Abstract
The Na+-dependent taurocholate cotransporting polypeptide (NTCP/SLC10A1) is a hepatocyte-specific solute carrier, which plays an important role in maintaining bile salt homeostasis in mammals. The absence of a hepatic Na+-dependent bile salt transport system in marine skate and rainbow trout raises a question regarding the function of the Slc10a1 gene in these species. Here, we have characterized the Slc10a1 gene in the marine skate, Leucoraja erinacea. The transcript of skate Slc10a1 (skSlc10a1) encodes 319 amino acids and shares 46% identity to human NTCP (hNTCP) with similar topology to mammalian NTCP. SkSlc10a1 mRNA was mostly confined to the brain and testes with minimal expression in the liver. An FXR-bile salt reporter assay indicated that skSlc10a1 transported taurocholic acid (TCA) and scymnol sulfate, but not as effectively as hNTCP. An [3H]TCA uptake assay revealed that skSlc10a1 functioned as a Na+-dependent transporter, but with low affinity for TCA (Km = 92.4 µM) and scymnol sulfate (Ki = 31 µM), compared with hNTCP (TCA, Km = 5.4 µM; Scymnol sulfate, Ki = 3.5 µM). In contrast, the bile salt concentration in skate plasma was 2 µM, similar to levels seen in mammals. Interestingly, skSlc10a1 demonstrated transport activity for the neurosteroids dehydroepiandrosterone sulfate and estrone-3-sulfate at physiological concentration, similar to hNTCP. Together, our findings indicate that skSlc10a1 is not a physiological bile salt transporter, providing a molecular explanation for the absence of a hepatic Na+-dependent bile salt uptake system in skate. We speculate that Slc10a1 is a neurosteroid transporter in skate that gained its substrate specificity for bile salts later in vertebrate evolution.
Keywords: sodium-dependent transporter, solute carrier, evolution, neuroactive steroid
a sodium (na+)-dependent bile salt transporter plays an essential role in hepatic uptake of bile acids in humans and rodents, where the functional determinant is the Na+-dependent taurocholate-cotransporting polypeptide (NTCP/SLC10A1) (2, 9, 19, 24). NTCP is a member of solute carrier family 10 A (SLC10A). There are seven members in this gene family in humans and rodents (3). Because three of them transport bile salts, this family is also called sodium-bile salt cotransporters (1, 3). NTCP/SLC10A1 is predominantly expressed in hepatocytes and is localized at the basolateral membrane, where it functions to clear conjugated bile salts from blood to the liver in humans and rodents (10, 19, 22). After entering hepatocytes, bile salts are excreted into bile canalicular lumen by another hepatocyte-specific transporter, the bile salt export pump (BSEP/ABCB11), which maintains the forward flow of bile (7). Dysfunction of NTCP/SLC10A1 or BSEP results in elevated levels of serum bile salts in humans and rodents, emphasizing the functional importance of this pair of transporters in maintaining bile salt homeostasis (23, 24, 26). In vitro studies indicate that NTCP can also transport some steroids and thyroid hormones and drugs, including estrone-3-sulfate, dehydroepiandrosterone sulfate (DHEAS), thyroxine, and statins (1, 18). However, very little is known about its orthologs in nonmammalian vertebrate species. In fact, previous functional studies indicate that a Na+-dependent bile salt transport system is absent in the marine skate (Leucoraja erinacea) and rainbow trout (4, 5, 15), two representative species for cartilaginous fish and teleost, respectively. These findings provide support for the notion that an NTCP-based bile salt uptake system is not present in early vertebrate evolution and suggest that Slc10a1 gene either does not exist or has not evolved as a bile salt transporter in these species.
Very recently, Murashita et al. (14) identified an Slc10a1 gene in rainbow trout and found that its expression was most abundant in the brain. However, it is not known whether this protein can transport bile salts. In addition, Ensembl genome annotations suggest that the Slc10a1 gene has evolved in evolutionarily primitive vertebrates, e.g., the sea lamprey. Our in silico analysis of unassembled skate genome also suggests that there is a Slc10a1 gene in this species. However, its transcript has not been identified, nor do we know whether this gene encodes a functional bile salt transporter.
In this report, we have characterized the Slc10a1 gene in the marine skate (Leucoraja erinacea) and examined its tissue expression and substrate specificity. Our data indicate that skate Slc10a1 (skSlc10a1) is not a physiological bile salt transporter, suggesting that it may rather be a neurosteroid transporter.
MATERIALS AND METHODS
Chemicals.
All chemicals were purchased from Sigma-Aldrich (St. Louis, MO), unless stated otherwise. [3H]taurocholic acid ([3H]TCA, 5.0 Ci/mmol), [3H]DHEAS (50 Ci/mmol), and [3H]estrone-3-sulfate (57.3 Ci/mmol) were purchased from PerkinElmer (Waltham, MA). Oligonucleotide synthesis and DNA sequencing services were provided by the Keck Biotechnology Resource Laboratory at Yale University. Scymnol sulfate was kindly provided by Dr. Lee Hagey (University of California, San Diego).
Animal tissue collection, RNA extraction, and quantification.
All animal-related experiments were approved by the local Institutional Animal Care and Use Committee. Male skates were caught in June–July 2010 from Gulf of Maine, and the tissues were collected at the Mount Desert Island Biological Laboratory in Salisbury Cove, ME, and stored in −80°C freezer, as described in our previous report (13). Mouse tissues were sampled from C57BL/6 males purchased from Jackson Laboratories (Bar Harbor, ME). Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA) and was purified using a kit (RNeasy clean-up kit, Qiagen, Valencia, CA). Two micrograms of total RNA from each sample was reverse transcribed into cDNA using a kit from Agilent (Santa Clara, CA). Real-time quantitative PCR was performed on a Roche LightCycler 480 Systems (Roche, Indianapolis, IN). The specific primers and probes are listed in Table 1. mRNA expression was normalized to 2 µg of total RNA. Results were expressed in copy number, where the cloned constructs or purified PCR fragment were used to establish a standard curve.
Table 1.
