Abstract
Background
The neonatal innate immune system differs to microbial infection both quantitatively and qualitatively when compared to adults. Here, we provide the first genome-wide ex-vivo expression profile of umbilical cord blood (UCB) neutrophils from full-term infants prior to and in response to whole-blood LPS stimulation. Additionally, we provide cytokine expression prior to and following LPS stimulation. The genomic expression and cytokine profile are compared to LPS-stimulated whole blood from healthy adult subjects (HC).
Methods
Whole blood from UCB (n=6) and HC (n=6) was studied at baseline or was stimulated for 24 hours with 100 ngs/ml of lipopolysaccharide (LPS). CD66b+ neutrophils were subsequently isolated with microfluidic techniques and genome-wide expression analyses were performed. Ingenuity Pathway Analysis (IPA) software was utilized to predict downstream functional effects. Additionally, cytokine concentrations in whole blood prior to and after 24 hours of LPS incubation were determined.
Results
LPS stimulated whole blood from UCB demonstrated significant differences in both ex-vivo cytokine production and PMN gene expression. Mixed-effect modeling identified 1153 genes whose expression changed significantly in UCB and HC after exposure to LPS (p<0.001 with a minimum 1.5-fold change). IPA downstream predictions suggest that PMNs from UCB fail to effectively upregulate genes associated with activation, phagocytosis, and chemotaxis in response to LPS stimulation. Furthermore, whole blood from UCB showed increased IL-10 production to LPS, but failed to significantly increase several pro-inflammatory cytokines.
Conclusions
LPS-stimulated whole blood from UCB exhibited a markedly suppressed inflammatory cytokine production and PMN innate immune genome response. These differences in gene expression and cytokine production may be an adaptive response to a prior fetal environment, but may also explain their increased susceptibility to infections. Characterization of these deficits is the first step towards developing prophylactic and therapeutic interventions.
Keywords: neonatal sepsis, innate immunity, inflammation, phagocytosis, immune activation, chemotaxis
Introduction
Infections are responsible for nearly half of the three million neonatal deaths that occur annually (1) and sepsis is the number one cause of neonatal morbidity worldwide (2). Additionally, the annual costs of treating neonatal infection and sepsis are estimated to be $1.7 billion in the US alone in 2005 (3). The prevention of infection remains the most promising approach to reducing mortality and the morbidity in this highly vulnerable population.
Prior murine research and early human studies suggest that the innate and not the adaptive immune system is most crucial in preventing infection in the newborn, and that the neonatal innate immune system has both quantitative and qualitative deficits (4–8). By validating prior murine findings, and working to fully characterize the deficits of the human neonate’s innate immune system, we can work to better identify neonates at risk of infectious complications and begin to develop therapies aimed at correcting these deficits potentially providing prophylactic interventions to infants most at risk.
With the advent of intra-partum antibiotic use to combat Group B streptococcus, E. coli has emerged as the primary causative pathogen within 72 hours of life in preterm infants, and as the second most common cause in full-term infants (9). Here, we utilized lipopolysaccharide (LPS), an endotoxin released from gram-negative microbes such as E. coli, to delineate how the innate immune system, specifically the neutrophil, of the neonate responds to an infectious insult in an ex-vivo model.
Investigation of the neonatal immune system has two significant limitations: 1) the inability to collect an adequate volume of blood from neonates weighing as little as 500 grams, and 2) concerns regarding the risk:benefit of human investigations in neonates as a vulnerable population. By using umbilical cord as a source of neonatal blood, we have noninvasively obtained blood samples from newborns without posing any additional risk to the neonate. We recognize that cord blood samples contain some maternal contamination, but the quantities are general minimal (~1%) and are usually discounted (10). In addition, by using microfluidic techniques to isolate neutrophils from miniscule quantities of blood (11), we have minimized volume requirements for future studies with direct neonatal blood sampling.
Here, we present the first genome-wide ex-vivo expression profile of cord blood neutrophils in response to ex-vivo whole blood LPS stimulation. The transcriptomic profile of neutrophils obtained from full-term infant umbilical cord blood (UCB) and from adult healthy control subjects (HC) at both rest and 24 hours following LPS stimulation are compared. Ex-vivo whole blood cytokine expression and neutrophil genome-wide expression analyses were performed, and down-stream signaling pathways were extrapolated from the change of individual gene transcription of neutrophils in response to ex-vivo LPS stimulation. This analysis required less than 1 mL of blood.
Materials and Methods
Data and Sample Collection
UCB sample collection from full term infants was approved by the University of Florida Institutional Review Board (IRB) with a waiver of consent. UCB is considered a waste product; blood was collected by laboratory personnel from isolated segments of umbilical cord via needle puncture in the delivery room shortly after birth. A brief questionnaire was filled out by nursing staff that included gestational age, gender, weight, mechanism of birth, and presence of any known perinatal infectious concerns. The cord blood came from full-term infants (>38 week gestation) undergoing normal vaginal delivery with no documented complications. Cord blood samples were not based on race, age or ethnicity of the parents, or the sex of the infant. Study staff were not permitted access to electronic medical records or have patient contact due to waiver of consent. Blood from HC were collected by venipuncture from healthy young adults (25–45 years old), after informed consent was obtained. Healthy adult subjects claimed no recent medical history, no history of autoimmune disease or cancer, and were not taking any medications, including nonsteroidal anti-inflammatory agents.
Within an hour of blood draw, neutrophil genomics and whole blood cytokine concentrations were determined on aliquots of blood obtained from UCB and HC. Remaining UCB and HC whole blood aliquots were incubated at 37° C in a CO2 controlled incubator with highly purified LPS (100 ng per mL of blood; ultrapure via ion exchange chromatography Escherichia coli O26:B6 (Sigma-Aldrich, St. Louis, MO)) overnight. Preliminary studies were conducted on cord blood purchased from the New York Blood Bank, and was stimulated with increasing quantities of LPS (1, 10, 100, and 1000 ngs/ml). One-hundred ngs/ml was selected based on the greatest ex-vivo cytokine production (data not shown). Unstimulated controls were performed at baseline rather than after 24 hours of no stimulation for two reasons. First, the null hypothesis was that the change in PMN gene expression in response to LPS from neonates was not different than from adults. Second, measuring this change after 24 hours of no stimulation is difficult because of the relative short half-lives of non-stimulated human neutrophils. Estimates of the half-life of neutrophils in the circulation have varied from six hours to five days and whether the half-lives differ in neonates versus adults is unknown (12, 13).
