Abstract
Background:
Several studies concerning the association between glutathione S-transferase P1 (GSTP1) Ile105Val polymorphism and male infertility risk have reported controversial findings. The present study was aimed to explore this association using a meta-analysis.
Methods:
The PubMed, EMBASE, China National Knowledge Infrastructure (CNKI), and Wanfang databases were searched. Odds ratios (ORs) with 95% confidence intervals (CIs) were calculated to estimate the strength of the association.
Results:
A total of 3282 cases and 3268 controls in nine case-control studies were included. There was no significant association between GSTP1 Ile105Val polymorphism and male infertility in the overall population, but significant associations were found under the dominant (OR = 1.23, 95% CI = 1.04–1.46, I2 = 32.2%) and heterozygote (OR = 1.29, 95% CI = 1.08–1.53, I2 = 26.8%) models after excluding studies for which the data did not satisfy Hardy-Weinberg equilibrium (HWE). Similarly, subgroup analyses revealed no significant association in Asians or Chinese population although a significant association was apparent among Chinese population in studies with HWE under the heterozygote model (OR = 1.25, 95% CI = 1.03–1.52, I2 = 44.1%). Significant heterogeneity could be observed in some genetic models, but this heterogeneity was not significant when stratified by HWE. No evidence for publication bias was found.
Conclusions:
The GSTP1 Ile105Val polymorphism might not be associated with male infertility risk, and thus additional well-designed studies with larger sample size are warranted.
Keywords: Glutathione S-transferase P1, Male Infertility, Polymorphism
Introduction
Male infertility is a multifactorial and heterogeneous condition that affects approximately 10–15% of the male adult population worldwide. For nearly half of cases, the cause of infertility is unknown (idiopathic male infertility).[1,2] It is widely accepted that many genetic and environmental factors interact and are implicated in the impairment of spermatogenesis and consequent infertility.[3] Approximately, 30% of male infertility is due to genetic abnormalities including chromosome aberrations, DNA damage, and single gene mutations.[4] Genome-wide association studies (GWAS) have identified several susceptibility loci and >200 single nucleotide polymorphisms (SNPs) that have been associated with oligozoospermia and/or azoospermia.[5,6,7] Genetic polymorphisms can lead to different levels of individual susceptibility to the potential adverse effects of environmental factors (e.g., exposure to chemicals) on reproductive function.[8]
Glutathione S-transferases (GSTs) are an important superfamily of Phase II multifunctional enzymes that contribute to the detoxification of a wide variety of natural and artificial compounds including chemotherapeutic drugs, carcinogens, and various xenobiotics.[9] GSTs play a central role in protecting cells against reactive oxygen species (ROS), which are produced under oxidative and electrophile stresses.[10] Numerous epidemiological studies have demonstrated that increased exposure to harmful environmental substances is associated with decreased semen quality and quantity and altered reproductive hormone levels that contribute to male infertility.[11,12] It has also been shown that levels of ROS in spermatozoa and seminal plasma are significantly higher in patients with idiopathic infertility compared to that in healthy controls.[13] Excessive ROS can cause substantial oxidative damage to sperm through enzyme inactivation, protein degradation, DNA fragmentation, and lipid peroxidation, which results in defective spermatogenesis and the consequent negative impact on fertility.[14,15] Genetic-based alterations in GSTs activities might alter their ability to detoxify potentially damaging agents and hence increase the risk of disease development.[16] The three GSTs GSTM1, GSTT1, and GSTP1 are the predominant members of the GST family, and deletion of the gene encoding GSTM1 or GSTT1 or the polymorphism Ile105Val (rs1695) of GSTP1 can impact the binding affinity of these enzymes and/or reduce or eliminate cellular GST activity.[17,18] The gene encoding GSTP1 is located at 11q13, spanning nearly 2.8 kb and containing seven exons.[19] GSTP1 plays a critical role in the biotransformation and inactivation of toxic and carcinogenetic electrophiles, especially those in cigarette smoke[20] and also inhibits apoptosis and promotes cellular proliferation through interacting with Jun N-terminal kinase (JNK) pathway.[21,22] The polymorphism Ile105Val in exon 5 of GSTP1 significantly alters the activity and heat stability of the encoded enzyme, decreasing its ability to detoxify environmental mutagens and protect against oxidative damage to DNA.[23] A number of studies have demonstrated that this polymorphism is an important risk factor for individual susceptibility to various diseases[23,24] and is a biomarker to predict the efficacy of chemotherapeutics for certain diseases including cancer.[25]
To date, several case-control studies have investigated the association between the GSTP1 Ile105Val polymorphism and risk of male infertility,[21,26,27,28,29,30,31,32,33] but the results have been inconclusive owing to the limitation of individual studies, for example, relatively small sample size and/or differences in patient populations, ethnicity, or genotyping methods. Therefore, we carried out a systematic review and meta-analysis of all available data to better assess the overall effects of this polymorphism on male infertility risk.
