Abstract
Clinical management of prostate cancer remains a significant challenge due to the lack of available tests for guiding treatment decisions. The blood prostate-specific antigen test has facilitated early detection and intervention of prostate cancer. However, blood prostate-specific antigen levels are less effective in distinguishing aggressive from indolent prostate cancers and other benign prostatic diseases. Thus, the development of novel approaches specific for prostate cancer that can differentiate aggressive from indolent disease remains an urgent medical need. In the current study, we evaluated urine specimens from prostate cancer patients using LC-MS/MS, with the aim of identifying effective urinary prostate cancer biomarkers. Glycoproteins from urine samples of prostate cancer patients with different Gleason scores were characterized via solid phase extraction of N-linked glycosite-containing peptides and LC-MS/MS. A total of 2923 unique glycosite-containing peptides were identified. Glycoproteomic comparison on urine and tissues from aggressive and non-aggressive prostate cancers as well as sera from prostate cancer patients revealed that the majority of AG prostate cancer associated glycoproteins were more readily detected in patient’s urine than serum samples. Our data collectively indicate that urine provides a potential source for biomarker testing in patients with AG prostate cancer.
Keywords: Aggressive, Glycoprotein, Glycoproteomics, Prostate cancer, Urine
1 Introduction
Prostate cancer is the most commonly diagnosed cancer type among American men, remaining the leading cause of new cancer cases and the second highest cause of death in men in the United States in 2015 [1]. Determination of serum prostate-specific antigen (PSA) levels and its isoforms (together with digital rectal examination) for prostate cancer screening [2] has facilitated early prostate cancer detection and intervention [3,4]. However, the major limitation of PSA as a surrogate serum marker for prostate cancer is its low specificity and high prevalence of detecting indolent prostate cancer [5]. Biopsy-based Gleason score is currently the optimal method to detect aggressive (AG) prostate cancer [6], but it generates sampling errors [7].
Proteomics provides a powerful means to examine the whole proteome or subproteomes in different disease conditions [8]. The majority of extracellular proteins, including cell surface, transmembrane, and secreted proteins, are modified by glycans, and therefore referred to as glycoproteins. Owing to their accessibility in body fluids for cancer diagnostics, glycoproteins constitute the major biochemical class of clinical biomarkers [9]. Most cancer biomarkers approved by the U.S. Food and Drug Administration to date are glycoproteins, for instance, PSA for prostate cancer, CA15-3 for breast cancer, and CA125 for ovarian cancer [10]. Protein glycosylation plays crucial roles in several diseases. For instance, glycosylation is reported to define cancer malignancy [11], changes in glycosylation are often a hallmark of disease states [12], and core fucosylation is related to AG prostate cancer [13].
Prostate cancer biomarker discovery and translation remains a challenging task, and there is currently a critical need for new markers that can discriminate indolent from AG prostate cancer [14]. Several different specimens have been analyzed for discovery of novel biomarkers, including prostatic tissue [15,16,19], serum [20–22], and urine [8,23–26], in recent years. To identify prostate cancer specific changes, biomarkers are preferably identified in cancer tissues, and subsequently detected and analyzed in body fluids [27]. Detection of biomarkers in serum or urine has advantages over tissue markers, since acquisition of cancer tissues involves invasive biopsy procedure that itself is prone to sampling errors [25]. Urine has potential advantages over serum, since urinary proteins are stable and do not undergo significant proteolysis for several hours after collection, compared with blood [8]. Moreover, due to its proximity to prostate relative to peripheral blood, urine-based biomarkers may outperform blood-based tests for early detection of aggressive prostate cancer.
Our laboratory has successfully developed and optimized effective glycoproteomic technologies for analysis of glycoproteins [28, 29], which have been used in marker discovery from prostate tumor tissues to distinguish aggressive from indolent tumors [15–18]. Using the SPEG approach, we previously identified tissue-derived glycoprotein changes associated with aggressive prostate cancer. In the current study, glycoproteins from urine and serum samples of prostate cancer patients were analyzed and compared with those obtained from prostate cancer tissues to determine whether the tissue proteins associated with aggresive prostate cancer are detectable in urine or serum, aiming for developing potential surrogate markers applicable for diagnosis of aggressive disease.
2 Materials and methods
2.1 Chemicals
Hydrazide resin was purchased from Bio-Rad (Hercules, CA), PNGase F from New England Biolabs (Ipswich, MA), C18 columns from Waters (Milford, MA), sequencing-grade trypsin from Promega (Madison, WI), and tris(2-chloroethyl) phosphate and the bicinchoninic acid assay (BCA) kit from Thermo Scientific (Waltham, MA). Centrifugal filter units were acquired from EMD Millipore (Billerica, MA), and ammonium bicarbonate, sodium periodate, iodoacetamide, and other chemicals were obtained from Sigma–Aldrich (St. Louis, MO).
2.2 Clinical specimens
Clinical specimens and medical information were obtained with informed consent, and experiments were performed with the approval of the Institutional Review Board of The Johns Hopkins University (Baltimore, MD). Urine samples from prostate cancer patients (n = 40, Supporting Information Table S1) were collected during office visits in the Department of Urology. Serum samples from prostate cancer (n = 119) and nonprostate cancer (n = 48) subjects were collected at the Clinical Chemistry Laboratory (Supporting Information Table S2). Tissue samples were collected as described previously [15, 16]. Fresh urine samples were centrifuged at 3000 × g for 20 min at 4°C to remove cells and debris. Centrifuged urine samples and sera from all patients were stored at −80°C until use.
