Abstract
MicroRNAs are small, noncoding, RNAs known for their powerful modulation of molecular processes, making them a major focus for studying pathological mechanisms. The human miR-146 family of microRNAs consists of two member genes, MIR146A and MIR146B. These two microRNAs are located on different chromosomes and exhibit differential regulation in many cases. However, they are nearly identical in sequence, sharing a seed region, and are thus predicted to target the same set of genes. A large proportion of the microRNA (miR)-146 literature focuses on its role in regulating the innate immune response in the context of various pathologies by modulating two widely studied target genes in the toll-like receptor signaling cascade. A growing subset of the literature reports a role of miR-146 in cardiovascular and renal disease, and data suggest there is exciting potential for miR-146 as a diagnostic and therapeutic target. Nevertheless, the published literature is confounded by unclear and imprecise language concerning the specific effects of the two miR-146 family members. The present review will compare the genomic origin and regulation of miR-146a and miR-146b, discuss some approaches to overcome analytical and experimental challenges, and summarize findings in major areas of miR-146 research. Moving forward, careful evaluation of miR-146a/b specificity in analytical and experimental approaches will aid researchers in elucidating the functional relevance of differential regulation of the miR-146 family members in health and disease.
Keywords: cancer, cardiac, microRNA, renal
microRNAs (miRNAs) are a class of small, endogenous, noncoding RNAs known to regulate the expression of protein-coding genes. They are found throughout the genome and are transcribed much like protein-coding sequences (34, 49). miRNA biogenesis (46), target recognition (5), and function (13), reviewed extensively elsewhere, are described briefly here. Posttranscriptional processing of the primary miRNA transcript results in formation of a bioactive mature miRNA ~18–22 nucleotides in length (46). The mature miRNA combines with an RNA-induced silencing complex to exert its biological function via binding to the 3′-untranslated region (3′-UTR) of the mRNA transcript and suppressing translation or promoting the degradation of the message. They bind their targets via seed region complementarity, corresponding to nucleotides 2–8 in the mature miRNA sequence. Many human miRNAs and their target sequences share sequence homology across species, making them good candidates for study in animal models (27). Previous studies have shown that many miRNAs can target a single mRNA transcript, and a single miRNA can target many mRNA transcripts (32, 41, 62, 72). miRNAs are thus intriguing molecules to study for their ability to be master regulators of pathophysiology (13, 58) and their attractive potential as therapeutic targets (69).
The publicly available database of confirmed and suspected miRNAs (29) now contains information on nearly 36,000 mature miRNA products from 223 species; including mature sequences for 2,588 human (hsa-), 1,915 mouse (mmu-), and 765 rat (rno-) miRNA. Many miRNAs have been grouped into families based on shared seed sequences, compounding the difficulty of teasing apart biological effects of a specific miRNA, one such family being miR-146. First described in humans by Taganov et al. in 2006 (85), the miR-146 family consists of two miRNAs with nearly identical sequences, miR-146a-5p and miR-146b-5p. The present review will compare the genomic organization and regulation of the two miR-146 family members, discuss several obstacles and caveats in studying miR-146a and miR-146b, and summarize findings in major areas of miR-146 research.
The miR-146 Family
Genomic location and organization.
Mir146 was first identified in mouse cardiac tissue in a study published by Lagos-Quintana et al. in 2002 (51). Three years later, Cai et al. (12) prepared cDNA libraries from size-selected small RNAs (18–24 nt in length) from the BC-1 human B cell lymphoma cell line and classified their sequences based on analysis from the GenBank database (National Institutes of Health) and miRNA registry (Sanger). They found 34 cellular miRNAs, “several of which [had] been reported only in rodents,” including miR-146. The authors note that per the Sanger Institute RNA family database, Rfam, MIR146A was a predicted human miRNA based on analysis of rodent or zebrafish miRNA clones. Taganov et al. (85) are credited with confirming the presence of a human homolog by providing the first characterization of the genomic location and regulation of the human miR-146 family in 2006.
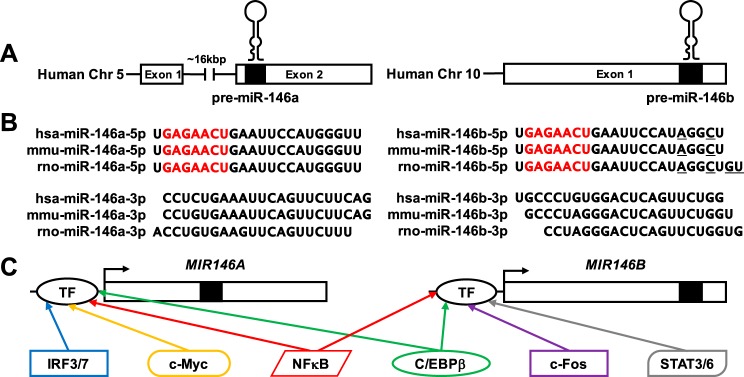
The primary transcripts (pri-miR) are transcribed in the nucleus from two genes, MIR146A and MIR146B. In the human, MIR146A is found within a larger long noncoding RNA host gene, MIR3142HG (chromosome 5q33.3), while MIR146B is found in an intergenic region of human chromosome 10 (10q24.32) (Fig. 1A). In the mouse, Mir146a is found in an intergenic region of chromosome 11 (band B1.1) and miR-146b is found within the first intron of the protein coding gene called major facilitator superfamily domain containing 13a (Mfsd13a or Tmem180) on chromosome 19 (band C3). In the rat, both Mir146a and Mir146b are found in intergenic regions of chromosomes 10q21 and 1q54, respectively. The precursor miRNA (pre-miR) sequences for these two miRNAs are nearly identical on the 5′-end and largely different on the 3′-end, regardless of species. Sequencing data compiled on the miRBase database provides evidence for the mature -5p strand to be the bioactive “guide strand” and the mature -3p strand to be the “passenger strand” for both miR-146a and miR-146b (Fig. 1B). Thus, for the remainder of this review the mention of miR-146a and miR-146b will refer to the -5p strands.
Fig. 1.
The microRNA (miR)-146 family consists of 2 members, miR-146a and miR-146b, found on human chromosomes 5 and 10, respectively (A). Basic exon structure for the human genes is shown, with black boxes and hairpin structures indicating the location of the precursor miR-146 sequences. B: sequences for the mature -5p and -3p strands are shown for both miR-146a and miR-146b in human, mouse, and rat. There is high sequence homology across species for miR-146a-5p and miR-146b-5p, respectively. Note that miR-146a-5p and miR-146b-5p differ by only a few bases on the 3′-end in each species (nucleotides differing between miR-146a-5p and miR-146b-5p are underlined). While the -3p sequences for both miR-146a and miR-146b, respectively, are largely homologous across species, within each species the miR-146a-3p and miR-146b-3p sequences are nonhomologous. C: a schematic of predicted and known transcription factor (TF) binding sites in the promoter regions of both MIR146A and MIR146B.
The mature sequences for miR-146a and miR-146b are highly conserved across species (Fig. 1B). Moreover, the two miRNAs differ only by two nucleotides on the 3′-end of the mature strand, not within the seed region. Research interest into the miR-146 family is growing, though published literature on miR-146a has outpaced that of miR-146b by nearly fivefold. It is common to find miRNA family members such as these presented as “miR-146a/b,” for example, because little was known about their function and regulation when they were first annotated. With advancements in miRNA research, i.e., the advent of miRNA deep sequencing (miR-seq), numerous miRNA profiling studies report differential expression of miRs in various pathologies. To that end, many publications report differential expression of the two miR-146 isoforms yet discuss the miR-146a/b family en bloc with no discrimination between isoform-specific effects. While they share homology and a miRNA family number, several characteristics highlighted in the reported literature suggest that they may have unique regulatory functions. The available evidence suggests that these two miRNAs are not transcribed in tandem; nor are they transcriptionally regulated by all the same factors and cofactors; nor is their expression temporally synchronized; nor do they have the same tissue-specific expression profiles. This strongly suggests that transcription and/or processing of miR-146a and miR-146b is intentionally specific and that they may even have unique regulatory functions determined by sequence characteristics outside of the seed region. The subsequent sections will summarize some of what is known of the miR-146 family; highlight the need for careful, precise discussion of the independence of these two miRNAs; and discuss limitations that should be considered when selecting approaches to discriminate the roles of miR-146a and miR-146b in pathology.
Transcriptional regulation.