Methods
Literature search strategy
The meta-analysis was performed in accordance with PRISMA guidelines.[34] We conducted a comprehensive literature search of the databases PubMed, EMBASE, China National Knowledge Infrastructure (CNKI), and Wanfang (updated as of September 10, 2016) for case-control studies evaluating the relationship between GSTP1 Ile105Val and male infertility, using a combination of the following search terms: (glutathione S-transferase P1 or GSTP1 or GSTP1 Ile105Val or A313G or rs1695) plus (polymorphism[s] or SNP[s] or variation or genotype[s]) plus (infertility or sterility or infecundity or sterile) plus (male or men). Data for other potentially relevant studies were also obtained by reviewing the references on this topic listed in retrieved original and review articles. No language or publication date restriction was applied in this meta-analysis.
Selection criteria
Eligible studies were selected using the following inclusion criteria: (1) they evaluated the association between GSTP1 Ile105Val and male infertility; (2) they were case-control studies; and (3) they provided sufficient data concerning genotypes and/or allele frequencies in both cases and controls to estimate odds ratios (ORs) with corresponding 95% confidence intervals (CIs).
Data extraction
Two reviewers independently extracted the relevant information from the identified studies, and disagreements were discussed and resolved with consensus. The following information was collected from each study: The first author's surname, year of publication, country in which the study was performed, ethnicity, total number of cases and controls, genotype distribution, allele frequencies, results of Hardy-Weinberg equilibrium (HWE) test, and method of genotyping test.
Quality score assessment
Two reviewers independently assessed the quality of the included case-control studies using the Newcastle-Ottawa Scale (NOS).[35] The quality of each study was evaluated by a “star” rating system (0–9 stars) based on three aspects of the study, namely, selection, comparability, and exposure. Studies with ≥7 stars were considered of high quality.
Statistical analysis
Deviation from HWE for each control group of the included studies was examined by Chi-square tests. OR values with 95% CIs were used to assess the strength of association between GSTP1 Ile105Val and male infertility susceptibility. The significance for pooled ORs was determined using the Z-test with P < 0.05 considered statistically significant. The ORs of the allele (Val vs. Ile), recessive (Val/Val vs. Ile/Val + Ile/Ile), dominant (Val/Val + Ile/Val vs. Ile/Ile), additive (Val/Val vs. Ile/Ile), and heterozygote (Ile/Val vs. Ile/Ile) models were calculated by the fixed effects model or random effects model according to the outcome of between-study heterogeneity detected by Chi-square test based on the Q test. The fixed effects model with Mantel-Haenszel method was used to estimate the pooled ORs when heterogeneity was indicated to be nonsignificant (P > 0.10 and I2 < 50%);[36] otherwise, the random effects model based on DerSimonian and Laird method was more appropriate.[37] Sensitivity was assessed by sequentially excluding individual studies to assess the stability and reliability of the results. A funnel plot and Egger's tests were applied to evaluate publication bias across studies (a value of P < 0.05 was considered statistically significant). Subgroup analyses were conducted based on ethnicity, HWE, and Chinese population subgroup. Data were analyzed mainly using STATA version 12.0 (StataCorp LP, College Station, Texas, USA).
Results
Characteristics of studies
As shown in Figure 1, 83 published studies that matched our predefined search strategy were identified after an initial search. After removing duplicate publications and screening titles and abstracts, 13 candidate studies were obtained. By reviewing full texts of these studies, four more were rejected (two were not case-control studies,[38,39] one did not focus on the polymorphism of rs1695,[40] and two reported on two similar populations).[32,41] No additional eligible studies were found through manual search of the reference lists. Finally, nine case-control studies were included in the meta-analysis, involving 3282 patients with male infertility and 3268 controls; eight of the included studies were conducted on the Asian population (two from Iran and six from China), and the remaining study was on the Caucasian population from Russia. The genotype distributions of the controls in four studies were consistent with HWE. Of not-in-HWE studies, three studies[26,32,33] did not mention HWE and two studies[28,31] stated that the genotype distributions were consistent with HWE but in fact are not. In addition, all not-in-HWE studies did not explain the effect of HWE on their conclusions. The results of the NOS assessment yielded an average score of 7.4 (range, 5–9); seven studies were regarded as high quality, whereas two other studies were of slightly low quality (scored only six stars for lack of detailed data for the study population selection). Genotyping methods included polymerase chain reaction-restriction fragment length polymorphism (PCR-RELP) which was most commonly used in these studies, MassArray, direct DNA sequencing, and TaqMan assay. Table 1 presents the characteristics of the included studies.
Figure 1.