2.3 N-Linked glycosite-containing peptide isolation from urine of prostate cancer patients
Urine samples were thawed at room temperature and vortexed for 2 min. Samples (15 mL) were concentrated to <1 mL using 10KD centrifugal devices. Protein concentrations of the urine samples were determined using the BCA assay. Proteins were denatured in 8 M urea/1 M ammonium bicarbonate buffer [30], reduced with 10 mM tris(2-chloroethyl) phosphate at 37°C for 1 h and alkylated with 15 mM iodoacetamide at room temperature in the dark for 30 min. Next, solutions were diluted fivefold with deionized water. Sequencing-grade trypsin (Promega, Madison, WI; protein–enzyme, 50:1, w/w) was added to samples and incubated at 37°C overnight with shaking. Samples were centrifuged at 13 000 × g for 10 min to remove precipitates and purified via C18 SPE. Peptides were eluted from the C18 column in 60% ACN/0.1% TFA, and concentrations were measured with the BCA assay.
N-Linked glycosite-containing peptides were extracted using SPEG (hydrazide chemistry) as described previously [15, 28], with minor modifications [31]. Briefly, peptides in the solution were oxidized with 10 mM sodium periodate solution at room temperature in the dark for 1 h. Samples were diluted with 0.1% TFA in 5% ACN and cleaned using the C18 column. The elution solution was collected directly into equilibrated hydrazide beads (300 μL of 50% slurry for each sample) and incubated with 100 mM aniline at room temperature for 1 h. Beads were washed three times each with 50% ACN, 1.5 M NaCl, water, and 100 mM ammonium bicarbonate buffer. Former N-glycopeptides were released via 3 μL PNGase F (New England Biolabs, Beverly, MA) in 25 mM NH4HCO3 buffer at 37°C overnight with shaking. The glycosite-containing peptides were collected in supernatant/wash solutions (200 μL of 50% ACN), dried via vacuum, and resuspended in 50 μL 0.2% formic acid (pH <3) for HPLC fractionation.
2.4 Isolation of N-linked glycosite-containing peptides from serum samples of prostate cancer patients
Serum N-linked glycosite-containing peptides were isolated using SPEG as described previously [32]. Briefly, 40 mL serum samples were diluted tenfold with 500 mM sodium acetate buffer and oxidized with 15 mM sodium periodate. After desalting, samples were coupled to hydrazide beads, followed by tryptic digestion and extensive washing. N-linked glycosite-containing peptides were released from hydrazide beads using PNGase-F.
2.5 Fractionation of N-linked glycosite-containing peptides
Equal amounts of N-linked glycosite-containing peptides isolated from urine or serum samples of prostate cancer patients were pooled. Fractionation of samples was performed using high pH RPLC on the 1220 Infinity LC system with a Zorbax Extended-C18 analytical column containing 1.8 μM particles (Agilent Technologies, Inc., Santa Clara, CA) at a flow rate of 0.2 mL/min. The mobile phase A consisted of 10 mM ammonium formate (pH 10) while B contained 10 mM ammonium formate and 90% ACN (pH 10). Sample separation was accomplished using the following linear gradient: 0–2% B in 10 min, 2–8% B in 5 min, 8–35% B in 85 min, 35–95% B in 5 min, and 95% B for an additional 15 min. Peptides were detected at 215 nm, and 96 fractions collected, along with the LC separation in a time-based mode from 16 to 112 min. Fractions were concatenated into 24 fractions by combining every 24 fractions (for instance, 1, 25, 49, 73; 2, 26, 50, and 74). Samples were dried in a speed vacuum and stored at −80°C until LC-MS/MS analysis.
2.6 LC-MS/MS analysis
Peptides (~1 μg) were separated through a Dionex Ultimate 3000 RSLC nanosystem (Thermo Scientific) with a 75 μm × 50 cm Acclaim PepMap100 RSLC column (Thermo Scientific). The mobile phase flow rate was 250 nL/min and included phase A (0.1% formic acid and 2% ACN in water) and B (0.1% formic acid and 95% ACN in water). The gradient profile was set as follows: 5–40% B for 68 min, 40–95% B for 5 min, 90% B for 10 min. MS analysis was performed using a Thermo Q Exactive mass spectrometer (Thermo Scientific). The spray voltage was set at 1.8 kV. The QE spectral (AGC) target for MS1 was set as 1 × 106 in 60 ms maximum time and AGC target for MS/MS as 1 × 105 (at a resolution of 17 500 and maximum IT 75 ms) of the 20 most abundant ions. Charge state screening was enabled to reject unassigned, singly charged, and equal or more than eight protonated ions. A dynamic exclusion time of 15 s was used to discriminate against previously selected ions.
