (suppl; abstr 3507). [Google Scholar]
- 12. Popovici V, Budinska E, Bosman FT. et al. Context-dependent interpretation of the prognostic value of BRAF and KRAS mutations in colorectal cancer. BMC Cancer 2013; 13: 439.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sinicrope FA, Mahoney MR, Yoon HH. et al. Analysis of molecular markers by anatomic tumor site in stage III colon carcinomas from adjuvant chemotherapy trial NCCTG N0147 (Alliance). Clin Cancer Res 2015; 21: 5294–5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lochhead P, Kuchiba A, Imamura Y. et al. Microsatellite instability and BRAF mutation testing in colorectal cancer prognostication. J Natl Cancer Inst 2013; 105: 1151–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Roth AD, Delorenzi M, Tejpar S. et al. Integrated analysis of molecular and clinical prognostic factors in stage II/III colon cancer. J Natl Cancer Inst 2012; 104: 1635–1646. [DOI] [PubMed] [Google Scholar]
- 16. Phipps AI, Limburg PJ, Baron JA. et al. Association between molecular subtypes of colorectal cancer and patient survival. Gastroenterology 2015; 148: 77–87.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guinney J, Dienstmann R, Wang X. et al. The consensus molecular subtypes of colorectal cancer. Nat Med 2015; 21: 1350–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Merok MA, Ahlquist T, Royrvik EC. et al. Microsatellite instability has a positive prognostic impact on stage II colorectal cancer after complete resection: results from a large, consecutive Norwegian series. Ann Oncol 2013; 24: 1274–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Marshall A, Altman DG, Royston P. et al. Comparison of techniques for handling missing covariate data within prognostic modelling studies: a simulation study. BMC Med Res Methodol 2010; 10: 17.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Blanche P, Dartigues JF, Jacqmin-Gadda H.. Estimating and comparing time-dependent areas under receiver operating characteristic curves for censored event times with competing risks. Stat Med 2013; 32: 5381–5397. [DOI] [PubMed] [Google Scholar]
- 21. Goodman SN. Toward evidence-based medical statistics. 2: the Bayes factor. Ann Intern Med 1999; 130: 1005–1013. [DOI] [PubMed] [Google Scholar]
- 22. R Development Core Team. R: A Language and Environment for Statistical Computing (http://www.R-project.org). Vienna: R Foundation for Statistical Computing 2008.
- 23. Missiaglia E, Jacobs B, D’Ario G. et al. Distal and proximal colon cancers differ in terms of molecular, pathological, and clinical features. Ann Oncol 2014; 25: 1995–2001. [DOI] [PubMed] [Google Scholar]
- 24. Venook A, Niedzwiecki D, Innocenti F. et al. Impact of primary tumor location on overall survival and progression-free survival in patients with metastatic colorectal cancer: analysis of CALGB/SWOG 80405 (Alliance). J Clin Oncol 2016; 34(Suppl): abstr 3504. [Google Scholar]
- 25. Domingo E, Freeman-Mills L, Rayner E. et al. Somatic POLE proofreading domain mutation, immune response, and prognosis in colorectal cancer: a retrospective, pooled biomarker study. Lancet Gastroenterol Hepatol 2016; 1: 207–216. [DOI] [PubMed] [Google Scholar]
- 26. Sinicrope F, Smyrk TC, Foster NR. et al. Association of tumor infiltrating lymphocytes (TILs) with molecular subtype and prognosis in stage III colon cancers (CC) from a FOLFOX-based adjuvant chemotherapy trial. J Clin Oncol 2016; 34(Suppl): abstr 3518. [Google Scholar]
- 27. Galon J, Mlecnik B, Marliot F. et al. Validation of the Immunoscore (IM) as a prognostic marker in stage I/II/III colon cancer: results of a worldwide consortium-based analysis of 1,336 patients. J Clin Oncol 2016; 34 (Suppl): abstr 3500. [Google Scholar]
- 28. Calon A, Lonardo E, Berenguer-Llergo A. et al. Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat Genet 2015; 47: 320–329. [DOI] [PubMed] [Google Scholar]
- 29. Berdiel-Acer M, Berenguer A, Sanz-Pamplona R. et al. A 5-gene classifier from the carcinoma-associated fibroblast transcriptomic profile and clinical outcome in colorectal cancer. Oncotarget 2014; 5: 6437–6452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Becht E, de Reynies A, Giraldo NA. et al. Immune and stromal classification of colorectal cancer is associated with molecular subtypes and relevant for precision immunotherapy. Clin Cancer Res 2016; 22: 4057–4066. [DOI] [PubMed] [Google Scholar]
- 31. Roepman P, Schlicker A, Tabernero J. et al. Colorectal cancer intrinsic subtypes predict chemotherapy benefit, deficient mismatch repair and epithelial-to-mesenchymal transition. Int J Cancer 2014; 134: 552–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Song N, Pogue-Geile KL, Gavin PG. et al. Clinical outcome from oxaliplatin treatment in stage II/III colon cancer according to intrinsic subtypes: secondary analysis of NSABP C-07/NRG Oncology Randomized Clinical Trial. JAMA Oncol 2016; 2: 1162–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.