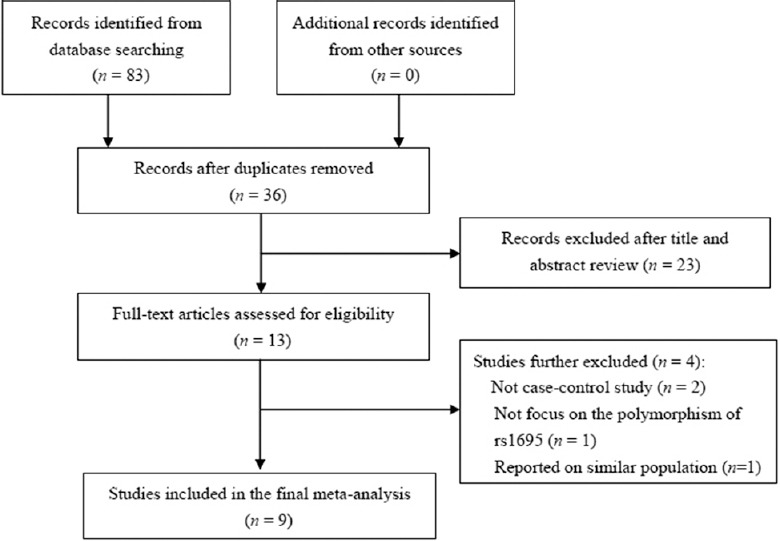
Flow chart of the study selection process.
Table 1.
Details of the individual studies included in the meta-analysis
| Author (year) | Country (ethnicity) | Case/control, n | Association of allele | HWE (χ2/P) | Score (NOS) | Genotyping method | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Numbers | Ile/Ile | Ile/Val | Val/Val | Val allele (%) | OR (95% CI) | P | |||||
| Yarosh et al. (2015)[30] | Russia (Caucasian) | 203/227 | 90/117 | 96/85 | 17/25 | 130/135 | 1.13 (0.83–1.49) | 0.467 | 2.451/0.117 | 8 | PCR-RFLP |
| Lakpour et al. (2013)[33] | Iran (Asian) | 95/26 | 95/26 | 0/0 | 0/0 | 0/0 | NR | NR | NA | 6 | PCR-RFLP |
| Safarinejad et al. (2010)[31] | Iran (Asian) | 166/166 | 102/86 | 59/76 | 5/4 | 69/84 | 0.78 (0.54–1.11) | 0.167 | 7.405/0.006 | 9 | PCR-RFLP |
| Zhou (2016)[27] | China (Asian) | 386/441 | 233/280 | 141/145 | 12/16 | 165/177 | 1.08 (0.85–1.37) | 0.513 | 0.273/0.601 | 8 | MassArray |
| Xiong et al. (2015)[21] | China (Asian) | 479/234 | 249/146 | 221/83 | 9/5 | 239/93 | 1.34 (1.02–1.76) | 0.034 | 3.031/0.082 | 8 | PCR-RFLP |
| Feng et al. (2015)[28] | China (Asian) | 216/198 | 137/141 | 47/43 | 32/14 | 111/71 | 1.58 (1.13–2.21) | 0.007 | 13.598/<0.001 | 7 | Sequencing |
| Tang et al. (2014)[32] | China (Asian) | 246/117 | 167/85 | 79/32* | NR | NR | NR | NA | 6 | PCR-RFLP | |
| Lu (2013)[26] | China (Asian) | 1255/717 | 728/393 | 78/56 | 449/268 | 976/592 | 0.91 (0.79–1.03) | 0.139 | 504.588/<0.001 | 8 | TaqMan |
| Li et al. (2013)[29] | China (Asian) | 236/142 | 156/90 | 73/45 | 7/7 | 87/59 | 0.86 (0.60–1.25) | 0.430 | 0.197/0.657 | 7 | PCR-RFLP |
| Total | 3282/3268 | 1957/1364 | 715/533 | 531/339 | 1777/1211 | 1.06 (0.97–1.15) | 0.216 | ||||
*The numbers of genotype distribution of Ile/Val + Val/Val in cases and controls. OR: Odds ratio; CI: Confidence interval; HWE: Hardy-Weinberg equilibrium; NOS: Newcastle-Ottawa Scale; PCR-RFLP: Polymerase chain reaction-restriction fragment length polymorphism; NR: Not report; NA: Not available.
Association of glutathione S-transferase P1 Ile105Val polymorphism with male infertility risk
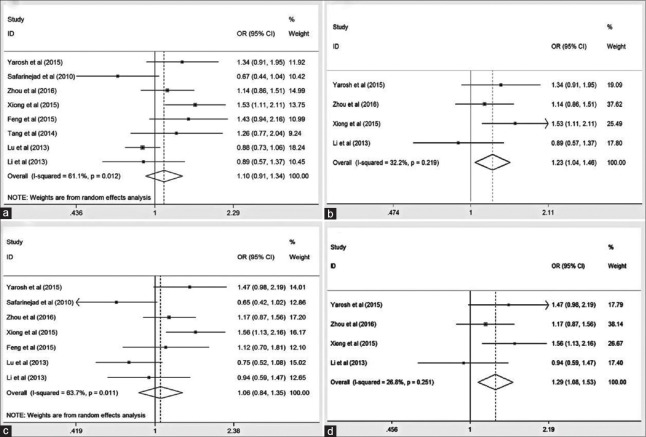
For the overall analyses, there was no significant association between GSTP1 Ile105Val polymorphism and male infertility in all the five genetic models [Table 2 and Figure 2a, 2c]; however, after excluding not-in-HWE studies, significant associations were detected under dominant (OR = 1.23, 95% CI = 1.04–1.46, I2 = 32.2%) and heterozygote (OR = 1.29, 95% CI = 1.08–1.53, I2 = 26.8%) models in the overall population [Table 2 and Figure 2b, 2d]. For subgroup analyses of Asians or Chinese population, the Ile105Val polymorphism showed no association with male infertility, whereas a significant association was found for the Chinese population in studies with HWE under heterozygote model (OR = 1.25, 95% CI = 1.03–1.52, I2 = 44.1%) [Table 2].
