Disruption of transcriptional gene expression is a hallmark of dilated cardiomyopathy; however, its etiology is not well understood. Cardiac-specific deletion of the transcriptional coactivator mediator subunit 1 (Med1) results in dilated cardiomyopathy, decreased cardiac function, and lethality. Med1 deletion disrupted cardiac mitochondrial and metabolic gene expression patterns.
Keywords: mediator complex, transcription, metabolism, cardiovascular disease
Abstract
The mediator complex, a multisubunit nuclear complex, plays an integral role in regulating gene expression by acting as a bridge between transcription factors and RNA polymerase II. Genetic deletion of mediator subunit 1 (Med1) results in embryonic lethality, due in large part to impaired cardiac development. We first established that Med1 is dynamically expressed in cardiac development and disease, with marked upregulation of Med1 in both human and murine failing hearts. To determine if Med1 deficiency protects against cardiac stress, we generated two cardiac-specific Med1 knockout mouse models in which Med1 is conditionally deleted (Med1cKO mice) or inducibly deleted in adult mice (Med1cKO-MCM mice). In both models, cardiac deletion of Med1 resulted in early lethality accompanied by pronounced changes in cardiac function, including left ventricular dilation, decreased ejection fraction, and pathological structural remodeling. We next defined how Med1 deficiency alters the cardiac transcriptional profile using RNA-sequencing analysis. Med1cKO mice demonstrated significant dysregulation of genes related to cardiac metabolism, in particular genes that are coordinated by the transcription factors Pgc1α, Pparα, and Errα. Consistent with the roles of these transcription factors in regulation of mitochondrial genes, we observed significant alterations in mitochondrial size, mitochondrial gene expression, complex activity, and electron transport chain expression under Med1 deficiency. Taken together, these data identify Med1 as an important regulator of vital cardiac gene expression and maintenance of normal heart function.
NEW & NOTEWORTHY Disruption of transcriptional gene expression is a hallmark of dilated cardiomyopathy; however, its etiology is not well understood. Cardiac-specific deletion of the transcriptional coactivator mediator subunit 1 (Med1) results in dilated cardiomyopathy, decreased cardiac function, and lethality. Med1 deletion disrupted cardiac mitochondrial and metabolic gene expression patterns.
the human heart consumes kilogram quantities of ATP daily to maintain cardiac contractility and homeostasis, and precise transcriptional control of metabolic gene expression is essential. Accordingly, disruption of metabolic gene expression leads to the dysregulation of cardiac energetics and heart disease (6). The mediator complex is an important director of cardiac transcription by integrating cellular signaling events. Mediator is a large, multisubunit complex consisting of 25–30 proteins. This complex regulates the transcription of select target genes by recruiting RNA polymerase II to DNA-bound transcription factors (27, 31).
Numerous signaling pathways responsible for cellular homeostasis, growth, differentiation, and metabolism converge on mediator through transcriptional coactivators and repressors that target one or more of the subunits of this complex (28). Interactions between individual mediator subunits and transcription factors or nuclear receptors provide specificity in the activation of different gene programs (4). Genetic studies of individual subunits of the mediator complex revealed the importance of these cofactors in regulating select gene expression patterns in different tissues during development and disease, including direct interaction with key metabolic transcription factors (3, 7). In highly metabolic organs, including liver, adipose, skeletal muscle, and myoblast cells, mediator subunit 1 (Med1) is a critical component for transcriptional regulation of metabolic genes (8, 9, 21, 37). In these organs, genetic deficiency of Med1 protects against metabolic dysfunction (1, 9, 22). Med1 directly interacts with nuclear receptors and coactivators in these tissues to regulate metabolic functions, providing a potential mechanistic link for how multiple transcription factors are able to act in a coordinated fashion. Genetic deficiency of Med1 results in early embryonic lethality at embryonic days 9.5–11.5, with evidence for significant impairment in cardiac development (18, 38). Despite the clear importance of metabolic function in cardiac tissue, the role of Med1 in the regulation of cardiac transcriptional gene expression and homeostasis is incompletely defined.
In the current study, we generated two mouse models with cardiomyocyte-specific deletion of Med1. We demonstrate a critical function for Med1 in regulation of key metabolic and mitochondrial gene expression profiles in the heart. Cardiac deletion of Med1 leads to dilated cardiomyopathy (DCM) and death. Together, these findings establish Med1 and mediator as key regulators of transcriptional responses essential for cardiac function in development and disease.
METHODS
Animals.
C57BL/6 mice were used for the developmental time period and transaortic banding studies. For embryonic day (E)18.5 and postnatal day (P)1 and P3 time periods, three hearts were pooled. For P7 and P14 time periods, two hearts were pooled. For all remaining time periods, only one heart was utilized. Mice deficient for Med1 in cardiomyocytes were generated as listed below. All animal procedures were conducted with the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health and were reviewed and approved by the Institutional Animal Care and Use Committee at the University of Iowa.
Transaortic constriction.
Male C57BL/6 mice between 9 and 10 wk of age were used for transaortic banding procedure. Mice were anesthetized using 2% isoflurane mixed with 0.5 l/min 100% O2. Partial thoracotomy was performed to access the transverse aorta. A Horison Titanium ligating clip (005200) was placed on the transverse aorta. The entire procedure is identical except for clipping in sham control mice. Echocardiography imaging was performed at 2 and 5 wk to confirm successful constriction. Ventricular tissues were harvested at 5 wk postconstriction and snap frozen in liquid nitrogen for Western blot processing.
Human explanted heart samples.
Human explanted heart samples were collected as part of the Human Explanted Heart Program (HELP) at the Mazankowski Alberta Heart Institute and the Human Organ Procurement and Exchange (HOPE) program at the University of Alberta Hospital. All experiments were performed in accordance with the institutional guidelines and were approved by the Institutional Ethics Committee. Informed consents were obtained from all participants. Explanted hearts with DCM (n = 3; 2 female/1 male; age = 35–60 yr; ejection fraction = 33.8 ± 2.4%) and nonfailing control hearts (n = 3; 1 female/2 male; age = 27–68 yr; ejection fraction ≥ 50%) were used. Our protocol enables dissection and snap-frozen of tissues in liquid nitrogen within 15 min of explantation.
Generation of Med1 conditional knockout mice.
The Med1 floxed mice (Med1fl/fl) were kindly provided by Dr. J. K. Reddy and Med1 cardiac-specific knockout (cKO) mice were generated by breeding Med1fl/fl mice with C57/BL6 transgenic mice containing the α-myosin heavy chain (α-MHC) promoter-driven cre recombinase and intercrossed to establish heterozygous Med1fl/+;αMHC-Cre mice. Male Med1fl/+-αMHC Cre+ mice were breed to female Med1fl/fl mice to establish homozygous Med1fl/fl mice. Studies were done using Cre-negative Med1fl/fl littermates as controls. Primers used for genotyping were as follows: Med1 forward 5′-GCA GGG AAA CCT GAG TGG AA-3′, Med1 reverse 5′-TGG GGC AAT TGG GAG TTT GT-3′; αMHC forward: 5′-CTA GCC CAC ACC AGA AAT GAC AGA C-3′; Cre reverse: 5′-GAA CCT CAT CAC TCG TTG CAT CGA C-3′. Inducible cardiomyocyte-specific deletion of Med1 was performed by crossing Med1fl/fl mice with Myh6-MerCreMer (MCM, tamoxifen-inducible Cre recombinase under the control of the αMHC promoter-Jackson Laboratories Stock: 005657). Homologous recombination was achieved by administration of tamoxifen chow (40 mg·kg−1·day−1; Harlan-Teklad Diets) for 2 wk with replacement to standard chow following the 2-wk period. Myh6-MCM mice were genotyped using the forward 5′-ATA CCG GAG ATC ATG CAA GC-3′ and reverse-5′-AGG TGG ACC TGA TCA TGG AG-3′ primers. Age-matched Med1fl/fl Cre negative and Myh6-MCM alone with or without tamoxifen treatment were used as controls. Animals were fed standard chow and water given ad libitum.
Transthoracic echocardiography.
Cardiac function was evaluated in conscious mice in the University of Iowa Cardiology animal phenotyping core laboratory. Left-sided chest hair was removed. Parasternal long and short axis views were obtained using a high-frequency echocardiography (30 MHz) linear array transducer (Vevo 2100; Visual Sonics). Measurements performed were done by a single experienced operator blinded to the mouse genotypes.
RNA isolation.