Oligonucleotide sequences used for molecular cloning and real-time RT-PCR detection of Ntcp/Slc10a1, Bsep (Abcb11), ActB (β-actin) genes from skate and mouse
| Name | Sequence (5′→3′) | Purpose | Gene |
|---|---|---|---|
| skSlc10a1-F9 | CGATGATCTCCTTGGGCTGCACC | Cloning | skate Slc10a1 |
| skSlc10a1-R10 | GAGCACCAATCTCCTCTCTGCAG | Cloning | skate Slc10a1 |
| skSlc10a1-F25 | GCATGGATCCTGGAGGGCATTAGCTTCTTGGCT | Cloning | skate Slc10a1 |
| skSlc10a1-R26 | AATGGGCATCAGTGCTGTGGTGA | Cloning | skate Slc10a1 |
| skSlc10a1-R29 | TCACCTTCTGCTTCGATAGATCAC | Cloning | skate Slc10a1 |
| skSlc10a1-F11 | GGACACTTTTCTCTCCTCACCTAATT | Cloning and Q-PCR | skate Slc10a1 |
| skSlc10a1-R13 | CGCAAGAGCATATCCTGAAGTGTA | Cloning and Q-PCR | skate Slc10a1 |
| skSlc10a1-P12 | Fam-TCACCACAGCACTGATGCCCATTATTG-BHQ | Q-PCR | skate Slc10a1 |
| skBsep-3854F | TTGCCATCGCACGAGCCAT | Q-PCR | skate Bsep |
| skBsep-3920R | AGCGCAGACGTGGCCTCAT | Q-PCR | skate Bsep |
| skBsep-3875P | Fam-TCCGTGACCCAAAGATCCTGCTCCTG-BHQ | Q-PCR | skate Bsep |
| skActB-564F | GAGTCACACTGTGCCCATCTACG | Q-PCR | skate ActB |
| skActB-630R | AGCCAGGTCCAGACGCAGG | Q-PCR | skate ActB |
| skActB-588P | Hex-AGGCTACGCCTTGCCACACGCC-BHQ | Q-PCR | skate ActB |
| mBsep TaqMan PP | Purchased from Life Technologies, Cat no. Mm00445168_m1 | Q-PCR | mouse Bsep |
| mNtcp TaqMan PP | Purchased from Life Technologies, Cat no. Mm00441421_m1 | Q-PCR | mouse Ntcp |
| mActB TaqMan PP | Purchased from Life Technologies, Cat no. Mm00607939_s1 | Q-PCR | mouse ActB |
Q-PCR, quantitative PCR; sk, skate; m, mouse.
Molecular cloning.
To clone skSlc10a1, we first retrieved sequence fragments from the skate genome by ortholog searching using human NTCP (hNTCP) protein sequence as a query. We then designed primers and amplified a 600-bp fragment from skate liver using RT-PCR. DNA sequencing and phylogenetic analysis confirmed that this fragment encoded a portion of skSlc10a1. A full-length skSlc10a1 was obtained by 5′- and 3′-RACE PCR using a kit from Clontech (Mountain View, CA). To functionally characterize skSlc10a1, its full-length coding region was subcloned into pcDNA3.1 vector. At least six clones were sequenced, and one clone with identical sequence to the skate genome or original RACE-PCR products was selected for further experiments. The primers are listed in Table 1.
Skate plasma total bile salt assay.
Total bile salt levels in skate plasma was assayed using an enzymatic reaction-based kit from Diazyme Laboratories (Poway, CA). It measures the concentration of total 3α-hydroxy bile salts, in which scymnol sulfate, the major endogenous bile salt in skate, was used for establishing a standard curve.
Phylogenetic analysis.
Phylogeny was inferred with Bayesian Markov Chain Monte Carlo (MCMC) analysis using MrBayes v3.2.0 software, as described previously (13). Briefly, protein sequences were obtained from NCBI and the Protein Data Bank. Sequence alignment was performed with Clustal Omega at standard settings on the EBI website (http://www.ebi.ac.uk/). The alignment was run in the MCMC analysis for 20,000 generations, with three heated chains and a single cold chain. Posterior probabilities were determined by sampling every 100 generations, where the initial 50 samples were discarded as “burn-in.”
Huh7 cell base [3H]TCA uptake assay.
Huh7 cell line was a gift from Dr. Yun-Chi Cheng’s laboratory (Yale University), which was originally acquired from Japanese Collection of Research Bioresources (Tokyo, Japan). The cells were maintained in monolayer culture at 37°C in an atmosphere of 5% CO2 in growth medium, DMEM with 10% FBS, 50 U/ml penicillin, and 50 µg/ml streptomycin, all from Invitrogen. Transfection was performed using Lipofectamine 2000 (Invitrogen) by following the manufacturer’s instruction. For uptake assays, cells (2 × 105/well) were seeded in 12-well plates 24 h before transfection. Either pcDNA3.1 (control), pcDNA3.1-hNTCP, or pcDNA3.1-skSlc10a1 was transfected into Huh7 cells. Twenty-four hours after transfection, cells were subjected to radioisotope-labeled substrate uptake assays, as previous described (17). All data presented in this report were from 10-min uptakes in a 37°C water bath, because a time-course experiment indicates that skSlc10a1 transport activity remains in the linear range for up to 10 min of incubation with substrate in these transfected Huh7 cells (data not shown). Transport activity was normalized to total cell protein. Uptake activity is presented as pmol TCA·mg protein−1·min−1. Kinetic constants are expressed as values ± SD and were calculated by nonlinear fitting of data to the Michaelis-Menten equation using least squares (GraphPad Prism 7, GraphPad Software).
Slc10a1-farnesoid X receptor (FXR/NR1H4) luciferase reporter assay for bile salt transport.
A dual-luciferase gene reporter assay (Promega, Madison, MI) was utilized to determine whether conjugated bile salts can be transported into cells by SLC10A1/Slc10a1, because intracellular bile salt can activate FXR and stimulate the expression of its target genes in a dose-dependent manner (13). Huh7 cells were maintained in growth medium. When cells reached 80% confluence, the culture medium was changed to DMEM supplemented with 0.5% charcoal-stripped FBS and cotransfected with 50 ng pcDNA3.1 (control) or 50 ng hNTCP or 50 ng skSlc10a1 along with 75 ng FXR, 50 ng RXR, 250 ng pGL3-hIBABP, and 10 ng Renilla using 3 μl Lipofectamine 2000 for each well in 24-well plates. Twenty-four hours after transfection, cells were treated with bile salts for an additional 24 h in 0.5% charcoal-stripped FBS DMEM. Passive lysis buffer (Promega) was used to prepare cell lysate, and luminescence was detected in a Synergy2 Microplate Reader (BioTek). Firefly luciferase readings were normalized to Renilla luciferase, the internal control.