Table 2.
Association between GSTP1 Ile105Val polymorphism and male infertility risk
| Comparison | Population | Number of studies | Test of association | Heterogeneity | |||
|---|---|---|---|---|---|---|---|
| OR (95% CI) | P | Model | P | I2 (%) | |||
| Val versus Ile (allele) | Overall | 7 | 1.06 (0.90–1.26) | 0.482 | R | 0.008 | 65.3 |
| Overall in HWE | 4 | 1.12 (0.97–1.29) | 0.116 | F | 0.294 | 19.2 | |
| Asian | 6 | 1.06 (0.87–1.29) | 0.588 | R | 0.005 | 70.3 | |
| Asian in HWE | 3 | 1.21 (0.96–1.32) | 0.162 | F | 0.156 | 46.1 | |
| China | 5 | 1.11 (0.90–1.38) | 0.343 | R | 0.005 | 72.8 | |
| China in HWE | 3 | 1.21 (0.96–1.32) | 0.162 | F | 0.156 | 46.1 | |
| Val/Val versus Ile/Val + Ile/Ile (recessive) | Overall | 7 | 0.97 (0.82–1.14) | 0.683 | F | 0.209 | 28.8 |
| Overall in HWE | 4 | 0.76 (0.51–1.16) | 0.203 | F | 0.945 | 0 | |
| Asian | 6 | 0.99 (0.83–1.17) | 0.860 | F | 0.171 | 35.4 | |
| Asian in HWE | 3 | 0.78 (0.46–1.34) | 0.373 | F | 0.836 | 0 | |
| China | 5 | 0.98 (0.83–1.17) | 0.824 | F | 0.107 | 47.4 | |
| China in HWE | 3 | 0.78 (0.46–1.34) | 0.373 | F | 0.836 | 0 | |
| Val/Val + Ile/Val versus Ile/Ile (dominant) | Overall | 8 | 1.10 (0.91–1.34) | 0.334 | R | 0.012 | 61.1 |
| Overall in HWE | 4 | 1.23 (1.04–1.46) | 0.015 | F | 0.219 | 32.2 | |
| Asian | 7 | 1.07 (0.87–1.33) | 0.517 | R | 0.012 | 63.4 | |
| Asian in HWE | 3 | 1.19 (0.89–1.58) | 0.235 | R | 0.122 | 52.5 | |
| China | 6 | 1.14 (0.92–1.41) | 0.229 | R | 0.029 | 59.8 | |
| China in HWE | 3 | 1.19 (0.89–1.58) | 0.235 | R | 0.122 | 52.5 | |
| Val/Val versus Ile/Ile (additive) | Overall | 7 | 0.96 (0.81–1.14) | 0.644 | F | 0.223 | 26.9 |
| Overall in HWE | 4 | 0.86 (0.56–1.31) | 0.472 | F | 0.879 | 0 | |
| Asian | 6 | 0.97 (0.81–1.15) | 0.701 | F | 0.148 | 38.7 | |
| Asian in HWE | 3 | 0.84 (0.49–1.44) | 0.525 | F | 0.719 | 0 | |
| China | 5 | 1.06 (0.70–1.63) | 0.773 | R | 0.087 | 50.8 | |
| China in HWE | 3 | 0.84 (0.49–1.44) | 0.525 | F | 0.719 | 0 | |
| Ile/Val versus Ile/Ile (heterozygote) | Overall | 7 | 1.06 (0.84–1.35) | 0.619 | R | 0.011 | 63.7 |
| Overall in HWE | 4 | 1.29 (1.08–1.53) | 0.005 | F | 0.251 | 26.8 | |
| Asian | 6 | 1.01 (0.78–1.31) | 0.951 | R | 0.015 | 64.6 | |
| Asian in HWE | 3 | 1.25 (1.03–1.52) | 0.026 | F | 0.167 | 44.1 | |
| China | 5 | 1.09 (0.85–1.41) | 0.499 | R | 0.054 | 57.0 | |
| China in HWE | 3 | 1.25 (1.03–1.52) | 0.026 | F | 0.167 | 44.1 | |
GSTP1: Glutathione S-transferase P1; OR: Odds ratio; CI: Confidence interval; HWE: Hardy-Weinberg equilibrium; R: Random effects model; F: Fixed effects model.
