Abstract
Combined fecal microbiota transfer and antibiotic treatment prevented recurrences of urinary tract infections with multidrug-resistant (MDR) Pseudomonas aeruginosa, but it failed to eradicate intestinal colonization with MDR Escherichia coli. Based on microbiota analysis, failure was not associated with distinct diminished microbiota diversity.
Keywords: fecal microbiota transplantation, multidrug resistance, microbiome, 16S analysis
Multidrug resistance (MDR) of Enterobacteriaceae is an increasing worldwide problem that challenges the treatment of common bacterial infections. Multidrug resistance has been declared one of the greatest challenges to global public health today, and innovative strategies for decolonization of MDR bacteria are urgently needed to reduce the use of reserve antibiotics and prevent transmission [1]. A few reports mention success with fecal microbiota transfer (FMT) to eliminate extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae. Failures have not been reported. We present a 34-year-old patient on peritoneal dialysis (PD), treated with FMT to eradicate a Verona integron-encoded metallo-β-lactamase (VIM)-positive Pseudomonas aeruginosa causing recurrent urinary tract infections (UTIs), which hampered planned kidney-pancreas transplantation. Microbiome analysis was performed before and after infusion of fecal microbiota.
CASE DESCRIPTION
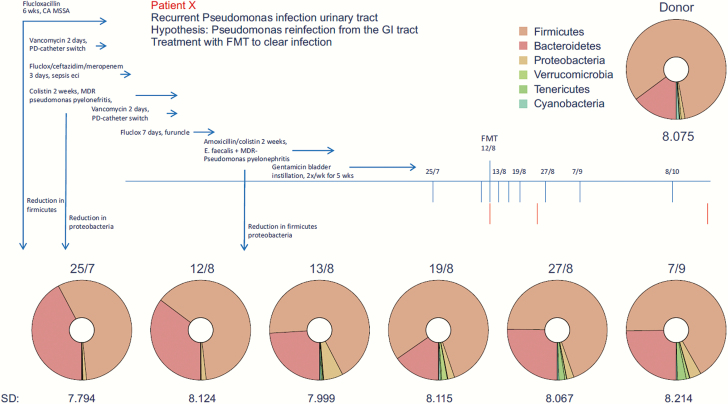
A 34-year-old male with type 1 diabetes mellitus was referred to our tertiary hospital because of diabetic nephropathy. Screening for combined kidney pancreas transplant started. Two months after starting hemodialysis, he was admitted because of bacteremia and catheter-related thrombophlebitis of the brachiocephalic vein by Staphylococcus aureus, which was treated with flucloxacillin for 6 weeks. Because the extensive thrombosis prohibited shunt or catheter placement, he was converted to PD. During admission, a transurethral catheter was placed because of neurogenic bladder dysfunction. Shortly after discharge, he returned to our hospital with a febrile catheter-related UTI and was treated empirically with ceftazidim. Urinary cultures were positive with a blaVIM carbapenemase-producing P aeruginosa, resistant to carbapenems, cephalosporins, quinolones, aminoglycosides, and fosfomycin, and only susceptible to colistin with a minimum inhibitory concentration of 4 mg/L. The same P aeruginosa was isolated from a rectal swab and the PD-catheter exit site. The patient received colistin intravenously (IV) for 2 weeks, and the urinary and PD catheter were replaced. In the following months, the patient suffered from recurrent febrile UTIs due to the MDR P aeruginosa (details on antibiotic use shown in Figure 1). Because of the high likelihood of recurrence of UTI caused by this MDR organism for which the only antibiotic was nephrotoxic, kidney transplantation was considered contraindicated and the patient was removed from the waiting list. During colistin treatment of the third episode, a plan for decolonization was developed. The transurethral catheter was removed, and intermittent catheterization with twice weekly prophylactic intravesical high-dose gentamicin instillations was started. Repeated negative cultures of urine, PD catheter-skin interface, skin, ears, and throat excluded chronic prostatitis or colonization at other sites than the gut. No oral selective digestive decontamination was given. After consultation with our ethics committee, informed consent was obtained from the patient for treatment with fecal microbiota: written informed consent was obtained from the patient for publication. Six weeks after the last IV course of colistin, the infusion of FMT was performed.
Figure 1.
Timeline of recurrent infections, antibiotic use, and microbiota diversity before and after fecal microbiota transfer. CA MSSA, catheter-related bacteremia with methicillin-sensitive Staphylococcus areus; FMT, fecal microbiotica transfer; GI, gastrointestinal; MDR, multidrug resistant; PD, peritoneal dialysis.