2.7 Glycoprotein and glycopeptide quantification
Acquired MS/MS spectra were searched against the RefSeq human protein database (downloaded on December 30, 2013) using the SEQUEST search engine (Thermo Proteome Discoverer v1.4; Thermo Fisher Scientific). Database search parameters were set as follows: a maximum of two missed cleavage sites permitted for trypsin digestion, 10 ppm precursor mass tolerance, 0.06 Da fragment mass tolerance, carbamidomethylation (C, +57.0215 Da) as a fixed modification, and oxidization (M, +15.9949 Da) and deamidation (N, +0.98 Da) as dynamic modifications. The results were filtered with 1% FDR. Spectral counting was used to quantify the peptides identified from LC-MS/MS data [33]. SIEVE software (Thermo Scientific, version 2.0) was used to conduct semiquantitative glycoproteomic analysis of urine and serum samples from patients with AG and non-AG (NAG) prostate cancer. To do this, raw files were imported into SIEVE software and the chromatograms were aligned. Frame parameters were set as follows: frame for all MS2 scans, retention time from 0 to 60 min, m/z from 350 to 1800, frame time retention time width 2.5 min, and frame m/z width 10 ppm. Following framing, the results from Proteome Discoverer search files were imported with a FDR of 1% and only peptides unique to one protein were used for quantification. Proteins with at least two spectral counts were reported. The integrated intensity of glycosite-containing peptides was normalized by the total ion chromatogram and used to calculate a P value. The average intensity of peptides was used to calculate fold change.
2.8 Statistics
Student’s t-test was used to calculate the P-value of differentially expressed glycoproteins. Data were considered significant at P-values ≤ 0.05.
3 Results
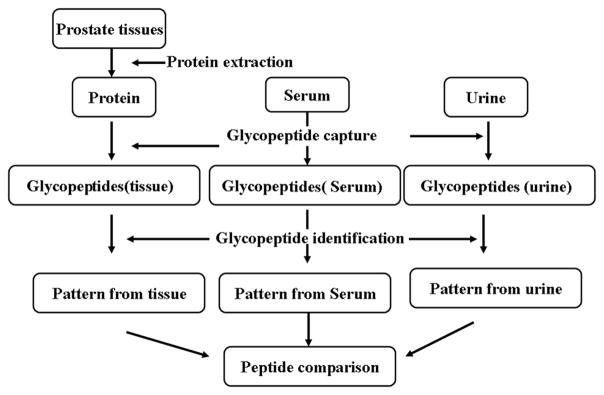
3.1 Workflow of detection of N-linked glycosite-containing peptides identified from prostate cancer tissues in serum and urine samples of prostate cancer patients
Due to the reduced complexity of MS-based analysis that focuses solely on N-linked glycosite-containing peptides from secreted or extracellular proteins, tissue-derived N-linked glycosite-containing peptides may be readily detected in body fluids using glycoproteomics. Accordingly, we analyzed N-linked glycosite-containing peptides from serum and urine of prostate cancer patients using the workflow described in Fig. 1. N-linked glycosite-containing peptides were extracted using the SPEG method [28, 34], analyzed via LC-MS/MS, and the database searched. Glycopeptides previously identified from tissues were compared with those detected in urine or serum samples from prostate cancer patients [16, 29].
Figure 1.
Workflow of detection of N-linked glycosite-containing peptides in tissue, serum, and urine samples from prostate cancer patients.
3.2 Glycoproteomic analysis of urine and serum samples from prostate cancer patients
To evaluate whether urine and serum specimens contain the signature glycopeptides detected in tissue samples of prostate cancer patients, we performed 2D-LC-MS/MS analyses of SPEG-isolated N-linked glycosite containing peptides from urine and serum samples. The peptides identified were filtered with 1% FDR. In total, 2923 and 2475 unique N-linked glycosite-containing peptides were identified in urine and serum, respectively (Supporting Information Table S3). We further compared the unique glycopeptides from urine and serum to those identified from tissues of prostate cancer patients (Supporting Information Table S3a). Overall, 40% of the glycopeptides from prostate cancer tissues were also detectable in urine, while only 13% were detected in serum (Fig. 2, Supporting Information Table 3a), suggesting that urine is a more favorable source than serum for detecting prostate tissue derived glycoproteins.
Figure 2.
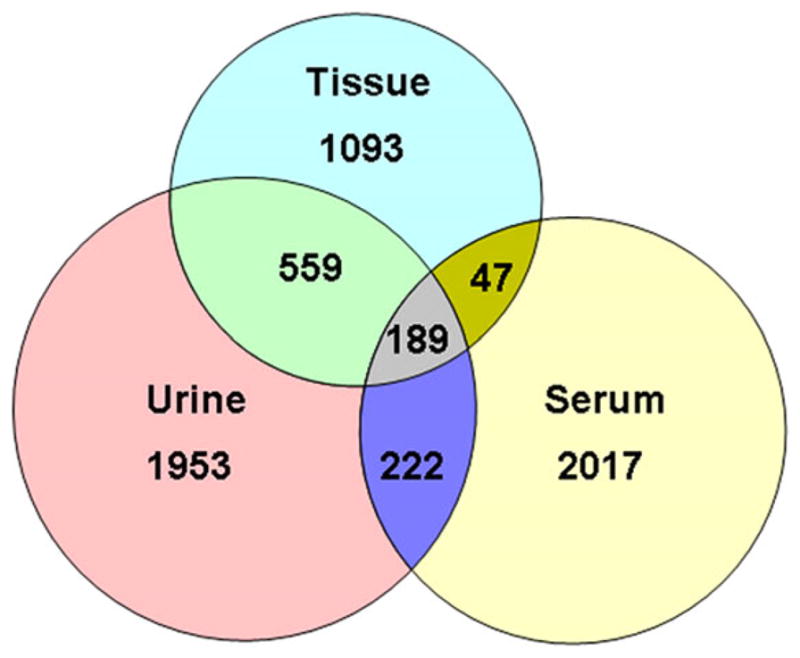
Identification of urine and serum N-linked glycosite-containing peptides and comparison with peptides identified in prostate tissues. In total, 1888, 2923 and 2475 glycopeptides were identified from tissue, urine, and serum, respectively.