Figure 2.
Forest plot of the glutathione S-transferase P1 Ile105Val polymorphism associated with male infertility susceptibility in (a) the overall population and (b) the overall population in studies with Hardy-Weinberg equilibrium under dominant model (Val/Val + Ile/Val vs. Ile/Ile), and (c) the overall population and (d) the overall population in studies with Hardy-Weinberg equilibrium under heterozygote model (Ile/Val vs. Ile/Ile). OR: Odds ratio; CI: Confidence interval.
Heterogeneity, sensitivity analysis, and publication bias
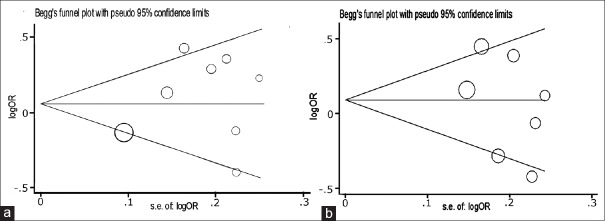
As shown in Table 2, significant between-study heterogeneity (P < 0.1 or I2 > 50%) was observed in some genetic comparison models, but this heterogeneity was not significant when stratified by HWE. Sensitivity analysis revealed that the corresponding pooled OR values were not significantly altered when one study at a time was removed from the overall pooled data (data not shown). Begg's funnel plot and Egger's test were conducted to assess the publication bias of literature. After combining the included studies, the results of Begg's funnel plot and Egger's test showed no apparent evidence of publication bias in any of the comparison models [Figure 3].
Figure 3.
Analysis of funnel plot asymmetry and publication bias for the associations between glutathione S-transferase P1 Ile105Val polymorphism and male infertility in the overall population under (a) dominant (Val/Val + Ile/Val vs. Ile/Ile; P = 0.399) and (b) heterozygote (Ile/Val vs. Ile/Ile; P = 0.367) models.
Discussion
Male infertility is increasingly common, especially in developed countries, mainly resulting from complicated interactions between genetic and environmental factors.[3] As a family of detoxifying and antioxidant enzymes, the dysfunctions of GSTs have been shown to be involved in the pathogenesis of various diseases.[16] Many studies have suggested that genetic mutations in GSTs genes might contribute to abnormal GST expression and are likely associated with male infertility risk[21,31,41] although other studies reported contradictory results. A recent meta-analysis conducted in 2012 showed that GSTM1 and GSTT1 null polymorphisms were associated with a significantly increased risk of male infertility, whereas the GSTP1 Ile105Val polymorphism has a protective effect against the development of male infertility.[42] For GSTP1 Ile105Val polymorphism, however, there were some limitations in this meta-analysis. First, only two case-control studies comprising 231 cases and 196 controls were included,[31,41] and thus the sample size was very small and the statistical power was relatively low. Second, of the two included case-control studies, only one indicated that the GSTP1 Ile105Val polymorphism might decrease the risk of male infertility,[31] and the genotype distribution of the control was not in agreement with HWE; for the other study, no significant relation was found, and this study which did not provide detailed data on genotyping methods and HWE.[41] Third, there was a significant between-study heterogeneity. Moreover, many recent studies have been published concerning the association between the GSTP1 Ile105Val polymorphism and male infertility risk.[21,26,27,28,29,30,32,33]
For the present meta-analysis, data for which were pooled from nine case-control studies including 3282 cases and 3268 controls, we systematically evaluated the association between the GSTP1 Ile105Val polymorphism and male infertility susceptibility. The results of the overall analysis indicated that this polymorphism was not significantly associated with male infertility although significant associations were found under the dominant and heterozygote models in the overall population after excluding not-in-HWE studies. Similarly, analysis of the Asian and Chinese population subgroups revealed a nonsignificant association, but there was an increased risk for the Chinese population in studies with HWE under the heterozygote model. These findings contradict the results of the recent meta-analysis that we discussed above.[42] Indeed, among the nine case-control studies in the current meta-analysis, five studies[26,27,29,32,33] showed no association between the Ile105Val polymorphism and male infertility, and three[21,28,30] reported an increased risk, whereas only one study[31] suggested a decrease risk. The present study revealed obvious between-study heterogeneity in several genetic models. Significant between-study heterogeneity was mild when not-in-HWE studies[26,28,31,32,33] were excluded in the overall and subgroup analysis, suggesting that not-in-HWE studies might contribute to the major source of heterogeneity. Nevertheless, the observed positive associations between this polymorphism and male infertility risk in the present study should be interpreted with caution, and further studies are needed to clarify these finding. This is a comprehensive meta-analysis investigating the association between the GSTP1 Ile105Val polymorphism and male infertility risk. Similar to other meta-analyses, some limitations should be noted. First, owing to the limited number of databases searched, other relevant published studies or unpublished studies with null results were not included, which might have caused publication bias even though it was not observed in the present study. Second, studies were conducted primarily in the Asian populations (six from China and two from Iran) and to a lesser extent in a single Caucasian population from Russia. More studies involving a wider spectrum of subjects should be carried out with different populations. Third, there were small sample sizes and relatively few eligible studies, particularly for the subgroup analysis. Although we detected significant associations in the overall population and Chinese population after excluding the not-in-HWE studies, only four and three studies were included in the statistical analysis, respectively. Furthermore, male infertility is a heterogeneous disease with multifactorial etiology. Yarosh et al.[30] observed that the genotype combination GSTP1 105Ile/Ile and GSTT1 null was associated with decreased risk of male infertility, whereas the genotype combination GSTP1 105 Ile/Val and GSTT1 was associated with increased risk. However, we could not further evaluate the gene-gene and gene-environment interactions or carry out a haplotype analysis of the development of male infertility because the original data for the eligible studies could not be obtained.
