Abstract
Background
Invasive group A Streptococcus (iGAS) disease caused by type emm89 strains has been increasing worldwide, driven by the emergence of an epidemic clonal variant (clade 3 emm89). The clinical characteristics of patients with emm89 iGAS disease, and in particular with clade 3 emm89 iGAS disease, are poorly described.
Methods
We used population-based iGAS surveillance data collected in metropolitan Toronto, Ontario, Canada during the period 2000–2014. We sequenced the genomes of 105 emm89 isolates representing all emm89 iGAS disease cases in the area during the period and 138 temporally matched emm89 iGAS isolates collected elsewhere in Ontario.
Results
Clades 1 and 2 and clade O, a newly discovered emm89 genetic variant, caused most cases of emm89 iGAS disease in metropolitan Toronto before 2008. After rapid emergence of new clade 3, previously circulating clades were purged from the population and the incidence of emm89 iGAS disease significantly increased from 0.14 per 100000 in 2000–2007 to 0.22 per 100000 in 2008–2014. Overall, emm89 organisms caused significantly more arthritis but less necrotizing fasciitis than strains of the more common type emm1. Other clinical presentations were soft tissue and severe respiratory tract infections. Clinical outcomes did not differ significantly between emm89 clades overall. However, clade 3 emm89 iGAS disease was more common in youth and middle-aged individuals.
Conclusions
The rapid shift in emm89 iGAS strain genetics in metropolitan Toronto has resulted in a significant increase in the incidence of emm89 iGAS disease, with noticeably higher rates of clade 3 disease in younger patients.
Keywords: emerging strain genotype, group A Streptococcus, invasive disease, populations at risk, whole-genome sequencing
Group A Streptococcus ([GAS] also known as Streptococcus pyogenes) causes a broad range of diseases, from pharyngitis and superficial skin infections to life-threatening necrotizing fasciitis and streptococcal toxic shock syndrome (STSS) [1]. After a decline in the mid-20th century, re-emergence of invasive GAS (iGAS) infections has led to disease incidences estimated at 1.5–4.3 cases per 100000 in high-income countries [2–4]. Group A Streptococcus isolates can be differentiated into more than 200 types by deoxyribonucleic acid (DNA) sequence analysis of the region of the emm gene encoding the variable amino terminal portion of the M protein (a virulence factor with antiphagocytic properties) [5–7]. Invasive GAS disease burden is associated with a relatively small number of emm types in many high- income countries [8–10].
Investigative approaches that include whole-genome sequencing (WGS) of population-based samples have resulted in several important discoveries that have enhanced our understanding of iGAS diseases [11–14]. For example, analysis of >3500 emm1 GAS genomes has uncovered allele variation due to recombination in the promoter spacer region of genes nga and slo encoding secreted toxins NAD+-glycohydrolase (NADase) and streptolysin O (SLO), respectively [15, 16]. Enhanced expression of these key virulence factors by a recombinant epidemic emm1 clone led to the global spread of severe emm1 iGAS infections in the 1980s [16, 17]. Similar WGS-based approaches have begun to identify contributors to the sudden increase in emm89 iGAS disease reported in recent years in several countries [16, 18–21]. Analysis of genome-wide single-nucleotide polymorphisms (SNPs) defined 3 genetically distinct emm89 clades, which differ in virulence in mouse and nonhuman primate models of infection [15, 16, 22]. The more virulent emm89 clonal clade (named “clade 3”) has the same polymorphisms in the promoter spacer region of the nga and slo gene cluster (also resulting in enhanced NADase and SLO toxin activity) as epidemic emm1 strains [16]. Furthermore, clade 3 emm89 strains lack the has genes required for the biosynthesis of the hyaluronic acid capsule typical of GAS [22]. Replacement of historic clades 1 and 2 by clade 3 temporally coincides with the increase in emm89 iGAS disease that has been documented in the United States and some European countries [16, 18].
Early reports described associations of emm89 GAS with skin infections [23], healthy children and young adult patients developing gastrointestinal symptoms and peritonitis [24, 25], and adults with necrotizing fasciitis and STSS [26]. Although our understanding of the biology of emm89 organisms has greatly advanced [15, 16, 18–20, 27], emm89 iGAS disease remains relatively poorly characterized clinically. Moreover, the clinical presentations and populations at risk for iGAS disease caused by the various emm89 clades have not been thoroughly investigated. To better understand these public health issues, we analyzed comprehensive population-based surveillance data for emm89 iGAS disease collected in metropolitan Toronto in the period 2000–2014, in combination with WGS analysis of the emm89 isolates responsible for these infections.