Cytokine Analysis
Whole blood was centrifuged at 1500 × g for 8 minutes at 4° C, and the supernatant was collected and stored in an −80° C freezer. At the completion of sample collection, concentrations of IL-1β, IL-4, IL-6, IL-10, IL-12, IFNɣ, TNFα, MIP-1α, MCP-1, GRO, and IP-10 and were determined using Milliplex kits (Millipore, Darmstadt Germany) on the Luminex® MAGpix Multiplex reader.
Genomic Analysis
Neutrophils were isolated from whole blood using anti-CD66b monoclonal antibody coated microfluidic devices as previously described (14). The cassettes capture a fixed number of PMNs (between 5 and 10 × 106 cells), so that the total number of PMNs captured were equivalent between UCB and HC samples, regardless of the total or differential counts. Isolated neutrophils were then lysed en bloc using RLT buffer (Qiagen, Valencia, CA), and the cell lysate was run through a QiaShredder™ column and samples were stored in an −80° C freezer. RNA was extracted from lysates using QIAGEN RNeasy™ Mini Kit (Qiagen). RNA integrity was assessed with an Agilent 2100 Bioanalyzer (Santa Clara, CA, USA). RNA was labeled using Nugen (NuGEN Technologies, Inc., San Carlos, CA) Ovation Pico WTA® System V2. Resulting cDNA was labeled, fragmented using Nugen Encore® Biotin Module, hybridized onto GeneChip® Human Transcriptome Array 2.0 (Affymetrix, Santa Clara, CA) and processed following manufacturer’s instructions.
Gene Expression Profile and Statistical Analysis
Microarray expression was normalized using RMA™ as implemented in Partek (Partek Inc., St Louis MO). Differences in gene expression between UCB and HC samples, both unstimulated and LPS-stimulated, were identified using both ANOVA and a linear mixed-effects model with a false discovery adjusted probability of p<0.001. The mixed effects model better handles related cohorts that have markedly different variances. The expression of genes significant at p<0.001 with a minimum 2-fold change to distinguish between UCB and HC patients, or between unstimulated and LPS stimulation, was confirmed using leave-one-out-cross-validation studies and Monte Carlo simulations with class prediction tools implemented in BRB-ArrayTools™ Version: 4.5.0 – Beta_1 Release, developed by Richard Simon & BRB-ArrayTools Development Team (http://linus.nci.nih.gov/BRB-ArrayTools.html).
Gene Ontology™ and BioCarta™ Pathway analysis was conducted using BRB ArrayTools. These genes were further analyzed with Ingenuity Pathway Analysis (IPA) software™. IPA software™ was employed to make downstream functional predictions from these groups of genes with a Z-score greater than two indicating significance.
Differences in cytokine concentration between UCB and HC, with and without LPS stimulation, were compared using Wilcoxon’s sign-rank test, as the cohorts failed tests of normality.
Results
Genomic Analysis
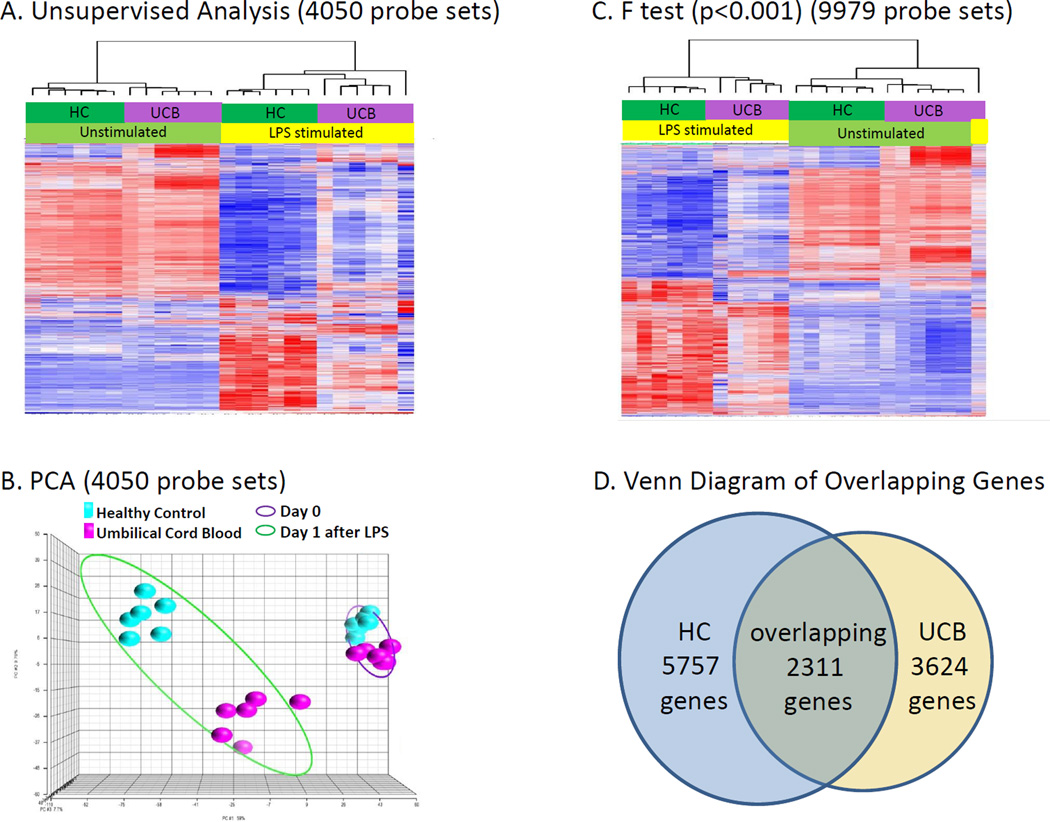
Initially, an unsupervised analysis was conducted using all 24 microarrays where the coefficient of variation for individual gene expression was set at 0.5. As shown in Figure 1A, 4,050 probe sets representing 3,762 genes had a variance of greater than 50%, and when hierarchically clustered, the gene expression patterns separated perfectly first between unstimulated samples and samples stimulated with LPS. Within each major subdivision, there was a further perfect separation between UCB and HC. When leave one out cross validation was used to predict the classification of samples into one of the four groups, the assignment was a perfect 100%, and a Monte Carlo simulation gave a probability of <0.001. The principal component analysis gives a better overall description of the sum of differences in gene expression between the four groups. As is evident, the differences in gene expression between UCB and HC were markedly greater after LPS stimulation that in unstimulated samples.