Statistical analysis.
Statistical analysis was performed with GraphPad Prism 7 (GraphPad software). All of the data were presented as the means ± SD. Paired two-tailed t-test was used to detect differences between two groups, with the level of significance set at 0.05.
RESULTS
Identification of skate Slc10a1.
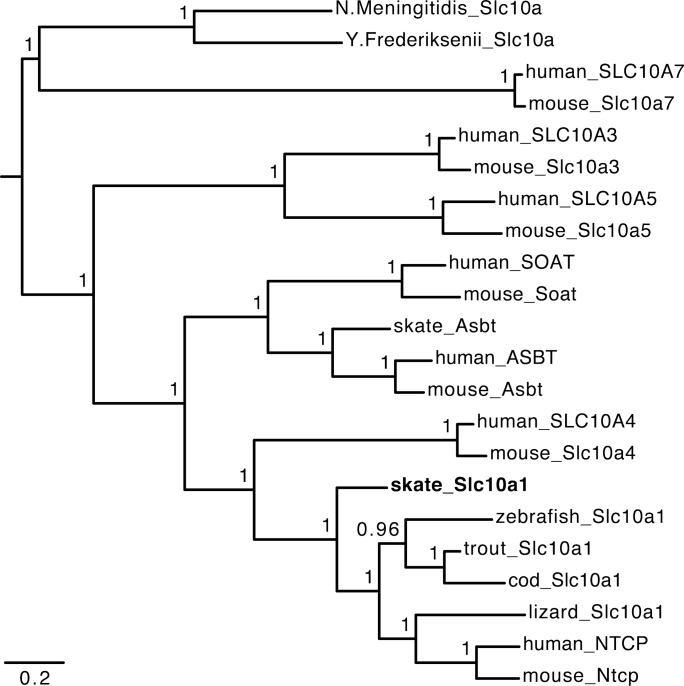
Two splicing isoforms of Slc10a1 were identified in the skate. The first one encodes 319 amino acids (GenBank accession no. KY245892). GenBank blasting revealed that it is most similar to NTCP/SLC10A1 from rainbow trout, humans, and rodents. Phylogenetic analysis confirmed that this protein is a Slc10a1 ortholog (Fig. 1). Therefore, we named it skate Slc10a1 (skSlc10a1). We also identified an alternative splicing isoform of skSlc10a1 gene (GenBank accession no.: KY245893), which contains multiple stop codons in the middle section of the transcript and is unlikely to encode any protein. Protein sequence alignment indicates that skSlc10a1 shares 46% identity to hNTCP and 56% identity to rainbow trout Ntcp-like (Slc10a1) protein. These amino acid sequence identities are much higher than two recently characterized bacterial Slc10a-like proteins, i.e., NmSlc10a (Protein ID: 3ZUY) and YfSlc10a (Protein ID: 4N7W), which only share ~22% identity to hNTCP and skSlc10a1. Interestingly, the residues directly binding Na+ in bacterial Slc10a are all conserved in skSlc10a1, as in other members of the vertebrate Slc10a1 family (Fig. 2), suggesting that skSlc10a1 is a Na+-dependent transporter. Protein structure predictions suggest that skSlc10a1 has nine transmembrane domains with its NH2 terminus having a glycosylation site (23NTIL) that is localized outside of cell plasma membrane, as is the predicted structure of NTCP/Ntcp in humans and rodents (Fig. 2).
Fig. 1.
Phylogeny of the Slc10a family shows that skate slc10a1 is the most primitive members of the NTCP/SLC10A1 subfamily. Phylogeny was inferred using Bayesian MCMC analysis. Posterior probabilities are indicated at nodes. Branch length is expressed as the number of expected substitutions per site. Protein names and accession numbers are human_NTCP, NP_003040; mouse_Ntcp, NP_001171032; lizard_Slc10a1, XP_016851911; zebrafish_Slc10a1, XP_690745; trout_Slc10a1, BAN81910; cod_Slc10a1, ENSGMOT00000017534; skate_Slc10a1, KY245892; human_ASBT, NP_000443; mouse_Asbt, NP_035518; skate_Asbt, AFM29856; human_SLC10A3, NP_062822; mouse_Slc10a3, NP_663381; human_SLC10A4, NP_689892; mouse_Slc10a4, NP_775579; human_SLC10A5, NP_001010893; mouse_Slc10a5, NP_001010834; human_SOAT, NP_932069; mouse_Soat, NP_083691; human_SLC10A7, NP_001025169; mouse_Slc10a7, NP_084012; N. Meningitidis_Slc10a, 3ZUY_A; Y. Frederiksenii_Slc10a, 4N7W.
Fig. 2.
Amino acid sequence alignment of SLC10A family members. The predicted nine transmembrane domains based on bacterial Slc10a-like genes are indicated in gray bars. Residues that directly interact with the two Na+ ions are highlighted. Residues whose mutations affect human NTCP binding activity with taurocholic acid appear in bold and are underlined (11). HuNTCP, human NTCP(NP_003040); RaNtcp, rat Ntcp(NP_058743); RtSlc10a1, rainbow trout Slc10a1(BAN81910); NmSlc10a, N. meningitidis Slc10a(3ZUY_A); YfSlc10a, Y. frederiksenii Slc10a(4N7W) are shown.