Unlike many other miRNA families, little detail of the transcriptional regulation of MIR146A and MIR146B has been published; however, those that have been identified are summarized in Fig. 1C. In Taganov’s original paper, both MIR146A and MIR146B were reported to be regulated by the transcription factor nuclear factor κ-light-chain-enhancer of activated B cells (NF-κB). They describe putative binding sites for the transcription factors interferon regulatory factor 3/7 (IRF3/7) and CCAAT-enhancer-binding-protein-β (C/EBPβ) upstream of the first exon of MIR146A. They also reported an NF-κB2 binding site ~34 kb upstream of the primary MIR146B transcript. Two years later Chang et al. (15) confirmed a human MIR146A transcription start site (TSS) and identified an additional transcription factor binding site for v-myc avian myelocytomatosis viral oncogene homolog (c-Myc). In 2011, Chien et al. (17) identified a TSS 813 bp upstream of the pre-miR-146b sequence. In 2014, Li et al. (53) reported two direct binding sites for C/EBPβ identified in the miR-146b promoter region ~2 kb upstream of the human precursor miR-146b sequence.
Caveats, Barriers, and Caution against Ambiguity
Target prediction.
The classic view of miRNA activity describes their biological function as inducing transcript degradation or causing translational repression after binding the 3′-UTR of the targeted mRNA transcript. As noted above, these processes are reviewed in depth elsewhere. We also now understand a variety of possible miRNA roles, including that of signaling molecule (6, 33) or biomarker (30) and noncanonical regulatory functions such as posttranscriptional upregulation of target mRNAs (89). Regardless of downstream effect, the miRNAs identify and bind their targets via complementary base pairing of the miRNA seed sequence with the mRNA transcript. Considering miR-146a and miR-146b have identical seed sequences, our ability to identify predicted miRNA target pairs based on sequence alone is limited. Many target prediction sites base prediction algorithms on seed sequence complementarity. Given the identical seed sequences of miR-146a and miR-146b, this poses an obvious problem. How does one differentiate activity of one miRNA over the other? In this section, we will discuss some of the technical limitations of different research approaches and various issues that should be considered when studying these two miRNAs with nearly identical sequences.
Predicted targets for miR-146a and miR-146b largely overlap based on 3′-UTR binding alone due to their identical seed sequences. Many times, these prediction sites have completely overlapping predicted targets. TargetScan v7.1 (1), a widely used target prediction site, identifies 3,887 human mRNA transcripts targeted by “miR-146-5p” totaling over 5,000 specific target sites, regardless of user input (i.e., “146a” or “146b”). Various prediction software suites will report a specific binding site for one or the other miRNA, but upon closer inspection both could potentially bind the target of interest due to seed sequence homology. There is a possibility that additional nucleotide binding sites that differ on the 3′-end of the mature miRNA sequences could preferentially stabilize the miRNA-mRNA complex and increase the probability that one miR-146 isoform is more effective at regulating a given target; however, this has not been described to date.
The gold standard for determining target binding is the 3′-UTR luciferase reporter assay, a cell-based assay that uses a reporter construct, generally transfected into a stable cell line, to investigate whether the mature miRNA sequence binds and regulates a predicted target sequence (82). For a variety of reasons, an investigator may choose to test only one of the miR-146 family members without making a clear distinction between the two miRNAs. The lack of this information limits our ability to understand the impact of differential pathophysiological regulation of shared predicted targets.
Investigators may rely on RNA sequencing data sets (including ncRNA data) to analyze gene expression profiles. The art is in the interpretation of these “big data.” One logical, rigorous, and highly accessible approach for miRNA research begins with comparing miRNA deep sequencing data with RNA sequencing data to identify potential miRNA-mRNA target pairs relevant to the disease model studied. miRNA activity can induce transcript degradation; therefore, target pairs showing reciprocal expression (i.e., increased miRNA abundance and decreased target mRNA abundance) can be passed through pathway analysis software to help identify potential mechanistic pathways on which to devote additional research efforts. miRNAs are also well known to repress translation without the transcript being degraded. For this reason, miRNA activity may correlate better with protein abundance than with message abundance, allowing researchers to detect target suppression induced by either mRNA degradation or translational repression. Recent efforts have attempted to analyze miRNA-sequencing data alongside large proteomics data sets (87). Given the high degree of difficulty associated with producing high-quality proteomics data, this approach will likely gain traction as quantitative techniques evolve (23, 52, 67). Careful and thorough investigation is required to support the selection of a miRNA target-of-interest, compiling all we know from published literature about potential target function in different tissues or organisms and pairing that with rigorous bench-top validation of meaningful miRNA-mRNA target interaction.
The method by which the presence and abundance of miRNA are detected or modulated is a key consideration as we progress in the field of miRNA research. Small RNA sequencing is a robust method that can provide single base resolution to identify differential expression of miRNA from the same family. However, the relatively high cost of such analysis and the burden of “big data” placed on an investigator may deter some from using this approach. Many investigators opt to incorporate smaller scale approaches to miRNA expression profiles, such as quantitative (q)PCR or qPCR-based microarrays; however, one must be careful to confirm the specificity of such techniques when studying miRNA families with nearly identical sequences. Specific information about the design and sequence of commercially available primers is generally unavailable to investigators, and specificity may not be guaranteed. Additionally, clear and accurate descriptions of research methods are important for proper understanding of the exact miRNA reported or studied.
Selective perturbation of miR-146a and/or miR-146b.
Recent published reports highlight various attempts to knock down and overexpress these miRNAs. Selective perturbation of the miR-146 family members helps shed light on the specific mechanisms of each miRNA. Current technology allows for knockdown of a mature miRNA via RNA silencing approaches and/or overexpression via delivery of exogenous precursor or mature miRNA. These provide the investigator tools to test the functional effects of modulating the abundance of a given miRNA in vitro or in vivo.
Anti-sense technologies utilize synthetic oligonucleotide sequences with complementarity to the mature miRNA with special chemical modifications that increases overall stability of the knock-down effect, and this can be successfully administered in vivo to knock down a specific miRNA (22, 25, 26, 47, 48). Because these anti-miRNAs work through sequence complementarity, it is important to test the specificity of the targeting oligonucleotides for a miRNA-of-interest as well as those of other highly similar miRNAs before specificity can be confirmed.
Precursor or mimic technologies function to increase the abundance of a mature sequence, either by providing an exogenous source of precursor miRNA for subsequent posttranscriptional processing to form mature miRNA or by supplying an exogenous mimic of the endogenous mature miRNA-of-interest. These technologies are a more robust way to ensure specific increase in abundance of the miRNA-of-interest. miR-146a and miR-146b have distinct primary and precursor sequences, and thus supplying an exogenous precursor containing a specific mature -5p sequence ensures a greater level of confidence in the specificity of the approach. These methods have been largely successful in upregulating target-miR expression in vitro, but genomic editing/transgenesis remains the dominant approach for studying miRNA upregulation in vivo.
Publication ambiguity.
For over a decade, miR-146a and miR-146b, as with many other miRNA families, have been reported together; sometimes presuming no difference in activity (often reported as “miR-146a/b”) and regularly making no distinction between the two (i.e., “miR-146”). Moreover, a complete description of the methods used is key to determining whether a distinction is possible using the selected approach. If there is any uncertainty, the author should make a statement identifying the factors limiting interpretation of the work. For example, one report highlights the finding that patients with coronary artery disease had increased abundance of both miR-146a and miR-146b. While in vitro treatment of cells with miR-146a and miR-146b mimics and inhibitors showed expected reciprocal expression of target gene expression, the in vivo data did not reveal this expected pattern, rather, showing a positive correlation between miR abundance and target expression (86). The authors of the report rightly address this discrepancy in the discussion section and suggest an explanation. Confusion ensues when a paper reports hsa-miR-146a to be upregulated in lung cancer (97) and is later cited as support that hsa-miR-146b “is known to be downregulated in lung cancer” (3). As will be discussed in the next section, a growing body of literature suggests that these two miRNAs have unique and important roles in pathophysiology. Therefore, it is vital that a clear distinction is made between miR-146a and miR-146b when data are reported. Moreover, a complete description of the methods used is key to determining whether a distinction can even be made with any accuracy using the selected approach. If there is any uncertainty, the author should make a statement identifying the factors limiting interpretation of the work. Publication of ambiguous or unclear results adds to the difficulty of determining miR-146a or miR-146b specific activity and limits understanding of their regulatory actions.
miR-146 Family in Pathophysiology
With the increasing accessibility to miRNA profiling data sets, there are a growing number of studies that report differential expression or correlation of the miR-146 family members with pathological conditions (Table 1). The studies that focus on these miRNAs fall mainly into two broad categories: cancer and inflammation. Many studies identifying miRNA-associated pathologies report expression profiles for a panel of miRNAs. As noted above, it is important to be aware of the method by which miRNA family members are reported, differentiated, and quantified; as these distinctions are vital to the proper interpretation of each study.