METHODS
Clinical Data, Isolates, and Laboratory Methods
The Toronto Invasive Bacterial Diseases Network (TIBDN) is a collaboration of all hospitals, microbiology laboratories, and public health units serving the city of Toronto and Peel region, Ontario, Canada (hereafter designated “metropolitan Toronto”; population 4.2 million in 2014). The TIBDN uses standardized methods and forms to collect clinical data including demographic information, disease manifestations, and underlying medical conditions from all patients with iGAS disease in the geographical area. Data used in this study are from January 1, 2000 to December 31, 2014; collection and usage was approved by the Research Ethics Boards of participating TIBDN institutions. Invasive GAS cases were defined as those in which acute illness occurred in association with the isolation of GAS from a normally sterile site (blood, cerebrospinal, pleural, peritoneal, pericardial, or joint fluid [including bursa], bone, aspirates, and tissue specimens or swabs obtained during surgery). Streptococcal toxic shock syndrome and necrotizing fasciitis were defined as previously described [27, 28].
We included 1 strain from each of the 105 emm89 iGAS disease cases recorded in metropolitan Toronto during the period (Table S1). We also analyzed clinical data and isolates from 138 additional emm89 iGAS cases that occurred in the province of Ontario outside of metropolitan Toronto (Table S1), recovered by hospitals that report and submit iGAS isolates to TIBDN on a voluntary basis. Clinical data were limited for these cases. Isolates were cultured at 37°C with 5% CO2 on Columbia blood agar plates containing 5% sheep blood or in Todd-Hewitt broth supplemented with 0.2% yeast extract. Isolates were confirmed to be GAS by β-hemolysis on sheep blood agar, grouping of carbohydrate antigen, large colony size, and bacitracin susceptibility [29].
Molecular Typing and Whole-Genome Sequencing of Group A Streptococcus Strains
Deoxyribonucleic acid was prepared from overnight cultures using the QIAamp DNA minikit (QIAGEN, Toronto, ON, Canada). emm typing was performed by polymerase chain reaction (PCR) and Sanger sequencing, as described previously [30]. We followed the procedures described by Nasser et al [17] to (1) prepare genomic libraries and perform paired-end Illumina genome sequencing of isolates, (2) identify polymorphisms against reference genomes, and (3) establish core-genome SNP-based phylogenies. We confirmed polymorphisms in the nga/slo promoter spacer region of all strains by PCR amplification and Sanger sequencing using previously reported primers [16]. Presence or absence of the 3-gene has locus was confirmed by PCR, as described previously [31].
Statistical Analysis
Statistical analysis was performed using SAS (version 9.3). Contingency tables were tested with 2-tailed χ2 or Fisher’s exact tests, as appropriate. Differences in disease incidence and length of hospitalization between emm89 clades were evaluated with Poisson regression analysis and the Kruskal-Wallis test, respectively. P values <.05 were considered statistically significant.
RESULTS
The Incidence of emm89 Invasive Group A Streptococcus Disease Significantly Increased in Metropolitan Toronto Commensurate With Emergence of Clade 3 Strains
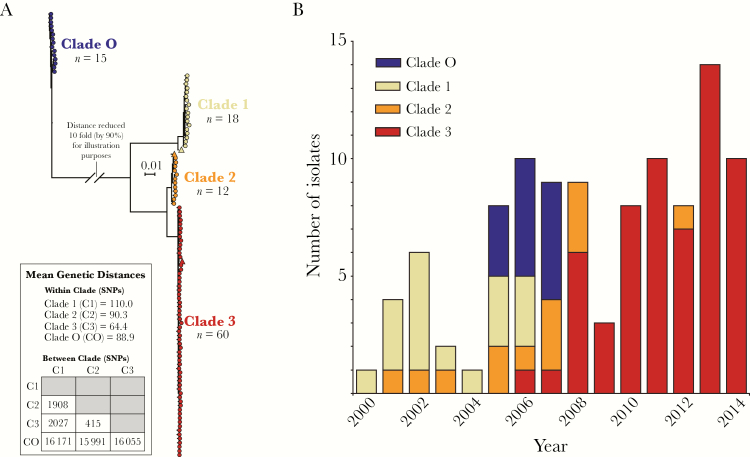
One hundred five of the 1596 (7%) iGAS disease cases in metropolitan Toronto between 2000 and 2014 were caused by emm89 strains. During this period, the overall incidence of iGAS infections remained relatively stable. In contrast, the incidence of emm89 GAS infections increased significantly from 0.14 per 100000 in 2000–2007 to 0.22 per 100000 in 2008–2014 (χ2, P = .021) (Table S2). To test the hypothesis that this increase in incidence was due to the emergence of clade 3 emm89 strains, we used core-genome-SNP analysis to determine the phylogenetic relationships of the 105 emm89 iGAS isolates. Results revealed a genetically diverse emm89 population, including strains belonging to previously described clades 1, 2 and 3, which were genetically closely related to reference strains of those clades isolated elsewhere (Figure 1A). We also identified a fourth, distinct phylogenetic group, which we have named clade O. Strains of this clade were genetically distantly related to clades 1, 2, or 3 strains (Figure 1A), further supporting the notion that genetic diversity among emm89 GAS is vastly higher than that observed in some other emm types [15]. We next studied the temporal distribution of the strains. Clade 1 strains (17% of the emm89 iGAS isolates) predominated in the earlier years of this investigation (Figure 1B). Clade 2 strains (11% of the emm89 iGAS isolates) were recovered primarily from 2005–2008. Clade O strains (14% of the total emm89 isolates) were found only in 2005–2007. The circulating emm89 strain population shifted dramatically in 2008, due to rapid expansion of clade 3 (57% of the total emm89 isolates, and 94% of those recovered from 2008–2014; Figure 1B), which effectively purged historic clades 1, 2, and O from the population. To investigate whether a similar replacement of historic clades by clade 3 occurred in the rest of Ontario, we sequenced the genomes of 138 emm89 isolates voluntarily submitted during the same time period (2000–2014) from areas of the province outside of metropolitan Toronto (Figure S1A). A similar increase in the number of emm89 iGAS cases caused by clade 3 strains was observed in the rest of the province beginning in 2007 (Figure S1B and S1C).