Figure 1. Umbilical Cord Blood and Healthy Control Subject Gene Expression.
A. When the 4,050 probe sets (representing 3,762 genes) with a CoV >0.5 in the unsupervised analysis were hierarchically clustered, defining differences in the patterns of gene expression was first the presence or absence of LPS and second, the source of the PMNs.
B. The unsupervised analysis of Figure 1A was processed using Principal Component Analysis (PCA) software which gives a better visual description of the sum of differences in gene expression between the four groups. Gene expression patterns from HC subjects (blue) and the UCB (pink) are tightly grouped to the right on Day 0; however, they respond differently to LPS exposure with the HC group clustering in the left upper corner and the UCB clustering in the lower center.
C. Using a false discovery adjusted probability of p<0.001 and a two-fold difference in expression, the temporal pattern of the expression of the LPS responsive genes differed between neutrophils from umbilical cord blood (UCB) (n=6) and healthy control (HC) subjects (n=6). There was a significant difference in the expression of 9,979 probe sets representing 9,296 unique genes between neutrophils isolated from UCB and HC subjects on prior to and overnight after LPS exposure.
D. Venn diagram showing the overlap between genes whose expression changed in response to LPS in the UCB and HC PMNs. 6,166 probe sets whose mRNA abundance changed significantly from HC (p < 0.001) in response to LPS stimulation which represented 5,757 unique genes, compared to only 3,741 probes sets representing 3,624 unique genes responding to LPS stimulation in PMNs from UCB. 2,389 probes sets representing 2,311 genes were identified as changing in common in both HC and UCB.
Figure 1C depicts an ANOVA on the four groups of data, UCB prior to and after LPS stimulation and HC prior to and after LPS stimulation which identified 9,979 probes sets as significant (p< 0.001 with a fold-change 2 cut-off) representing 9,296 unique genes. Figure 1D demonstrates the 6,166 probes sets whose mRNA abundance changed significantly from HC (p<0.001) in response to LPS stimulation which represented 5,757 unique genes, compared to only 3,741 probes sets representing 3,624 unique genes responding to LPS stimulation in PMNs from UCB. 2,389 probes sets representing 2,311 genes were identified as changing in common in both HC and UCB.
Mixed-effect modeling identified 1,790 genes whose magnitude of change was significantly different between PMNs obtained from UCB and HC (p<0.001; from prior to after LPS stimulation). These genes were further evaluated and 1,153 genes were identified as having a minimum 1.5-fold change.
Table 1 identifies the ten genes most upregulated and down regulated from UCB and HC in response to LPS stimulation. Five of the ten genes that are most up-regulated in adults and umbilical cord blood overlap (Panel A), but in every case, the magnitude of the increase is markedly less (from 85% to approximately 50%) in UCB samples. Four of the five genes are clearly involved in the inflammatory response, and even the fifth, the succinate receptor has been implicated as a coactivator of platelets (15). There was no significant overlap in the ten genes most down-regulated in response to LPS in PMNs from UCB and HC (Panel B). But, similar to that seen for upregulated genes, the magnitude of the changes in gene expression from UCB PMNs is markedly lower than from HC.
Table 1. Genes Whose Expression Increased and Decreased the Greatest in Response to Ex Vivo LPS Stimulation.
A. Top ten genes whose expression increased in response to ex-vivo LPS stimulation. Five of the ten genes are seen in both PMNs from UCB and HC; in every case, however, the increase was greater in PMNs from HC. Interestingly, only one gene on this list was expressed significantly greater in UCB: semenogelin (SEMG1). Degradation products of semenogelin are potent antimicrobials, including sgl29 (15).
B. Top ten genes whose expression is decreased in response to ex vivo LPS stimulation. In this case, there is no overlap between expression patterns in PMNs isolated from UCB and HC.
| Adult Healthy Control Subject | Full Term Umbilical Cord Blood | ||||
|---|---|---|---|---|---|