Ventricular tissue was isolated and flash frozen in liquid nitrogen and stored at −80°C until processed. Flash frozen ventricles were pulverized with a Bessman tissue pulverizer before homogenization. Total RNA was extracted in TRIzol reagent (Invitrogen) using a Postter- elvehjem tissue grinder. Reverse transcriptase-PCR was performed to generate cDNA using SuperScript-III (Invitrogen). For quantitative real-time PCR (qPCR), 50 ng of cDNA were used for each reaction with iTAQ Universal Sybergreen reagent (BioRad) using the QuantStudio 6 Flex system (Applied Biosystems). Gene expression was analyzed using the ΔΔCT method and relative expression was normalized to Rpl7l1. Sequences for the primers are as follows: Med1 (forward: 5′-CAT TAA ATG ACC CCC ACT CCG-3′; reverse: 5′-GTC CAG AGC TTG TAC AGC AGT T-3′), Myh6 (forward: 5′-ACA TTC TTC AGG ATT CTC TG-3′; reverse: 5′-CTC CTT GTC ATC AGG CAC), Myh7 (forward: 5′-TTC CTT ACT TGC TAC CCT C-3′; reverse: 5′-CTT CTC AGA CTT CCG CAG-3′), Nppa (forward: 5′-GTC TTG GCC TTT TGG CTT C-3′; reverse: 5′-TTC CTC AGT CTG CTC ACT C-3′), Nppb (forward-5′-GCT GCT TTG GGC ACA AGA TAA-3′; reverse: 5′-GCA GCC AGG AGG TCT TCC TA-3′), Col12a1 (forward: 5′-CTA TCC AGG TTC CGG CTA ACA-3′; reverse: 5′-CCT CCT GTA TGA TGC CGA CC-3′), Ctgf (forward: 5′-CTG CTG TGC ATC CTC CTA CC-3′; reverse: 5′-CAC GCT CCG TAC ACA GTT CT-3′), Tgfb2 (forward: 5′-CTG CCT TCG CCC TCT TTA CA-3′; reverse: 5′-GGC TGA GGA CTT TGG TGT GT), Tgfb3 (forward: 5′-CGC TGA ATG GCT GTC TTT CG-3′; reverse: 5′-GAC CCA AGT TGG ACT CTC TCC-3′), Fabp3 (forward: 5′-TGA CAG CAG ATG ACC GGA AG; reverse:-5′-CCA CAC TGC CAT GAG TGA GA-3′), Fatp (forward: 5′-CCA TCT GGG AGG AGT TCA CG-3′; reverse: 5′-ACA CAT GCG TGA GGA TAC GG-3′), Cpt1a (forward: 5′-CAC CAA CGG GCT CAT CTT CT-3′; reverse-5′-AGG GGA GGC CAC TCC ATT AT-3′), Cpt1b (forward: 5′-ATG ATG GCT ACG GGG TCT CT-3′; reverse: 5′-CCA CCC CTT ATG CCT GTG AG-3′), Cpt2 (forward: 5′-CAC AGC ATC GTA CCC ACC AT-3′; reverse: 5′-TCC TTC CCA ATG CCG TTC TC-3′), Acadm (forward: 5′-GCG ATG AAG GTT GAA CTC GC-3′; reverse: 5′-TCA GTG GCT AGC TGA TTG GC-3′), Acadvl (forward:5′-AAG ATT CGG GAT GGC TGC AA-3′; reverse: 5′-GCT GAT GGC GGC TTC TAT CT-3′), Cs (forward: 5′-CGG AAC AAG GGC TCA GGA AT-3′; reverse: 5′-ATG AGA AGA AGG CCC CAA GC-3′), Ckmt2 (forward: 5′-GCA GGA TCT GGA TAC GAC AGA-3′; reverse: 5′-GGC CAT CCT TCT TCT TGG TT-3′), Esrra (forward: 5′-TGC CCA GCA TAG GGT GTT AG-3′; reverse: 5′-CTG GCT TCT GAC AAT CCC CC-3′), Ppara (forward: 5′-CTG GTC TTA ACC GGC CCA AT-3′; reverse: 5′-TGC ACA TAG CCA GAA GGG TG-3′), Ppargc1a (forward: 5′-GTA AAT CTG CGG GAT GAT GG-3′; reverse: 5′-AGC AGG GTC AAA ATC GTC TG-3′), Ppargc1b (forward: 5′-GCC TTC CCA GAA CTG GAT GAA-3′; reverse: 5′-TCA GAG CTT GCT GTT GGG GA-3′).
RNA-seq.
The University of Iowa Institute of Human Genetics, Genomics Division, generated polyA-enriched stranded RNA libraries followed by RNAseq using the Illumina HiSeq platform. Raw sequence reads were analyzed with BaseSpace (www.illumina.com) by aligning reads to the Mus musculus mm10 genome using the TopHat Alignment app. Transcripts were assembled and significant differentially expressed genes were determined with the Cufflinks Assembly and DE app using a false discovery rate <0.05. Data have been deposited into GEO (GSE84160).
Panther gene ontology analysis.
RNA-seq analysis list of differentially regulated genes significantly changed between Med1fl/fl and Med1cKO mouse hearts was entered into the PANTHER classification system (Pantherdb.org) (29, 30). Classification of data was compiled via Biological Process and a Pie Chart was created. Additional viewing of the alterations in Biological Processes pertaining to Metabolic Processes (GO:0008152) was done to break down individual metabolic processes.
Ingenuity pathway analysis.
RNA-seq analysis data was uploaded to Qiagen’s Ingenuity Pathway Analysis (IPA; Valencia, CA) and biological and interaction networks were generated. The gene list was analyzed for biological function enrichment analysis, upstream regulator analysis, and networks of interaction between molecules algorithmically generated based on their connectivity.
DNA isolation and mitochondrial DNA quantification.
Twenty-five milligrams of ventricular tissue from all groups were pulverized and total DNA was prepared from Qiagen’s DNeasy blood and tissue kit (69054). The relative copy numbers of nuclear vs. mitochondrial genomes from 1 ng total DNA was determined by real-time PCR with primers specific to mitochondrial Cytb (forward: 5′-CAT TTA TTA TCG CGG CCC TA-3′; reverse: 5′-TGT TGG GTT GTT TGA TCC TG-3′) and mtCO1 (forward: 5′-TGC TAG CCG CAG GCA TTA CT-3′; reverse:5′-CGG GAT CAA AGA AAG TTG TGT-3′) and nuclear Gcg (forward: 5′ CAG GGC CAT CTC AGA ACC-3′; reverse: 5′-GCT ATT GGA AAG CCT CTT GC-3′) and Hbb (forward: 5′ GAA GCG ATT CTA GGG AGC AG-3′; reverse: 5′-GGA GCA GCG AT CTG AGT AGA-3′) genes.
Western blot analysis.
Ventricles were homogenized in RIPA lysis buffer containing protease/phosphatase inhibitors. Fifty nanograms of total protein were resolved using SDS-PAGE electrophoresis and transferred to nitrocellulose membranes. Membranes were blocked for 1 h in a 5% dry nonfat milk-Tris buffered Saline solution with tween. Primary antibodies were used at the following dilutions: Med1 [Bethyl A300-793A, 1:2,000 (mouse tissues) or Thermo PA5-37384, 1:1,000 (human tissues)], Pgc1-α (Santa Cruz Biotechnology Sc-13067, 1:3,000), Ppar-α (Santa Cruz Biotech Sc-9000, 1:1,000), ERR-α (Cell Signaling 13826, 1:1,000), and mitochondrial oxidative phosphorylation (OxPhos) complexes (Mitosciences MS604-300, 1:6,000). Appropriate secondary antibodies were than used (1:10,000). Antibody signals were detected using ECL. Gapdh (Cell Signaling 2118, 1:4,000) or sarcomeric α-actinin (Sigma A7811, 1:10,000) were used as loading controls. Quantification of Western blot was performed after scanning of film using ImageJ software.
Histology.
Hearts were isolated and fixed in 10% formalin and processed for paraffin embedment or transient electron microscopy. For paraffin-embedded sample staining 5-µm thick sections were cut and stained with hematoxylin and eosin, Masson’s trichrome, and terminal deoxynucleotidyl transferase dUTP-mediated nick-end labeling (TUNEL; in Situ Cell Death Detection Kit, Fluorescein; Roche) by the University of Iowa’s Central Microscopy Research Facility. All staining was performed on three to six hearts per group with three sections per heart. Five images were taken was each heart using an Olympus BX61. Fibrosis was quantified using ImageJ. For TUNEL analysis 10 images were taken per heart section and the number of TUNEL-positive cells was quantified using ImageJ. Each section was also stained with a negative control (no TUNEL) and positive control (TUNEL in the presence of a nuclease). For transient electron microscopy, ultrastructural examination of left ventricular tissue fixed with osmium tetroxide/uranyl acetate staining (90 nm) was performed with a Jeol electron microscope (JEM-1230) at ×1,500-30,000 direct magnifications (Jeol). Mitochondrial number and size was measured using ImageJ.
Citrate synthase activity.
Cellular citrate synthase activity was measured using the protocol previously described (2). Briefly, ventricular protein extracts were homogenized in DETAPAC buffer and sonicated three times for 10 s each. Each sample went through at least one freeze-thaw cycle before the assay. Protein was quantified using the Lowry assay (25). 50 μg protein was combined with NADP + oxaloacetic acid (0.5 mM) and dTNB (0.1 mM). Reaction was monitored at 412 nm for the formation of TNB formed every 30 min for 2 min. Rates were determined from slopes determined by regression analysis of the data.
Mitochondrial electron chain complex activities.
All electron transport chain (ETC) activity assays were adapted from previously described protocols (5). The assays were performed on a Beckman DU 800 spectrophotometer (Brea, CA). Complex I activity was assayed as the rate of rotenone-inhibitable NADH oxidation. Ventricular tissue protein extracts were assayed with or without 200 μg/ml rotenone in working buffer containing 25 mmol/l potassium phosphate buffer, 5 mmol/l magnesium chloride, 2 mmol/l potassium cyanide, 2.5 mg/ml BSA, 0.13 mmol/l NADH, 200 μg/ml antimycin A, and 7.5 mmol/l coenzyme Q1. Complex II activity was assayed as the rate of reduction of 2,6-dichloroindophenol by coenzyme Q in the presence and absence of 0.2 M succinate. Ventricular protein extracts were incubated with or without succinate in 25 mmol/l potassium phosphate buffer, 5 mmol/l magnesium chloride, 2 mmol/l potassium cyanide, 2.5 mg/ml BSA for 10 min at 30 C. After incubation, 200 μg/ml antimycin A, 200 μg/ml rotenone, 5 mmol/l DCIP, and 7.5 mmol/l coenzyme Q1 were added to each cuvette and incubated for 1 min before absorbance rates were read. Complex III activity was assayed as the rate of cytochrome c reduction by coenzyme Q2. Ventricular protein extracts were assayed in 25 mM potassium phosphate buffer, 5 mmol/l magnesium chloride, 2 mmol/l potassium cyanide, 2.5 mg/ml BSA, 0.5 mmol/l n-dodecyl β-maltoside, 200 μg/ml rotenone, 1.5 mmol/l cytochrome c, and 3.5 mmol/l coenzyme Q2.