In conclusion, the present meta-analysis suggests that the GSTP1 Ile105Val polymorphism might not contribute to risk of male infertility. Well-designed and larger studies of populations with different ethnicities should be performed to validate or refute the conclusions.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Footnotes
Edited by: Peng Lyu
References
- 1.Krausz C, Escamilla AR, Chianese C. Genetics of male infertility: From research to clinic. Reproduction. 2015;150:R159–74. doi: 10.1530/REP-15-0261. doi: 10.1530/REP-15-0261. [DOI] [PubMed] [Google Scholar]
- 2.Zang ZJ, Ji SY, Zhang YN, Gao Y, Zhang B. Effects of saikokaryukotsuboreito on spermatogenesis and fertility in aging male mice. Chin Med J. 2016;129:846–53. doi: 10.4103/0366-6999.178972. doi: 10.4103/0366-6999.178972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finotti AC, Costa E, Silva RC, Bordin BM, Silva CT, Moura KK. Glutathione S-transferase M1 and T1 polymorphism in men with idiopathic infertility. Genet Mol Res. 2009;8:1093–8. doi: 10.4238/vol8-3gmr642. doi: 10.4238/vol8-3gmr642. [DOI] [PubMed] [Google Scholar]
- 4.Chen P, Wang X, Xu C, Xiao H, Zhang WH, Wang XH, et al. Association of polymorphisms of A260G and A386G in DAZL gene with male infertility: A meta-analysis and systemic review. Asian J Androl. 2016;18:96–101. doi: 10.4103/1008-682X.153542. doi: 10.4103/1008-682X.153542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu B, Huang Z. Variations in antioxidant genes and male infertility. Biomed Res Int. 2015;2015:513196. doi: 10.1155/2015/513196. doi: 10.1155/2015/513196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aston KI, Krausz C, Laface I, Ruiz-Castané E, Carrell DT. Evaluation of 172 candidate polymorphisms for association with oligozoospermia or azoospermia in a large cohort of men of European descent. Hum Reprod. 2010;25:1383–97. doi: 10.1093/humrep/deq081. doi: 10.1093/humrep/deq081. [DOI] [PubMed] [Google Scholar]
- 7.Kosova G, Scott NM, Niederberger C, Prins GS, Ober C. Genome-wide association study identifies candidate genes for male fertility traits in humans. Am J Hum Genet. 2012;90:950–61. doi: 10.1016/j.ajhg.2012.04.016. doi: 10.1016/j.ajhg.2012.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cummings AM, Kavlock RJ. Gene-environment interactions: A review of effects on reproduction and development. Crit Rev Toxicol. 2004;34:461–85. doi: 10.1080/10408440490519786. [DOI] [PubMed] [Google Scholar]
- 9.Gorukmez O, Yakut T, Gorukmez O, Sag SO, Topak A, Sahinturk S, et al. Glutathione S-transferase T1, M1 and P1 genetic polymorphisms and susceptibility to colorectal cancer in Turkey. Asian Pac J Cancer Prev. 2016;17:3855–9. [PubMed] [Google Scholar]
- 10.Mann CL, Davies MB, Boggild MD, Alldersea J, Fryer AA, Jones PW, et al. Glutathione S-transferase polymorphisms in MS: Their relationship to disability. Neurology. 2000;8(54):552–7. doi: 10.1212/wnl.54.3.552. [DOI] [PubMed] [Google Scholar]
- 11.Bhatt RV. Environmental influence on reproductive health. Int J Gynaecol Obstet. 2000;70:69–75. doi: 10.1016/s0020-7292(00)00221-6. doi: 10.1016/S0020-7292(00)00221-6. [DOI] [PubMed] [Google Scholar]
- 12.Melgarejo M, Mendiola J, Koch HM, Moñino-García M, Noguera-Velasco JA, Torres-Cantero AM. Associations between urinary organophosphate pesticide metabolite levels and reproductive parameters in men from an infertility clinic. Environ Res. 2015;137:292–8. doi: 10.1016/j.envres.2015.01.004. doi: 10.1016/j.envres.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 13.Alkan I, Simsek F, Haklar G, Kervancioglu E, Ozveri H, Yalçin S, et al. Reactive oxygen species production by the spermatozoa of patients with idiopathic infertility: Relationship to seminal plasma antioxidants. J Urol. 1997;157:140–3. doi: 10.1097/00005392-199701000-00044. [PubMed] [Google Scholar]