| Fold Change |
Name | Symbol | Fold Change |
Name | Symbol |
| 100 | chemokine (C-C motif) ligand 2 |
CCL2 | 20.8 | interleukin 1, alpha | IL1A |
| 55.6 | SLAM family member 7 | SLAMF7 | 19.2 | semenogelin I | SEMG1 |
| 41.7 | interleukin 1, alpha | IL1A | 17.9 | tumor necrosis factor (ligand) superfamily, member 15 |
TNFSF15 |
| 40 | chemokine (C-X-C motif) ligand 10 |
CXCL10 | 17.2 | microRNA 147b | MIR147B |
| 37 | succinate receptor 1 | SUCNR1 | 16.7 | succinate receptor 1 | SUCNR1 |
| 35.7 | tumor necrosis factor (ligand) superfamily, member 15 |
TNFSF15 | 16.4 | chemokine (C-C motif) ligand 2 |
CCL2 |
| 34.5 | interleukin 23, alpha subunit p19 |
IL23A | 14.1 | SLAM family member 7 | SLAMF7 |
| 29.4 | microRNA 222 | MIR222 | 8.3 | chemokine (C-C motif) receptor-like 2 |
CCRL2 |
| 26.3 | small nucleolar RNA, H | SNORA20 | 8.3 | interleukin-1 receptor- associated kinase 2 |
IRAK2 |
| 25.6 | chemokine (C-C motif) ligand 20 |
CCL20 | 8.3 | interferon-induced protein 44 | IFI44 |
| Fold- change |
Name | Symbol | Fold- change |
Name | Symbol |
|---|---|---|---|---|---|
| −36.1 | platelet and endothelial cell adhesion molecule 1 |
PECAM1 | −16.7 | defensin, alpha 4, corticostatin |
DEFA4 |
| −22.7 | RNA, 5S ribosomal 498 | RN5S498 | −14.6 | cathepsin G | CTSG |
| −21.4 | complement component (3b/4b) receptor 1 |
CR1 | −10.5 | histone cluster 1, H1c | HIST1H1C |
| −20.1 | microRNA 644a | MIR644A | −8 | ribonuclease, RNase A family, 2 (liver, eosinophil- derived neuro |
RNASE2 |
| −17.7 | RBM5 antisense RNA | LUST | −7.8 | ribonuclease, RNase A family, 3 |
RNASE3 |
| −17.3 | adhesion molecule, interacts with CXADR antigen 1 |
AMICA1 | −7.2 | cathelicidin antimicrobial peptide |
CAMP |
| −17 | toll-like receptor 6 | TLR6 | −6.6 | latexin | LXN |
| −15.9 | ring finger protein 141 | RNF141 | −6.1 | RBM5 antisense RNA | LUST |
| −13.5 | family with sequence similarity 65, member B |
FAM65B | −5.8 | activating transcription factor 6 |
ATF6 |
| −12.8 | maltase-glucoamylase (alpha-glucosidase) |
MGAM | −5.7 | IQ motif containing GTPase activating protein 2 |
IQGAP2 |
Significantly Downregulated Umbilical Cord Blood Functional Predictions
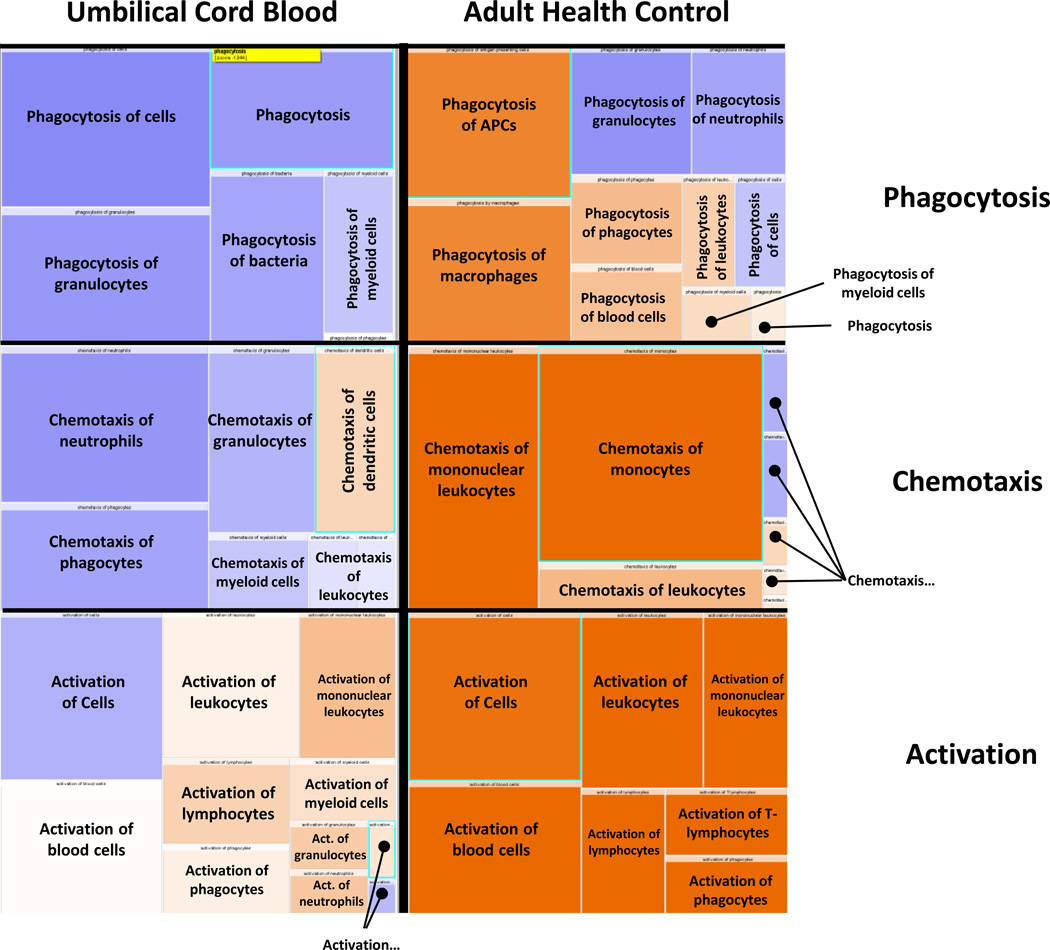
IPA software™ was utilized to predict downstream effects from this neutrophil transcriptomic profile and revealed a general propensity of HC to upregulate expression of ontologies of genes related to neutrophil activation, phagocytosis and chemotaxis, while UCB exhibited attenuated upregulation or downregulation of these same groups (Figure 2).
Figure 2. Ingenuity Pathway Analysis (IPA) Downstream Predictions.
IPA software™ was employed to analyze the neutrophil transcriptomic changes in umbilical cord blood (UCB) vs healthy control (HC) from Day 0 to Day 1 after lipopolysaccharide (LPS) incubation. The data shown represent 9,296 genes that are significantly different between the groups (p<0.001 with a fold-change 2 cut-off). Here, the orange represents over-representation of genes (upregulated) and the blue represents downregulated. The UCB shows almost global downregulation, or attenuated upregulation of genes involved in phagocytosis, chemotaxis and activation after LPS exposure. Whereas, the adult favors a more upregulated transcriptomic profile.
Downstream predictions for these genes identified significant preponderance of genes involved in “cytotoxicity”, “toxicity of cells”, and “killing of cells” (Z= −2.408; −2.136; −2.058, respectively) that were down-regulated in UCB; conversely in PMNs from HC, LPS stimulation showed a trend towards upregulation (Table 2). Additionally, gene expression profiles involved in “cell movement of neutrophils, granulocytes, and phagocytes” (Z= −3.011; −2.718; −2.081, respectively), “apoptosis of hematopoietic cell lines” (Z= −2.027), “phagocytosis of granulocytes” (Z= −2.191), “synthesis of lipids” (Z= −2.504), “synthesis and metabolism of eicosanoids” (Z= −2.150; −2.182) and “metabolism of reactive oxygen species” (Z= −2.088) were all significantly downregulated in UCB after LPS stimulation whereas HC response was not significantly downregulated.
Table 2. Predicted Functional Outcomes from Transcriptomic Expression Changes after LPS Exposure in Umbilical Cord Blood vs. Healthy Control.