Statistical analysis.
Results are expressed as means ± SE. An unpaired Student’s t test was used to determine statistical significance of all samples where there were two groups, and ANOVA analysis was performed for groups of three or more with a Tukey correction. Kaplan Meier curve analysis was performed using GraphPad Prism. A log rank test (Mantel-Cox) was performed for survival curve analysis. Significance was defined as indicated.
RESULTS
Dynamic expression of Med1 during development and disease.
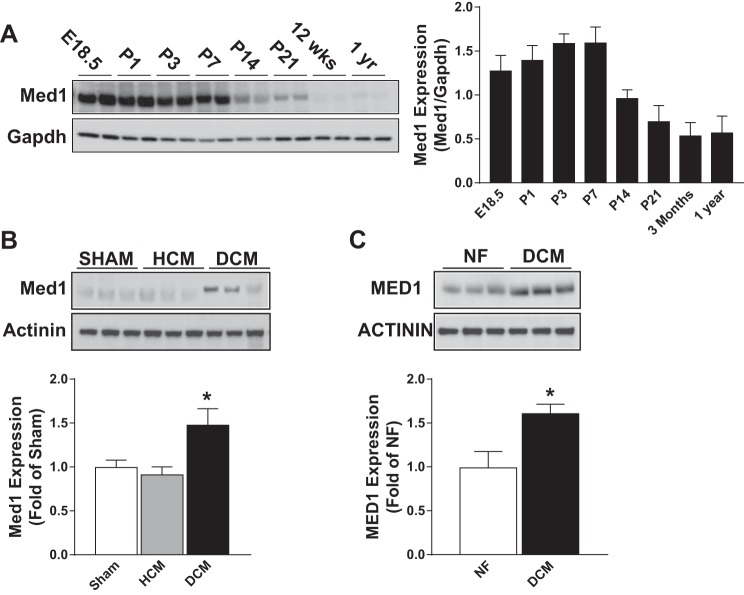
In the developing Mus musculus heart, Med1 expression was highly expressed until postnatal day 7. Expression decreased at postnatal days 14 and 21 and further decreased into adulthood (Fig. 1A). Med1 expression was assessed in ventricular tissue form mice that underwent a sham operation (sham), a transaortic constriction where there was compensative hypertrophic cardiomyopathy (HCM) or a transaortic constriction where the hearts developed a dilated cardiomyopathy (DCM). The expression of Med1 was unchanged in ventricular tissue from HCM relative to sham-operated hearts but was elevated in DCM tissue (Fig. 1B). In the human heart, MED1 expression was elevated in left ventricular tissue from patients with a history of DCM as compared with tissue from a nonfailing heart (Fig. 1C). Together, these results demonstrate that Med1 is dynamically expressed during both development and disease.
Fig. 1.
Cardiac mediator subunit 1 (Med1) expression is dynamic during development and disease. A: immunoblotting of Med1 and Gapdh in mouse heart tissues from time periods embroynic day (E)18.5, postnatal day (P)1, P3, P7, P14, P21, 12 wk, and 1 yr. Left: representative immunoblot. Right: quantitative analysis (right panel). (n = 4/time period). B: immunoblotting of Med1 and α-actinin in mouse ventricular tissue from mice that underwent a sham operation [sham: heart weight (HW; mg): 119 ± 6.8: ejection fraction (EF; %): 72 ± 5.2], a transaortic constriction where there was a compensated hypertrophic cardiomyopathy (HCM: HW: 196 ± 5.6; EF:50 ± 6.1), and a transaortic constriction where the hearts developed a dilated cardiomyopathy (DCM: HW:261 ± 20; EF: 19 ± 4.9). Top: representative immunoblot. Bottom: quantitative analysis. *P < 0.05 vs. sham control; n = 4/group. C: immunoblotting of Med1 and α-Actinin in human ventricular tissue samples from patients with nonfailing hearts (NF) or dilated cardiomyopathy (DCM). Top: representative immunoblot. Bottom: quantitative analysis. *P < 0.05 vs. NF control; n = 3/group.
Cardiac Med1 is necessary for survival in mice.
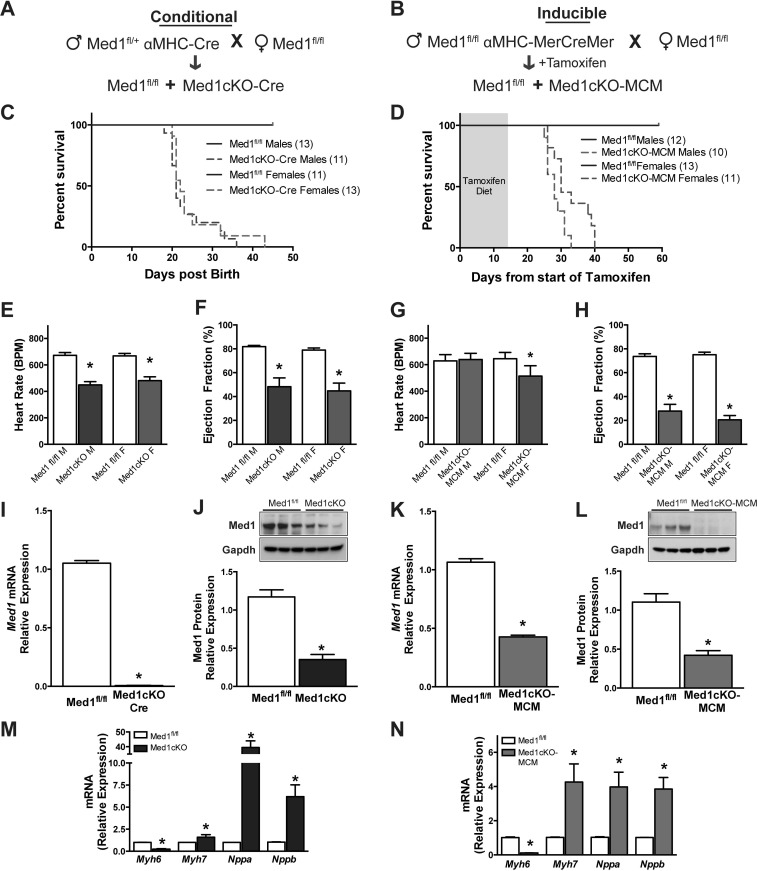
The developmental regulation and potential link to DCM prompted us to investigate whether loss of Med1 in cardiomyocytes protects against cardiac stress. Germline deletion of Med1 results in embryonic lethality, presumably due to defects in cardiac development (18, 38). To generate cardiomyocytes deficient in Med1, we utilized a constitutive cardiomyocyte-specific α-myosin heavy chain (αMHC)-promotor driven Cre recombinase bred to conditional Med1 floxed mice (Med1fl/fl) (38) (denoted as “Med1cKO mice,” Fig. 2A). In this model, αMHC expression is present as early as E7.5, decreases until E16, and by P7 becomes the dominant myosin heavy chain isoform expressed in the heart (26). To inducibly delete Med1 in cardiomyocytes in the adult heart in vivo, we crossed Med1fl/fl mice with the inducible modified estrogen receptor MerCreMer (MCM)-αMHC-driven Cre recombinase mice (denoted as “Med1cKO-MCM mice”). Med1 was deleted by administration of tamoxifen in chow beginning at 6–8 wk of age (Fig. 2B).
Fig. 2.
Conditional deletion of cardiac Med1 results in heart failure and death. A: schematic of conditional αMHC-Cre Med1 (Med1cKO) gene deletion. B: schematic of adult tamoxifen-inducible αMHC-MerCreMer (MCM) mediated Med1 gene deletion (Med1cKO-MCM). C and D: survival analysis of male and female Med1cKO offspring and adult Med1cKO-MCM mice following tamoxifen treatment (2 wk, 40 mg·kg−1·day−1). In C, P < 0.0001 in survival curves between Med1cKO mice and Med1fl/fl mice. P = 0.4915 in survival curves was found between sexes. In D, P < 0.0001 in survival curves between Med1cKO mice and Med1fl/fl mice. P = 0.031 between male and female Med1cKO-MCM mice. E–H: echocardiographic analysis showing heart rate [beats per minutes (bpm)] and ejection fraction (%) in male (M) and female (F) Med1fl/fl and Med1cKO (E and F, respectively) and Med1fl/fl and Med1cKO-MCM (G and H, respectively) mice. Data are means ± SE (n = 4–14); *P < 0.05 vs. sex-matched Med1fl/fl controls. See also Table 1. I and K: mRNA expression of Med1 gene in 3-wk-old Med1cKO and 21–28 days after the start of tamoxifen administration Med1cKO-MCM hearts. Data are means ± SE (n = 5–6); *P < 0.05 vs. Med1fl/fl controls. J and L: immunoblotting of Med1 in ventricular tissue at 3 wk of age in Med1cKO mice and 21–28 days after the start of tamoxifen administration in Med1cKO-MEM hearts. Top: representative immunoblot. Bottom: quantitative analysis. Data are means ± SE; n = 6/group; *P < 0.05 vs. Med1fl/fl controls. M and N: mRNA expression of markers of cardiac failure Myh6, Myh7, Nppa, and Nppb in ventricular tissue from 3-wk-old Med1cKO hearts (M) and Med1cKO-MCM hearts 21–28 days after the start of tamoxifen administration (N) compared with Med1fl/fl controls. Data are means ± SE; n = 5–6; *P < 0.05 vs. Med1fl/fl controls.