The GOASVM algorithm is generally used to predict subcellular location [35]. The distribution data indicated that the majority of identified glycoproteins in prostate tissue, urine, and serum reside either in the extracellular compartment or plasma membrane. These results were as same as expected, since N-linked glycoproteins are most frequently secretory or plasma membrane proteins. The cancer-associated extracellular proteins from prostate cancer tissues are likely to be secreted by cancer cells or shed from cell surfaces, thus serving as potential biomarkers in body fluids [36].
3.3 Detection of tissue-specific AG prostate cancer associated glycoproteins in urine and serum
To further determine whether the glycoproteins associated with AG prostate cancer identified in tissues are detectable in urine and serum, we compared the prostate cancer tissue-derived candidate glycoproteins previously discovered by our group as candidate markers for AG disease to those detected in urine and serum samples. Almost all candidates (except Serpin H1, Ectonucleoside triphosphate diphosphohydrolase 1 and protein FAM3B) were readily detected in urine, but not serum samples (Table 1, Supporting Information Table 4). Moreover, clinically used prostate cancer specific proteins, including PSA and prostatic acid phosphatase, which were not identified in patient serum in this study without targeted analysis, were readily identified in urine samples from prostate cancer patients using the same shotgun glycoproteomics approach. Our results further confirmed that urine provides an abundant source of proteins associated with AG prostate cancer.
Table 1.
Detection of aggressive prostate cancer associated glycoproteins in urine and serum
| Protein (human) | UniProt | Protein name | Detected in urine | Detected in serum |
|---|---|---|---|---|
| POSTN | Q15063 | Periostin | Yes | No |
| ASPN | Q9BXN1 | Asporin | Yes | No |
| LAMB2 | P55268 | Laminin subunit beta-2 | Yes | No |
| SERPH | P50454 | Serpin H1 | No | No |
| CSPG2 | P13611 | Versican core protein | Yes | No |
| ENTP1 | P49961 | Ectonucleoside triphosphate diphosphohydrolase 1 | No | No |
| SE1L1 | Q9UBV2 | Protein sel-1 homolog 1 | Yes | No |
| ITAV | P06756 | Integrin alpha-V | Yes | No |
| FOLH1 | Q04609 | Glutamate carboxypeptidase 2 | Yes | No |
| STIM1 | Q13586 | Stromal interaction molecule 1 | Yes | No |
| PTK7 | Q13308 | Inactive tyrosine-protein kinase 7 | Yes | No |
| ICOSL | O75144 | ICOS ligand | Yes | Yes |
| SPP2A | Q8TCT8 | Signal peptide peptidase-like 2A | Yes | No |
| CD276 | Q5ZPR3 | CD276 antigen | Yes | No |
| KLK3 | P07288 | Prostate-specific antigen | Yes | No |
| ZA2G | P25311 | Zinc-alpha-2-glycoprotein | Yes | Yes |
| FBN1 | P35555 | Fibrillin-1 | Yes | No |
| CD38 | P28907 | ADP-ribosyl cyclase 1 | Yes | No |
| CNTP2 | Q9UHC6 | Contactin-associated protein-like 2 | Yes | No |
| AMPN | P15144 | Aminopeptidase N | Yes | Yes |
| PAM3B | P58499 | Protein FAM3B | No | No |
| RNT2 | O00584 | Ribonuclease T2 | Yes | No |
| NAAA | Q02083 | N-acylethanolamine-hydrolyzing acid amidase | Yes | No |
| PPAP | P15309 | Prostatic acid phosphatase | Yes | No |
| GSLG1 | Q92896 | Golgi apparatus protein 1 | Yes | No |
| TSN1 | O60635 | Tetraspanin-1 | Yes | No |
| DPP4 | P27487 | Dipeptidyl peptidase 4 | Yes | No |
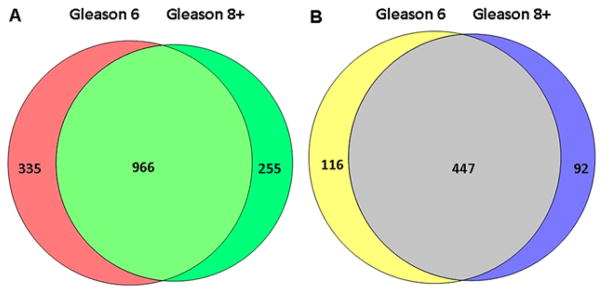
3.4 Quantitative glycoproteomic analysis of urine samples from patients with AG and NAG prostate cancer
To determine whether aggressive prostate cancer associated tissue glycoproteins detected in urine are samples also differentially expressed in urine samples from between AG and NAG groups, we separately analyzed 20 additional urine samples from ten men with low-grade (Gleason score of 3 + 3) and ten men with high-grade (Gleason score of at least 4 + 4) prostate cancer using the glycoproteomics approach. LC-MS/MS analyses of individual samples led to the identification of 1556 N-linked glycosite-containing peptides representing 655 unique glycoproteins (Fig. 3, Supporting Information Table S5). Peak area integration was performed for quantification of identified peptides using SIEVE software [37]. The most abundant glycosite-containing peptide was selected from each glycoprotein for quantification of fold changes in AG and NAG groups (Supporting Information Table S6). The levels of five out of 26 glycoproteins, specifically, protein tyrosine kinase 7 (PTK7), ICOS ligand (ICOSL), zinc alpha 2 glycoprotein (AZGP1), fibrillin-1 and Golgi apparatus protein 1, were significantly decreased in urine samples of patients with AG disease.
