Abstract
Multidrug-resistant Neisseria gonorrhoeae infections have been declared one of the top three urgent threats to public health. Approaches to combat resistance include targeted therapy with antibiotics previously thought to be ineffective, made possible by rapid molecular assays to predict susceptibility. Previous studies have associated the gyrase A (gyrA) gene of Neisseria gonorrhoeae with in vitro resistance to ciprofloxacin. We conducted a systematic review of studies comparing Neisseria gonorrhoeae gyrA genotype results with conventional antimicrobial susceptibility test results. We identified 31 studies meeting inclusion criteria, among which seven different loci for mutations in the gyrA gene were identified, from sixteen countries between the years of 1996 and 2016. We then performed a meta-analysis among those studies stratifying by use of real-time PCR or non-real-time PCR technique, and compared the summary receiver operating characteristic curves (sROC) between the two PCR methods. Among studies using real-time PCR the pooled estimate of sensitivity and specificity of gyrA genotype results for the prediction of Neisseria gonorrhoeae susceptibility to ciprofloxacin were 98.2% (95% CI 96.5%-99.1%) and 98.6% (95% CI 97.0%-99.3%), respectively. The sROCs for studies using real-time PCR techniques were well separated from those using non-real-time PCR techniques, with only slight overlap in the confidence intervals, suggesting that real-time PCR techniques were a more accurate approach. GyrA genotype testing is a novel approach to combating the emergence of multi drug-resistant Neisseria gonorrhoeae, and is a sensitive and specific method to predict in vitro ciprofloxacin susceptibility.
Keywords: Neisseria gonorrhoeae Resistance, Gyrase A, GyrA, Ciprofloxacin
Introduction
Multidrug-resistant Neisseria gonorrhoeae infections have been declared one of the top three urgent threats to public health [1]. Neisseria gonorrhoeae has developed resistance to all antimicrobials currently available [2, 3], and there are only a few novel antibiotics in development [4-6], therefore new approaches are urgently needed. A recently described approach calls for targeted therapy with antibiotics previously thought to be ineffective [7, 8], which has been made possible by the development of rapid molecular assays that predict in vitro antimicrobial susceptibility [9].
In the United States, it is estimated that approximately 80% of Neisseria gonorrhoeae infections are susceptible to ciprofloxacin [10]. Previous work has shown that mutation of the gyrase A (gyrA) gene of Neisseria species is associated with ciprofloxacin resistance, particularly mutation at the Ser91 codon [9] [30s]. Mutations in other loci have been shown to confer resistance to ciprofloxacin, such as parC, however prior studies have demonstrated that mutations in other locations, which confer resistance, usually occur in parallel with the gyrA mutation; prediction of ciprofloxacin susceptibility, therefore, may be done solely by detection of mutation in the gyrA gene [30s-34s]. In response, real-time polymerase chain reaction (PCR) –based molecular assays have been developed to predict gonococcal susceptibility to ciprofloxacin [33s].
Those rapid molecular genotypic assays have been studied extensively to determine how they correlate with standard agar dilution susceptibility methods [32s-43s]. Here we review the previously published reports describing the sensitivity, specificity, negative and positive predictive values of the gyrA genotypic assays for the prediction of Neisseria gonorrhoeae susceptibility to ciprofloxacin.
Methods
In September 2016, we conducted a literature search using the following text-word terms: “GYRA” OR “CIPROFLOXACIN” OR “QUINOLONE” AND “GONORRH*” with PubMed as a search platform. Our search resulted in over 900 studies which was further restricted to 831 in the English language. Our only inclusion criterion was the use of gyrA genotypic determination techniques and the comparison of genotype results with conventional methods for determining ciprofloxacin susceptibility. We excluded articles that only evaluated resistant or susceptible strains of N. gonorrhoeae because there would be no comparison of susceptibility results. Similarly, we excluded articles reporting five or fewer isolates with resistant or susceptible phenotypes. Wild-type is defined as the most prevalent sequence of the gyrase A gene, which predicts full ciprofloxacin susceptibility. One article was excluded as a cluster analysis was performed and the results provided were insufficient to distinguish which mutant or wild-type strains were associated with which susceptibility results. In total we identified 30 articles meeting inclusion criteria. We were also aware of one article that was not available on PubMed, which fulfilled inclusion criteria. Here we report sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) of the wild-type gyrA gene for predicting susceptibility to ciprofloxacin from those 31 reports.
Statistical Analysis
Sensitivity, specificity and their corresponding 95% confidence intervals of individual studies were calculated and presented in forest plots (Figures 1 and 2). For studies with zero event cell count, continuity correction of 0.5 was added to each cell to enable calculations. Heterogeneities of sensitivities and specificities among included studies were assessed by Cochran Q and chi-squared test [11], respectively. Given the strong evidence of heterogeneities (p-values of Chi-squared test <0.05) and known correlation between the pair of sensitivity and specificity, we obtained the summary statistics of sensitivity and specificity by using a bivariate random effects modeling approach [12]. The summary operating characteristic curves (sROC) [13] were constructed and compared between studies using real-time PCR techniques and non-real-time PCR techniques (Figure 3). All the statistical analyses were conducted using Package MADA (Meta-Analysis of Diagnostic Accuracy) in the R statistical software environment version 3.3.1 [14].