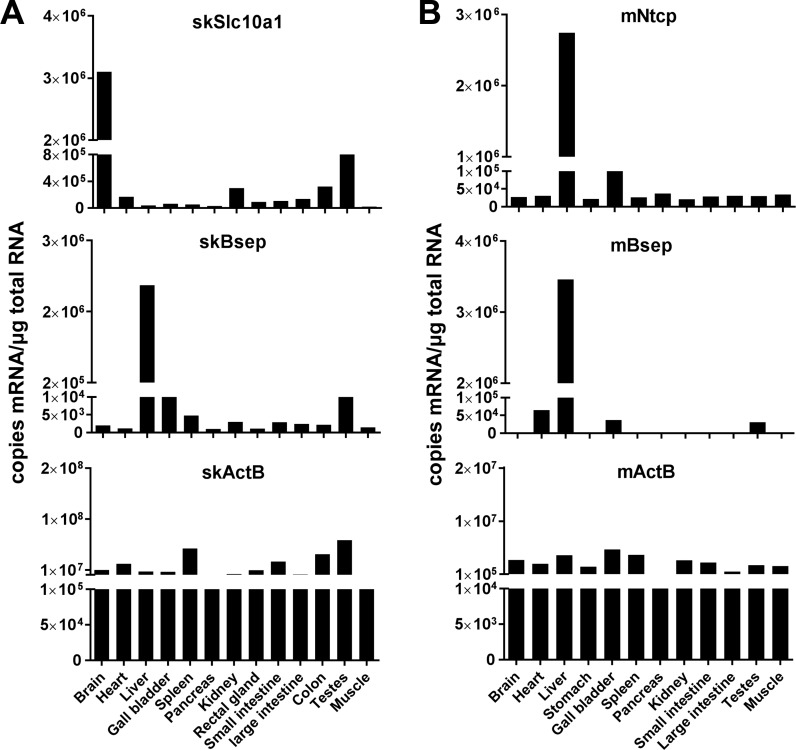
SkSlc10a1 is primarily expressed in the brain and testes but not the liver.
To check whether the tissue distribution of Slc10a1 in skate is the same as its mammalian orthologs, i.e., liver specific, we performed quantitative real-time PCR to assess its mRNA expression in a variety of skate tissues. In parallel, β-actin mRNA levels were quantified to ensure RNA integrity (Fig. 3). To our surprise, skSlc10a1 was most abundant in the brain and testes, while its expression was relatively low in the liver and other tissues (Fig. 3A). This expression pattern is similar to Slc10a1 in rainbow trout but strikingly different from NTCP/SLC10A1 in humans and mice (10, 14, 22) (Fig. 3B). In contrast, the expression of Bsep/Abcb11 was liver specific in both skate and mouse, as is mouse Ntcp (Fig. 3). The low expression of skSlc10a1 in the liver raises the question of whether this gene is a bile salt transporter in skate.
Fig. 3.
mRNA tissue distribution of Slc10a1, and Bsep in skate and mouse. skBsep, mNtcp, and mBsep appeared to be liver-specific genes, whereas skSlc10a1 was abundantly expressed in the brain and testes with low levels in the liver. β-Actin (ActB) served as a control. Expression below 1,000 copies of mRNA per microgram of total RNA was considered background. All values are expressed as means from triplicate measurements.
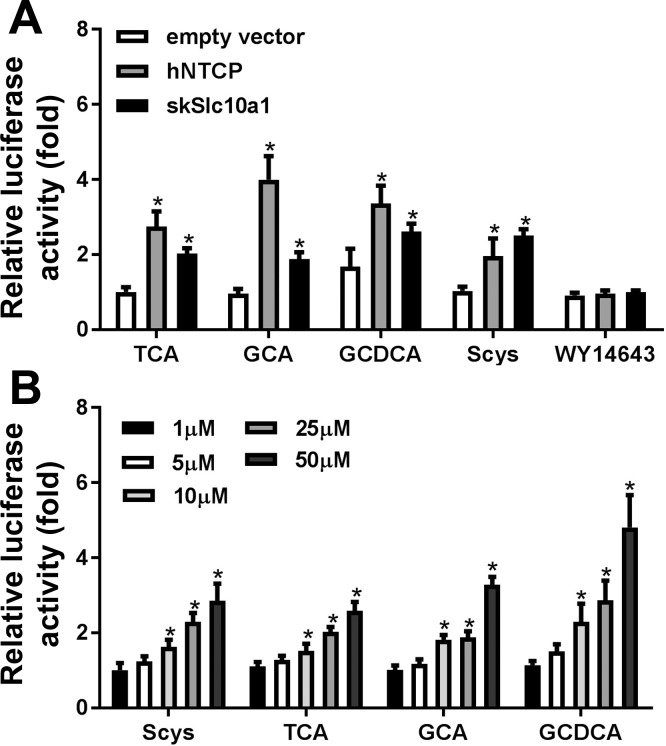
SkSlc10a1 can transport TCA and scymnol sulfate in an FXR-bile acid reporter assay.
To test whether skSlc10a1 can transport bile salts, we first performed an FXR-bile salt reporter assay in transfected Huh7 cells. As demonstrated in Fig. 4A, cotransfection of skSlc10a1 significantly increased FXR activity when cells were treated with 25 µM bile salts but not WY14643 (a PPARα agonist). This indicates that skSlc10a1 can transport conjugated bile salts across cell membranes, although it is much less effective than hNTCP. To assess skSlc10a1’s transport efficiency for bile salts, we treated transfected Huh7 cells with different concentrations of bile salts. As shown in Fig. 4B, to activate FXR, at least 10 µM of bile salt was needed. In contrast, 5 µM of TCA or scymnol sulfate significantly induced FXR activity by 2.5-fold in hNTCP-transfected cells, suggesting that the affinity of skSlc10a1 for bile salts is low.
Fig. 4.
NTCP-FXR luciferase reporter assay in cotransfected Huh7 cells. Cells were cotransfected with FXR reporter constructs plus either empty vector, hNTCP or skSlc10a1 for 24 h and subjected to bile salt treatment for an additional 24 h. A: cells transfected with empty vector, hNTCP, or skSlc10a1 were treated with 25 µM TCA, GCA, GCDCA, scymnol sulfate (Scys), or WY14643. B: cells transfected with skSlc10a1 were treated with five different concentrations of Scys, TCA, GCA, or GCDCA. Luciferase readings of this analysis are relative to cells transfected with pcDNA3.1 empty vector control. *P < 0.05 vs. medium control. All values represent at least three independent experiments and are expressed as means ± SD.