Table 1.
Summary of the findings of miR-146a and miR-146b activity in associated pathologies
| Associated Pathology or Process | miRNA | Target | Tissue/Cell Type | Reference | |
|---|---|---|---|---|---|
| Cancer, inflammation, and innate immune response | expression profile of regulatory T cells compared with naïve T cells | 146 ↑ | N/A | regulatory T cells | Cobb (19) |
| inhibition of NF-κB signaling | 146a/b ↑ | IRAK1/TRAF6 ↓ | leukemic monocyte cell line | Taganov (85) | |
| Treg treatment of graft-vs.-host disease | 146b ↓ | TRAF6 ↑ | regulatory T cells | Lu (64) | |
| breast cancer malignancy, increased patient survival | 146b ↑ | IRAK1/TRAF6 ↓ | breast cancer cell | Xiang (93) | |
| 5q- syndrome | 146a ↓ | TRAF6 ↑ | hematopoietic stem and progenitor cells | Starczynowski (83) | |
| FOXP3-induced cellular apoptosis | 146a ↑ | IRAK1/TRAF6 ↓ | breast cancer cell | Liu (60) | |
| PDGF-stimulated tumor growth | 146b ↑ | EGFR ↓ | glioblastoma cells | Shao (80) | |
| Cardiovascular disease | coronary artery disease | 146a/b ↑ | IRAK1/TRAF6 ↓ (in vitro) | peripheral blood mononuclear cells | Takahashi (86) |
| atherosclerotic plaque formation | 146a/b ↑ | IGSF1, SORT1, and NOVA1 ↓ | patient arteries | Raitoharju (75) | |
| angiopoietin-1 mediates suppression of LPS-induced inflammatory response | 146b ↑ | IRAK1/TRAF6 ↓ | human umbilical vein endothelial cells | Echavarria (24) | |
| cardioprotection in microbial sepsis | 146a ↑ | IRAK1/TRAF6 ↓ | cardiac tissue; cardiac myocytes and monocytic cells | Gao (28) | |
| risk for coronary artery disease positively correlated with presence of SNP | 146a ↓ | N/A | N/A; SNP typing | Jazdzewski (39), Xiong (95) | |
| left-ventricular remodeling after myocardial infarction | 146a ↑ | N/A | circulating plasma | Liu (61) | |
| cyanotic congenital heart disease and chronic hypoxia | 146b ↑ | RNase L ↓ | myocardial tissue, rat cardiomyocytes | Li (54) | |
| attenuation of proinflammatory stress in hyperlipidemia | 146a ↑ | IRAK1/TRAF6 ↓ | monocytes and macrophages | Li (55) | |
| Acute and chronic kidney disease | CKD patients on hemodialysis | 146b ↑ and 146a ↓ | N/A | peripheral blood mononuclear cells | Zawada (99) |
| acute kidney injury and kidney fibrosis | 146b ↑ | N/A | renal cortex tissue | Pellegrini (73) | |
| unilateral ureteral obstruction model of kidney fibrosis | 146b ↑ | Smad4 ↓ | renal cortex tissue | Morishita (70) | |
| TGF-β1 induced renal interstitial fibrosis | 146b ↓ | N/A | renal epithelial cells | Kriegel (42) | |
| cisplatin-induced acute kidney injury | 146b ↑ | ErbB4 ↓ | renal tissue and renal tubular epithelial cells | Zhu (101) | |
miR, miRNA: microRNA. N/A, not available.
Cancer, inflammation, and the innate immune response.
The involvement of miR-146a and miR-146b in inflammation is the most advanced field of miR-146 research. Beginning with Taganov’s seminal paper in 2006, which spearheaded interest in miR-146 research, the role of this miRNA family in toll-like receptor (TLR) signaling and the innate immune response has been widely studied (16, 21, 66, 74, 78, 81). The same year, Cobb et al. (19) reported the important role of “miR-146” in the development of mature regulatory T cells (Tregs), including miR-146 in a characteristic subset of miRNAs uniquely expressed in Tregs distinct from naïve T cells. By 2008, Sheedy et al. (81) published a review that made special note of the growing knowledge of the activity of this miRNA family in the innate immune response, a common pathway suggested by early miR-146 publications (i.e., TLR-4 signaling). A large proportion of the reported studies identify interleukin-1 receptor-associated kinase 1 (IRAK1) and tumor necrosis factor receptor-associated factor 6 (TRAF6) as the key targets of the miR-146 family, as summarized in Fig. 2 (9, 36, 93). IRAK1 and TRAF6 have long been known to play a vital role in proper TLR signaling, one mechanism by which the innate immune system senses a pathogenic stimulus. Bacterial lipopolysaccharide (LPS, also: endotoxin) stimulates TLR-4 and sets off a signaling cascade acting through IRAK1/TRAF6 culminating in the activation of NF-κB and AP-1 transcription factors and expression of immune response genes (2). While several classes of negative feedback controllers of TLR-4 signaling had been previously reported, Taganov et al. (85) were the first to suggest that the miR-146 family acted as such. THP-1 cells (derived from an acute monocytic leukemia patient) were stimulated with LPS, and the expression profile of miRNAs was analyzed by microarray. The investigators found that both MIR146A and MIR146B are endotoxin-responsive genes, and promotor analysis identified MIR146A upregulation as NF-κB dependent. After validating IRAK1 and TRAF6 as targets of miR-146a and miR-146b with luciferase reporter constructs, they proposed that the upregulation of these miRNAs serves as negative feedback regulation of TLR-4 signaling (85). At the same time confirming miR-146a/b are endotoxin responsive, Kutty et al. (50) also reported preferential upregulation of miR-146a or miR-146b depends on the inflammatory stimulus. In vitro treatment of human retinal epithelial cells with various cytokines revealed maximal miR-146a upregulation is dependent on interleukin-1β and miR-146b dependent on interferon-γ. These data highlight the unique role the individual miR-146 family members may play in mediating the immune response.
Fig. 2.
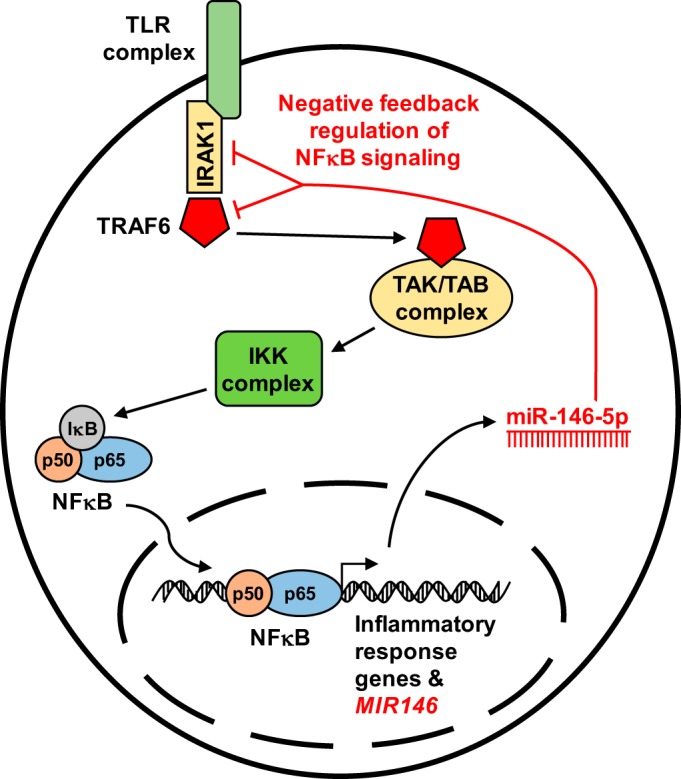
A large majority of miR-146 literature focuses on the innate immune and inflammatory responses mediated by NF-κB signaling. miR-146 has been shown to act as a negative feedback regulator of NF-κB signaling by suppressing the translation of 2 targets: interleukin-1 receptor-associated kinase 1 (IRAK1) and tumor necrosis factor receptor-associated factor 6 (TRAF6). Stimulation of the toll-like receptor complex (TLR) leads to the association of IRAK1 and TRAF6 with the TAK/TAB complex, resulting in the subsequent activation of the IKK complex and translocation of NF-κB to the nucleus where it induces expression of inflammatory/immune response genes and induces expression of miR-146. miR-146 then may act as a molecular brake on NF-κB signaling by suppressing IRAK1 and TRAF6 expression.