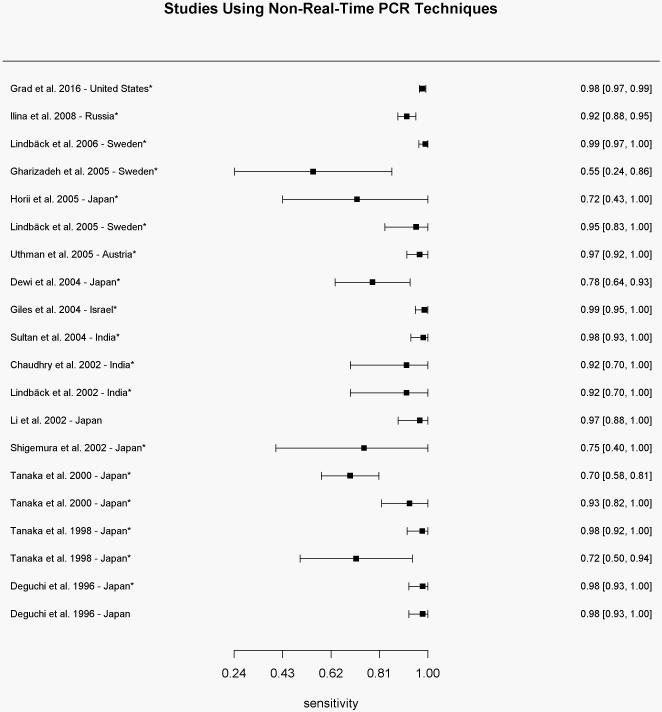
Figure 1.
Sensitivity results for each study using non-real-time PCR-based techniques for determination of the gyrase A genotype compared to conventional methods of susceptibility testing.
* Sensitivity values were calculated based on reported results
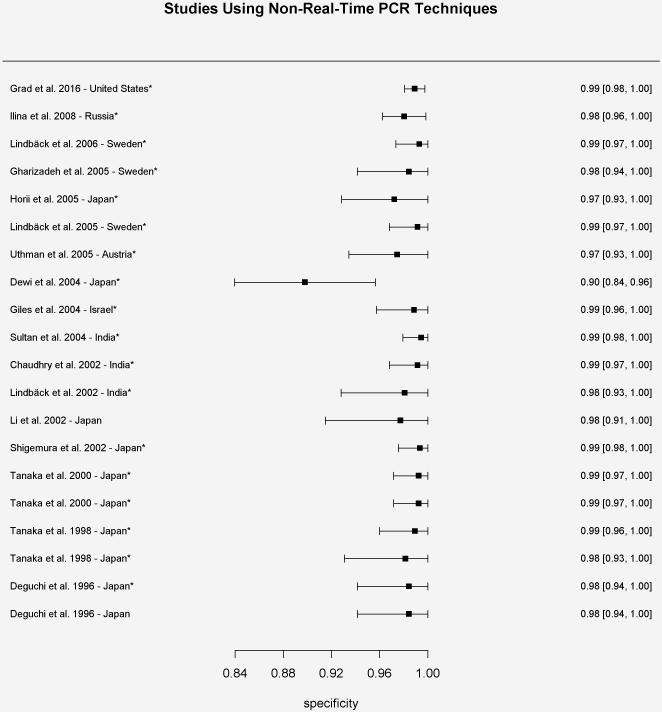
Specificity results for each study using non-real-time PCR-based techniques for determination of the gyrase A genotype compared to conventional methods of susceptibility testing.
*Specificity values were calculated based on reported results
Figure 2.
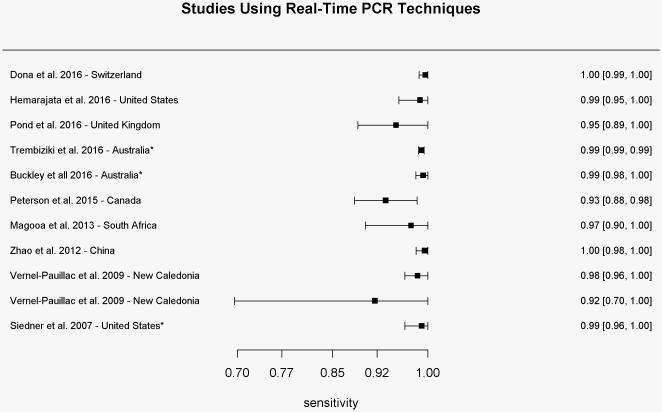
Sensitivity results for each study using real-time PCR-based techniques for determination of the gyrase A genotype compared to conventional methods of susceptibility testing.