Figure 3.
Identification of N-linked glycosite-containing peptides in urine samples from prostate cancer patients with different Gleason scores. (A) Identification of N-linked glycosite-containing peptides in urine samples from prostate cancer patients with Gleason scores 6 and 8+. (B) Identification of glycoproteins from prostate cancer patients with Gleason scores 6 and 8+.
4 Discussion
Proteomic technologies have been widely applied to identify cancer biomarkers in recent years. However, to our knowledge, this is the first study to utilize urine samples for analysis of glycoproteins. Our group has identified a number of N-linked glycoproteins associated with AG prostate cancer in tissues [13, 15, 16]. In the current study, we compared the glycoproteins identified in urine samples to those detected in sera and tissues of prostate cancer patients. The majority of the glycoproteins identified in prostate tissue were readily detected in patient urine samples, indicating that urine may serve as an abundant source for prostate cancer biomarker discovery.
Although the proteins themselves are common, ubiquitous, abundant, and familiar, earlier studies indicated that glycoproteins produced by cancer cells have altered glycan structures [38]. Similar to normal cells during embryogenesis, tumor cells undergo activation and rapid growth, adhere to a variety of other cell types and cell matrices, and invade tissues. Embryonic development and cellular activation in vertebrates are typically accompanied by changes in cellular glycosylation profiles [39]. Thus, it is not surprising that glycoprotein changes represent a universal feature of malignant transformation and tumor progression.
Analysis of urine is relevant in view of its proximity to the prostate, compared to peripheral blood. In blood, prostate cancer specific proteins become diluted upon mixing with proteins secreted from other tissues, resulting in decreased sensitivity and specificity of detection. The use of urine-based biomarkers as a surrogate for the early detection of AG prostate cancer is an ideal approach, since urine presents a rapid, inexpensive, reliable, noninvasive, and easy-to-use specimen for diagnosis that requires little or no training and no specialized instrumentation. Due to its proximity to the prostate and relatively low exposure to organs other than bladder and kidney, enhanced sensitivity and specificity of detecting glycoproteins released from prostate cancer patients may be achieved.
Furthermore, urine contains vesicles derived from the prostate and other urological organs that are released into preurine via direct shedding, budding from the plasma membrane, or fusion of intracellular multivesicular bodies with the plasma membrane [40]. Interestingly, the majority of surface plasma proteins are glycoproteins. Glycoproteomic methods have been applied to efficiently enrich and analyze exosome-like vesicles from human urine [41–44], further facilitating the identification of potential candidates in urine specimens that contribute to AG prostate disease.
Analysis of the urine glycoproteome revealed that the glycoproteins identified from prostate cancer tissues are also readily detectable in urine samples, including those already used in the clinic or for research, including PSA, PAP [45], and kallikrein-11 [46]. A number of glycoproteins have been identified as potential markers effective in distinguishing between AG and NAG prostate cancers, including zinc alpha 2 glycoprotein (AZGP1) [47], CD276 antigen [5], and metalloproteinase inhibitor 1 [48]. Earlier, our group showed that an increase in PTK7 and periostin and a decrease in N-acylethanolamine acid amidase in prostate tissue are potentially associated with AG prostate cancer [15, 16]. In addition, cathepsin F and acid sphingomyelinase-like phosphodiesterase 3b are reported to be associated with cervical cancer [49] and ovarian tumor, respectively [36]. However, all previous studies have been based on tissue or serum samples. The current approach focusing on identification of glycoproteins in urine samples should aid in the discovery of effective noninvasive biomarkers for prostate cancer.
We selected an additional 20 random urine samples from prostate cancer patients to determine the relative abundance of prostate cancer tissue-derived glycoproteins in urine of patients with AG and NAG disease. Glycoproteomic results indicated a decrease in the majority of the identified glycoproteins in AG urine, except ribonuclease T2. Interestingly, PTK7 and ICOSL levels were decreased in urine samples, but increased in AG prostate tissue. On the other hand, similar to findings in tissue, three glycoproteins, including AZGP1, fibrillin-1, and Golgi apparatus protein 1, were decreased in urine samples from the AG group. Expression of AZGP1 has been used as a predictor of metastatic prostate cancer following radical prostatectomy [47]. Data from the current study showed for the first time that AZGP1 expression appear to decrease in urine samples of AG prostate cancer patients. However, the quantitative analysis results in this study were based on a small sample size of 10 individuals in aggressive and non-aggressive groups. A large number urine samples in each group would be necessary to determine the changes of identified aggressive prostate cancer associated glycoproteins from tissues in urine samples.