Dysregulation of the innate immune response has been shown to be oncogenic, and miR-146a/b have been implicated in this process, as outlined in Fig. 3 (76). In 2008, Bhaumik et al. (8) reported increased expression of miR-146a/b in metastatic human breast cancer cells negatively regulated NF-κB signaling and reduced the cells’ metastatic phenotype, providing the first clear functional connection of miR-146a/b to cancer pathology. In 2010, Starczynowski et al. (83) published a study in Nature Medicine in which they reported the loss of MIR146A in 5q- syndrome (a hematological disorder and subtype of myelodysplastic syndrome in which there is a loss of the long arm of chromosome 5q33.1) accounted for, in part, patients’ increased susceptibility to cancer. The data showed a clear, inverse correlation between a reduction in miR-146a abundance and increased expression of the target gene TRAF6, which leads to inappropriate activation of innate immune signaling in hematopoietic stem and progenitor cells (83). Derepression of TRAF6 signaling in a mouse culture model altered hematopoiesis and bone marrow failure, suggesting the importance of miR-146a in relation to the pathogenesis of malignancy (88). Clinical data support this, showing 5q- syndrome associates with poorer prognoses in patients with acute myeloid leukemia (10, 68). Xiang et al. (93) highlighted the tumor suppressor role of miR-146b in modulating negative feedback regulation of NF-κB signaling, suggesting that upregulation of miR-146b may be a molecular brake on tumorigenesis.
Fig. 3.
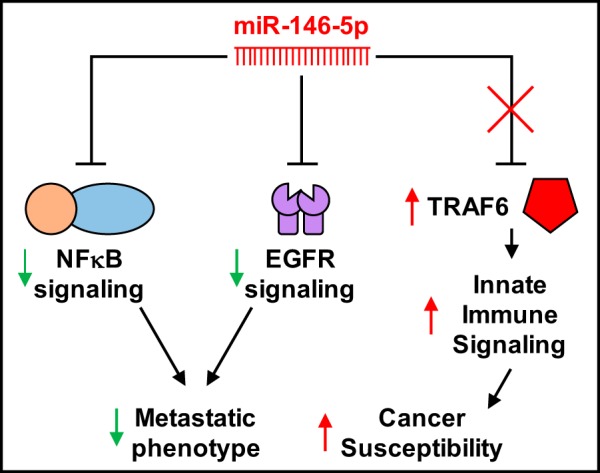
Diagram depicting 3 simple pathways of miR-146-5p modulation of cancer phenotypes. The previously discussed NF-κB signaling pathway has been shown to be induce the metastatic phenotype of various breast cancer cells. Increased expression of miR-146a/b acts to block NF-κB signaling and thus reduces this metastatic potential. miR-146b also fine-tunes the cell’s response to growth factor stimulus in a cancer setting. Platelet-derived growth factor increases expression of miR-146b, which subsequently blocks epidermal growth factor receptor (EGFR) signaling thereby halting metastatic growth. When miR-146a is lost due to a chromosomal abnormality (5q syndrome), aberrant expression of tumor necrosis factor receptor-associated factor 6 (TRAF6) induces NF-κB signaling and increases cancer susceptibility.
Aberrant signaling by platelet-derived growth factor (PDGF) is another important oncogenic mechanism through which the miR-146 family is involved. In 2011, Shao et al. (80) identified a negative feedback regulatory role for miR-146b in human glioblastoma cells. PDGF-BB (a known ligand of PDGF) induced the expression of miR-146b via a c-fos-dependent mechanism. Increased miR-146b inversely correlated with abundance of the epidermal growth factor receptor, fine-tuning the cells’ response to growth factor stimulus in a cancer setting (80), supporting a previous study that suggested therapeutic potential for miR-146a or miR-146b in controlling breast cancer metastasis (38). Liu et al. (60) reported interesting data suggesting a specific role for miR-146a and not miR-146b in FOXP3-induced apoptosis of breast cancer cells. The authors showed miR-146a upregulated by FOXP3+ T-regulatory cells inhibited NF-κB signaling through suppression of IRAK1 and TRAF6, leading to increased cellular apoptosis, suggesting a new mechanism for FOXP3-mediated tumor suppression critically supported by the miR-146 family (60).
The role of the miR-146 family in the immune response has focused on functions outside of cancer biology as well (35, 64, 90). In a recent study investigating the role of Tregs in the maintenance of proper immune response balance, Lu et al. (64) proposes knockdown of miR-146b (and not miR-146a) helps to prolong the immune-suppressive effect of Tregs in graft vs. host disease, a disease in which adoptive Treg transfer is effective treatment. The data showed knockdown of miR-146b in thymic-derived Tregs (tTregs) led to increased nuclear localization and activation of NF-κB via increased expression of TRAF6. The anti-miR-146b-treated tTregs showed enhanced prosurvival pathway activation and greater efficacy to suppress inflammatory responses (64). In another example, miR-146a has been implicated in the pathogenesis of Alzheimer’s disease (56, 65). miR-146a was shown to be upregulated in brain regions exhibiting neuroinflammation, supporting the role of miR-146a in modulating the immune response in neuropathology. These studies, and others, highlight the complexity and great potential of miRNA research and point to the multitude of cell-specific or condition-specific effects miRNAs may have in pathology.
Relevance to cardiovascular pathophysiology.
While the innate immune and inflammatory responses account for a clear majority of the miR-146 literature, a substantial subsegment of the reported literature regarding the pathophysiological role of the miR-146 family has focused on cardiovascular inflammatory diseases. An early report from Takahashi et al. (86) examined the abundance of miR-146a/b by real-time PCR in peripheral blood mononuclear cells in patients with coronary artery disease (CAD) compared with non-CAD control patients, finding increased abundance of both miR-146a and miR-146b in CAD patients compared with controls. They suggested the importance of the miR-146 family in modulating TLR4 signaling through IRAK1 and TRAF6 inhibition, thereby mediating the development and progression of atherosclerotic disease. Additional evidence suggesting the miR-146 family is modulated in vascular inflammatory disease was reported in a study examining miRNA profiles in atherosclerotic plaques in arteries from patients enrolled in the Tampere Vascular Study (75). The study analyzed miRNA expression data by microarray (Agilent) after which differentially expressed target miRNAs were verified by qRT-PCR. The report showed miR-146a and miR-146b-5p, among others, to be upregulated in the atherosclerotic arteries compared with control arteries. No further mechanism was examined, though these findings suggest this to be an intriguing area of future study. Apolipoprotein E (ApoE) is known to display anti-inflammatory properties and protect from atherosclerosis and inflammatory disease. Loss of ApoE can enhance NF-κB signaling and lead to proinflammatory stress responses. Li et al. (55) reported delivery of miR-146a mimetic to ApoE knockout mice (ApoE−/−) attenuated macrophage activation, atherosclerosis, and proinflammatory responses in hyperlipidemia. This study is one, among many, to suggest the great therapeutic potential of new miRNA drug delivery techniques (57, 91, 98).
There is also evidence of a direct link between miR-146a and myocardial infarction (MI). Zidar et al. (102) correlated miR-146a levels in autopsied heart tissue with severity of infarction in patients who died from complications following MI, including ventricular rupture. The authors suggest that miR-146a is upregulated in response to NF-κB activation in the early innate immune response to severe MI. However, the mechanisms leading to, or resulting from, miR-146a modulation were not identified. It was not determined whether its role is protective or damaging to the heart. In 2012, a published report showed increased expression of circulating miR-146a, among other miRNAs, in the serum of patients diagnosed with acute coronary syndrome (ACS) in an emergency setting (71). The authors suggest their data point to a diagnostic role of circulating miR-146a in early assessment of suspected ACS patients. One study by Liu et al. (61) examined the potential to use circulating miR-146a levels as a biomarker to predict left-ventricular remodeling (LVR) after MI. The authors reported abundance of circulating miR-146a in patients with LVR to be significantly higher than those patients without LVR, subsequently showing miR-146a abundance was an independent predictor of LVR development (odds ratio 2.127). Other studies directly link miR-146b with cardiac pathology. Li et al. (54) showed chronic upregulation of miR-146b-5p in the heart tissue of patients with cyanotic congenital heart disease and chronic hypoxia and suggest a protective role for miR-146b in chronic cardiac hypoxia.