Figure 1.
The increase in incidence of emm89 invasive group A Streptococcus (iGAS) disease in metropolitan Toronto correlated with emergence of clade 3 emm89 strains. (A) shows the inferred genetic relationships among emm89 isolates causing iGAS disease in metropolitan Toronto. The neighbor-joining phylogenetic tree was constructed using 18925 concatenated single nucleotide polymorphism (SNP) loci identified relative to the genome of reference strain MGAS23530 (clade 2; GenBank accession number CP013839). The analysis identified 4 distinct clades (1, 2, 3, and O) among isolates from metropolitan Toronto. The genetic and virulence differences between clades 1, 2, and 3 are shown and have been previously described [15]. Clade 1, 2, and 3 isolates from metropolitan Toronto were closely related to their respective reference strains (indicated in the tree by the triangles). Clade 1 strains, including reference strain MGAS11027 (GenBank accession number CP013838), differed pairwise on average by 110 SNPs. Clade 2 strains including reference strain MGAS23530 differed on average by 90 SNPs. Clade 3 strains including reference strain MGAS27061 (GenBank accession number CP013840) differed on average by 64 SNPs. Clade O strains differed on average by 89 SNPs. Clade O strains were distantly related to clades 1, 2, and 3 strains, differing on average by 16171, 15991, and 16055 SNPs, respectively. Clade O strains were also distantly related to previously described emm89 “distant and near outliers” [15] (see also Figure S1B). (B) shows the yearly distribution of emm89 iGAS disease cases in metropolitan Toronto over the period 2000–2014. The different colors identify the genetic makeup of the emm89 isolates causing each disease case. In earlier years, most cases were caused by clades 1 and 2. Clade O strains were only identified in years 2005 to 2007. Clade 3 strains were first observed in 2006. Clade 3 infections become more prevalent in 2008 and have since essentially replaced all historic clades. Clade 3 strains accounted for all but 1 of the 53 emm89 iGAS isolates recovered after 2008.
Patients With Invasive Group A Streptococcus Disease Caused by Strains of the Different emm89 Clades Differed in Comorbidities and Risk Factors
At least 1 underlying illness potentially predisposing to iGAS disease was present in 76% of emm89-infected patients, and 37% of patients had more than 1 underlying illness. Although there was no significant difference in the frequency of underlying illness between the different emm89 clades, the distribution of underlying illness and risk factors was slightly different. Patients infected with clade 2 had higher rates of diabetes and cancer than patients infected with the other 3 clades (Table 1). This might be correlated with the observation that clade 2-infected patients had higher rates of STSS, intensive care unit (ICU) admission, and case fatality (Table 2). In contrast, we did not observe statistically significant differences in the length of hospitalization (median, 10 days) between patients infected with the different clades. More patients infected with clade 1 were admitted from nursing homes, whereas more patients infected with clade 2 and clade 3 strains were admitted from home than other clades (Table 1). Intravenous (IV) drug use and alcohol abuse was reported by 4% and 11% of emm89-infected patients, respectively, and all cases of self- reported IV drug use were associated with clades 1 or O strains (Table 1). None of these cases were clustered in space or time or associated with homelessness, although, as a group, clade O strains occurred more frequently among homeless patients (Table 1). Although we did not observe clade 3 emm89 infections in metropolitan Toronto among IV drug users, 14 clade 3 patients from the rest of Ontario had a history of IV drug use (Table S3).