In conclusion, direct LC-MS/MS analysis of glycopeptides from patient urine samples yielded abundant information, with detection of a large number of peptides also identified in prostate cancer tissue. This study is mainly focused on the detection of proteins from prostate tissues in urine, not on the quantitative changes due to the small sample size. Our on-going project will further focus on AG-specific markers (Supporting Information Table 7) as well as on those that show a difference between the AG and NAG samples with larger sample sizes.
Supplementary Material
Significance of the study.
Biopsy in prostate cancer diagnosis is invasive and serum PSA levels are less effective in distinguishing aggressive prostate cancer from non-aggressive prostate cancer and other benign prostatic diseases, highlighting the urgent medical need for novel biomarkers that allow differentiation of aggressivity from indolent disease. Over the years, our group has developed and optimized solid phase extraction of N-linked glycosite-containing peptides (SPEG) for the analysis of glycoproteins. Here, we applied the SPEG methodology to analyze glycoproteins from urine and serum samples of prostate cancer patients. The results were compared with glycoproteins identified from prostate cancer tissues and serum samples to determine whether proteins from prostate cancer tissues can be detected in urine or serum samples. The majority of cancer-associated glycoproteins displayed higher abundance in urine compared to serum samples, indicating that urine is a highly reliable source of prostate cancer specific proteins. To our knowledge, this is the first documented analysis of the urinary glycoproteome from prostate cancer patients and comparison to glycoproteins associated with aggressive prostate tumor in tissues.
Abbreviations
- AG
aggressive
- BCA
bicinchoninic acid assay
- NAG
non-AG
- PSA
prostate-specific antigen
- PTK7
protein tyrosine kinase 7
- SPEG
solid phase extraction of glycosite-containing peptides
Footnotes
The authors have declared no conflict of interest.
Additional supporting information may be found in the online version of this article at the publisher’s website
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Chan D, Bruzek D, Oesterling J, Rock R, Walsh P. Prostate-specific antigen as a marker for prostatic cancer: a monoclonal and a polyclonal immunoassay compared. Clin Chem. 1987;33:1916–1920. [PubMed] [Google Scholar]
- 3.Presti JC. Prostate biopsy strategies. Nat Clin Pract Urol. 2007;4:505–511. doi: 10.1038/ncpuro0887. [DOI] [PubMed] [Google Scholar]
- 4.Garcia M, Jemal A, Ward EM, Center MM, Hao Y, Siegel RL, Thun MJ. Global cancer facts and figures 2007. American Cancer Society; Atlanta, GA: 2007. [Google Scholar]
- 5.Velonas VM, Woo HH, dos Remedios CG, Assinder SJ. Current status of biomarkers for prostate cancer. Int J Cancer. 2013;14:11034–11060. doi: 10.3390/ijms140611034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muniyan S, Ingersoll MA, Batra SK, Lin MF. Cellular prostatic acid phosphatase, a PTEN-functional homologue in prostate epithelia, functions as a prostate-specific tumor suppressor. Biochim Biophys Acta. 2014;1846:88–98. doi: 10.1016/j.bbcan.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gonzalgo ML, Bastian PJ, Mangold LA, Trock BJ, et al. Relationship between primary Gleason pattern on needle biopsy and clinicopathologic outcomes among men with Gleason score 7 adenocarcinoma of the prostate. Urology. 2006;67:115–119. doi: 10.1016/j.urology.2005.07.037. [DOI] [PubMed] [Google Scholar]
- 8.Haj-Ahmad TA, Abdalla MA, Haj-Ahmad Y. Potential urinary protein biomarker candidates for the accurate detection of prostate cancer among benign prostatic hyperplasia patients. J Cancer. 2014;5:103–114. doi: 10.7150/jca.6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sokoll LJ, Wang YH, Feng ZD, Kagan J, et al. [-2]Proenzyme prostate specific antigen for prostate cancer detection: a National Cancer Institute Early Detection Research Network validation study. J Urol. 2008;180:539–543. doi: 10.1016/j.juro.2008.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meany DL, Chan DW. Aberrant glycosylation associated with enzymes as cancer biomarkers. Clin Proteomics. 2011;8:7. doi: 10.1186/1559-0275-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hakomori S. Glycosylation defining cancer malignancy: New wine in an old bottle. Proc Natl Acad Sci USA. 2002;99:10231–10233. doi: 10.1073/pnas.172380699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dube DH, Bertozzi CR. Glycans in cancer and inflammation—potential for therapeutics and diagnostics. Nat Rev Drug Discov. 2005;4:477–488. doi: 10.1038/nrd1751. [DOI] [PubMed] [Google Scholar]