A common minor C allele single nucleotide variation (rs2910164, MAF C = 0.2792) found within the passenger strand of pre-miR-146a was reported to affect the processing of the mature miR-146a sequence by altering transcription factor binding efficiency, reducing the abundance of both pre-miR-146a and miR-146a-5p by nearly twofold and affecting the efficiency of translation repression of target genes (39). This common genetic variation at the miR-146a locus within the Chinese Han population correlates with an increased risk for CAD (95). After genotyping 295 CAD patients and 283 controls by restriction fragment length polymorphism PCR, logistic regression analysis revealed heterozygous patients (CG) or patients homozygous for the minor allele (CC) to have significantly increased risk for CAD compared with patients that did not carry the minor allele. A recent meta-analysis of 10 studies examined the correlation of this common SNP with coronary heart disease, adding support for the pathological relevance of miR-146a regulation in cardiovascular disease (94).
Some investigators have focused on the role of miR-146b in regulating cardiovascular responses to experimentally induced inflammatory processes. Echavarria et al. (24) suggest a role for miR-146b-5p in mediating the LPS-induced inflammatory response in culture human endothelial cells. The study reported angiopoietin-1 mediated the upregulation of miR-146b-5p and subsequently inhibited LPS-induced TLR-4 signaling through suppression of IRAK1 and TRAF6. In 2015, Gao et al. (28) suggested miR-146a is cardioprotective, attenuating cardiac dysfunction in microbial sepsis. Mice subjected to a sepsis-inducing cecal puncture procedure were treated with lenti-viral miR-146a, and cardiac function was monitored via echocardiography. Fractional shortening and ejection fraction were significantly greater in miR-146a-treated animals than untransfected or scramble transfected controls. The authors propose miR-146a upregulation inhibited NF-κB-mediated expression of inflammatory cytokines and infiltration of proinflammatory cells into the myocardium. NF-κB has been reported to play a role in pathological cardiac hypertrophy and heart failure, suggesting it may be an important link between miR-146 function and heart failure (31, 37, 92).
Relevance to renal pathophysiology.
There is recent movement toward understanding the genetic and epigenetic dysregulation of genes involved in miRNA regulation of renal physiology in both chronic and acute pathophysiology (7, 40, 43–45, 59, 77, 96). Zawada et al. (99) published a paper in which they examined miRNA-sequencing data from peripheral blood mononuclear cells in chronic kidney disease (CKD) patients on hemodialysis and compared expression profiles to healthy controls. They identified 182 differentially expressed miRNAs, finding that miR-146b was upregulated and miR-146a was downregulated in this patient population. These data suggest and support the differential regulation of miR-146 family members in CKD. The miR-146 family has also been associated with acute kidney injury (AKI) in humans. Pellegrini et al. (73) published data in 2016 showing miR-146b was highly upregulated in the renal cortex in patients with AKI, noting that miR-146b was maximally upregulated at the peak of fibrosis. The upregulation of miR-146b was recapitulated in a unilateral ureteral obstruction model of kidney fibrosis in the rat. The authors highlight the use of small-RNA sequencing technology to survey expression profiles in a temporal manner throughout distinct phases of kidney injury. Additionally, in an in vitro model of renal interstitial fibrosis, Kriegel et al. (42) examined the change in miRNA expression following TGF-β1 (transforming growth factor-β 1) treatment of human renal epithelial cells. This study reported the downregulation of miR-146b-5p by nearly 1.5-fold in the treated cells compared with vehicle-treated controls, confirmed by qRT-PCR analysis. In a mouse model of unilateral ureteral obstruction, treatment with exogenous miR-146a attenuated renal fibrosis by inhibiting the proinflammatory and profibrotic NF-κB and TGF-β1 signaling cascades (70), suggesting a protective role for miR-146a. In contrast, a recent report published by Zhu et al. (101) suggests a deleterious effect of miR-146b-5p upregulation in cisplatin-induced AKI. Knocking down miR-146b-5p in rat renal tubule epithelial cells using miR-146b inhibitors resulted in protection from cisplatin-induced apoptosis in vitro. This study is one case, among many, where there was clear evidence that the knockdown of miR-146b-5p was effective, but there was no mention of off-target effects on miR-146a expression or compensatory upregulation of miR-146a.
Ongoing Considerations and Future Directions
There are many questions yet unanswered in miRNA research, especially with regard to the miR-146 family. The advent of small noncoding RNA sequencing technologies allows unprecedented precision for researchers to investigate expression profiles of many noncoding RNAs, including miRNAs. Moreover, anti-sense and precursor technologies are new tools at our disposal to study the effects of differential miRNA expression in vivo and in vitro. New evidence shows increased support for “noncanonical” miRNA activity, for example, non-3′-UTR target binding or translational activation of an mRNA transcript (11, 18, 89, 100). Genomic editing technologies that allow researchers to pinpoint knockdown or knock-in of a sequence of interest are one potential avenue on which to find these answers. CRISPR-mediated genomic editing is becoming the new standard for stable knockdown of miRNAs in vitro and in vivo (14). Many studies are identifying miRNAs as exciting biomarkers in pathology (4, 30, 84), and new strategies are in development for the effective delivery of miRNA mimics, pre-miRNAs, or anti-sense oligos, with dozens tested in cancer therapies and some new technologies even reaching clinical trials. The potential therapeutic benefit of in vivo miRNA modulation is only beginning to be uncovered (6, 20, 63, 79). Rigorous experimental approaches will aid us in understanding how and why these miRNAs are differentially regulated in health and disease as well as ways in which we might exploit this knowledge to identify potential therapeutic pathways.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
M.R.P. prepared figures; M.R.P. drafted manuscript; M.R.P. and A.J.K. edited and revised manuscript; A.J.K. approved final version of manuscript.