Table 1.
Underlying Conditions Associated With emm89 and emm1 iGAS Infections, and Location From Which iGAS Patients From Metropolitan Toronto Were Admitted to Hospital, 2000–2014
| Underlying Condition | Clade O No. (%) |
Clade 1 No. (%) |
Clade 2 No. (%) |
Clade 3 No. (%) |
Total emm89 No. (%) |
emm1
No. (%) |
|---|---|---|---|---|---|---|
| Total | 15 (100) | 18 (100) | 12 (100) | 60 (100) | 105 (100) | 397 (100)d |
| Diabetes mellitus | 1 (7) | 4 (22) | 4 (33) | 17 (25) | 26 (25) | 54 (14)g |
| Cardiac diseasea | 0 (0) | 5 (28) | 0 (0) | 7 (12) | 12 (11) | 55 (14) |
| Pulmonary diseaseb | 1 (7) | 2 (11) | 2 (17) | 9 (15) | 14 (13) | 62 (16) |
| Renal disease | 0 (0) | 0 (0) | 0 (0) | 3 (5) | 3 (3) | 16 (4.1) |
| Liver disease | 3 (20) | 1 (6) | 0 (0) | 1 (2) | 5 (5) | 10 (2.6) |
| Immunodeficiencyc | 1 (7) | 6 (33) | 5 (42) | 9 (15) | 21 (20) | 41 (11)e |
| Alcohol abuse | 6 (40) | 0 (0) | 1 (8) | 5 (8) | 12 (11) | 22 (5.7)g |
| IV drug use | 3 (20)f | 1 (6)f | 0 (0)f | 0 (0)f | 4 (4) | 6 (1.5) |
| Admission from | ||||||
| Total | 15 (100) | 18 (100) | 12 (100) | 59 (100) | 104 (100)h | 393 (100)i |
| Home | 10 (67) | 12 (67) | 11 (92) | 48 (81) | 81 (78) | 342 (87) |
| Nursing home | 0 (0) | 4 (22) | 0 (0) | 5 (8) | 9 (9) | 16 (4.1) |
| Hospital | 0 (0) | 1 (6) | 0 (0) | 4 (7) | 5 (5) | 27 (6.9) |
| Retirement home/group home | 1 (7) | 1 (6) | 1 (8) | 2 (3) | 5 (5) | 6 (2) |
| Homeless | 4 (27) | 0 (0) | 0 (0) | 0 (0) | 4 (4) | 2 (1) |
Abbreviations: HIV, human immunodeficiency virus; iGAS, invasive group A Streptococcus; IV, intravenous; SLE systemic lupus erythematosus.
aIncludes cardiac disease and congestive heart failure.
bIncludes asthma, chronic bronchitis, and other respiratory conditions such as interstitial lung disease and bronchiectasis.
cIncludes previous organ/stem cell transplant, SLE, HIV infection, and cancer.
dData available for 389 of 397 cases.
eData for organ/stem cell transplant and SLE not available for emm1. Statistical analysis performed only for the comparison of HIV infection and cancer. No statistical difference found between emm89 and emm1.
f P < .05 for the comparison between emm89 clades clade O, clade 1, and clade 2, and the emerging clade 3.
g P < .05 for the comparison with all emm89 patients.
hData available for 104 of 105 patients.
iData available for 393 of 397 patients.
Table 2.
Clinical Presentation and Outcomes of Patients With emm89 and emm1 iGAS Infections in Metropolitan Toronto, 2000–2014
| Clinical Presentation/Outcome | Clade O No. (%) |
Clade 1 No. (%) |
Clade 2 No. (%) |
Clade 3 No. (%) |
emm89 Total No. (%) | emm1 No. (%) |
|---|---|---|---|---|---|---|
| Total | 15 (100) | 18 (100) | 12 (100) | 60 (100) | 105 (100) | 397 (100) |
| Arthritis | 2 (13) | 4 (22) | 2 (17) | 9 (15) | 17 (16) | 32 (8)c |
| Bacteremia without focus | 0 (0) | 3 (17) | 3 (25) | 7 (12) | 13 (12) | 61 (15) |
| Soft Tissue Infection | ||||||
| Necrotizing fasciitis | 0 (0) | 0 (0) | 0 (0) | 2 (3) | 2 (2) | 36 (9)c |
| Other soft tissue | 10 (67) | 4 (22) | 1 (8) | 20 (33) | 35 (33) | 138 (35) |
| Respiratory Tract Infection | ||||||
| Lower respiratory | 2 (13) | 3 (17) | 5 (42) | 8 (13) | 18 (17) | 79 (20) |
| Upper respiratory | 1 (6) | 1 (6) | 1 (8) | 7 (12) | 10 (9) | 20 (5) |
| Peripartum infection | 0 (0) | 1 (6) | 0 (0) | 4 (7) | 5 (5) | 8 (2) |
| Othera | 0 (0) | 2 (11) | 0 (0) | 3 (5) | 5 (5) | 23 (6) |
| STSS | 1 (7) | 5 (28) | 5 (42) | 14 (23) | 25 (24) | 117 (29) |
| Case fatalityb | 1 (7) | 4 (22) | 5 (42) | 10 (17) | 20 (19) | 81 (20) |
| ICU admission | 4 (27) | 6 (33) | 5 (42) | 16 (27) | 31 (30) | 158 (40) |
Abbreviations: ICU, intensive care unit; iGAS, invasive group A Streptococcus; STSS, streptococcal toxic shock syndrome.