The transcriptomic upregulation or downregulation of 1,153 genes, identified by mixed-effect modeling (p<0.001; with 1.5 fold-change cut-off) were analyzed by IPA software™ and significant downstream functional predictions (defined as −2.00< Z >2.00) were identified for umbilical cord blood (UCB) and healthy control (HC). Here is a comparison of Z-scores (from Day 0 vs Day 1 after LPS) between UCB and HC. UCB demonstrated significant downregulation of cell movement, phagocytosis, apoptosis, cytotoxicity, and synthesis of lipids and eicosanoids. UCB had significant upregulation of genes responsible for quantity of neutrophils, as well as organismal death and morbidity and mortality. Although HC showed similar trends in these categories, the change was not significant. Conversely, HC demonstrated significant downregulation of adhesion and binding genes and significant upregulation of activation of immune cells and signaling pathways. Both UCB and HC significantly upregulated quantity of phagocytes, antigen presentation, and response of antigen presenting cells.
| Significantly Downregulated in Umbilical Cord Blood | Umbilical Cord Blood Z-Score |
Healthy Control Z-Score |
|---|---|---|
| Cell Movement | ||
| Cell movement of neutrophils | −3.011 | −1.876 |
| Cell movement of granulocytes | −2.718 | −1.089 |
| Cell movement of phagocytes | −2.081 | −1.067 |
| Phagocytosis | ||
| Phagocytosis of granulocytes | −2.191 | −1.195 |
| Cell Death and Survival | ||
| Apoptosis of Hematopoietic Cell Lines | −2.027 | −1.977 |
| Cytotoxicity | −2.408 | 0.129 |
| Cytotoxicity of Cells | −2.006 | 0.025 |
| Toxicity of Cells | −2.136 | 0.182 |
| Killing of Cells | −2.058 | 0.018 |
| Lipid Metabolism | ||
| Synthesis of Lipids | −2.504 | −0.918 |
| Synthesis of Eicosanoid | −2.15 | −0.225 |
| Metabolism of Eicosanoid | −2.182 | −0.174 |
| Free Radical Scavenging | ||
| Metabolism of Reactive Oxygen Species | −2.088 | −0.902 |
| Significantly Upregulated in Umbilical Cord Blood | ||
| Hematological System Development and Function | ||
| Quantity of Blood Cells | 2.142 | 1.615 |
| Quantity of Leukocytes | 2.24 | 1.126 |
| Quantity of Neutrophils | 2.034 | 0.973 |
| Organism Survival | ||
| Organismal Death | 3.621 | 1.04 |
| Morbidity and Mortality | 3.472 | 0.847 |
| Significantly Downregulated in Healthy Control | ||
| Hematological System Development and Function | ||
| Adhesion of Neutrophils | −1.08 | −2.178 |
| Adhesion of Granulocytes | −1.108 | −2.337 |
| Adhesion of Immune Cells | −0.771 | −2.223 |
| Binding of Granulocytes | −0.489 | −2.56 |
| Binding of Professional Phagocytic Cells | −1.782 | −2.302 |
| Binding of Leukocytes | −0.156 | −2.171 |
| Significantly Upregulated in Healthy Control | ||
| Cell-to-Cell Signaling and Interaction | ||
| Activation of Antigen Presenting Cells | 0.6 | 2.634 |
| Activation of Leukocytes | 0.152 | 2.407 |
| Activation of Phagocytes | 0.313 | 2.37 |
| Activation of Blood Cells | −0.055 | 2.15 |
| Immune Response of Antigen Presenting Cells | 1.868 | 2.417 |
| Cell Signaling | ||
| Protein Kinase Cascade | 0.923 | 2.884 |
| I-kappaB kinase/NF-kappaB Cascade | 0.423 | 2.262 |
| Significantly Upregulated in Healthy Control and Umbilical Cord Blood | ||
| Quantity of Phagocytes | 3.147 | 2.062 |
| Antigen Presentation | 2.903 | 3.39 |
| Response of Antigen Presentation Cells | 2.213 | 2.197 |
Significantly Upregulated Umbilical Cord Blood Functional Predictions
PMNs from UCB had overexpression of genes significantly upregulated in pathways linked to “organismal death (Z= 3.621) and morbidity and mortality” (Z= 3.472) as well as “quantity of blood cells, leukocytes, and neutrophils” (Z= 2.142; 2.240; 2.034, respectively) following LPS incubation; these were not significantly upregulated in HC.
Significant Downregulated Healthy Control Functional Predictions
In contrast, PMNs from HC had overexpression of genes that were significantly downregulated involved in “adhesion of neutrophils, granulocytes, and immune cells” (Z= −2.178; −2.337; −2.223) as well as “binding of granulocytes, professional phagocytic cells, and leukocytes” (Z= −2.560; −2.302; −2.171, respectively); while expression of PMNs from UCB was downregulated but not significantly.
Significant Upregulated Healthy Control Functional Predictions
PMNs from HC demonstrated overexpression of genes involved in the upregulation of “activation of blood cells” (Z= 2.150) while PMNs from UCB were not significantly downregulated. Additionally, PMNs from HC showed upregulation of “activation of antigen presenting cells, leukocytes, and phagocytes” (Z= 2.634; 2.407; 2.370, respectively), as well as “immune response of antigen presenting cells” (Z= 2.417); UCB demonstrated upregulation that was attenuated and not significant.
Significantly Upregulated Healthy Control Signaling Predictions
In addition to predicted functional effects, HC also demonstrated significantly increased “signaling through I-κB kinase/NF-κB cascade and protein kinase cascade” (Z= 2.262; 2.884) while UCB demonstrated an attenuated non-significant upregulation.
Cytokine Analysis
(Figure 3). In response to LPS, whole blood from HC significantly increased IFNƔ concentrations (p<0.01) while UCB demonstrated a non-significant decrease. IL-12 (p<0.01), MIP-1α (p<0.0001), TNF-α (p<0.005), IP-10 (p<0.0001) concentrations were all increased by LPS stimulation in HC, while whole blood stimulation from UCB had an attenuated non-significant increase in these cytokine concentrations. IL-1β and IL-6 were significantly upregulated by LPS stimulation in both HC (p<0.005; p<0.01) and UCB (p<0.05; p<0.001) with a trend toward more robust response from HC; however, the fold change post LPS incubation between HC and UCB were not statistically different. Whole blood from UCB had a significant increase in IL-10 (p<0.0001) following LPS stimulation, while HC showed an attenuated non-significant increase. Whole blood from UCB also had a trend towards greater increases in MCP-1 post-LPS stimulation (p<0.0001) when compared to HC; however, the fold change (Day 0 vs Day 1 post LPS incubation) difference between HC and UCB stimulated whole blood was not statistically different and HC also had significant increase in MCP-1 (p<0.01).
Figure 3. Cytokine Responses Following Overnight Incubation with Lipopolysaccharide.