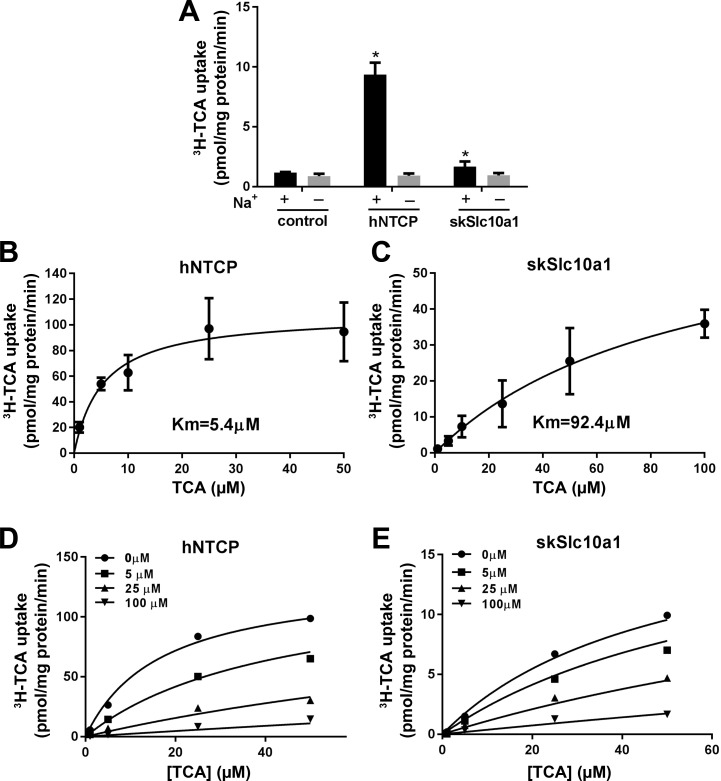
SkSlc10a1 is a Na+-dependent transporter but with very low affinity for bile salts.
To determine whether the transport activity of skSlc10a1 for bile salt is Na+-dependent, and also to assess the affinity of skSlc10a1 for bile salts, we carried out an uptake assay using [3H]TCA in transfected Huh7 cells. In this experiment, both hNTCP and skSlc10a1 again demonstrated transport activity for TCA. However, when Na+ was replaced by choline, the transport activities of both transporters were abolished (Fig. 5A), indicating that they are Na+-dependent transporters. When different concentrations of TCA were assayed in these uptake experiments, the results confirmed that hNTCP has high affinity for TCA with a Km = 5.4 µM (Fig. 5B), a value comparable to previous reports (10). In contrast, the Km of skSlc10a1 for TCA was 92.4 µM, a concentration that is more than 10-fold above normal physiological levels of serum bile salt seen in humans and rodents (Fig. 5C). Since the major bile salt in skate is scymnol sulfate, we assessed whether scymnol sulfate can effectively compete with 3H-TCA. This experiment yielded a Ki of 31 µM for scymnol sulfate (Fig. 5E), indicating that skSlc10a1 has a similarly low affinity for the endogenous skate bile salt scymnol sulfate as for TCA. However, skSlc10a1 may still be a physiological bile salt transporter if the plasma bile salt concentrations were sufficiently high. Because there is no published data on the level of serum bile salts in skate, we assayed the concentration of total bile salt in skate plasma, which was 2 ± 2 µM (n = 6), a value comparable to that seen in other vertebrates. These findings are consistent with the conclusion that skSlc10a1 is not an effective bile salt transporter in skate.
Fig. 5.
[3H]taurocholic acid (TCA) uptake assay in transfected Huh7 cells. A: uptake of 10 µM [3H]TCA in cells transiently transfected with empty vector (control), hNTCP, or skSlc10a1. Cells were incubated for 10 min in medium containing Na+ (+) or medium where Na+ was substituted with choline (–). *P < 0.05 vs. choline. Kinetics of [3H]TCA uptake in cells transfected with hNTCP (B) or skSlc10a1 (C). Cells were incubated for 10 min, and background uptake levels derived from cells transfected with vector control were subtracted. Data were normalized to total cell protein. Kinetic parameters were obtained by nonlinear curve fitting using the Michaelis-Menten equation. Competition assays for uptake of [3H]TCA and scymnol sulfate in hNTCP (D) and skSlc10a1 (E) in transfected Huh7 cells. Cells were incubated with indicated concentrations of TCA and scymnol sulfate for 10 min. The calculated Ki of scymnol sulfate for hNTCP and skSlc10a1 are 3.5 ± 0.57 µM (mean ± SD) and 31 ± 5 µM, respectively from three independent experiments. A representative plot is presented.
SkSlc10a1 transports neuroactive steroids at physiological concentration.
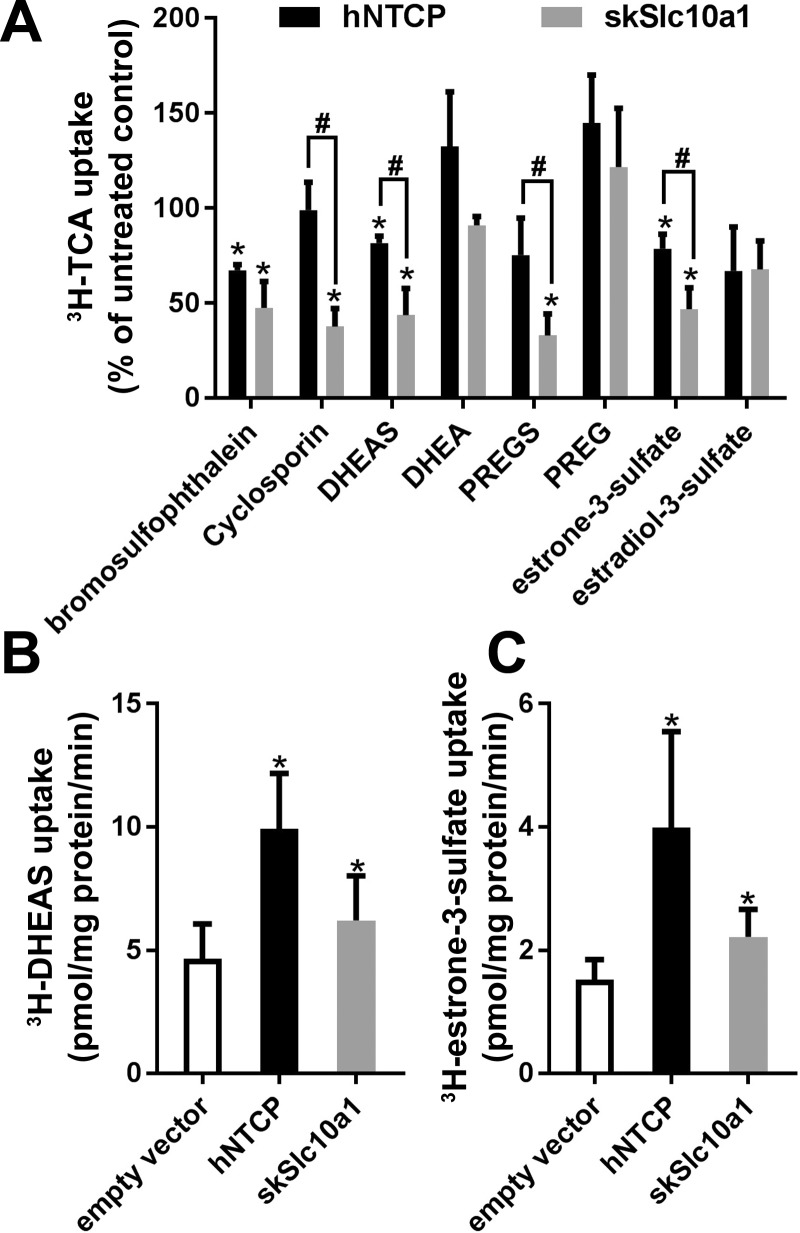
Because skSlc10a1 was abundantly expressed in the brain and testes and also because mammalian NTCP/Ntcps are capable of transporting certain steroids (6, 17, 18, 25), we asked whether steroids could be substrates of skSlc10a1. To address this question, we initially tested several neuroactive steroids using a competitive uptake assay. As demonstrated in Fig. 6A, 2 µM [a physiologically relevant concentration (16)] of DHEAS, pregnenolone sulfate (PREGS), or estrone-3-sulfate significantly reduced skSlc10a1 uptake of [3H]TCA by 55%, 65%, and 52%, respectively. DHEAS and estrone-3-sulfate also significantly reduced hNTCP uptake of TCA but to a lesser extent. In contrast, even at 10 µM concentration, the unconjugated form of these neurosteroids, dehydroepiandrosterone (DHEA), and pregnenolone (PREG) did not compete with TCA uptake by skSlc10a1 or hNTCP. As controls, bromosulfophthalein (2 µM) inhibited the uptake of TCA by both skSlc10a1 and hNTCP, whereas 1 µM cyclosporine A only inhibited skSlc10a1 (Fig. 6A).
Fig. 6.