aOther includes peritoneal infection, gynecological infection not associated with pregnancy.
bCase fatality was defined as death that could be attributed to GAS infection within 30 days of positive culture.
c P < .05 for the comparison with all emm89 patients.
Overall, Clade 3 emm89 Strains Caused Invasive Group A Streptococcus Disease in Younger Patients
The 105 emm89 iGAS disease cases from metropolitan Toronto occurred in patients aged 8 months to 96 years old, 53% of whom were female. The median age at infection was 51 years old for females and 48 years old for males. Males and females and patients of different ages had slightly different disease manifestations (Figure 2). Adult males had more arthritis, and soft tissue infections were more common among the elderly. The greatest incidence of infection occurred in those patients >75 years old, and this age group had the highest case fatality rate (Table S4). The median age of patients infected with historic clades 1, 2, and O was 51.4 years (range 6–96 years), whereas among clade 3-infected patients the median age was 49.3 years (8 months–90 years). Relative to the historic clades, the incidence of clade 3 infections was higher across all age groups, and particularly among those <19 years old, and middle-aged patients (those in their 30s and 40s) (Table S4). Although our data do not allow the calculation of precise incidence rates in areas of Ontario outside of metropolitan Toronto, we observed that in these areas the largest proportion of patients infected with clade 3 strains was in the 30–39 age group. Relative to the historic clades, significantly more clade 3 strains were isolated from patients aged 20–49 (Figure S1D). Taking all isolates from Ontario together, clade 3 strains infected proportionally more patients aged 20–49 and fewer patients aged 75+, relative to historic clades (Table S5).
Figure 2.
Clinical presentation of emm89 invasive group A Streptococcus (iGAS) disease cases in metropolitan Toronto (2000–2014) by sex, age group, and emm89 clade. The different clinical presentations are depicted in different colors as per the legend. Soft tissue and respiratory infections predominated among both female (top panel) and male patients (bottom panel) across all age groups and particularly in the elderly (75 years old or greater). Overall, more cases of clade 3 emm89 iGAS occurred among children and younger adults. Arthritis was more common among adult males aged 30–49. “Other” includes peritoneal infections and gynecological infections not associated with pregnancy.
Clinical Characteristics of emm89 Invasive Group A Streptococcus Infections
The most common manifestation of emm89 iGAS disease was soft tissue infection, followed by lower respiratory infections and arthritis (Table 2). Streptococcal toxic shock syndrome occurred in 24% of emm89-infected patients. The overall case fatality rate was 19%. To assess the features of emm89 disease, we compared the cohort of emm89 patients to that of patients with emm1 iGAS disease in the same population (Table 2). The most common disease manifestation among emm1 strains was also soft tissue infection, followed by lower respiratory infections and bacteremia without focus. Significantly more cases of arthritis and significantly fewer cases of necrotizing fasciitis were observed among patients with emm89 iGAS disease compared with those with emm1 disease (Table 2). However, STSS, ICU admission, and case fatality rates were not significantly different between emm89 and emm1 iGAS cases (Table 2). There were no significant differences between clades with respect to the sites of isolation, although emm89 iGAS strains were less frequently isolated from blood than emm1 (Table 3). Overall, 76% of emm89-infected patients and 70% of emm1-infected patients in metropolitan Toronto had at least 1 underlying illness. A significantly greater proportion of emm89-infected patients had diabetes compared with emm1-infected patients (Table 1).
Table 3.