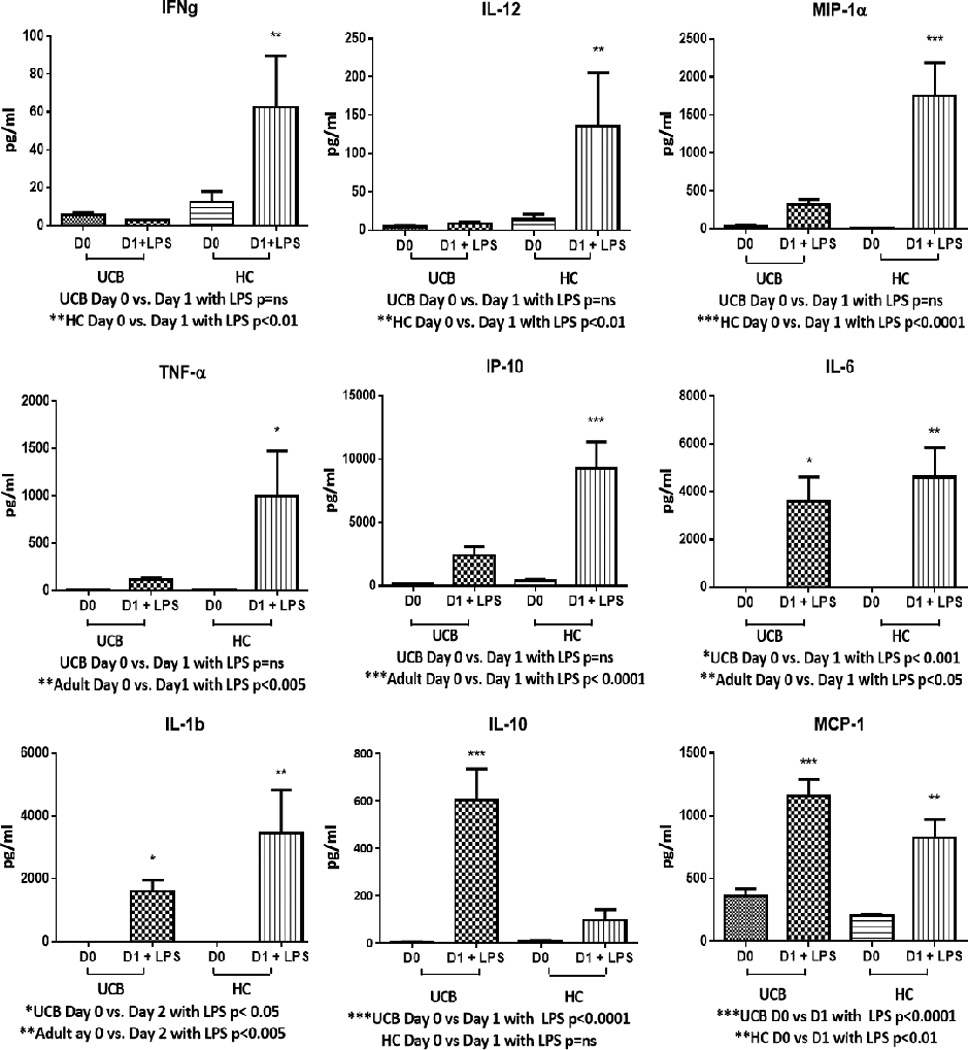
In response to overnight whole-blood stimulation with lipopolysaccharide (LPS), healthy control (HC) significantly upregulated IFNƔ (p<0.01), IL-12 (p<0.01), MIP-1α (p<0.0001), TNFα (p<0.005), IP-10 (p<0.0001) while umbilical cord blood (UCB) had attenuated non-significant responses. IL-1β and IL-6 concentrations were significantly increased by both HC (p<0.005; p<0.01) and UCB (p<0.05; p<0.001). UCB had a significant increase in IL-10 (p<0.0001), while HC showed an attenuated non-significant increase. UCB also had a trend towards greater increase in MCP-1 post-LPS stimulation (p<0.0001) when compared to HC (p<0.01).
Discussion
Infection continues to be a significant cause of morbidity and mortality in the neonatal population. Previous research suggests that it is primarily the innate and not the adaptive immune system that plays the most crucial role in combating bacterial infection (4, 5). While the innate immune system is a complex network, neutrophil function plays a predominant role in combating infections (16). LPS, an endotoxin, has been shown to be a major mediator of neutrophil activation via TLR4 receptor signaling. Here, we compared neutrophil genomic and whole blood responses from full-term neonate UCB and compared it to HC volunteers in order to highlight the differences in neutrophil responsiveness following overnight LPS stimulation ex-vivo. Surprisingly, gene expression patterns at baseline were surprisingly similar between PMNs obtained from full-term infants and healthy adult subjects; however, their responses to LPS were significantly different. To our knowledge, this is the first comparison of the genome-wide ex-vivo expression profile of neutrophils prior to and in response to LPS stimulation from cord blood. Prior microarray analysis has only been conducted on infected or septic children (17–20) or in response to peptidoglycan (21).
Transcriptomic analysis of the PMN genome suggests that in response to LPS stimulation, PMNs from UCB have a reduced capacity to upregulate transcription of classes of genes related to neutrophil activation, phagocytosis and chemotaxis when compared to PMNs from HC. Neutrophil activation has been shown to be influenced by length of labor (22), but has not been well characterized at the level of genomic expression. Our findings concerning chemotaxis and phagocytosis are supported by prior functional studies that found a 60% decrease in neonatal neutrophil chemotaxis to the chemoattractant fMLP (23), as well as a reduced phagocytic capacity (24, 25). We are reluctant to define these differences in genomic and functional responses as being ‘defective’ in the newborn, as they obviously reflect an evolutionary response to the fetal state and the transition to independence. However, a better understanding of these differences provides a potential target for future therapies aimed at facilitating the transition to a more adult-like state. Additionally, transcriptomic signatures may enable risk stratification and facilitate closer surveillance and more aggressive clinical management of high risk neonates, while at the same time, minimizing interventional therapies and broad spectrum antibiotic use in low risk neonates.