- 14.Aydemir B, Onaran I, Kiziler AR, Alici B, Akyolcu MC. Increased oxidative damage of sperm and seminal plasma in men with idiopathic infertility is higher in patients with glutathione S-transferase Mu-1 null genotype. Asian J Androl. 2007;9:108–15. doi: 10.1111/j.1745-7262.2007.00237.x. doi: 10.1111/j.1745-7262.2007.00237.x. [DOI] [PubMed] [Google Scholar]
- 15.Tang M, Wang S, Wang W, Cao Q, Qin C, Liu B, et al. The glutathione-S-transferase gene polymorphisms (GSTM1 and GSTT1) and idiopathic male infertility risk: A meta-analysis. Gene. 2012;511:218–23. doi: 10.1016/j.gene.2012.09.054. doi: 10.1016/j.gene.2012.09.054. [DOI] [PubMed] [Google Scholar]
- 16.Hollman AL, Tchounwou PB, Huang HC. The Association between Gene-environment interactions and diseases involving the human GST superfamily with SNP variants. Int J Environ Res Public Health. 2016;13:379. doi: 10.3390/ijerph13040379. doi: 10.3390/ijerph13040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kadam PD, Chuan HH. Erratum to: Rectocutaneous fistula with transmigration of the suture: A rare delayed complication of vault fixation with the sacrospinous ligament. Int Urogynecol J. 2016;27:505. doi: 10.1007/s00192-016-2952-5. doi: 10.1007/s00192-016-2952-5. [DOI] [PubMed] [Google Scholar]
- 18.Weich N, Ferri C, Moiraghi B, Bengió R, Giere I, Pavlovsky C, et al. GSTM1 and GSTP1, but not GSTT1 genetic polymorphisms are associated with chronic myeloid leukemia risk and treatment response. Cancer Epidemiol. 2016;44:16–21. doi: 10.1016/j.canep.2016.07.008. doi: 10.1016/j.canep.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 19.Cowell IG, Dixon KH, Pemble SE, Ketterer B, Taylor JB. The structure of the human glutathione S-transferase pi gene. Biochem J. 1988;255:79–83. doi: 10.1042/bj2550079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hengstler JG, Arand M, Herrero ME, Oesch F. Polymorphisms of N-acetyltransferases, glutathione S-transferases, microsomal epoxide hydrolase and sulfotransferases: Influence on cancer susceptibility. Recent Results Cancer Res. 1998;154:47–85. doi: 10.1007/978-3-642-46870-4_4. [DOI] [PubMed] [Google Scholar]
- 21.Xiong DK, Chen HH, Ding XP, Zhang SH, Zhang JH. Association of polymorphisms in glutathione S-transferase genes (GSTM1, GSTT1, GSTP1) with idiopathic azoospermia or oligospermia in Sichuan, China. Asian J Androl. 2015;17:481–6. doi: 10.4103/1008-682X.143737. doi: 10.4103/1008-682X.143737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Luca A, Federici L, De Canio M, Stella L, Caccuri AM. New insights into the mechanism of JNK1 inhibition by glutathione transferase P1-1. Biochemistry. 2012;51:7304–12. doi: 10.1021/bi300559m. doi: 10.1021/bi300559m. [DOI] [PubMed] [Google Scholar]
- 23.Salimi S, Nakhaee A, Jafari M, Jahantigh D, Sandooghi M, Zakeri Z, et al. Combination effect of GSTM1, GSTT1 and GSTP1 polymorphisms and risk of systemic lupus erythematosus. Iran J Public Health. 2015;44:814–21. [PMC free article] [PubMed] [Google Scholar]
- 24.Vasieva O. The many faces of glutathione transferase pi. Curr Mol Med. 2011;11:129–39. doi: 10.2174/156652411794859278. doi: 10.2174/156652411794859278. [DOI] [PubMed] [Google Scholar]
- 25.Shen X, Wang J, Yan X, Ren X, Wang F, Chen X, et al. Predictive value of GSTP1 Ile105Val polymorphism in clinical outcomes of chemotherapy in gastric and colorectal cancers: A systematic review and meta-analysis. Cancer Chemother Pharmacol. 2016;77:1285–302. doi: 10.1007/s00280-016-3047-1. doi: 10.1007/s00280-016-3047-1. [DOI] [PubMed] [Google Scholar]
- 26.Lu J. GST Gene Polymorphisms and Idiopathic Male Factor Infertility (in Chinese) Jiangsu: Nanjing Medical University; 2013. [Google Scholar]
- 27.Zhou Q. Study on the Associations between the CYP1A1 and GST Gene Polymorphisms and Male Infertility (in Chinese) Jiangsu: Jiangsu University; 2016. [Google Scholar]