MATERIAL AND METHODS
Donor feces infusion was performed using the support of the National Donor Feces Bank (http://www.ndfb.nl/) according to the FECAL trial protocol with minor modifications [2]. In summary, donor feces was obtained from an unrelated healthy volunteer. Donor serum and feces were extensively screened for fecal and blood transmitted diseases including MDR bacteria. Seventy-five grams of feces was homogenized with saline and sieved (300-μm mesh) to remove undigested food fragments. Within 8 hours after defecation of the donor, 300-mL fecal suspension was infused in the duodenum of the patient through a nasoduodenal tube, after full colon lavage. Stool samples were collected before infusion, after 1 week, 2 weeks, 1 month, 2 months, and 3 months and screened for MDR presence using selective enrichment media, as described previously [3]. A portion of the feces was stored within 4 hours after delivery at −80°C for microbiome research. To assess the relatedness of bacterial strains, Amplified Fragment Length Polymorphism (AFLP) technique was performed as described previously [4].
MICROBIOTA ANALYSIS
Bacterial deoxyribonucleic acid (DNA) was isolated from the fecal samples using the ZR Fecal DNA MiniPrep kit (Zymo Research). Library preparation and amplification of the V4 hypervariable region 16S ribosomal ribonucleic acid gene was performed using NEXTflex 16S V4 Ampliconseq kit, version 2.0. High-throughput sequencing was executed at ServiceXS (Leiden, the Netherlands) on the Illumina HiSeq 2500 platform (Illumina, San Diego, CA) in rapid run mode paired-end 250 base pairs read length. Raw sequences were processed and analyzed using the open-source bioinformatics pipeline QIIME 1.9.1 (http://qiime.org/), and the Operational Taxonomic Unit was picked using the open-reference protocol. Subsequently, microbiota profiles were reported at phylum level and visualized using the visualization tool Krona [5].
RESULTS
No adverse event occurred during or after the infusion of microbiota, other than loose stools for 3 days. The stool culture taken before FMT was negative for the MDR P aeruginosa, but it did contain an ESBL-producing Escherichia coli, susceptible to carbapenems, gentamicin, piperacillin/tazobactam, and colistin. Subsequently, 5 stool cultures up to 3 months of follow up remained negative for P aeruginosa but persisted in containing the ESBL-producing E coli. The E coli post-FMT was identical to the E coli found before FMT, using AFLP. No infectious complications caused by P aeruginosa were noted during 18 months of follow up. However, the patient was treated once with trimethoprim-sulfamethoxazole for cystitis caused by an ESBL-positive E coli 8 months after FMT. Unfortunately, this strain was not available for AFLP analysis.
16S analysis of the patient’s stool 19 and 1 days before FMT revealed a diverse microbiota composition, ie, high Shannon diversity index of 7.8 and 8.1, respectively. No significant changes in microbiota diversity of the recipient were observed after the FMT (Figure 1). At phyla level, a high similarity of donor and recipient microbiota was observed with respect to the Firmicutes and Bacteroidetes as the expected main phyla of the microbiota (Figure 1).
DISCUSSION
A 34-year-old patient on PD and recurrent UTIs with a VIM-positive P aeruginosa was treated with infusion of fecal microbiota to eradicate P aeruginosa from the intestinal tract. A clinical success was observed, because at a follow-up period of 18 months no recurrent infections by P aeruginosa were diagnosed. Fecal microbiota transfer may have contributed to clinical success, but it cannot be excluded that MDR P aeruginosa was already eradicated from the gut before FMT, because the P aeruginosa could not be cultured the day before FMT.
A remarkable observation is the persistence of an ESBL-positive E coli after FMT. The E coli was presumably acquired after eradication treatment for P aeruginosa, because it had not been detected in earlier cultures. It is possible that the incomplete eradication of the MDR E coli is the result of coexistence of donor and patient E coli strains after FMT. A recent study showed this coexistence of donor and recipient strains, which persisted for at least 3 months after FMT for treatment of patients with metabolic syndrome [6]. This suggests that novel strains, acquired via FMT, can colonize the gut without replacing the indigenous strain population of the recipient.
In contrast to the diminished microbiota of recurrent Clostridium difficile infection (CDI) patients, our patient had an intact microbiota diversity and composition at phylum level before FMT. Previous antibiotic treatment (Figure 1) had not resulted in a distinct disturbance of the intestinal flora. Only minor changes of the microbiota composition were observed after FMT with a slight increase of cyanobacteria and tenericutes. We suggest that diminished diversity appears not to play a role in MDR carriership as opposed to recurrent CDI [7]. Therefore, one might question the efficacy of fecal transplantation in patients with a normal microbiota diversity. The disturbed microbiota and its recovery after FMT might explain the positive results of MDR eradication in patients with recurrent CDI [8, 9]. It is interesting to note that a recent paper showed that infusion of fecal microbiota in patients with recurrent CDI decreased the number and diversity of antimicrobial resistance genes, particularly by restoring dysbiosis and reducing the number of Proteobacteria [10]. Furthermore, beneficial effect of microbiota transfer has been shown in mice colonized with vancomycin-resistant Enterococcus (VRE) [11]. It is clear that more research on FMT for eradication of colonization of different MDR bacterial species is required.