REFERENCES
- 1.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. eLife 4: e05005, 2015. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol 4: 499–511, 2004. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 3.Backes C, Meese E, Lenhof HP, Keller A. A dictionary on microRNAs and their putative target pathways. Nucleic Acids Res 38: 4476–4486, 2010. doi: 10.1093/nar/gkq167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker M. RNA interference: microRNAs as biomarkers. Nature 464: 1225–1228, 2010. doi: 10.1038/4641227a. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233, 2009. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev 96: 1297–1325, 2016. doi: 10.1152/physrev.00041.2015. [DOI] [PubMed] [Google Scholar]
- 7.Bhatt K, Kato M, Natarajan R. Mini-review: emerging roles of microRNAs in the pathophysiology of renal diseases. Am J Physiol Renal Physiol 310: F109–F118, 2016. doi: 10.1152/ajprenal.00387.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhaumik D, Scott GK, Schokrpur S, Patil CK, Campisi J, Benz CC. Expression of microRNA-146 suppresses NF-kappaB activity with reduction of metastatic potential in breast cancer cells. Oncogene 27: 5643–5647, 2008. doi: 10.1038/onc.2008.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhaumik D, Scott GK, Schokrpur S, Patil CK, Orjalo AV, Rodier F, Lithgow GJ, Campisi J. MicroRNAs miR-146a/b negatively modulate the senescence-associated inflammatory mediators IL-6 and IL-8. Aging (Albany NY) 1: 402–411, 2009. doi: 10.18632/aging.100042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boultwood J, Pellagatti A, McKenzie AN, Wainscoat JS. Advances in the 5q- syndrome. Blood 116: 5803–5811, 2010. doi: 10.1182/blood-2010-04-273771. [DOI] [PubMed] [Google Scholar]
- 11.Brümmer A, Hausser J. MicroRNA binding sites in the coding region of mRNAs: extending the repertoire of post-transcriptional gene regulation. BioEssays 36: 617–626, 2014. doi: 10.1002/bies.201300104. [DOI] [PubMed] [Google Scholar]
- 12.Cai X, Lu S, Zhang Z, Gonzalez CM, Damania B, Cullen BR. Kaposi’s sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc Natl Acad Sci USA 102: 5570–5575, 2005. doi: 10.1073/pnas.0408192102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell 136: 642–655, 2009. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang H, Yi B, Ma R, Zhang X, Zhao H, Xi Y. CRISPR/cas9, a novel genomic tool to knock down microRNA in vitro and in vivo. Sci Rep 6: 22312, 2016. doi: 10.1038/srep22312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, Dang CV, Thomas-Tikhonenko A, Mendell JT. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet 40: 43–50, 2008. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chassin C, Kocur M, Pott J, Duerr CU, Gütle D, Lotz M, Hornef MW. miR-146a mediates protective innate immune tolerance in the neonate intestine. Cell Host Microbe 8: 358–368, 2010. doi: 10.1016/j.chom.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 17.Chien CH, Sun YM, Chang WC, Chiang-Hsieh PY, Lee TY, Tsai WC, Horng JT, Tsou AP, Huang HD. Identifying transcriptional start sites of human microRNAs based on high-throughput sequencing data. Nucleic Acids Res 39: 9345–9356, 2011. doi: 10.1093/nar/gkr604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cipolla GA. A non-canonical landscape of the microRNA system. Front Genet 5: 337, 2014. doi: 10.3389/fgene.2014.00337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cobb BS, Hertweck A, Smith J, O’Connor E, Graf D, Cook T, Smale ST, Sakaguchi S, Livesey FJ, Fisher AG, Merkenschlager M. A role for Dicer in immune regulation. J Exp Med 203: 2519–2527, 2006. doi: 10.1084/jem.20061692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Creemers EE, van Rooij E. Function and therapeutic potential of noncoding RNAs in cardiac fibrosis. Circ Res 118: 108–118, 2016. doi: 10.1161/CIRCRESAHA.115.305242. [DOI] [PubMed] [Google Scholar]
- 21.Curtale G, Mirolo M, Renzi TA, Rossato M, Bazzoni F, Locati M. Negative regulation of Toll-like receptor 4 signaling by IL-10-dependent microRNA-146b. Proc Natl Acad Sci USA 110: 11499–11504, 2013. doi: 10.1073/pnas.1219852110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davis S, Lollo B, Freier S, Esau C. Improved targeting of miRNA with antisense oligonucleotides. Nucleic Acids Res 34: 2294–2304, 2006. doi: 10.1093/nar/gkl183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duque-Guimarães DE, de Almeida-Faria J, Ong TP, Ozanne SE. Pulsed SILAC as a approach for miRNA targets identification in cell culture. Methods Mol Biol 1546: 149–159, 2017. doi: 10.1007/978-1-4939-6730-8_11. [DOI] [PubMed] [Google Scholar]
- 24.Echavarria R, Mayaki D, Neel JC, Harel S, Sanchez V, Hussain SN. Angiopoietin-1 inhibits toll-like receptor 4 signalling in cultured endothelial cells: role of miR-146b-5p. Cardiovasc Res 106: 465–477, 2015. doi: 10.1093/cvr/cvv120. [DOI] [PubMed] [Google Scholar]
- 25.Elmén J, Lindow M, Schütz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjärn M, Hansen HF, Berger U, Gullans S, Kearney P, Sarnow P, Straarup EM, Kauppinen S. LNA-mediated microRNA silencing in non-human primates. Nature 452: 896–899, 2008. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 26.Elmén J, Thonberg H, Ljungberg K, Frieden M, Westergaard M, Xu Y, Wahren B, Liang Z, Ørum H, Koch T, Wahlestedt C. Locked nucleic acid (LNA) mediated improvements in siRNA stability and functionality. Nucleic Acids Res 33: 439–447, 2005. doi: 10.1093/nar/gki193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19: 92–105, 2009. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao M, Wang X, Zhang X, Ha T, Ma H, Liu L, Kalbfleisch JH, Gao X, Kao RL, Williams DL, Li C. Attenuation of cardiac dysfunction in polymicrobial sepsis by microRNA-146a is mediated via targeting of IRAK1 and TRAF6 expression. J Immunol 195: 672–682, 2015. doi: 10.4049/jimmunol.1403155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 34: D140–D144, 2006. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Halkein J, Tabruyn SP, Ricke-Hoch M, Haghikia A, Nguyen NQ, Scherr M, Castermans K, Malvaux L, Lambert V, Thiry M, Sliwa K, Noel A, Martial JA, Hilfiker-Kleiner D, Struman I. MicroRNA-146a is a therapeutic target and biomarker for peripartum cardiomyopathy. J Clin Invest 123: 2143–2154, 2013. doi: 10.1172/JCI64365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hamid T, Guo SZ, Kingery JR, Xiang X, Dawn B, Prabhu SD. Cardiomyocyte NF-κB p65 promotes adverse remodelling, apoptosis, and endoplasmic reticulum stress in heart failure. Cardiovasc Res 89: 129–138, 2011. doi: 10.1093/cvr/cvq274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hausser J, Zavolan M. Identification and consequences of miRNA-target interactions–beyond repression of gene expression. Nat Rev Genet 15: 599–612, 2014. doi: 10.1038/nrg3765. [DOI] [PubMed] [Google Scholar]
- 33.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 5: 522–531, 2004. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 34.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA, Hannon GJ. A microRNA component of the p53 tumour suppressor network. Nature 447: 1130–1134, 2007. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hessam S, Sand M, Skrygan M, Gambichler T, Bechara FG. Expression of miRNA-155, miRNA-223, miRNA-31, miRNA-21, miRNA-125b, and miRNA-146a in the inflammatory pathway of hidradenitis suppurativa. Inflammation, 2016. [Epub ahead of print]. doi: 10.1007/s10753-016-0492-2. [DOI] [PubMed] [Google Scholar]
- 36.Hou J, Wang P, Lin L, Liu X, Ma F, An H, Wang Z, Cao X. MicroRNA-146a feedback inhibits RIG-I-dependent Type I IFN production in macrophages by targeting TRAF6, IRAK1, and IRAK2. J Immunol 183: 2150–2158, 2009. doi: 10.4049/jimmunol.0900707. [DOI] [PubMed] [Google Scholar]