Whole-blood from HC stimulated with LPS exhibited significant increases in pro-inflammatory cytokine concentrations (INFƔ, IL-12, MIP-1α, TNFα, IL-1β and IL-6) while whole blood from UCB showed only significantly increased IL-6 concentrations. Additionally, LPS stimulated HC blood exhibited a significant increase in IP-10 concentrations, a chemoattractant for monocytes, macrophages, T-cells, NK cells, and dendritic cells. Conversely, LPS-stimulated whole blood from UCB had significantly increased levels of the anti-inflammatory cytokine IL-10, which is known to downregulate Th1 polarization of T-cells, decrease MHC II complexes on antigen presenting cells (APC) and block NF-κB translocation (26). This cytokine profile in response to LPS incubation suggests that whole blood from UCB is unable to mount an inflammatory response to TLR4 stimulation which is likely to result in less Th1 T-cell polarization, less immune cell activation and recruitment and poor antigen presentation (Figure 3). These results largely correlate with similar prior studies; however, these studies were limited by lack of comparison to HC or included a limited number of cytokines (27–29). Our study uniquely demonstrates a significant increase in UCB IL-10 expression and fails to support a significant increase in UCB TNFα as reported by prior studies.
The shortcomings of this study are the low number of enrolled study participants which may be underpowered to detect all significant differences between UCB and HC. Our neonate study population is also limited to full-term infants due to the frequent use of cord blood ‘milking’ at the time of delivery for preterm infants whereby the umbilical cord blood is pushed back towards the infant and into the infant’s circulatory system. Therefore, our study doesn’t represent the population with the highest incidence of sepsis. However, we do believe that the shortcomings of the neonate’s immune system are a continuum and continued study in preterm neonates will likely include the differences noted here in term infant UCB as well as additional differences. Finally, cord blood contains some maternal cells and plasma, so it is not a true reflection of the infant per se. However, the degree of maternal contamination is generally considered modest, and efforts to obtain sufficient quantities of blood for genomic and cytokine measurements directly from the newborn are often difficult. Due to the waiver of consent, we were limited in our ability to review the medical history of the mother and infant for infection; however, the absence of any known pre-natal infectious complications was confirmed at the time of birth. Additionally, the information presented here reflects neutrophil transcription and either protein production or functional responses are needed to confirm whether these transcriptional changes translate into differences in PMN anti-microbial activity.
Future study will need to focus on neonatal blood collected at or around the time of birth from all viable gestational ages in order to fully characterize the gradation of unique changes exhibited by preterm and low-birth weight neonatal immune systems. Once the immune system is characterized, interventions aimed at reducing infections can be undertaken.
Acknowledgments
This work was supported by R01 GM097531, awarded by the National Institute of General Medical Sciences. BM and JM were supported by a National Institute of General Medical Sciences post-doctoral training grant in burns and trauma (T32 GM-008621-15).
Footnotes
Conflict of Interest and Financial Disclosure Statement: No conflict of or competing interests have been declared.
References
- 1.Liu L, Johnson HL, Cousens S, Perin J, Scott S, Lawn JE, Rudan I, Campbell H, Cibulskis R, Li M, Mathers C, Black RE W. H. O. Child Health Epidemiology Reference Group of and Unicef. Global, regional, and national causes of child mortality: an updated systematic analysis for 2010 with time trends since 2000. Lancet. 2012;379(9832):2151–2161. doi: 10.1016/S0140-6736(12)60560-1. [DOI] [PubMed] [Google Scholar]
- 2.Camacho-Gonzalez A, Spearman PW, Stoll BJ. Neonatal infectious diseases: evaluation of neonatal sepsis. Pediatr Clin North Am. 2013;60(2):367–389. doi: 10.1016/j.pcl.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watson RS, Carcillo JA. Scope and epidemiology of pediatric sepsis. Pediatr Crit Care Med. 2005;6(3 Suppl):S3–S5. doi: 10.1097/01.PCC.0000161289.22464.C3. [DOI] [PubMed] [Google Scholar]
- 4.Cuenca AG, Wynn JL, Moldawer LL, Levy O. Role of innate immunity in neonatal infection. Am J Perinatol. 2013;30(2):105–112. doi: 10.1055/s-0032-1333412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gentile LF, Nacionales DC, Lopez MC, Vanzant E, Cuenca A, Cuenca AG, Ungaro R, Szpila BE, Larson S, Joseph A, Moore FA, Leeuwenburgh C, Baker HV, Moldawer LL, Efron PA. Protective immunity and defects in the neonatal and elderly immune response to sepsis. J Immunol. 2014;192(7):3156–3165. doi: 10.4049/jimmunol.1301726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Melvan JN, Bagby GJ, Welsh DA, Nelson S, Zhang P. Neonatal sepsis and neutrophil insufficiencies. Int Rev Immunol. 2010;29(3):315–348. doi: 10.3109/08830181003792803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Erdman SH, Christensen RD, Bradley PP, Rothstein G. Supply and release of storage neutrophils. A developmental study. Biol Neonate. 1982;41(3–4):132–137. doi: 10.1159/000241541. [DOI] [PubMed] [Google Scholar]
- 8.Wynn JL, Scumpia PO, Winfield RD, Delano MJ, Kelly-Scumpia K, Barker T, Ungaro R, Levy O, Moldawer LL. Defective innate immunity predisposes murine neonates to poor sepsis outcome but is reversed by TLR agonists. Blood. 2008;112(5):1750–1758. doi: 10.1182/blood-2008-01-130500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stoll BJ, Hansen NI, Sanchez PJ, Faix RG, Poindexter BB, Van Meurs KP, Bizzarro MJ, Goldberg RN, Frantz ID, 3rd, Hale EC, Shankaran S, Kennedy K, Carlo WA, Watterberg KL, Bell EF, Walsh MC, Schibler K, Laptook AR, Shane AL, Schrag SJ, Das A, Higgins RD H. Eunice Kennedy Shriver National Institute of Child and N. Human Development Neonatal Research: Early onset neonatal sepsis: the burden of group B Streptococcal and E. coli disease continues. Pediatrics. 2011;127(5):817–826. doi: 10.1542/peds.2010-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poli F, Crespiatico L, Lecchi L, Sirchia G, Scalamogna M, Sirchia SM, Garagiola I, Pedranzini L. Highly sensitive chemiluminescent method for the detection of maternal cell contamination in human cord blood stored for allotransplantation: the experience of the Milano cord blood bank. Blood. 1997;89(8):3061–3062. [PubMed] [Google Scholar]