- 13.Wang X, Chen J, Li QK, Peskoe SB, et al. Overexpression of α (1, 6) fucosyltransferase associated with aggressive prostate cancer. Glycobiology. 2014;24:935–944. doi: 10.1093/glycob/cwu051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parekh DJ, Ankerst DP, Troyer D, Srivastava S, Thompson IM. Biomarkers for prostate cancer detection. J Urol. 2007;178:2252–2259. doi: 10.1016/j.juro.2007.08.055. [DOI] [PubMed] [Google Scholar]
- 15.Chen J, Xi J, Tian Y, Bova GS, Zhang H. Identification, prioritization, and evaluation of glycoproteins for aggressive prostate cancer using quantitative glycoproteomics and antibody-based assays on tissue specimens. Proteomics. 2013;13:2268–2277. doi: 10.1002/pmic.201200541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y, Chen J, Sethi A, Li QK, et al. Glycoproteomic analysis of prostate cancer tissues by SWATH mass spectrometry discovers N-acylethanolamine acid amidase and protein tyrosine kinase 7 as signatures for tumor aggressiveness. Mol Cell Proteomics. 2014;13:1753–1768. doi: 10.1074/mcp.M114.038273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tian Y, Bova GS, Zhang H. Quantitative glycoproteomic analysis of optimal cutting temperature-embedded frozen tissues identifying glycoproteins associated with aggressive prostate cancer. Anal Chem. 2011;83:7013–7019. doi: 10.1021/ac200815q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shah P, Wang X, Yang W, Toghi Eshghi S, et al. Integrated proteomic and glycoproteomic analyses of prostate cancer cells reveals glycoprotein alteration in protein abundance and glycosylation. Mol Cell Proteomics. 2015;14:2753–2763. doi: 10.1074/mcp.M115.047928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rubin MA, Zhou M, Dhanasekaran SM, Varambally S, et al. α-Methylacyl coenzyme A racemase as a tissue biomarker for prostate cancer. JAMA. 2002;287:1662–1670. doi: 10.1001/jama.287.13.1662. [DOI] [PubMed] [Google Scholar]
- 20.Petricoin EF, Ornstein DK, Paweletz CP, Ardekani A, et al. Serum proteomic patterns for detection of prostate cancer. J Natl Cancer Inst. 2002;94:1576–1578. doi: 10.1093/jnci/94.20.1576. [DOI] [PubMed] [Google Scholar]
- 21.Diamandis EP, Okui A, Mitsui S, Luo LY, et al. Human kallikrein 11 a new biomarker of prostate and ovarian carcinoma. Cancer Res. 2002;62:295–300. [PubMed] [Google Scholar]
- 22.Adam BL, Qu Y, Davis JW, Ward MD, et al. Serum protein fingerprinting coupled with a pattern-matching algorithm distinguishes prostate cancer from benign prostate hyperplasia and healthy men. Cancer Res. 2002;62:3609–3614. [PubMed] [Google Scholar]
- 23.Pang JX, Ginanni N, Dongre AR, Hefta SA, Opiteck GJ. Biomarker discovery in urine by proteomics. J Proteome Res. 2002;1:161–169. doi: 10.1021/pr015518w. [DOI] [PubMed] [Google Scholar]
- 24.Nilsson J, Skog J, Nordstrand A, Baranov V, et al. Prostate cancer-derived urine exosomes: a novel approach to biomarkers for prostate cancer. Br J Cancer. 2009;100:1603–1607. doi: 10.1038/sj.bjc.6605058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ploussard G, de la Taille A. Urine biomarkers in prostate cancer. Nat Rev Urol. 2010;7:101–109. doi: 10.1038/nrurol.2009.261. [DOI] [PubMed] [Google Scholar]
- 26.Struck-Lewicka W, Kordalewska M, Bujak R, Mpanga AY, et al. Urine metabolic fingerprinting using LC-MS and GC-MS reveals metabolite changes in prostate cancer: a pilot study. J Pharm Biomed Anal. 2015;111:351–361. doi: 10.1016/j.jpba.2014.12.026. [DOI] [PubMed] [Google Scholar]
- 27.Zhang H, Chan DW. Cancer biomarker discovery in plasma using a tissue-targeted proteomic approach. Caner Epidemiol Biomarkers Prev. 2007;16:1915–1917. doi: 10.1158/1055-9965.EPI-07-0420. [DOI] [PubMed] [Google Scholar]
- 28.Zhang H, Li X-j, Martin DB, Aebersold R. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat Biotechnol. 2003;21:660–666. doi: 10.1038/nbt827. [DOI] [PubMed] [Google Scholar]
- 29.Sun S, Shah P, Toghi Eshghi S, Yang W, et al. Comprehensive analysis of protein glycosylation by solid-phase extraction of N-linked glycans and glycosite-containing peptides. Nat Biotechnol. 2016;34:84–88. doi: 10.1038/nbt.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun S, Zhou JY, Yang W, Zhang H. Inhibition of protein carbamylation in urea solution using ammonium-containing buffers. Anal Biochem. 2014;446:76–81. doi: 10.1016/j.ab.2013.