- 37.Huang H, Joseph LC, Gurin MI, Thorp EB, Morrow JP. Extracellular signal-regulated kinase activation during cardiac hypertrophy reduces sarcoplasmic/endoplasmic reticulum calcium ATPase 2 (SERCA2) transcription. J Mol Cell Cardiol 75: 58–63, 2014. doi: 10.1016/j.yjmcc.2014.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hurst DR, Edmonds MD, Scott GK, Benz CC, Vaidya KS, Welch DR. Breast cancer metastasis suppressor 1 up-regulates miR-146, which suppresses breast cancer metastasis. Cancer Res 69: 1279–1283, 2009. doi: 10.1158/0008-5472.CAN-08-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jazdzewski K, Murray EL, Franssila K, Jarzab B, Schoenberg DR, de la Chapelle A. Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proc Natl Acad Sci USA 105: 7269–7274, 2008. doi: 10.1073/pnas.0802682105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kato M, Wang M, Chen Z, Bhatt K, Oh HJ, Lanting L, Deshpande S, Jia Y, Lai JY, O’Connor CL, Wu Y, Hodgin JB, Nelson RG, Bitzer M, Natarajan R. An endoplasmic reticulum stress-regulated lncRNA hosting a microRNA megacluster induces early features of diabetic nephropathy. Nat Commun 7: 12864, 2016. doi: 10.1038/ncomms12864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kriegel AJ, Baker MA, Liu Y, Liu P, Cowley AW Jr, Liang M. Endogenous microRNAs in human microvascular endothelial cells regulate mRNAs encoded by hypertension-related genes. Hypertension 66: 793–799, 2015. doi: 10.1161/HYPERTENSIONAHA.115.05645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kriegel AJ, Fang Y, Liu Y, Tian Z, Mladinov D, Matus IR, Ding X, Greene AS, Liang M. MicroRNA-target pairs in human renal epithelial cells treated with transforming growth factor beta 1: a novel role of miR-382. Nucleic Acids Res 38: 8338–8347, 2010. doi: 10.1093/nar/gkq718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kriegel AJ, Liu Y, Cohen B, Usa K, Liu Y, Liang M. MiR-382 targeting of kallikrein 5 contributes to renal inner medullary interstitial fibrosis. Physiol Genomics 44: 259–267, 2012. doi: 10.1152/physiolgenomics.00173.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kriegel AJ, Liu Y, Fang Y, Ding X, Liang M. The miR-29 family: genomics, cell biology, and relevance to renal and cardiovascular injury. Physiol Genomics 44: 237–244, 2012. doi: 10.1152/physiolgenomics.00141.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kriegel AJ, Mladinov D, Liang M. Translational study of microRNAs and its application in kidney disease and hypertension research. Clin Sci (Lond) 122: 439–447, 2012. doi: 10.1042/CS20110159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 11: 597–610, 2010. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 47.Krützfeldt J, Kuwajima S, Braich R, Rajeev KG, Pena J, Tuschl T, Manoharan M, Stoffel M. Specificity, duplex degradation and subcellular localization of antagomirs. Nucleic Acids Res 35: 2885–2892, 2007. doi: 10.1093/nar/gkm024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 438: 685–689, 2005. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 49.Kulshreshtha R, Davuluri RV, Calin GA, Ivan M. A microRNA component of the hypoxic response. Cell Death Differ 15: 667–671, 2008. doi: 10.1038/sj.cdd.4402310. [DOI] [PubMed] [Google Scholar]
- 50.Kutty RK, Nagineni CN, Samuel W, Vijayasarathy C, Jaworski C, Duncan T, Cameron JE, Flemington EK, Hooks JJ, Redmond TM. Differential regulation of microRNA-146a and microRNA-146b-5p in human retinal pigment epithelial cells by interleukin-1β, tumor necrosis factor-α, and interferon-γ. Mol Vis 19: 737–750, 2013. [PMC free article] [PubMed] [Google Scholar]
- 51.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol 12: 735–739, 2002. doi: 10.1016/S0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 52.Lam MP, Lau E, Ng DC, Wang D, Ping P. Cardiovascular proteomics in the era of big data: experimental and computational advances. Clin Proteomics 13: 23, 2016. doi: 10.1186/s12014-016-9124-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li J, Shan F, Xiong G, Wang JM, Wang WL, Xu X, Bai Y. Transcriptional regulation of miR-146b by C/EBPβ LAP2 in esophageal cancer cells. Biochem Biophys Res Commun 446: 267–271, 2014. doi: 10.1016/j.bbrc.2014.02.096. [DOI] [PubMed] [Google Scholar]
- 54.Li JW, He SY, Feng ZZ, Zhao L, Jia WK, Liu P, Zhu Y, Jian Z, Xiao YB. MicroRNA-146b inhibition augments hypoxia-induced cardiomyocyte apoptosis. Mol Med Rep 12: 6903–6910, 2015. doi: 10.3892/mmr.2015.4333. [DOI] [PubMed] [Google Scholar]
- 55.Li K, Ching D, Luk FS, Raffai RL. Apolipoprotein E enhances microRNA-146a in monocytes and macrophages to suppress nuclear factor-κB-driven inflammation and atherosclerosis. Circ Res 117: e1–e11, 2015. doi: 10.1161/CIRCRESAHA.117.305844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li YY, Cui JG, Hill JM, Bhattacharjee S, Zhao Y, Lukiw WJ. Increased expression of miRNA-146a in Alzheimer’s disease transgenic mouse models. Neurosci Lett 487: 94–98, 2011. doi: 10.1016/j.neulet.2010.09.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li Z, Rana TM. Therapeutic targeting of microRNAs: current status and future challenges. Nat Rev Drug Discov 13: 622–638, 2014. doi: 10.1038/nrd4359. [DOI] [PubMed] [Google Scholar]
- 58.Liang M. MicroRNA: a new entrance to the broad paradigm of systems molecular medicine. Physiol Genomics 38: 113–115, 2009. doi: 10.1152/physiolgenomics.00080.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liang M, Liu Y, Mladinov D, Cowley AW Jr, Trivedi H, Fang Y, Xu X, Ding X, Tian Z. MicroRNA: a new frontier in kidney and blood pressure research. Am J Physiol Renal Physiol 297: F553–F558, 2009. doi: 10.1152/ajprenal.00045.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu R, Liu C, Chen D, Yang WH, Liu X, Liu CG, Dugas CM, Tang F, Zheng P, Liu Y, Wang L. FOXP3 controls an miR-146/NF-κB negative feedback loop that inhibits apoptosis in breast cancer cells. Cancer Res 75: 1703–1713, 2015. doi: 10.1158/0008-5472.CAN-14-2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu X, Dong Y, Chen S, Zhang G, Zhang M, Gong Y, Li X. Circulating microRNA-146a and microRNA-21 predict left ventricular remodeling after ST-elevation myocardial infarction. Cardiology 132: 233–241, 2015. doi: 10.1159/000437090. [DOI] [PubMed] [Google Scholar]
- 62.Liu Y, Taylor NE, Lu L, Usa K, Cowley AW Jr, Ferreri NR, Yeo NC, Liang M. Renal medullary microRNAs in Dahl salt-sensitive rats: miR-29b regulates several collagens and related genes. Hypertension 55: 974–982, 2010. doi: 10.1161/HYPERTENSIONAHA.109.144428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lorenzen JM, Haller H, Thum T. MicroRNAs as mediators and therapeutic targets in chronic kidney disease. Nat Rev Nephrol 7: 286–294, 2011. doi: 10.1038/nrneph.2011.26. [DOI] [PubMed] [Google Scholar]
- 64.Lu Y, Hippen KL, Lemire AL, Gu J, Wang W, Ni X, Ranganathan P, Levine BL, Riley JL, June CH, Turka LA, Munn DH, Garzon R, Lu L, Blazar BR. miR-146b antagomir-treated human Tregs acquire increased GVHD inhibitory potency. Blood 128: 1424–1435, 2016. doi: 10.1182/blood-2016-05-714535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lukiw WJ, Zhao Y, Cui JG. An NF-kappaB-sensitive micro RNA-146a-mediated inflammatory circuit in Alzheimer disease and in stressed human brain cells. J Biol Chem 283: 31315–31322, 2008. doi: 10.1074/jbc.M805371200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ma X, Becker Buscaglia LE, Barker JR, Li Y. MicroRNAs in NF-kappaB signaling. J Mol Cell Biol 3: 159–166, 2011. doi: 10.1093/jmcb/mjr007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ma X, Zhu Y, Huang Y, Tegeler T, Gao SJ, Zhang J. Quantitative proteomic approach for microRNA target prediction based on (18)O/(16)O labeling. Cancer Inform 14, Suppl 5: 163–173, 2016. doi: 10.4137/CIN.S30563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mauritzson N, Albin M, Rylander L, Billström R, Ahlgren T, Mikoczy Z, Björk J, Strömberg U, Nilsson PG, Mitelman F, Hagmar L, Johansson B. Pooled analysis of clinical and cytogenetic features in treatment-related and de novo adult acute myeloid leukemia and myelodysplastic syndromes based on a consecutive series of 761 patients analyzed 1976-1993 and on 5098 unselected cases reported in the literature 1974-2001. Leukemia 16: 2366–2378, 2002. doi: 10.1038/sj.leu.2402713. [DOI] [PubMed] [Google Scholar]
- 69.Montgomery RL, van Rooij E. Therapeutic advances in microRNA targeting. J Cardiovasc Pharmacol 57: 1–7, 2011. doi: 10.1097/FJC.0b013e3181f603d0. [DOI] [PubMed] [Google Scholar]