A: competition assay for [3H]TCA transport. Huh7 cells transfected with skSlc10a1, hNTCP, or empty vector were incubated for 10 min in Na+ medium supplemented with 10 µM [3H]TCA with or without competitor. Cells were treated with 1 µM cyclosporin A, 10 µM dehydroepiandrosterone (DHEA), 10 µM pregnenolone (PREG), and all the other competitors were 2 µM. Background levels derived from cells transfected with vector control were subtracted, and uptake measurements were normalized to protein concentration. Transport activity from a noncompetitor control was set at 100%. All values represent at least three independent experiments and are expressed as means ± SD. *P < 0.05, noncompetitor control vs. competitor; #P < 0.05, skSlc10a1 vs. hNTCP; n ≥ 3. Uptake assays for [3H]DHEAS (B) and [3H]estrone-3-sulfate (C) in Huh7 cells transfected with expression vector for hNTCP, skSlc10a1, or empty vector were incubated for 10 min in Na+ medium with 0.5 µM DHEAS or estrone-3-sulfate. Values are expressed as means ± SD; n = 3. *P < 0.05, compared with empty vector in paired analysis.
To further confirm that DHEAS and estrone-3-sulfate are substrates of skSlc10a1, we performed direct uptake assay in transfected Huh7 cells using radioactive isotope-labeled compounds. As shown in Fig. 6, B and C, both skSlc10a1 and hNTCP demonstrated significant higher transport activity for DHEAS and estrone-3-sulfate than vector control even at 0.5 µM concentrations.
DISCUSSION
NTCP/SLC10A1 has been characterized as a Na+-dependent hepatocyte-specific bile salt transporter in humans and rodents and plays an important role in maintaining bile salt homeostasis. Since the enterohepatic circulation of bile salts has evolved in all vertebrates, one might expect that Slc10a1 would have evolved as a conserved hepatic Na+-dependent bile salt transporter as well. However, our molecular characterization of skSlc10a1 revealed that 1) skSlc10a1 transcript was most abundant in the brain and testes, with minimal expression in the liver, in striking contrast to human and rodent NTCP/Ntcp/Slc10a1 (Fig. 3) (10, 22); 2) although skSlc10a1 demonstrated Na+-dependent transport activity for TCA and scymnol sulfate in both the [3H]TCA uptake assay and FXR-bile salt-reporter assay (Figs. 4 and 5), its affinity for these bile salts was very low (hNTCP: Km = 5.4 µM for TCA vs. skSlc10a1: Km = 92.4 µM for TCA; scymnol sulfate Ki = 31 µM for skSlc10a1); and 3) the physiological bile salt concentration in skate plasma is ~2 µM, as seen in other vertebrates, which is much lower than the concentration that skSlc10a1 would require for an efficient uptake. Therefore, we conclude that Slc10a1 is not a physiological bile salt transporter in the marine skate. These findings provide a molecular explanation for prior physiological studies that failed to demonstrate a Na+-dependent hepatic bile salt transport system in isolated skate hepatocytes and perfused skate liver preparations (4, 5, 20). We speculate that the SLC10A1/Slc10a1 transporter gains substrate specificity for bile salts later in vertebrate evolution.
Consistent with previous reports (17, 18), we confirmed that hNTCP transported estrone-3-sulfate and DHEAS (Fig. 6). Most importantly, we demonstrated that skSlc10a1 transport these neurosteroid conjugates at physiological relevant concentrations. Therefore, we propose that neurosteroid conjugates are evolutionarily conserved endogenous substrates for the SLC10A1/Slc10a1 transporter. This speculation is also supported by its tissue distribution in skate and rainbow trout, where it is mainly expressed in the brain and testis (14) (Fig. 2). It is known that these neurosteroids play an important role in brain and sex organs in regulating the expression of genes involved in development, physiology, and sex behavior. However, SLC10A1/Slc10a1 in humans and rodents appears to be expressed primarily in the liver. It is possible that the functional role of SLC10A1 in the brain in these species has been substituted by the later evolution of two paralog genes, i.e., SLC10A4 and SLC10A6, as they are abundantly expressed in brain and also transport neurosteroid conjugates (8, 21). SLC10A1 has then further evolved, shifting its tissue distribution from brain to liver and gaining its substrate specificity for bile salts, which are also steroid-like metabolites of cholesterol. Future studies may explore these speculations.