Site of Isolation of emm89 and emm1 Isolates Causing iGAS Disease in Metropolitan Toronto, 2000–2014
| Site | Clade O No. (%)a |
Clade 1 No. (%) |
Clade 2 No. (%) |
Clade 3 No. (%) |
Total emm89 No. (%) |
emm1
No. (%) |
|---|---|---|---|---|---|---|
| Total | 15 (100) | 18 (100) | 12 (100) | 60 (100) | 105 (100) | 397 (100) |
| Blood and CSFb | 10 (67) | 9 (50) | 9 (75) | 44 (73) | 72 (69) | 312 (79)d |
| Otherc | 4 (27) | 5 (28) | 2 (17) | 6 (10) | 17 (16) | 48 (12) |
| Synovial fluid | 1 (7) | 2 (11) | 1 (8) | 6 (10) | 10 (10) | 21 (5) |
| Peritoneal fluid | 0 (0) | 1 (6) | 0 (0) | 1 (2) | 2 (2) | 4 (1) |
| Pleural fluid | 0 (0) | 1 (6) | 0 (0) | 3 (5) | 4 (4) | 12 (3) |
Abbreviations: CSF, cerebrospinal fluid; iGAS, invasive group A Streptococcus.
aPercentages may not add up to 100 due to rounding.
bOne single CSF isolate was obtained from an emm1-infected patient.
cOther includes isolates obtained from abscesses, aspirates, and specimens obtained during surgical procedures.
d P < .05 for the comparison with all emm89 patients.
DISCUSSION
Systems biology approaches combining in-depth genomic strain characterization with in vitro testing and experimental infection of nonhuman primates has unambiguously demonstrated that the recently emerged genetic clade 3 emm89 GAS, which overexpresses the cytolytic toxins NADase and streptolysin O, is the main driver of the rapid worldwide increase in emm89 iGAS disease [15, 16, 18, 22]. Here, using similar WGS-based approaches and temporal analysis, we show that in metropolitan Toronto the emergence and rapid expansion of clade 3 clonal progeny is responsible for the significant increase in incidence of emm89 iGAS disease observed since 2008 in a context of relatively stable total iGAS disease burden. Emergence and rapid expansion of clade 3 in metropolitan Toronto was concurrent with rapid decline and apparent extinction of previously circulating emm89 clades 1, and 2, and O.
The paucity of reports examining the clinical features of emm89 iGAS disease is at odds with the magnitude of the increase in emm89 iGAS disease reported here and in several other countries [15, 16, 18, 20, 32]. To begin to address this circumstance, we assessed comprehensively the clinical features of emm89 iGAS disease using population-based surveillance data collected over 14 years. One of our findings was that emm89 iGAS disease is characterized by the very frequent occurrence of soft tissue, severe respiratory infections, and arthritis. The prevalence of arthritis among emm89 patients was significantly higher than among type emm1 strains in metropolitan Toronto and similar to that reported previously in Europe [23, 33]. In contrast, emm89 organisms caused significantly less necrotizing fasciitis than emm1 strains. We next investigated the hypothesis that, similar to what has been described in animal models [16, 22], clade 3 emm89 causes more severe disease in human patients than previously circulating clades. Although clade 3 caused slightly more necrotizing fasciitis, there were no significant differences in clinical diagnosis between patients infected with the different clades. Similar to a previous report [18], we also did not observe significant differences in 7- or 30-day mortality between clade 3 and other emm89 clade types. We did not detect significant differences between emm89 clades for predisposing conditions such as chronic illness or immune suppression. Although significantly fewer clade 3-infected patients were IV drug users in metropolitan Toronto, data from the rest of Ontario showed that IV drug usage was significantly associated with clade 3 strains.
We next examined whether clade 3 emergence and rapid expansion correlated with enhanced ability of the strains to cause disease in different groups of individuals than previously circulating clades, and we made several interesting observations. First, although studies of iGAS incidence by age are frequently bimodal, with rates peaking in children <2 years old and highest rates in those >85 years of age [26, 27, 34], we identified very few cases of historic clades 1, 2, and O in children <5 years old and in those aged 5–19, age groups in which clade 3 emm89 iGAS disease was more frequent. In addition, clade 3 cases were significantly more common in patients aged 20–49 years old, who less frequently reported underlying comorbidities. Thus, since the emergence of clade 3, there is a trend towards a profile of emm89 GAS infected patient that is younger, slightly healthier, and with a lower rate of underlying illness.