Mating of heterozygous male Med1fl/+-αMHC cre mice to female Med1fl/fl yielded offspring whose genotypes met Mendelian ratios of inheritance at birth, but no homozygous cardiac-specific Med1 knockout (Med1cKO) mice survived beyond 6 wk. Kaplan-Meier analysis demonstrated that Med1cKO mice died between 3 and 6 wk of age. Survival analysis results in a P < 0.0001 indicating a significant difference in survival curves between Med1cKO mice and Med1fl/fl mice. No significant difference in survival curves was found between sexes (P = 0.4915; Fig. 2C). Inducible deletion of Med1 in adult mice in the Med1cKO-MCM model resulted in death on average 24 days from the start of tamoxifen treatment. In the Med1cKO-MCM group, P < 0.0001 was found indicating a significant difference in survival curves between Med1cKO mice and Med1fl/fl mice. A significant difference was found between male and female Med1cKO-MCM mice survival curves (P = 0.031) with females living longer than males; however, there are no mice remaining by 40 days (Fig. 2D). In all of the following studies, we focused our analysis at a time period in which we observed ~50% survival after Med1 deletion (3 wk of age in the Med1cKO group and 21–28 days after tamoxifen administration in the Med1cKO-MCM mice).
Med1 is necessary for normal cardiac function.
Echocardiography revealed that deletion of Med1 in the heart caused a reduction in both heart rate (~200 beats/min decrease) and ejection fraction (~50% decrease) in the Med1cKO mice among both sexes when compared with sex-matched Med1fl/fl controls (Fig. 2, E and F, and Table 1). A significant reduction in heart rate was observed only in female Med1cKO-MCM mice, while ejection fraction was reduced by 80% in both males and females when compared with sex-matched tamoxifen-treated Med1fl/fl controls (Fig. 2, G and H, and Table 1). There was an increase in both end diastolic volume and end systolic volume following Med1 deletion in both male and female Med1cKO and Med1cKO-MCM mice (Table 1). As expected, no changes in gross cardiac function were observed in adult Med1fl/fl, Med1cKO-MCM male mice maintained on a standard diet or wild-type C57BL/6-MCM males 28 days after tamoxifen administration (data not shown). These data verify that the decrease in cardiac function in the Med1cKO-MCM mice is attributable to Med1 deletion rather than an effect of tamoxifen treatment. Ventricular cardiac samples harvested from mice at 3 wk of age in the Med1cKO group and 21–28 days posttamoxifen administration in the Med1cKO-MCM mice revealed significant reductions in both Med1 mRNA and protein expression in Med1cKO mice (Fig. 2, I and J) and Med1cKO-MCM mice (Fig. 2, K and L) as compared with Med1fl/fl controls. The low level of cardiac Med1 protein expression is likely attributable to noncardiomyocyte cell types within the ventricles lacking the recombinase. As no striking evidence of sex differences have emerged, equal use of both male and females in each group is represented in subsequent experiments.
Table 1.
Echocardiographic analysis of Med1cKO mice
| Genotype | n | Sex | HR, Beats/min | EDV, μl | ESV, μl | Mass, mg | SV, μl | CO, μl/m | EF, % |
|---|---|---|---|---|---|---|---|---|---|
| Postnatal | |||||||||
| Med1fl/fl | 11 | M | 672 ± 22 | 13.8 ± 0.94 | 2.6 ± 0.26 | 42.2 ± 4.3 | 11.3 ± 0.7 | 7437 ± 657 | 82.0 ± 0.01 |
| Med1cKO | 8 | M | 449 ± 24* | 47.8 ± 14.3* | 32.4 ± 13.4* | 66.2 ± 11.4* | 15.3 ± 1.7** | 6639 ± 513 | 48.2 ± 0.08* |
| Med1fl/fl | 6 | F | 668 ± 17 | 11.3 ± 0.07 | 2.3 ± 0.19 | 43.5 ± 4.1 | 9.0 ± 0.7 | 5434 ± 638 | 79.0 ± 0.02 |
| Med1cKO | 7 | F | 481 ± 29** | 48.5 ± 15.8** | 32.6 ± 12.2** | 69.4 ± 6.6** | 16.0 ± 1.7** | 7823 ± 1202 | 44.8 ± 0.08** |
| Adult | |||||||||
| Med1fl/fl | 6 | M | 629 ± 47 | 32.0 ± 5.9 | 9.3 ± 2.9 | 76.2 ± 3.3 | 22.6 ± 3.4 | 13673 ± 1579 | 72 ± 0.03 |
| Med1cKO-MCM | 9 | M | 640 ± 46 | 86.5 ± 9.9* | 67.6 ± 10.4* | 102.9 ± 6.9* | 18.9 ± 2.8 | 12439 ± 2144 | 26 ± 0.06* |
| Med1fl/fl | 4 | F | 645 ± 47 | 41.0 ± 10.5 | 11.0 ± 3.2 | 88.5 ± 11.1 | 30.0 ± 7.4 | 9233 ± 3984 | 74 ± 0.02 |
| Med1cKO-MCM | 5 | F | 513 ± 78 | 106.9 ± 12.6** | 79.6 ± 14.7** | 122.4 ± 11.3** | 27.4 ± 2.8 | 13879 ± 2326 | 28 ± 0.06** |
Values are means ± SE. Echocardiographic analysis of 3-wk-old Med1fl/fl and Med1cKO male (M) and female (F) mice and Med1fl/fl and Med1cKO-MCM male and female mice 21–28 days after the start of tamoxifen treatment. HR, heart rate; EDV, end diastolic volume; ESV, end systolic volume; SV, stroke volume; CO, cardiac output; EF, ejection fraction.
P < 0.05 compared with Med1fl/fl M mice;
P < 0.05 compared with Med1fl/fl F mice.
During cardiac development, expression of a major contractile component of the sarcomere β-myosin heavy chain (β-MHC, myh7) is high, but at birth βMHC expression is decreased and expression of α-myosin heavy chain (αMHC, myh6) dominates. During heart failure, there is a reversion to an increase in expression of β-MHC and reduction in the α-MHC (32). In line with the changes in gross cardiac function, expression of myh6 was reduced while expression of myh7 was elevated in Med1cKO (Fig. 2M) and Med1cKO-MCM (Fig. 2N) hearts. Additionally, as the heart begins to fail, it transcribes and translates natriuretic peptides, atrial natriuretic peptide (ANP) and brain natriuretic peptide (BNP), to be released into the circulation. ANP and BNP are often used as prognostic markers for cardiovascular disease (11, 14). Expression of ANP and BNP genes Nppa and Nppb was increased in both Med1cKO and Med1cKO-MCM hearts (Fig. 2, M and N). Together these data demonstrate that mice with cardiac deficiency of Med1 exhibit early lethality and cardiac failure.
Med1 deletion alters cardiac structure and leads to DCM.
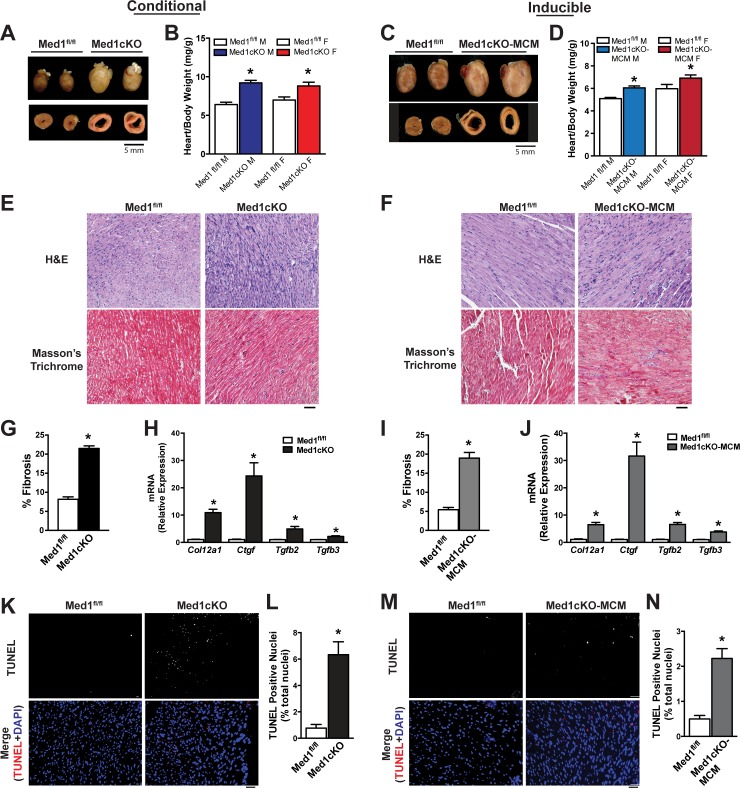
Consistent with the observed reduction in cardiac function, structural analysis of hearts from Med1cKO (Fig. 3A) and Med1cKO-MCM mice (Fig. 3C) revealed hypertrophic hearts with ventricular dilation as observed in transverse sections. Heart weights were increased in both male (44%) and female (27%) Med1cKO as compared with Med1fl/fl controls (Fig. 3B). Adult cardiac deletion of Med1 resulted in a 20% increase in heart weight in both male and females as compared with Med1fl/fl controls (Fig. 3D). No difference in body weight was observed between groups [Med1cKO (g): Med1fl/fl-M-9.3 ± 0.5; Med1cKO-M-8.7 ± 0.2; Med1fl/fl-F-9.8 ± 0.5; Med1cKO-F-8.9 ± 0.3; Med1cKO-MCM (g): Med1fl/fl-M-23.6 ± 0.6; Med1cKO-MCM-M-22.4 ± 0.6; Med1fl/fl-F-16.9 ± 0.4; Med1cKO-MCM-F-16.1 ± 0.4]. Histological examination revealed increased nuclei staining and increased Masson’s trichrome intensity, indicative of fibrosis (Fig. 3, E and F, and G and I, respectively). Expression of profibrotic genes (Col12a1, Ctgf, Tgfb2, and Tgfb3) was increased in ventricle samples from Med1cKO (Fig. 3H) and Med1cKO-MCM (Fig. 3J) as compared with Med1fl/fl controls. Deletion of Med1 also promoted a significant increase in apoptosis (Fig. 3, K–N). These findings identify pathological remodeling as an underlying feature of cardiac dysfunction under Med1 deficiency.