Sequence alignment of SLC10A1 members demonstrate that the Na+-binding sites identified in NmSlc10a and YfSlc10a in bacteria are all conserved among these SLC10A proteins (Fig. 2), indicating that they all are Na+-dependent transporters. This was experimentally confirmed in the [3H]TCA uptake assay, as replacing Na+ with choline abolished skSlc10a1 transport activity (Fig. 5A). However, although skSlc10a1 shares 46% identity to human NTCP, its affinity for TCA was very low compared with hNTCP (92.4 µM for skate vs. 5.4 µM for human). In contrast, both NmSlc10a and YfSlc10a only share 22% identity to hNTCP, but demonstrate a higher affinity for TCA (Km = 50 µM) than skSlc10a1 (12, 27). Yet the residues interacting with TCA in NmSlc10a and YfSlc10a are conserved between hNTCP and skSlc10a1, raising questions as to whether the substrate-binding residues identified in NmSlc10a and YfSlc10a represent how bile salts interact with mammalian NTCP. It is also possible that other residues may effect binding of TCA. This speculation is supported by the findings for a human NTCP variant, i.e., S267F that was identified and characterized in vitro (11). Although this residue is conserved between skSlc10a1 and hNTCP, the human S267F variant reduced the transport of TCA but not estrone-3-sulfate (11). Future studies of additional mutants or X-ray crystal structures of SLC10A family members are needed to resolve this issue.
Perspectives and Significance
Although it is well accepted that NTCP/SLC10A1 is a liver-specific bile salt transporter in humans and rodents, characterization of its ortholog in skate indicates that skSlc10a1 is unlikely a physiological bile salt transporter in evolutionarily primitive vertebrates. Rather, Slc10a1 is most likely a neurosteroid conjugate transporter in these species, indicating that substrate specificity for bile salts and tissue specificity for the liver occurred later in vertebrate evolution. Given the difference in bile salt hepatic clearance between lower vertebrates and mammals, we can also speculate that bile salt may have different functional roles in vertebrate evolution. Characterization of Slc10a1 from additional species could shed light on these questions.
GRANTS
This study was supported in part by National Institutes of Health Grants DK-34989 (Yale Liver Center) and DK-25636 (to J. L. Boyer) and by the Ensign Endowment Fund (to J. L. Boyer).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
D.Y., H.Z., D.A.L., and S.-Y.C. performed experiments; D.Y., H.Z., D.A.L., and S.-Y.C. analyzed data; D.Y., D.A.L., and S.-Y.C. prepared figures; D.Y. and S.-Y.C. drafted manuscript; D.Y., H.Z., D.A.L., J.L.B., and S.-Y.C. approved final version of manuscript; D.A.L., J.L.B., and S.-Y.C. edited and revised manuscript; J.L.B. and S.-Y.C. interpreted results of experiments; S.-Y.C. conceived and designed research.
ACKNOWLEDGMENTS
Present addresses: H. Zhang, Dept. of Dermatology, Peking University People's Hospital, Beijing, 100044, China; D. A. Lionarons, Oncogene Biology Laboratory, The Francis Crick Institute, London WC2A 3LY, United Kingdom.
REFERENCES
- 1.Anwer MS, Stieger B. Sodium-dependent bile salt transporters of the SLC10A transporter family: more than solute transporters. Pflügers Arch 466: 77–89, 2014. doi: 10.1007/s00424-013-1367-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyer JL. Bile formation and secretion. Compr Physiol 3: 1035–1078, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Döring B, Lütteke T, Geyer J, Petzinger E. The SLC10 carrier family: transport functions and molecular structure. Curr Top Membr 70: 105–168, 2012. doi: 10.1016/B978-0-12-394316-3.00004-1. [DOI] [PubMed] [Google Scholar]
- 4.Fricker G, Dubost V, Finsterwald K, Boyer JL. Characteristics of bile salt uptake into skate hepatocytes. Biochem J 299: 665–670, 1994. doi: 10.1042/bj2990665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fricker G, Hugentobler G, Meier PJ, Kurz G, Boyer JL. Identification of a single sinusoidal bile salt uptake system in skate liver. Am J Physiol Gastrointest Liver Physiol 253: G816–G822, 1987. [DOI] [PubMed] [Google Scholar]
- 6.Friesema EC, Docter R, Moerings EP, Stieger B, Hagenbuch B, Meier PJ, Krenning EP, Hennemann G, Visser TJ. Identification of thyroid hormone transporters. Biochem Biophys Res Commun 254: 497–501, 1999. doi: 10.1006/bbrc.1998.9974. [DOI] [PubMed] [Google Scholar]
- 7.Gerloff T, Stieger B, Hagenbuch B, Madon J, Landmann L, Roth J, Hofmann AF, Meier PJ. The sister of P-glycoprotein represents the canalicular bile salt export pump of mammalian liver. J Biol Chem 273: 10046–10050, 1998. doi: 10.1074/jbc.273.16.10046. [DOI] [PubMed] [Google Scholar]
- 8.Geyer J, Döring B, Meerkamp K, Ugele B, Bakhiya N, Fernandes CF, Godoy JR, Glatt H, Petzinger E. Cloning and functional characterization of human sodium-dependent organic anion transporter (SLC10A6). J Biol Chem 282: 19728–19741, 2007. doi: 10.1074/jbc.M702663200. [DOI] [PubMed] [Google Scholar]
- 9.Hagenbuch B, Dawson P. The sodium bile salt cotransport family SLC10. Pflügers Arch 447: 566–570, 2004. doi: 10.1007/s00424-003-1130-z. [DOI] [PubMed] [Google Scholar]