CONCLUSIONS
Clonal replacement among iGAS is a well described phenomenon [35, 36]. Given that the replacement of historic emm89 clades by clade 3 has occurred simultaneously in several unrelated countries, which have different models of access to healthcare, and whose populations are dissimilar in many social and human factors [16, 18], we speculate that emergence of this clone is dependent primarily on bacterial factors such as enhanced ability to persist and infect naive hosts rather than on host factors. The use of nonhuman primate models of infection has shown that enhanced NADase and SLO toxin activity has endowed clade 3 strains with increased fitness in the upper respiratory tract [15, 16]. It is interesting to note that emm89 pharyngitis also strikingly increased in frequency in 2007 in Ontario [21], likely related to the emergence of clade 3. We speculate that this enhanced ability to persist in the upper respiratory tract may be one key contributor to the rapid dissemination of clade 3 and its ability to readily infect younger, healthier patients. Data presented here, data from the Centers for Disease Control and Prevention Active Bacterial Core surveillance, and our unpublished data for Ontario for the period 2015–2016 suggest that emm89 iGAS cases continue to occur in high numbers in North America. Very recent reports from Finland have shown that diversifying clonal variants (subclades) of clade 3 emm89 have appeared that are associated with increased mortality [37]. Continued monitoring of changes in emm89 genetic diversity and associated iGAS disease is warranted.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Acknowledgments
We thank Chin-Yu (Amanda) Hong, Agron Plevneshi, and Wallis Rudnick (Toronto Invasive Bacterial Diseases Network) for database curation. We are grateful to Kelsie Jagt and Adriana Peci (Public Health Ontario) for statistical consultation. We also thank John McLaughlin (Public Health Ontario) for critical reading of an earlier version of this manuscript.
Financial support. This work was funded by the Fondren Foundation (to J. M. M.) and Public Health Ontario (internal grant PIF-2015-007; to N. F.).
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Cole JN, Barnett TC, Nizet V, Walker MJ. Molecular insight into invasive group A streptococcal disease. Nat Rev Microbiol 2011; 9:724–36. [DOI] [PubMed] [Google Scholar]
- 2. Hoge CW, Schwartz B, Talkington DF, et al. The changing epidemiology of invasive group A streptococcal infections and the emergence of streptococcal toxic shock-like syndrome. A retrospective population-based study. JAMA 1993; 269:384–9. [PubMed] [Google Scholar]
- 3. Ralph AP, Carapetis JR. Group a streptococcal diseases and their global burden. Curr Top Microbiol Immunol 2013; 368:1–27. [DOI] [PubMed] [Google Scholar]
- 4. Laupland KB, Ross T, Church DL, Gregson DB. Population-based surveillance of invasive pyogenic streptococcal infection in a large Canadian region. Clin Microbiol Infect 2006; 12:224–30. [DOI] [PubMed] [Google Scholar]
- 5. Lancefield RC. The antigenic complex of Streptococcus haemolyticus: I. Demonstration of a type-specific substance in extracts of Streptococcus haemolyticus. J Exp Med 1928; 47:91–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Scott JR, Pulliam WM, Hollingshead SK, Fischetti VA. Relationship of M protein genes in group A streptococci. Proc Natl Acad Sci U S A 1985; 82:1822–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Manjula BN, Acharya AS, Fairwell T, Fischetti VA. Antigenic domains of the streptococcal Pep M5 protein. Localization of epitopes crossreactive with type 6 M protein and identification of a hypervariable region of the M molecule. J Exp Med 1986; 163:129–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Walker MJ, Barnett TC, McArthur JD, et al. Disease manifestations and pathogenic mechanisms of group A Streptococcus. Clin Microbiol Rev 2014; 27:264–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Steer AC, Law I, Matatolu L, et al. Global emm type distribution of group A streptococci: systematic review and implications for vaccine development. Lancet Infect Dis 2009; 9:611–6. [DOI] [PubMed] [Google Scholar]
- 10. Smeesters PR, McMillan DJ, Sriprakash KS, Georgousakis MM. Differences among group A Streptococcus epidemiological landscapes: consequences for M protein-based vaccines? Expert Rev Vaccines 2009; 8:1705–20. [DOI] [PubMed] [Google Scholar]
- 11. Musser JM, Shelburne SA., 3rd A decade of molecular pathogenomic analysis of group A Streptococcus. J Clin Invest 2009; 119:2455–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Carroll RK, Shelburne SA, 3rd, Olsen RJ, et al. Naturally occurring single amino acid replacements in a regulatory protein alter streptococcal gene expression and virulence in mice. J Clin Invest 2011; 121:1956–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Olsen RJ, Sitkiewicz I, Ayeras AA, et al. Decreased necrotizing fasciitis capacity caused by a single nucleotide mutation that alters a multiple gene virulence axis. Proc Natl Acad Sci U S A 2010; 107:888–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bessen DE, McShan WM, Nguyen SV, et al. Molecular epidemiology and genomics of group A Streptococcus. Infect Genet Evol 2015; 