Fig. 3.
Med1 deletion in the heart induces pathological cardiac remodeling. Data were acquired from 3-wk-old Med1cKO hearts and Med1cKO-MCM hearts 21–28 days after the start of tamoxifen administration. A and C: whole heart and transverse sections of Med1cKO hearts (A) and Med1cKO-MCM hearts (C) compared with age-matched Med1fl/fl controls. B and D: whole heart weights of Med1cKO (B) and Med1cKO-MCM (D) hearts vs. Med1fl/fl controls. Data are means ± SE (n = 12–35); *P < 0.05 vs. Med1fl/fl controls. E and F: histological images of paraffin embedded samples stained using hematoxylin and eosin (H&E) and Masson’s trichrome in Med1cKO (E) and Med1cKO-MCM (F) samples with Med1fl/fl controls. G and I: quantification of fibrosis in Med1cKO (G) and Med1cKO-MCM (I) samples with Med1fl/fl controls. *P < 0.05 vs. Med1fl/fl controls; n = 3–6/group. H and J: mRNA expression of profibrotic genes Col12a1, Ctgf, Tgfb2, and Tgfb3 from ventricular samples in Med1cKO (H) and Med1cKO-MCM (J) groups compared with Med1fl/fl controls. Data are means ± SE; n = 6; *P < 0.05 vs. Med1fl/fl controls. K and M: terminal deoxynucleotidyl transferase (TUNEL) staining images in ventricular tissue from Med1cKO (K) and Med1cKO-MCM (M) vs. Med1fl/fl controls. Image magnification: ×20; scale bar = 50 pixels. L and N: quantitative analysis of TUNEL stain images (K and M, respectively). Quantified as %TUNEL-positive cells/field of view analyzed in blinded fashion. Data are means ± SE; n = 3/group; *P < 0.05 vs. Med1fl/fl controls.
Med1 regulates cardiac metabolic and mitochondrial genes.
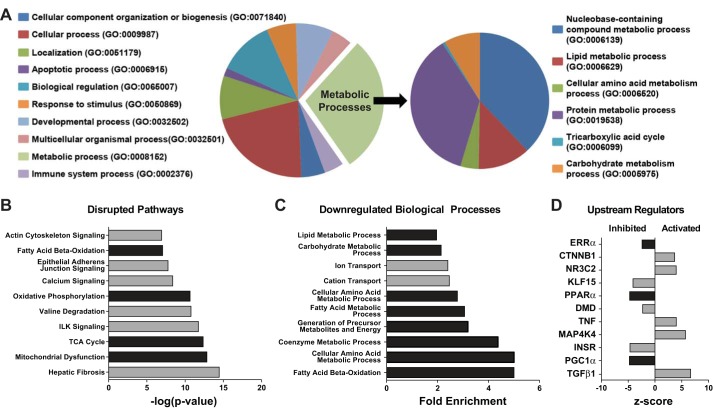
To determine the genetic basis for cardiac failure in Med1cKO mice, we performed RNA-seq analysis from Med1cKO hearts. A total of 5,568 genes were shown to be significantly altered in Med1cKO hearts, with 2,814 upregulated and 2,754 downregulated (GEO GSE84160). Gene Ontology (GO) Panther analysis was performed to identify cellular biological processes that were changed between groups. The leading biological category was metabolic processes, with nearly 40% of all genes associated with this process (GO:0008152, Fig. 4A). A subgroup analysis of the metabolic processes implicated multiple cellular metabolic pathways, including: tricarboxylic acid cycle (TCA), protein metabolic process, lipid metabolic process, and carbohydrate metabolic process.
Fig. 4.
Med1 deletion in the heart results in altered in metabolic gene expression. Molecular RNA-seq analysis of differential gene expression in 3-wk-old Med1fl/fl and Med1cKO hearts using Gene Ontology (GO) Panther analysis classification system (A) and Ingenuity Pathway Analysis (IPA; B, C, and D). n = 4/group. A: Panther GO analysis of Med1cKO RNA-seq differential gene expression. Pie chart of the top GO biological processes predicted to be altered. Smaller pie chart representing the predicted metabolic processes altered in the Med1cKO hearts. B; IPA of predicted disrupted pathways in the Med1cKO mouse heart. C: IPA of downregulated biological processes in the Med1cKO mouse heart. D: IPA of upstream biological regulators predicted activation or inhibition state in the Med1cKO mouse heart.
IPA was used to further stratify the differential changes in gene expression. The top disrupted biological pathways included mitochondrial dysfunction, metabolic [fatty acid oxidation (FAO), OxPhos, TCA] and contractile pathways (Fig. 4B). Of note, 93 genes were significantly downregulated and 7 upregulated related to mitochondrial function (see Supplemental Material, Supplemental Table S1; Supplemental Material for this article can be found at the American Journal of Physiology-Heart and Circulatory Physiology website). Importantly, the top downregulated biological processes by IPA mirrored the GO Panther analysis, substantiating the predominant downregulation metabolic cellular processes (Fig. 4C).
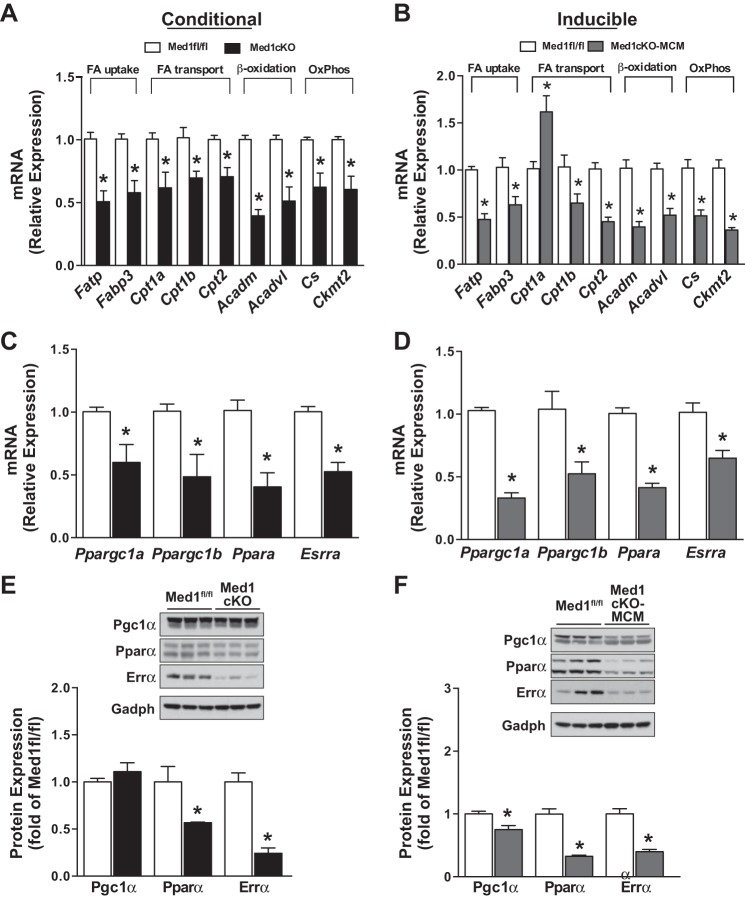
We also applied the upstream regulator analysis tool to identify the putative transcriptional regulators of the differentially expressed genes (Fig. 4D). Many canonical metabolic regulators (e.g., Esrrα, Pparα, and Ppargc1α/Pgc1-α) were predicted to be inhibited, while pathological cardiac remodeling pathways were predicted to be activated (e.g., CTNNB1, TNF, MAP3K4, and TGFβ1). Building on these data and to verify the RNA-seq data, we used qRT-PCR to measure expression of select metabolic genes that are regulated by these predicted transcription factors. Expression of fatty acid (FA) uptake (Fatp and Fabp3), FA transport (Cpta, Cptb, and Cpt2), β-oxidation (Acadm and Acadvl), and OxPhos (Cs and Ckmt2) genes were decreased in hearts from Med1cKO (Fig. 5A) and Med1cKO-MCM (Fig. 5B) mice. The only exception was an observed increase in Cpt1a expression in Med1cKO-MCM hearts. Interestingly, Cpt1a is being currently explored as a biomarker for cardiovascular disease risk (17).
Fig. 5.
Med1 deletion leads to changes in metabolic gene and transcriptional regulators in the heart. Data were acquired from 3-wk-old Med1cKO hearts and Med1cKO-MCM hearts 21–28 days after the start of tamoxifen administration. A–D: RT-qPCR validation of mRNA expression for genes encoding enzymes involved in cardiac metabolism (A and B) or regulatory transcription factors (C and D) in Med1cKO and Med1cKO-MCM ventricular samples. Data are means ± SE; n = 5–6. *P < 0.05 vs. Med1fl/fl controls. Fatty acids uptake genes: Fatp and Fatpb3. Fatty acid transport Cpt1a, Cpt1b, and Cpt2. β-Oxidation genes: Acadm and Acadvl. Oxidative Phosphorylation genes: Cs and Ckmt2. Transcriptional regulators: Essra, Ppara, Pargc1a, and Ppargc1b. E and F: immunoblotting of transcriptional regulatory proteins Pgc1α, Pparα and Errα in Med1cKO and Med1cKO-MCM ventricular samples. Top: representative immunoblot. Bottom: quantitative analysis. Data are means ± SE (n = 6/group); *P < 0.05 vs. Med1fl/fl controls.