- 10.Hagenbuch B, Meier PJ. Molecular cloning, chromosomal localization, and functional characterization of a human liver Na+/bile acid cotransporter. J Clin Invest 93: 1326–1331, 1994. doi: 10.1172/JCI117091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ho RH, Leake BF, Roberts RL, Lee W, Kim RB. Ethnicity-dependent polymorphism in Na+-taurocholate cotransporting polypeptide (SLC10A1) reveals a domain critical for bile acid substrate recognition. J Biol Chem 279: 7213–7222, 2004. doi: 10.1074/jbc.M305782200. [DOI] [PubMed] [Google Scholar]
- 12.Hu NJ, Iwata S, Cameron AD, Drew D. Crystal structure of a bacterial homologue of the bile acid sodium symporter ASBT. Nature 478: 408–411, 2011. doi: 10.1038/nature10450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lionarons DA, Boyer JL, Cai SY. Evolution of substrate specificity for the bile salt transporter ASBT (SLC10A2). J Lipid Res 53: 1535–1542, 2012. doi: 10.1194/jlr.M025726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murashita K, Yoshiura Y, Chisada S, Furuita H, Sugita T, Matsunari H, Iwashita Y, Yamamoto T. Homologue gene of bile acid transporters ntcp, asbt, and ost-α in rainbow trout Oncorhynchus mykiss: tissue expression, effect of fasting, and response to bile acid administration. Fish Physiol Biochem 40: 511–525, 2014. doi: 10.1007/s10695-013-9862-y. [DOI] [PubMed] [Google Scholar]
- 15.Råbergh CM, Ziegler K, Isomaa B, Lipsky MM, Eriksson JE. Uptake of taurocholic acid and cholic acid in isolated hepatocytes from rainbow trout. Am J Physiol Gastrointest Liver Physiol 267: G380–G386, 1994. [DOI] [PubMed] [Google Scholar]
- 16.Rainey WE, Nakamura Y. Regulation of the adrenal androgen biosynthesis. J Steroid Biochem Mol Biol 108: 281–286, 2008. doi: 10.1016/j.jsbmb.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schroeder A, Eckhardt U, Stieger B, Tynes R, Schteingart CD, Hofmann AF, Meier PJ, Hagenbuch B. Substrate specificity of the rat liver Na+-bile salt cotransporter in Xenopus laevis oocytes and in CHO cells. Am J Physiol Gastrointest Liver Physiol 274: G370–G375, 1998. [DOI] [PubMed] [Google Scholar]
- 18.Schweigmann H, Sánchez-Guijo A, Ugele B, Hartmann K, Hartmann MF, Bergmann M, Pfarrer C, Döring B, Wudy SA, Petzinger E, Geyer J, Grosser G. Transport of the placental estriol precursor 16α-hydroxy-dehydroepiandrosterone sulfate (16α-OH-DHEAS) by stably transfected OAT4-, SOAT-, and NTCP-HEK293 cells. J Steroid Biochem Mol Biol 143: 259–265, 2014. doi: 10.1016/j.jsbmb.2014.03.013. [DOI] [PubMed] [Google Scholar]
- 19.Slijepcevic D, Kaufman C, Wichers CG, Gilglioni EH, Lempp FA, Duijst S, de Waart DR, Elferink RP, Mier W, Stieger B, Beuers U, Urban S, van de Graaf SF. Impaired uptake of conjugated bile acids and hepatitis b virus pres1-binding in Na+-taurocholate cotransporting polypeptide knockout mice. Hepatology 62: 207–219, 2015. doi: 10.1002/hep.27694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith DJ, Grossbard M, Gordon ER, Boyer JL. Taurocholate uptake by isolated skate hepatocytes: effect of albumin. Am J Physiol Gastrointest Liver Physiol 252: G479–G484, 1987. [DOI] [PubMed] [Google Scholar]
- 21.Splinter PL, Lazaridis KN, Dawson PA, LaRusso NF. Cloning and expression of SLC10A4, a putative organic anion transport protein. World J Gastroenterol 12: 6797–6805, 2006. doi: 10.3748/wjg.v12.i42.6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stieger B, Hagenbuch B, Landmann L, Höchli M, Schroeder A, Meier PJ. In situ localization of the hepatocytic Na+/Taurocholate cotransporting polypeptide in rat liver. Gastroenterology 107: 1781–1787, 1994. doi: 10.1016/0016-5085(94)90821-4. [DOI] [PubMed] [Google Scholar]
- 23.Strautnieks SS, Bull LN, Knisely AS, Kocoshis SA, Dahl N, Arnell H, Sokal E, Dahan K, Childs S, Ling V, Tanner MS, Kagalwalla AF, Németh A, Pawlowska J, Baker A, Mieli-Vergani G, Freimer NB, Gardiner RM, Thompson RJ. A gene encoding a liver-specific ABC transporter is mutated in progressive familial intrahepatic cholestasis. Nat Genet 20: 233–238, 1998. doi: 10.1038/3034. [DOI] [PubMed] [Google Scholar]
- 24.Vaz FM, Paulusma CC, Huidekoper H, de Ru M, Lim C, Koster J, Ho-Mok K, Bootsma AH, Groen AK, Schaap FG, Oude Elferink RP, Waterham HR, Wanders RJ. Sodium taurocholate cotransporting polypeptide (SLC10A1) deficiency: conjugated hypercholanemia without a clear clinical phenotype. Hepatology 61: 260–267, 2015. doi: 10.1002/hep.27240. [DOI] [PubMed] [Google Scholar]
- 25.Visser WE, Wong WS, van Mullem AA, Friesema EC, Geyer J, Visser TJ. Study of the transport of thyroid hormone by transporters of the SLC10 family. Mol Cell Endocrinol 315: 138–145, 2010. doi: 10.1016/j.mce.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 26.Wang R, Salem M, Yousef IM, Tuchweber B, Lam P, Childs SJ, Helgason CD, Ackerley C, Phillips MJ, Ling V. Targeted inactivation of sister of P-glycoprotein gene (spgp) in mice results in nonprogressive but persistent intrahepatic cholestasis. Proc Natl Acad Sci USA 98: 2011–2016, 2001. doi: 10.1073/pnas.98.4.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou X, Levin EJ, Pan Y, McCoy JG, Sharma R, Kloss B, Bruni R, Quick M, Zhou M. Structural basis of the alternating-access mechanism in a bile acid transporter. Nature 505: 569–573, 2014. doi: 10.1038/nature12811. [DOI] [PMC free article] [PubMed] [Google Scholar]