33:393–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Beres SB, Kachroo P, Nasser W, et al. Transcriptome remodeling contributes to epidemic disease caused by the human pathogen Streptococcus pyogenes. MBio 2016; 7 pii: e00403-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhu L, Olsen RJ, Nasser W, et al. A molecular trigger for intercontinental epidemics of group A Streptococcus. J Clin Invest 2015; 125:3545–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nasser W, Beres SB, Olsen RJ, et al. Evolutionary pathway to increased virulence and epidemic group A Streptococcus disease derived from 3615 genome sequences. Proc Natl Acad Sci U S A 2014; 111:1768–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Turner CE, Abbott J, Lamagni T, et al. Emergence of a new highly successful acapsular group a Streptococcus clade of genotype emm89 in the United Kingdom. MBio 2015; 6:e00622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ikebe T, Tominaga K, Shima T, et al. Increased prevalence of group A Streptococcus isolates in streptococcal toxic shock syndrome cases in Japan from 2010 to 2012. Epidemiol Infect 2015; 143:864–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Friães A, Machado MP, Pato C, et al. Emergence of the same successful clade among distinct populations of emm89 Streptococcus pyogenes in multiple geographic regions. MBio 2015; 6:e01780–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shea PR, Ewbank AL, Gonzalez-Lugo JH, et al. Group A Streptococcusemm gene types in pharyngeal isolates, Ontario, Canada, 2002–2010. Emerg Infect Dis 2011; 17:2010–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhu L, Olsen RJ, Nasser W, et al. Trading capsule for increased cytotoxin production: contribution to virulence of a newly emerged clade of emm89 Streptococcus pyogenes. MBio 2015; 6:e01378–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Luca-Harari B, Darenberg J, Neal S, et al. Clinical and microbiological characteristics of severe Streptococcus pyogenes disease in Europe. J Clin Microbiol 2009; 47:1155–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gillespie RS, Hauger SB, Holt RM. Primary group A streptococcal peritonitis in a previously healthy child. Scand J Infect Dis 2002; 34:847–8. [DOI] [PubMed] [Google Scholar]
- 25. Ekelund K, Lemcke A, Konradsen HB. Evaluation of gastrointestinal symptoms as primary sign of severe invasive group A streptococcal infections. Indian J Med Res 2004; 119:179–82. [PubMed] [Google Scholar]
- 26. Plainvert C, Doloy A, Loubinoux J, et al. Invasive group A streptococcal infections in adults, France (2006–2010). Clin Microbiol Infect 2012; 18:702–10. [DOI] [PubMed] [Google Scholar]
- 27. Davies HD, McGeer A, Schwartz B, et al. Invasive group A streptococcal infections in Ontario, Canada. Ontario Group A Streptococcal Study Group. N Engl J Med 1996; 335:547–54. [DOI] [PubMed] [Google Scholar]
- 28. Defining the group A streptococcal toxic shock syndrome. Rationale and consensus definition. The Working Group on Severe Streptococcal Infections. JAMA 1993; 269:390–1. [PubMed] [Google Scholar]
- 29. Facklam RR, Washington JA II. Streptococcus and related catalase-negative Gram-positive cocci. In: Balows A, Hausler WJ, Jr, Herrmann KLet al. (eds). Shadomy Manual of Clinical Microbiology, 5th ed Washington, D.C: American Society for Microbiology; 1991: pp 238–57. [Google Scholar]
- 30. Beall B, Gherardi G, Lovgren M, et al. emm and sof gene sequence variation in relation to serological typing of opacity-factor-positive group A streptococci. Microbiology 2000; 146:1195–209. [DOI] [PubMed] [Google Scholar]
- 31. Flores AR, Jewell BE, Fittipaldi N, et al. Human disease isolates of serotype M4 and M22 group a streptococcus lack genes required for hyaluronic acid capsule biosynthesis. MBio 2012; 3:e00413-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Latronico F, Nasser W, Puhakainen K, et al. Genomic tracks behind spread of bacteremic Group A Streptococcus type emm89 in Finland, 2004–2014. J Infect Dis 2016; 214:1987–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Olafsdottir LB, Erlendsdottir H, Melo-Cristino J, et al. Invasive infections due to Streptococcus pyogenes: seasonal variation of severity and clinical characteristics, Iceland, 1975 to 2012. Euro Surveill 2014; 19:5–14. [PubMed] [Google Scholar]
- 34. Nelson GE, Pondo T, Toews KA, et al. Epidemiology of invasive group A streptococcal infections in the United States, 2005–2012. Clin Infect Dis 2016; 63:478–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Athey TB, Teatero S, Sieswerda LE, et al. High incidence of invasive group A Streptococcus disease caused by strains of uncommon emm types in Thunder Bay, Ontario, Canada. J Clin Microbiol 2016; 54:83–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fittipaldi N, Beres SB, Olsen RJ, et al. Full-genome dissection of an epidemic of severe invasive disease caused by a hypervirulent, recently emerged clone of group A Streptococcus. Am J Pathol 2012; 180:1522–34. [DOI] [PubMed] [Google Scholar]
- 37. Latronico F, Nasser W, Puhakainen K, et al. Genomic tracks behind spread of bacteremic group A Streptococcus type emm89 in Finland, 2004–2014. J Infect Dis 2016; 214:1987–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.