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun S, Zhang B, Aiyetan P, Zhou JY, et al. Analysis of N-glycoproteins using genomic N-glycosite prediction. J Proteome Res. 2013;12:5609–5615. doi: 10.1021/pr400575f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen J, Shah P, Zhang H. Solid phase extraction of N-linked glycopeptides using hydrazide tip. Anal Chem. 2013;85:10670–10674. doi: 10.1021/ac401812b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tian Y, Tan AC, Sun X, Olson MT, et al. Quantitative proteomic analysis of ovarian cancer cells identified mitochondrial proteins associated with paclitaxel resistance. Proteomics Clin Appl. 2009;3:1288–1295. doi: 10.1002/prca.200900005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang H, Liu AY, Loriaux P, Wollscheid B, et al. Mass spectrometric detection of tissue proteins in plasma. Mol Cell Proteomics. 2007;6:64–71. doi: 10.1074/mcp.M600160-MCP200. [DOI] [PubMed] [Google Scholar]
- 35.Wan S, Mak MW, Kung SY. GOASVM: a subcellular location predictor by incorporating term-frequency gene ontology into the general form of Chou’s pseudo-amino acid composition. J Theor Biol. 2013;323:40–48. doi: 10.1016/j.jtbi.2013.01.012. [DOI] [PubMed] [Google Scholar]
- 36.Tian Y, Yao Z, Roden R, Zhang H. Identification of glycoproteins associated with different histological subtypes of ovarian tumors using quantitative glycoproteomics. Proteomics. 2011;11:4677–4687. doi: 10.1002/pmic.201000811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neilson KA, Ali NA, Muralidharan S, Mirzaei M, et al. Less label, more free: approaches in label-free quantitative mass spectrometry. Proteomics. 2011;11:535–553. doi: 10.1002/pmic.201000553. [DOI] [PubMed] [Google Scholar]
- 38.Narimatsu H, Sawaki H, Kuno A, Kaji H, et al. A strategy for discovery of cancer glycobiomarkers in serum using newly developed technologies for glycoproteomics. FEBS J. 2010;277:95–105. doi: 10.1111/j.1742-4658.2009.07430.x. [DOI] [PubMed] [Google Scholar]
- 39.Varki A, Kannagi R, Toole BP. In: Glycans in Acquired Human Diseases. Varki A, Cummings R, Esko J, editors. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press, New York; 2009. pp. 1–14. [Google Scholar]
- 40.Benito-Martin A, Ucero AC, Zubiri I, Posada-Ayala M, et al. Osteoprotegerin in exosome-like vesicles from human cultured tubular cells and urine. PloS One. 2013;8:e72387. doi: 10.1371/journal.pone.0072387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kosanović M, Janković M. Isolation of urinary extracellular vesicles from Tamm-Horsfall protein-depleted urine and their application in the development of a lectin-exosome-binding assay. BioTechniques. 2014;57:143–149. doi: 10.2144/000114208. [DOI] [PubMed] [Google Scholar]
- 42.Hogan MC, Johnson KL, Zenka RM, Charlesworth MC, et al. Subfractionation, characterization, and in-depth proteomic analysis of glomerular membrane vesicles in human urine. Kidney Int. 2014;85:1225–1237. doi: 10.1038/ki.2013.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Echevarria J, Royo F, Pazos R, Salazar L, et al. Microarray-based identification of lectins for the purification of human urinary extracellular vesicles directly from urine samples. ChemBioChem. 2014;15:1621–1626. doi: 10.1002/cbic.201402058. [DOI] [PubMed] [Google Scholar]
- 44.Gerlach JQ, Krüger A, Gallogly S, Hanley SA, et al. Surface glycosylation profiles of urine extracellular vesicles. PLoS One. 2013;8:e74801. doi: 10.1371/journal.pone.0074801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ercole C, Lange PH, Mathisen M, Chiou R, et al. Prostatic specific antigen and prostatic acid phosphatase in the monitoring and staging of patients with prostatic cancer. J Urol. 1987;138:1181–1184. doi: 10.1016/s0022-5347(17)43543-9. [DOI] [PubMed] [Google Scholar]
- 46.Nakamura T, Scorilas A, Stephan C, Jung K, et al. The usefulness of serum human kallikrein 11 for discriminating between prostate cancer and benign prostatic hyperplasia. Cancer Res. 2003;63:6543–6546. [PubMed] [Google Scholar]
- 47.Henshall SM, Horvath LG, Quinn DI, Eggleton SA, et al. Zinc-alpha2-glycoprotein expression as a predictor of metastatic prostate cancer following radical prostatectomy. J Natl Cancer Inst. 2006;98:1420–1424. doi: 10.1093/jnci/djj378. [DOI] [PubMed] [Google Scholar]
- 48.Jung K, Nowak L, Lein M, Priem F, et al. Matrix metalloproteinases 1 and 3, tissue inhibitor of metalloproteinase-1 and the complex of metalloproteinase-1/tissue inhibitor in plasma of patients with prostate cancer. Int J Cancer. 1997;74:220–223. doi: 10.1002/(sici)1097-0215(19970422)74:2<220::aid-ijc14>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 49.Vazquez-Ortiz G, Pina-Sanchez P, Vazquez K, Duenas A, et al. Overexpression of cathepsin F, matrix metalloproteinases 11 and 12 in cervical cancer. BMC Cancer. 2005;5:68. doi: 10.1186/1471-2407-5-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.