To elucidate changes in major transcriptional metabolic players, we next measured expression of predicted upstream metabolic transcriptional regulators. Esrra, Ppara, Ppargc1a, and Ppargc1b were downregulated at the mRNA in both Med1cKO (Fig. 5C) and Med1cKO-MCM (Fig. 5D) hearts. Similarly, Pparα and Errα protein levels were lower Med1cKO and Med1cKO-MCM mouse hearts as compared with control (Fig. 5, E and F). Protein expression of Pgc1α was not altered in Med1cKO hearts but was decreased in Med1cKO-MCM hearts. Together, these data identify a role for Med1 as a transcriptional cofactor in coordinating the cardiac metabolic gene expression profile.
Cardiac mitochondrial function is altered in Med1cKO mice.
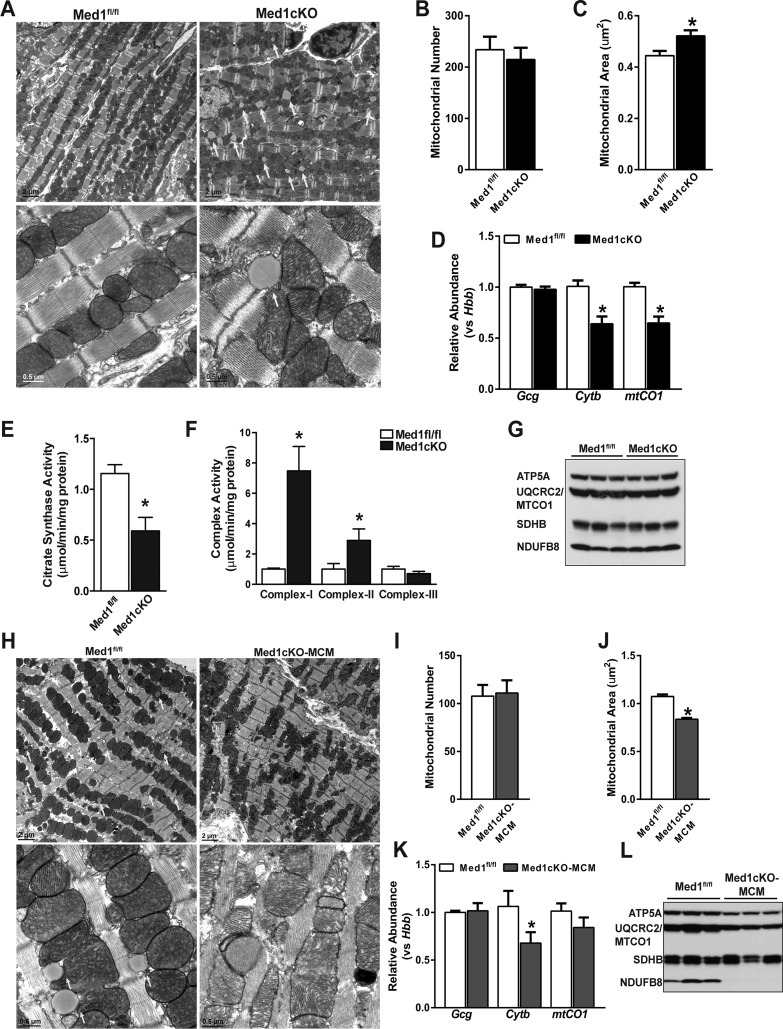
Since mitochondrial dysfunction is associated with cardiac failure and DCM, and our data indicate that Med1 is essential for normal cardiac function (33), we examined cardiac mitochondrial integrity. In 3-wk-old Med1cKO hearts, transmission electron microscopy studies revealed striking muscle fiber and mitochondrial damage, including mitochondrial disarray, myofibril disorganization, and mitochondrial size heterogeneity (Fig. 6A). Myocardial lipid content was increased in the Med1cKO hearts (yellow arrows), which is indicative of decreased mitochondrial metabolism in damaged tissue. Mitochondria number was not different in left ventricle sections between Med1fl/fl and Med1cKO hearts (Fig. 6B). However, relative mitochondrial area was increased (Fig. 6C) and mtDNA content was decreased under Med1 deficiency (Fig. 6D).
Fig. 6.
Alterations in cardiac mitochondria following Med1 deletion. Data were acquired from 3-wk-old Med1cKO hearts and Med1cKO-MCM hearts 21–28 days after the start of tamoxifen administration. A: transmission electron microscopy (TEM) images of Med1cKO vs. Med1fl/fl left ventricle samples. White arrows indicate lipid droplets. B and C: quantitative analysis of TEM images of mitochondrial number (B) and mitochondrial area (C). D: mitochondrial DNA analysis using mitochondrial gene Cytb and mtCO1 and nuclear gene Gcg compared with nuclear gene Hbb from Med1cKO ventricular samples. Data are means ± SE; n = 6/group; *P < 0.05 vs. Med1fl/fl controls. E: citrate synthase analysis in Med1cKO hearts. Data are means ± SE; n = 4/group; *P < 0.05 vs. Med1fl/fl controls. F: complex I, II, and III activities normalized to protein and citrate synthase activity in Med1cKO hearts. Data are means ± SE; n = 4/group; *P < 0.05 vs. Med1fl/fl controls. G: immunoblot for electron transport chain complex expression in ventricular tissue from Med1cKO and Med1fl/fl controls; n = 3. H: TEM images of Med1cKO-MCM vs. Med1fl/fl left ventricle samples. Yellow arrows indicate lipid droplets. I and J: Quantitative analysis of TEM images of mitochondrial number (I) and mitochondrial area (J); n = 4/group; *P < 0.05 vs. Med1fl/fl controls. K: mitochondrial DNA analysis using mitochondrial gene Cytb and mtCO1 and nuclear gene Gcg compared with nuclear gene Hbb from Med1cKO-MCM ventricular samples. Data are means ± SE; n = 6/group; *P < 0.05 vs. Med1fl/fl controls. L: immunoblot for electron transport chain complex expression in ventricular tissue from Med1cKO-MCM and Med1fl/fl controls; n = 3.
Given these changes, we assessed TCA cycle and ETC activity in Med1cKO hearts. First, citrate synthase activity was decreased in the Med1cKO heart (Fig. 6E). However, there was an increase in ETC complex I and II activity, with no change in complex III activity (Fig. 6F). Interestingly, no difference was observed in protein expression of ETC complexes in Med1cKO using an antibody cocktail (Fig. 6G, CI: Ndufb8, CII: Sdhb, CIII: Uqrc2, CIV: Mtco1, and CV: Atp5a).
In transient electron microscopy images from Med1cKO-MCM hearts taken 21–25 days after the start of tamoxifen treatment, similar changes were observed including mitochondrial disarray, myofibril disorganization, and mitochondrial size heterogeneity (Fig. 6H). In healthy Med1fl/fl control mice, structures resembling lipid droplets were present coupled next to healthy mitochondria. In sharp contrast, these structures were absent in myofibrils from cMed1KO-MCM heart tissue. While there was no change in mitochondrial number (Fig. 6I), there was a decrease in mitochondrial area (Fig. 6J) and mtDNA content (Fig. 6K). Expression of ETC complex I (NDUFB8), IV (MTCO1), and V(ATP5A) subunits were decreased or absent in Med1cKO-MCM hearts (Fig. 6L).
DISCUSSION
The progression to heart failure results from transcriptional and metabolic remodeling in cardiomyocytes. The mediator complex plays a key role in integrating the cellular signaling events leading to transcriptional remodeling and the changes in metabolic gene expression, yet few studies have directly tested the function of mediator in the heart. In an effort to elucidate the role of mediator, specifically Med1, we deleted Med1 in cardiomyocytes, which resulted in progressive DCM, cardiac failure, and death. Following cardiac Med1 deletion dramatic changes occurred in the transcriptional architecture and cardiac function. These changes are at least, in part, due to the overarching role of Med1 in mediating transcriptional gene programs of key nuclear metabolic transcription factors, Pgc1α, Pparα, and Errα. Together, our results uncover a new cardiac player in the pathogenesis of DCM and heart failure and provide insight into how changes in the metabolic gene expression profile can lead to deleterious effects on cardiac function.
DCM is defined as left ventricular dilation and systolic dysfunction (12). Consistent with our observed increase in Med1 protein levels in human heart failure and an animal model of DCM, Med1 m RNA expression has been reported to be upregulated in isolated cardiomyocytes from a genetic mouse model that models a human mutation leading to DCM (35) arising from a missense mutation in phospholamban (p.Arg9Cys). Interestingly, in our investigation loss of Med1 leads to DCM and is essential for normal cardiac function. Echocardiographic analysis revealed decreased ejection fraction and increased end systolic volumes in both the postnatal and adult Med1cKO animals. Med1cKO and Med1cKO-MCM hearts displayed cardiac remodeling typical of what is observed during DCM including increased heart mass, ventricular dilation, increased expression of profibrotic genes, and apoptosis. Together with our data in Med1cKO mice, we speculate that the increased Med1 expression in failing hearts is a compensatory response to try to combat the pathological cardiac remodeling through transcriptional alterations rather than a key driver of the phenotype. Furthermore, loss of Med1 disrupts homeostatic transcriptional responses, therefore leading to a pathological cardiac remodeling and DCM phenotype.