- 70.Morishita Y, Imai T, Yoshizawa H, Watanabe M, Ishibashi K, Muto S, Nagata D. Delivery of microRNA-146a with polyethylenimine nanoparticles inhibits renal fibrosis in vivo. Int J Nanomedicine 10: 3475–3488, 2015. doi: 10.2147/IJN.S82587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Oerlemans MI, Mosterd A, Dekker MS, de Vrey EA, van Mil A, Pasterkamp G, Doevendans PA, Hoes AW, Sluijter JP. Early assessment of acute coronary syndromes in the emergency department: the potential diagnostic value of circulating microRNAs. EMBO Mol Med 4: 1176–1185, 2012. doi: 10.1002/emmm.201201749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pasquinelli AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet 13: 271–282, 2012. doi: 10.1038/nrg3162. [DOI] [PubMed] [Google Scholar]
- 73.Pellegrini KL, Gerlach CV, Craciun FL, Ramachandran K, Bijol V, Kissick HT, Vaidya VS. Application of small RNA sequencing to identify microRNAs in acute kidney injury and fibrosis. Toxicol Appl Pharmacol 312: 42–52, 2016. doi: 10.1016/j.taap.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Perry MM, Moschos SA, Williams AE, Shepherd NJ, Larner-Svensson HM, Lindsay MA. Rapid changes in microRNA-146a expression negatively regulate the IL-1beta-induced inflammatory response in human lung alveolar epithelial cells. J Immunol 180: 5689–5698, 2008. doi: 10.4049/jimmunol.180.8.5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Raitoharju E, Lyytikäinen LP, Levula M, Oksala N, Mennander A, Tarkka M, Klopp N, Illig T, Kähönen M, Karhunen PJ, Laaksonen R, Lehtimäki T. miR-21, miR-210, miR-34a, and miR-146a/b are up-regulated in human atherosclerotic plaques in the Tampere Vascular Study. Atherosclerosis 219: 211–217, 2011. doi: 10.1016/j.atherosclerosis.2011.07.020. [DOI] [PubMed] [Google Scholar]
- 76.Rakoff-Nahoum S, Medzhitov R. Regulation of spontaneous intestinal tumorigenesis through the adaptor protein MyD88. Science 317: 124–127, 2007. doi: 10.1126/science.1140488. [DOI] [PubMed] [Google Scholar]
- 77.Reddy MA, Natarajan R. Recent developments in epigenetics of acute and chronic kidney diseases. Kidney Int 88: 250–261, 2015. doi: 10.1038/ki.2015.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Saba R, Sorensen DL, Booth SA. MicroRNA-146a: a dominant, negative regulator of the innate immune response. Front Immunol 5: 578, 2014. doi: 10.3389/fimmu.2014.00578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schmidt MF. miRNA targeting drugs: the next blockbusters? Methods Mol Biol 1517: 3–22, 2017. doi: 10.1007/978-1-4939-6563-2_1. [DOI] [PubMed] [Google Scholar]
- 80.Shao M, Rossi S, Chelladurai B, Shimizu M, Ntukogu O, Ivan M, Calin GA, Matei D. PDGF induced microRNA alterations in cancer cells. Nucleic Acids Res 39: 4035–4047, 2011. doi: 10.1093/nar/gkq1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sheedy FJ, O’Neill LA. Adding fuel to fire: microRNAs as a new class of mediators of inflammation. Ann Rheum Dis 67, Suppl 3: iii50–iii55, 2008. doi: 10.1136/ard.2008.100289. [DOI] [PubMed] [Google Scholar]
- 82.Smale ST. Luciferase assay. Cold Spring Harb Protoc 2010: pdb.prot5421, 2010. doi: 10.1101/pdb.prot5421. [DOI] [PubMed] [Google Scholar]
- 83.Starczynowski DT, Kuchenbauer F, Argiropoulos B, Sung S, Morin R, Muranyi A, Hirst M, Hogge D, Marra M, Wells RA, Buckstein R, Lam W, Humphries RK, Karsan A. Identification of miR-145 and miR-146a as mediators of the 5q- syndrome phenotype. Nat Med 16: 49–58, 2010. doi: 10.1038/nm.2054. [DOI] [PubMed] [Google Scholar]
- 84.Sun M, Fang S, Li W, Li C, Wang L, Wang F, Wang Y. Associations of miR-146a and miR-146b expression and clinical characteristics in papillary thyroid carcinoma. Cancer Biomark 15: 33–40, 2015. doi: 10.3233/CBM-140431. [DOI] [PubMed] [Google Scholar]
- 85.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA 103: 12481–12486, 2006. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Takahashi Y, Satoh M, Minami Y, Tabuchi T, Itoh T, Nakamura M. Expression of miR-146a/b is associated with the Toll-like receptor 4 signal in coronary artery disease: effect of renin-angiotensin system blockade and statins on miRNA-146a/b and Toll-like receptor 4 levels. Clin Sci (Lond) 119: 395–405, 2010. doi: 10.1042/CS20100003. [DOI] [PubMed] [Google Scholar]
- 87.Torres S, Garcia-Palmero I, Bartolomé RA, Fernandez-Aceñero MJ, Molina E, Calviño E, Segura MF, Ignacio Casal J. Combined miRNA profiling and proteomics demonstrates that different miRNAs target a common set of proteins to promote colorectal cancer metastasis. J Pathol [Epub ahead of print]. doi: 10.1002/path.4874. [DOI] [PubMed] [Google Scholar]
- 88.Varney ME, Niederkorn M, Konno H, Matsumura T, Gohda J, Yoshida N, Akiyama T, Christie S, Fang J, Miller D, Jerez A, Karsan A, Maciejewski JP, Meetei RA, Inoue J, Starczynowski DT. Loss of Tifab, a del(5q) MDS gene, alters hematopoiesis through derepression of Toll-like receptor-TRAF6 signaling. J Exp Med 212: 1967–1985, 2015. doi: 10.1084/jem.20141898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vasudevan S. Posttranscriptional upregulation by microRNAs. Wiley Interdiscip Rev RNA 3: 311–330, 2012. doi: 10.1002/wrna.121. [DOI] [PubMed] [Google Scholar]
- 90.Wang LL, Huang Y, Wang G, Chen SD. The potential role of microRNA-146 in Alzheimer’s disease: biomarker or therapeutic target? Med Hypotheses 78: 398–401, 2012. doi: 10.1016/j.mehy.2011.11.019. [DOI] [PubMed] [Google Scholar]
- 91.Wen D, Danquah M, Chaudhary AK, Mahato RI. Small molecules targeting microRNA for cancer therapy: Promises and obstacles. J Control Release 219: 237–247, 2015. doi: 10.1016/j.jconrel.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wong SC, Fukuchi M, Melnyk P, Rodger I, Giaid A. Induction of cyclooxygenase-2 and activation of nuclear factor-kappaB in myocardium of patients with congestive heart failure. Circulation 98: 100–103, 1998. doi: 10.1161/01.CIR.98.2.100. [DOI] [PubMed] [Google Scholar]
- 93.Xiang M, Birkbak NJ, Vafaizadeh V, Walker SR, Yeh JE, Liu S, Kroll Y, Boldin M, Taganov K, Groner B, Richardson AL, Frank DA. STAT3 induction of miR-146b forms a feedback loop to inhibit the NF-κB to IL-6 signaling axis and STAT3-driven cancer phenotypes. Sci Signal 7: ra11, 2014. doi: 10.1126/scisignal.2004497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Xie X, Shi X, Xun X, Rao L. Association between microRNA polymorphisms and coronary heart disease: A meta-analysis. Herz, 2016. [Epub ahead of print]. doi: 10.1007/s00059-016-4495-4. [DOI] [PubMed] [Google Scholar]
- 95.Xiong XD, Cho M, Cai XP, Cheng J, Jing X, Cen JM, Liu X, Yang XL, Suh Y. A common variant in pre-miR-146 is associated with coronary artery disease risk and its mature miRNA expression. Mutat Res 761: 15–20, 2014. doi: 10.1016/j.mrfmmm.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 96.Xu X, Kriegel AJ, Liu Y, Usa K, Mladinov D, Liu H, Fang Y, Ding X, Liang M. Delayed ischemic preconditioning contributes to renal protection by upregulation of miR-21. Kidney Int 82: 1167–1175, 2012. doi: 10.1038/ki.2012.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, Calin GA, Liu CG, Croce CM, Harris CC. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 9: 189–198, 2006. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 98.Yin H, Kanasty RL, Eltoukhy AA, Vegas AJ, Dorkin JR, Anderson DG. Non-viral vectors for gene-based therapy. Nat Rev Genet 15: 541–555, 2014. doi: 10.1038/nrg3763. [DOI] [PubMed] [Google Scholar]
- 99.Zawada AM, Rogacev KS, Müller S, Rotter B, Winter P, Fliser D, Heine GH. Massive analysis of cDNA Ends (MACE) and miRNA expression profiling identifies proatherogenic pathways in chronic kidney disease. Epigenetics 9: 161–172, 2014. doi: 10.4161/epi.26931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang Y, Wang Z, Gemeinhart RA. Progress in microRNA delivery. J Control Release 172: 962–974, 2013. doi: 10.1016/j.jconrel.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhu Y, Yu J, Yin L, Zhou Y, Sun Z, Jia H, Tao Y, Liu W, Zhang B, Zhang J, Wang M, Zhang X, Yan Y, Xue J, Gu H, Mao F, Xu W, Qian H. MicroRNA-146b, a Sensitive Indicator of Mesenchymal Stem Cell Repair of Acute Renal Injury. Stem Cells Transl Med 5: 1406–1415, 2016. doi: 10.5966/sctm.2015-0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zidar N, Boštjančič E, Glavač D, Stajer D. MicroRNAs, innate immunity and ventricular rupture in human myocardial infarction. Dis Markers 31: 259–265, 2011. doi: 10.1155/2011/247654. [DOI] [PMC free article] [PubMed] [Google Scholar]