- 28.Feng ZQ, Jing ZA, Liu HY, Liao SS, Guo LJ, Mao CQ, et al. Association of SPOI1 and GST gene polymorphisms with idiopathic male infertility in ethnic Han Chinese (in Chinese) Chin J Med Genet. 2015;32:866–70. doi: 10.3760/cma.j.issn.1003-9406.2015.06.025. doi: 10.3760/cma.j.issn.1003-9406.2015.06.025. [DOI] [PubMed] [Google Scholar]
- 29.Li C, Ding XP, Fu L, Chen L. Association between glatathione-S-transferase gene polymorphisms (GSTM, GSTT, GSTPl) and idiopathic azoospermia (in Chinese) Chin J Med Genet. 2013;30:102–5. doi: 10.3760/cma.j.issn.1003-9406.2013.01.025. doi: 10.3760/cma.j.issn.1003-9406.2013.01.025. [DOI] [PubMed] [Google Scholar]
- 30.Yarosh SL, Kokhtenko EV, Churnosov MI, Solodilova MA, Polonikov AV. Joint effect of glutathione S-transferase genotypes and cigarette smoking on idiopathic male infertility. Andrologia. 2015;47:980–6. doi: 10.1111/and.12367. doi: 10.1111/and.12367. [DOI] [PubMed] [Google Scholar]
- 31.Safarinejad MR, Shafiei N, Safarinejad S. The association of glutathione-S-transferase gene polymorphisms (GSTM1, GSTT1, GSTP1) with idiopathic male infertility. J Hum Genet. 2010;55:565–70. doi: 10.1038/jhg.2010.59. doi: 10.1038/jhg.2010.59. [DOI] [PubMed] [Google Scholar]
- 32.Tang KF, Xu SY, Zou TJ, Ding SS, Liu RZ, Wang XY, et al. Correlative analysis of between glutathione S-transferase polymorphisms and idiopathic male infertility (in Chinese) Chin J Androl. 2014;28:3–7. [Google Scholar]
- 33.Lakpour N, Mirfeizollahi A, Farivar S, Akhondi MM, Hashemi SB, Amirjannati N, et al. The association of seminal plasma antioxidant levels and sperm chromatin status with genetic variants of GSTM1 and GSTP1 (Ile105Val and Ala114Val) in infertile men with oligoasthenoteratozoospermia. Dis Markers. 2013;34:205–10. doi: 10.3233/DMA-120954. doi: 10.3233/DMA-120954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moher D, Liberati A, Tetzlaff J, Altman DG PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6:e1000097. doi: 10.1371/journal.pmed.1000097. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wells GA, Shea B, O’Connell D, Peterson J, Welch V, Losos M, et al. The Newcastle–Ottawa Scale (NOS) for Assessing the Quality of Nonrandomised Studies in Meta-analyses. Ottawa Health Research Institute. [Last accessed on 2017 Jan 10]. Available from: http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp .
- 36.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–48. [PubMed] [Google Scholar]
- 37.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–88. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 38.Wang H, Sun F, Xing JP, Ding SS, Sun C, Wang XY, et al. Correlation between glutathione S-transferase polymorphisms and sperm DNA integrity in male patients with idiopathic infertile (in Chinese) J China Med Univ. 2015;44:1075–8. [Google Scholar]
- 39.Messaros BM, Rossano MG, Liu G, Diamond MP, Friderici K, Nummy-Jernigan K, et al. Negative effects of serum p, p’-DDE on sperm parameters and modification by genetic polymorphisms. Environ Res. 2009;109:457–64. doi: 10.1016/j.envres.2009.02.009. doi: 10.1016/j.envres.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 40.Sun L, Mipam TD, Zhao F, Liu W, Zhao W, Wu S, et al. Comparative testis proteome of cattleyak from different developmental stages. Animal. 2017;11:101–11. doi: 10.1017/S1751731116001191. doi: 10.1017/S1751731116001191. [DOI] [PubMed] [Google Scholar]
- 41.Tang K, Xue W, Xing Y, Xu S, Wu Q, Liu R, et al. Genetic polymorphisms of glutathione S-transferase M1, T1, and P1, and the assessment of oxidative damage in infertile men with varicoceles from northwestern China. J Androl. 2012;33:257–63. doi: 10.2164/jandrol.110.012468. doi: 10.2164/jandrol.110.012468. [DOI] [PubMed] [Google Scholar]
- 42.Safarinejad MR, Dadkhah F, Ali Asgari M, Hosseini SY, Kolahi AA, Iran-Pour E. Glutathione S-transferase polymorphisms (GSTM1, GSTT1, GSTP1) and male factor infertility risk: A pooled analysis of studies. Urol J. 2012;9:541–8. [PubMed] [Google Scholar]