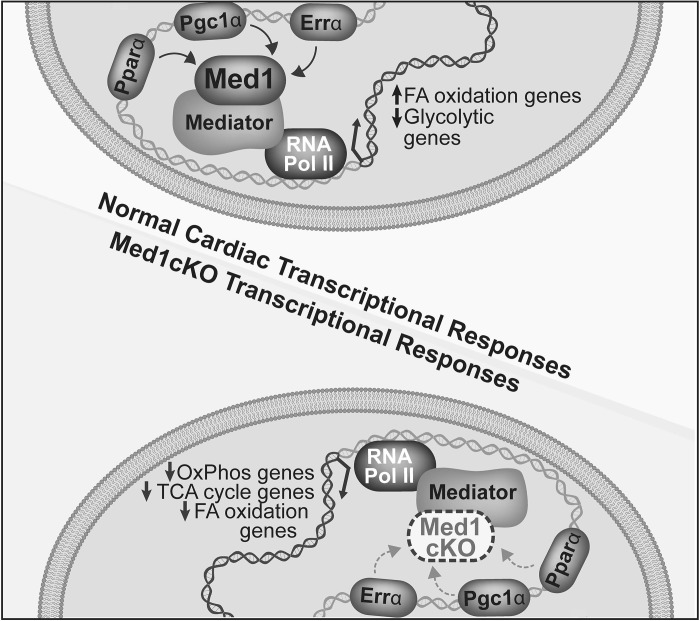
In the heart, nuclear receptor families, including Ppars and Errs, and their coactivator Pgc1 are important regulators of cardiac metabolism and mitochondrial homeostasis during development and disease (24, 34). For example, direct transcriptional control of key metabolic genes occurs via an interaction between Pgc1α and Pparα, guiding cardiac FAO enzyme transcription (13, 36). Furthermore, the interaction between Errα and Pgc1α regulates genes involved in cardiac energetics, mitochondrial biogenesis, dynamics, and OxPhos (16). While Pgc1α is commonly thought of as a master regulator of cellular metabolism, it lacks DNA binding activity and acts through Ppars and Errs to mediate molecular signaling cascades. How these transcriptional signaling responses are integrated with DNA binding mediators resulting in an overall large scale change in metabolic and mitochondrial gene expressions is unclear. Our results address this question by identifying Med1 as an integral regulator of these transcription factors and thereby metabolic processes (Fig. 7). Importantly, Med1 regulates metabolic transcriptional pathways involved in FAO, TCA cycle, ETC complexes, and OxPhos. Ingenuity pathway analysis identified mitochondrial dysfunction as a major pathway associated with the differentially regulated genes in the Med1cKO hearts. Given that Med1 regulates many vital cellular processes, the phenotype of Med1cKO hearts cannot be ascribed to small changes in gene expression but rather a large scale imbalance in transcriptional signaling underlying cellular metabolism and mitochondrial function.
Fig. 7.
Schematic depicting the role of Med1 in regulating cardiac transcription. Expression of Med1 in cardiomyocytes is required for normal transcriptional metabolic adaptations regulating expression of metabolic transcriptional pathways involved in fatty acid oxidation (FAO), TCA cycle, and oxidative phosphorylation.
In addition to the DCM phenotype observed in both the conditional and inducible cardiac Med1 deletion models, similarities were found between important players in cardiac mitochondrial and metabolic homeostasis. Expressions of Pparα and Esrra/Errα both at the gene and protein level were decreased when Med1 was deleted in both groups. Deletion of Med1 in liver parenchymal cells results in the attenuation of Pparα target gene transcription, and overexpression of Med1 produces the converse effect (20, 21). We found attenuation of gene expression levels of key players of FAO, β-oxidation, and OxPhos pathways were decreased following Med1 deletion in the heart. These data likewise follow the pattern observed when Errα is knocked out of the mouse heart (15).
While there were many similarities between the conditional and inducible Med1 deletion models, several differences have been identified. While Ppargc1α mRNA expression was reduced in Med1cKO and Med1cKO-MCM hearts, Pgc1α protein expression was maintained in Med1cKO but decreased in Med1cKO-MCM hearts. Stabilization and sustained activation of Pgc1α is undesirable as it can lead to uncontrollable mitochondrial biogenesis and respiration, disrupting sarcomere structure within the heart and decreasing cardiac function (23). Cpt1α expression was higher in the Med1cKO-MCM heart compared with the Med1cKO heart. Expression of Cpt1α is thought to be beneficial in the developing heart to aid in the utilization of fatty acids. In Med1cKO hearts, Cpt1α expression is lost, possibly contributing in part to the early lethality. It has also been observed that in the rat heart Cpt1α expression can be induced due to diabetic and fasting stress (10). In the Med1cKO-MCM heart, the increased expression of Cpt1α could likewise be due to the increase stress the heart is incurring. Transient electron microscopy images of myocytes in the Med1cKO hearts showed an increase in lipid droplet-like structures compared with floxed control myocytes. In contrast, these structures were absent in the Med1cKO-MCM myocytes. Mitochondrial area was increased in the cMed1KO mice yet decreased in cMed1KO-MCM mice relative to littermates. Individual expression of the ETC proteins in the hearts of Med1cKO was not changed; however, in the Med1cKO-MCM hearts, expression of ETC complex I, III, and V subunits was decreased or absent. Further investigations will help identify mitochondrial alterations between these two groups.
The importance of cardiac Med1 has similarly been reported by the Reddy laboratory (19). Their work likewise demonstrates a DCM phenotype consistent with our findings in which Med1 has been deleted from the heart either conditionally or in adulthood. Our study is the first to identify cardiac protein expression patterns of Med1 through development as well as disease. During transaortic constriction, Med1 expression increases only when hearts are failing vs. undergoing compensated hypertrophy. Of clinical importance, we are the first to report increased Med1 expression in human ventricular samples from patients with DCM. Notably, in our study both males and females were used equally to provide evidence of nongender-biased role of Med1 as a vital integrator of cellular signaling and cardiac homeostasis. Interestingly, adult female mice with induced cardiac deletion of Med1 lived longer compared with males. Differential gene analysis between these two studies identified similar themes of disrupted biological processes; nonetheless, the strength of our study was by using all the significantly differentially regulated genes rather than those of a selected cut-off of fold change difference allowed for a broader and global understanding of Med1 function in the heart. Furthermore, the RNA-seq analysis performed here identified not only significant alterations in metabolic and mitochondrial signaling pathways but upstream regulators that are vital to cardiac contractility and health.
A lingering question is whether the transcriptional reprogramming observed in Med1-deficient mice is a direct cause or consequence of the DCM observed in these mice. This question is difficult to address in the Med1cKO mice, as the study of earlier time periods might be confounded by developmental transcriptional changes that occur early in postnatal life. While the tamoxifen-inducible Med1cKO model is ideally suited to address this question through time-dependent Med1 deletion, a limitation of our studies is the early lethality of the Med1cKO-MCM model.
DCM and heart failure is an increasing burden on society. The transcriptional reprogramming of metabolic and mitochondrial processes is a hallmark of the pathological changes that lead to DCM and heart failure. Our finding that Med1 promotes metabolic and mitochondrial gene transcriptional downregulation implicates Med1 as an important transcriptional coactivator in the heart. Given that mitochondrial dysfunction underlies the pathogenesis of a wide range of cardiovascular diseases, the Med1 transcriptional regulatory pathway is a novel candidate for the investigation of experimental and possibly therapeutic strategies. Future studies aimed at unraveling the molecular role of Med1 in cardiac disease states associated with metabolic and mitochondrial dysfunction may identify specific therapeutic targets and signaling interactions.
GRANTS
This work was support by generous research support from National Institutes of Health (NIH) Grants R01-HL-125436-02, S10-OD-019941-01, and S10-RR-026293-01, Fraternal Order of Eagles Diabetes Research Center and the University of Iowa Carver College of Medicine (to C. E. Grueter), and NIH Grant T32 Postdoctoral Fellowship 5-T32-HL-007121-38 (to K. M. Spitler).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
K.M.S. and C.E.G. conceived and designed research; K.M.S. and J.M.P. performed experiments; K.M.S., J.M.P., D.D.H., and C.E.G. analyzed data; K.M.S., J.M.P., G.Y.O., D.D.H., and C.E.G. interpreted results of experiments; K.M.S., D.D.H., and C.E.G. prepared figures; K.M.S. and C.E.G. drafted manuscript; K.M.S., J.M.P., G.Y.O., D.D.H., and C.E.G. edited and revised manuscript; K.M.S., J.M.P., G.Y.O., D.D.H., and C.E.G. approved final version of manuscript.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge use of the University of Iowa Central Microscopy Research Facility, a core resource supported by the Vice President for Research and Economic Development, the Holden Comprehensive Cancer Center, and the Carver College of Medicine. This work used the JEOL JEM-1230 Transmission Electron Microscope in the University of Iowa Central Microscopy Research Facilities, which was purchased with funding from National Institutes of Health SIG Grant-1 S10-RR-018998-01. This work used the Vevo 2100 in the University of Iowa Mouse Cardiovascular Imaging Core which was purchased using National Institutes of Health Grant S10 OD019941-01 and S10 RR026293-01. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We thank the University of Iowa Free Radical and Radiation Biology Research Core for assistance with ETC assays. We thank Dr. J. K. Reddy for the Med1-floxed mice. We acknowledge Dr. Eric Olson for intellectual expertise and reading of the manuscript. We thank Dr. Colleen Johnson for critical reading of the manuscript.