- 11.Warner EA, Kotz KT, Ungaro RF, Abouhamze AS, Lopez MC, Cuenca AG, Kelly-Scumpia KM, Moreno C, O'Malley KA, Lanz JD, Baker HV, Martin LC, Toner M, Tompkins RG, Efron PA, Moldawer LL. Microfluidics-based capture of human neutrophils for expression analysis in blood and bronchoalveolar lavage. Lab Invest. 2011;91(12):1787–1795. doi: 10.1038/labinvest.2011.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dancey JT, Deubelbeiss KA, Harker LA, Finch CA. Neutrophil kinetics in man. J Clin Invest. 1976;58(3):705–715. doi: 10.1172/JCI108517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pillay J, den Braber I, Vrisekoop N, Kwast LM, de Boer RJ, Borghans JA, Tesselaar K, Koenderman L. In vivo labeling with 2H2O reveals a human neutrophil lifespan of 5.4 days. Blood. 2010;116(4):625–627. doi: 10.1182/blood-2010-01-259028. [DOI] [PubMed] [Google Scholar]
- 14.Kotz KT, Xiao W, Miller-Graziano C, Qian WJ, Russom A, Warner EA, Moldawer LL, De A, Bankey PE, Petritis BO, Camp DG, 2nd, Rosenbach AE, Goverman J, Fagan SP, Brownstein BH, Irimia D, Xu W, Wilhelmy J, Mindrinos MN, Smith RD, Davis RW, Tompkins RG, Toner M Inflammation and P. the Host Response to Injury Collaborative Research. Clinical microfluidics for neutrophil genomics and proteomics. Nat Med. 2010;16(9):1042–1047. doi: 10.1038/nm.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Macaulay IC, Tijssen MR, Thijssen-Timmer DC, Gusnanto A, Steward M, Burns P, Langford CF, Ellis PD, Dudbridge F, Zwaginga JJ, Watkins NA, van der Schoot CE, Ouwehand WH. Comparative gene expression profiling of in vitro differentiated megakaryocytes and erythroblasts identifies novel activatory and inhibitory platelet membrane proteins. Blood. 2007;109(8):3260–3269. doi: 10.1182/blood-2006-07-036269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kobayashi SD, DeLeo FR. Role of neutrophils in innate immunity: a systems biology-level approach. Wiley Interdiscip Rev Syst Biol Med. 2009;1(3):309–333. doi: 10.1002/wsbm.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dickinson P, Smith CL, Forster T, Craigon M, Ross AJ, Khondoker MR, Ivens A, Lynn DJ, Orme J, Jackson A, Lacaze P, Flanagan KL, Stenson BJ, Ghazal P. Whole blood gene expression profiling of neonates with confirmed bacterial sepsis. Genom Data. 2015;3:41–48. doi: 10.1016/j.gdata.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cernada M, Serna E, Bauerl C, Collado MC, Perez-Martinez G, Vento M. Genome-wide expression profiles in very low birth weight infants with neonatal sepsis. Pediatrics. 2014;133(5):e1203–e1211. doi: 10.1542/peds.2013-2552. [DOI] [PubMed] [Google Scholar]
- 19.Smith CL, Dickinson P, Forster T, Craigon M, Ross A, Khondoker MR, France R, Ivens A, Lynn DJ, Orme J, Jackson A, Lacaze P, Flanagan KL, Stenson BJ, Ghazal P. Identification of a human neonatal immune-metabolic network associated with bacterial infection. Nat Commun. 2014;5:4649. doi: 10.1038/ncomms5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong HR. Genome-wide expression profiling in pediatric septic shock. Pediatr Res. 2013;73(4 Pt 2):564–569. doi: 10.1038/pr.2013.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fong ON, Chan KY, Leung KT, Lam HS, Cheung HM, Leung TY, Li K, Ng PC. Expression profile of cord blood neutrophils and dysregulation of HSPA1A and OLR1 upon challenge by bacterial peptidoglycan. J Leukoc Biol. 2014;95(1):169–178. doi: 10.1189/jlb.0413219. [DOI] [PubMed] [Google Scholar]
- 22.Weinschenk NP, Farina A, Bianchi DW. Neonatal neutrophil activation is a function of labor length in preterm infants. Pediatr Res. 1998;44(6):942–945. doi: 10.1203/00006450-199812000-00020. [DOI] [PubMed] [Google Scholar]
- 23.Weinberger B, Laskin DL, Mariano TM, Sunil VR, DeCoste CJ, Heck DE, Gardner CR, Laskin JD. Mechanisms underlying reduced responsiveness of neonatal neutrophils to distinct chemoattractants. J Leukoc Biol. 2001;70(6):969–976. [PMC free article] [PubMed] [Google Scholar]
- 24.Miller ME. Phagocyte function in the neonate: selected aspects. Pediatrics. 1979;64(5 Pt 2 Suppl):709–712. [PubMed] [Google Scholar]
- 25.Filias A, Theodorou GL, Mouzopoulou S, Varvarigou AA, Mantagos S, Karakantza M. Phagocytic ability of neutrophils and monocytes in neonates. BMC Pediatr. 2011;11:29. doi: 10.1186/1471-2431-11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Driessler F, Venstrom K, Sabat R, Asadullah K, Schottelius AJ. Molecular mechanisms of interleukin-10-mediated inhibition of NF-kappaB activity: a role for p50. Clin Exp Immunol. 2004;135(1):64–73. doi: 10.1111/j.1365-2249.2004.02342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Powell C, Orsi N, Simpson N, Levene M. Characterisation of the cytokine inflammatory response in LPS stimulated full-term cord blood. J Perinat Med. 2004;32(5):440–445. doi: 10.1515/JPM.2004.144. [DOI] [PubMed] [Google Scholar]
- 28.Seghaye MC, Heyl W, Grabitz RG, Schumacher K, von Bernuth G, Rath W, Duchateau J. The production of pro- and anti-inflammatory cytokines in neonates assessed by stimulated whole cord blood culture and by plasma levels at birth. Biol Neonate. 1998;73(4):220–227. doi: 10.1159/000013980. [DOI] [PubMed] [Google Scholar]
- 29.Kollmann TR, Crabtree J, Rein-Weston A, Blimkie D, Thommai F, Wang XY, Lavoie PM, Furlong J, Fortuno ES, 3rd, Hajjar AM, Hawkins NR, Self SG, Wilson CB. Neonatal innate TLR-mediated responses are distinct from those of adults. J Immunol. 2009;183(11):7150–7160. doi: 10.4049/jimmunol.0901481. [DOI] [PMC free article] [PubMed] [Google Scholar]