Only 8 case reports have been published showing that FMT resulted in intestinal decolonization of ESBL- and carbapenemase-producing Enterobacteriaceae, VRE, or methicillin-resistant S aureus [12–15]. It is unfortunate that no information has been provided on microbiota composition before and after transplantation. Five trials are currently underway regarding the use of FMT for MDR bacterial decolonization, which should provide more insight on the role of the microbiota on colonization with specific microorganisms [12].
A limitation of our analysis is that the microbiota was determined by 16S analysis. Although very useful in bacterial taxonomic classification, it lacks the required resolution to track transmission of bacterial strains in the microbiota using single-nucleotide variants in metagenomes [6]. Therefore, it was not possible to compare the composition of the microbiota at strain level, allowing a comparison between the donor and patient P aeruginosa strains. However, no VIM gene was detected by polymerase chain reaction on DNA from 3 feces samples after FMT.
CONCLUSIONS
In conclusion, combined FMT and antibiotic treatment prevented recurrence of UTI with MDR P aeruginosa. Intestinal colonization with ESBL-producing E coli persisted in the presence of a microbiota with intact diversity, suggesting that eradication of E coli requires perhaps other specific strain(s) of microbes. More detailed analysis, such as metagenomics, could identify specific strains that add to decolonization and should be applied in current studies on FMT for intestinal eradication of different MDR bacterial species.
Acknowledgments
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. World Health Organization. Worldwide Country Situation Analysis: Response to Antimicrobial Resistance. Geneva: World Health Organization; 2015. [Google Scholar]
- 2. van Nood E, Vrieze A, Nieuwdorp M, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med 2013; 368:407–15. [DOI] [PubMed] [Google Scholar]
- 3. Paltansing S, Vlot JA, Kraakman ME, et al. Extended-spectrum β-lactamase-producing Enterobacteriaceae among travelers from the Netherlands. Emerg Infect Dis 2013; 19:1206–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vos P, Hogers R, Bleeker M, et al. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 1995; 23:4407–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ondov BD, Bergman NH, Phillippy AM. Interactive metagenomic visualization in a web browser. BMC Bioinformatics 2011; 12:385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li SS, Zhu A, Benes V, et al. Durable coexistence of donor and recipient strains after fecal microbiota transplantation. Science 2016; 352:586–9. [DOI] [PubMed] [Google Scholar]
- 7. Chang JY, Antonopoulos DA, Kalra A, et al. Decreased diversity of the fecal microbiome in recurrent Clostridium difficile-associated diarrhea. J Infect Dis 2008; 197:435–8. [DOI] [PubMed] [Google Scholar]
- 8. García-Fernández S, Morosini MI, Cobo M, et al. Gut eradication of VIM-1 producing ST9 Klebsiella oxytoca after fecal microbiota transplantation for diarrhea caused by a Clostridium difficile hypervirulent R027 strain. Diagn Microbiol Infect Dis 2016; 86:470–1. [DOI] [PubMed] [Google Scholar]
- 9. Dubberke ER, Mullane KM, Gerding DN, et al. Clearance of vancomycin-resistant Enterococcus concomitant with administration of a microbiota-based drug targeted at recurrent Clostridium difficile infection. Open Forum Infect Dis 2016; 3:ofw133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Millan B, Park H, Hotte N, et al. Fecal microbial transplants reduce antibiotic-resistant genes in patients with recurrent Clostridium difficile infection. Clin Infect Dis 2016; 62:1479–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ubeda C, Taur Y, Jenq RR, et al. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J Clin Invest 2010; 120:4332–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Manges AR, Steiner TS, Wright AJ. Fecal microbiota transplantation for the intestinal decolonization of extensively antimicrobial-resistant opportunistic pathogens: a review. Infect Dis (Lond) 2016; 48:587–92. [DOI] [PubMed] [Google Scholar]
- 13. Singh R, van Nood E, Nieuwdorp M, et al. Donor feces infusion for eradication of extended spectrum beta-lactamase producing Escherichia coli in a patient with end stage renal disease. Clin Microbiol Infect 2014; 20:O977–8. [DOI] [PubMed] [Google Scholar]
- 14. Crum-Cianflone NF, Sullivan E, Ballon-Landa G. Fecal microbiota transplantation and successful resolution of multidrug-resistant-organism colonization. J Clin Microbiol 2015; 53:1986–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lagier JC, Million M, Fournier PE, et al. Faecal microbiota transplantation for stool decolonization of OXA-48 carbapenemase-producing Klebsiella pneumoniae. J Hosp Infect 2015; 90:173–4. [DOI] [PubMed] [Google Scholar]