REFERENCES
- 1.Bai L, Jia Y, Viswakarma N, Huang J, Vluggens A, Wolins NE, Jafari N, Rao MS, Borensztajn J, Yang G, Reddy JK. Transcription coactivator mediator subunit MED1 is required for the development of fatty liver in the mouse. Hepatology 53: 1164–1174, 2011. doi: 10.1002/hep.24155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrientos A, Marín C, Miró O, Casademont J, Gómez M, Nunes V, Tolosa E, Urbano-Márquez A, Cardellach F. Biochemical and molecular effects of chronic haloperidol administration on brain and muscle mitochondria of rats. J Neurosci Res 53: 475–481, 1998. doi:. [DOI] [PubMed] [Google Scholar]
- 3.Baskin KK, Winders BR, Olson EN. Muscle as a “mediator” of systemic metabolism. Cell Metab 21: 237–248, 2015. doi: 10.1016/j.cmet.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belakavadi M, Fondell JD. Role of the mediator complex in nuclear hormone receptor signaling. Rev Physiol Biochem Pharmacol 156: 23–43, 2006. [DOI] [PubMed] [Google Scholar]
- 5.Birch-Machin MA, Briggs HL, Saborido AA, Bindoff LA, Turnbull DM. An evaluation of the measurement of the activities of complexes I–IV in the respiratory chain of human skeletal muscle mitochondria. Biochem Med Metab Biol 51: 35–42, 1994. doi: 10.1006/bmmb.1994.1004. [DOI] [PubMed] [Google Scholar]
- 6.Carvajal K, Moreno-Sánchez R. Heart metabolic disturbances in cardiovascular diseases. Arch Med Res 34: 89–99, 2003. doi: 10.1016/S0188-4409(03)00004-3. [DOI] [PubMed] [Google Scholar]
- 7.Chen W, Roeder RG. Mediator-dependent nuclear receptor function. Semin Cell Dev Biol 22: 749–758, 2011. doi: 10.1016/j.semcdb.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen W, Yang Q, Roeder RG. Dynamic interactions and cooperative functions of PGC-1alpha and MED1 in TRalpha-mediated activation of the brown-fat-specific UCP-1 gene. Mol Cell 35: 755–768, 2009. doi: 10.1016/j.molcel.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen W, Zhang X, Birsoy K, Roeder RG. A muscle-specific knockout implicates nuclear receptor coactivator MED1 in the regulation of glucose and energy metabolism. Proc Natl Acad Sci USA 107: 10196–10201, 2010. doi: 10.1073/pnas.1005626107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cook GA, Edwards TL, Jansen MS, Bahouth SW, Wilcox HG, Park EA. Differential regulation of carnitine palmitoyltransferase-I gene isoforms (CPT-I alpha and CPT-I beta) in the rat heart. J Mol Cell Cardiol 33: 317–329, 2001. doi: 10.1006/jmcc.2000.1304. [DOI] [PubMed] [Google Scholar]
- 11.Dewey CM, Spitler KM, Ponce JM, Hall DD, Grueter CE. Cardiac-secreted factors as peripheral metabolic regulators and potential disease biomarkers. J Am Heart Assoc 5: e003101, 2016. doi: 10.1161/JAHA.115.003101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elliott P. Cardiomyopathy. Diagnosis and management of dilated cardiomyopathy. Heart 84: 106–112, 2000. doi: 10.1136/heart.84.1.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gulick T, Cresci S, Caira T, Moore DD, Kelly DP. The peroxisome proliferator-activated receptor regulates mitochondrial fatty acid oxidative enzyme gene expression. Proc Natl Acad Sci USA 91: 11012–11016, 1994. doi: 10.1073/pnas.91.23.11012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hasegawa K, Fujiwara H, Doyama K, Mukoyama M, Nakao K, Fujiwara T, Imura H, Kawai C. Ventricular expression of atrial and brain natriuretic peptides in dilated cardiomyopathy. An immunohistocytochemical study of the endomyocardial biopsy specimens using specific monoclonal antibodies. Am J Pathol 142: 107–116, 1993. [PMC free article] [PubMed] [Google Scholar]
- 15.Huss JM, Imahashi K, Dufour CR, Weinheimer CJ, Courtois M, Kovacs A, Giguère V, Murphy E, Kelly DP. The nuclear receptor ERRalpha is required for the bioenergetic and functional adaptation to cardiac pressure overload. Cell Metab 6: 25–37, 2007. doi: 10.1016/j.cmet.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 16.Huss JM, Torra IP, Staels B, Giguère V, Kelly DP. Estrogen-related receptor alpha directs peroxisome proliferator-activated receptor alpha signaling in the transcriptional control of energy metabolism in cardiac and skeletal muscle. Mol Cell Biol 24: 9079–9091, 2004. doi: 10.1128/MCB.24.20.9079-9091.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Irvin MR, Aslibekyan S, Hidalgo B, Arnett D. CPT1A: the future of heart disease detection and personalized medicine? Clin Lipidol 9: 9–12, 2014. doi: 10.2217/clp.13.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ito M, Yuan CX, Okano HJ, Darnell RB, Roeder RG. Involvement of the TRAP220 component of the TRAP/SMCC coactivator complex in embryonic development and thyroid hormone action. Mol Cell 5: 683–693, 2000. doi: 10.1016/S1097-2765(00)80247-6. [DOI] [PubMed] [Google Scholar]
- 19.Jia Y, Chang HC, Schipma MJ, Liu J, Shete V, Liu N, Sato T, Thorp EB, Barger PM, Zhu YJ, Viswakarma N, Kanwar YS, Ardehali H, Thimmapaya B, Reddy JK. Cardiomyocyte-specific ablation of med1 subunit of the mediator complex causes lethal dilated cardiomyopathy in mice. PLoS One 11: e0160755, 2016. doi: 10.1371/journal.pone.0160755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jia Y, Guo GL, Surapureddi S, Sarkar J, Qi C, Guo D, Xia J, Kashireddi P, Yu S, Cho YW, Rao MS, Kemper B, Ge K, Gonzalez FJ, Reddy JK. Transcription coactivator peroxisome proliferator-activated receptor-binding protein/mediator 1 deficiency abrogates acetaminophen hepatotoxicity. Proc Natl Acad Sci USA 102: 12531–12536, 2005. doi: 10.1073/pnas.0506000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jia Y, Qi C, Kashireddi P, Surapureddi S, Zhu YJ, Rao MS, Le Roith D, Chambon P, Gonzalez FJ, Reddy JK. Transcription coactivator PBP, the peroxisome proliferator-activated receptor (PPAR)-binding protein, is required for PPARalpha-regulated gene expression in liver. J Biol Chem 279: 24427–24434, 2004. doi: 10.1074/jbc.M402391200. [DOI] [PubMed] [Google Scholar]
- 22.Jia Y, Viswakarma N, Fu T, Yu S, Rao MS, Borensztajn J, Reddy JK. Conditional ablation of mediator subunit MED1 (MED1/PPARBP) gene in mouse liver attenuates glucocorticoid receptor agonist dexamethasone-induced hepatic steatosis. Gene Expr 14: 291–306, 2009. doi: 10.3727/105221609788681213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lehman JJ, Barger PM, Kovacs A, Saffitz JE, Medeiros DM, Kelly DP. Peroxisome proliferator-activated receptor gamma coactivator-1 promotes cardiac mitochondrial biogenesis. J Clin Invest 106: 847–856, 2000. doi: 10.1172/JCI10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab 1: 361–370, 2005. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 25.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem 193: 265–275, 1951. [PubMed] [Google Scholar]
- 26.Lyons GE, Schiaffino S, Sassoon D, Barton P, Buckingham M. Developmental regulation of myosin gene expression in mouse cardiac muscle. J Cell Biol 111: 2427–2436, 1990. doi: 10.1083/jcb.111.6.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malik S, Roeder RG. Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem Sci 30: 256–263, 2005. doi: 10.1016/j.tibs.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 28.Malik S, Roeder RG. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet 11: 761–772, 2010. doi: 10.1038/nrg2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc 8: 1551–1566, 2013. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mi H, Poudel S, Muruganujan A, Casagrande JT, Thomas PD. PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic Acids Res 44: D336–D342, 2016. doi: 10.1093/nar/gkv1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mittler G, Kremmer E, Timmers HT, Meisterernst M. Novel critical role of a human Mediator complex for basal RNA polymerase II transcription. EMBO Rep 2: 808–813, 2001. doi: 10.1093/embo-reports/kve186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morkin E. Control of cardiac myosin heavy chain gene expression. Microsc Res Tech 50: 522–531, 2000. doi:. [DOI] [PubMed] [Google Scholar]
- 33.Rosca MG, Hoppel CL. Mitochondria in heart failure. Cardiovasc Res 88: 40–50, 2010. doi: 10.1093/cvr/cvq240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scarpulla RC. Metabolic control of mitochondrial biogenesis through the PGC-1 family regulatory network. Biochim Biophys Acta 1813: 1269–1278, 2011. doi: 10.1016/j.bbamcr.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Truszkowska GT, Bilińska ZT, Kosińska J, Śleszycka J, Rydzanicz M, Sobieszczańska-Małek M, Franaszczyk M, Bilińska M, Stawiński P, Michalak E, Małek LA, Chmielewski P, Foss-Nieradko B, Machnicki MM, Stokłosa T, Ponińska J, Szumowski Ł, Grzybowski J, Piwoński J, Drygas W, Zieliński T, Płoski R. A study in Polish patients with cardiomyopathy emphasizes pathogenicity of phospholamban (PLN) mutations at amino acid position 9 and low penetrance of heterozygous null PLN mutations. BMC Med Genet 16: 21, 2015. doi: 10.1186/s12881-015-0167-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vega RB, Huss JM, Kelly DP. The coactivator PGC-1 cooperates with peroxisome proliferator-activated receptor alpha in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Mol Cell Biol 20: 1868–1876, 2000. doi: 10.1128/MCB.20.5.1868-1876.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu J, Xiao Y, Liu J, Ji Y, Liu H, Xu J, Jin X, Liu L, Guan MX, Jiang P. Loss of MED1 triggers mitochondrial biogenesis in C2C12 cells. Mitochondrion 14: 18–25, 2014. doi: 10.1016/j.mito.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 38.Zhu Y, Qi C, Jia Y, Nye JS, Rao MS, Reddy JK. Deletion of PBP/PPARBP, the gene for nuclear receptor coactivator peroxisome proliferator-activated receptor-binding protein, results in embryonic lethality. J Biol Chem 275: 14779–14782, 2000. doi: 10.1074/jbc.C000121200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.