* Sensitivity values were calculated based on reported results
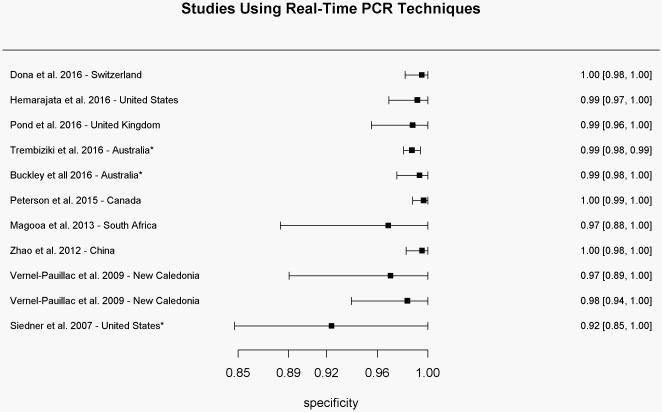
Specificity results for each study using real-time PCR-based techniques for determination of the gyrase A genotype compared to conventional methods of susceptibility testing.
*Specificity values were calculated based on reported results
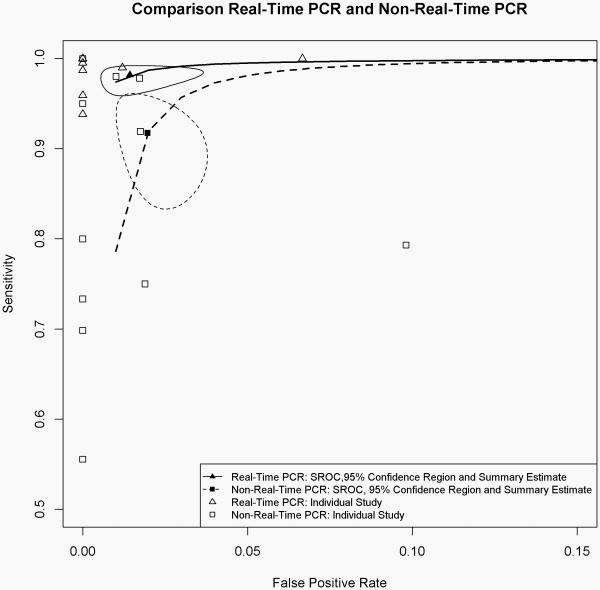
Figure 3.
Summary operating characteristic curves for sensitivity and specificity comparing studies using non-real-time PCR and real-time PCR techniques for the determination of the gyrase A genotype. Solid and dashed circled areas represent the 95% confidence regions for studies using real-time PCR and non-real-time PCR techniques, respectively.
Results
Of the 31 articles meeting inclusion criteria, seven different loci for mutations in the gyrA gene were identified from samples collected in sixteen countries between the years of 1996 and 2016 [30s-60s]. Eleven studies used real-time polymerase chain reaction techniques for determination of the gyrA genotype, while 20 were conducted using other methods such as pyrosequencingTM, probe hybridization, isothermic chimeric primer-initiated amplification, or gel electrophoresis to determine the gyrA genotype.
All studies compared genotype results with minimum inhibitory concentration (MIC) assay results using agar dilution techniques, except one study, which used available phenotypic susceptibility data without MIC assay results [40s]. The cutoff MIC for reduced susceptibility and resistance varied slightly among studies with cutoff values ranging from ≥ 0.06-0.125 μg/ml and ≥ 0.05-1.0 μg/ml, respectively.
Studies Using Non-Real-Time Polymerase Chain Reaction Methods for Determination of GyrA Genotype
We identified twenty studies (N=2931 isolates) from seven countries using non-real-time PCR techniques for the determination of the gyrA genotype. The range of sensitivities and specificities of wild-type gyrA genotype for predicting ciprofloxacin susceptibility reported by those studies was 58.0-100% (mean of 91.2%) and 90.5-100% (mean of 99.2%), respectively (Figure 1a and 1b). Positive predictive values ranged from 76.3-100% (mean of 97.9%), while negative predictive values ranged from 72.4-100% (mean of 95.4%). The pooled estimate of sensitivity and specificity based on the bivariate mixed-effect modeling approach were 91.7% (95% CI 85.4%-95.5%) and 98.0% (95% CI 96.7%-98.8%), respectively.
Shigemura et al. used denaturing high-performance liquid chromatography and DNA sequencing in 2002 among samples collected in Japan and found 83.3% sensitivity of gyrA mutation for predicting reduced ciprofloxacin susceptibility, however results improved to 100% when only evaluating mutations at codon 91 of the gyrA gene. Similarly, Dewi et al. found 80.6% sensitivity, which improved to 93.5%; and Tanaka et al. found 75.0% sensitivity, which improved to 100% when analyzing only mutations in codon 91.
Gharizadeh et al. found 10 isolates with mutations in the gyrA gene (two in region Asp95 and eight in region Ser91) that showed phenotypic susceptibility to ciprofloxacin, however the median MIC was higher than for wild-type strains (0.21 μg/ml and 0.03 μg/ml, respectively). Conversely, Grad et al. report five isolates with phenotypic resistance to ciprofloxacin (MIC values > 1 μg/ml) that had no identifiable mutation in gyrA or parC.
Fourteen of the twenty studies included sequence analysis of parC as well as gyrA. Uthman et al. found slightly higher sensitivity of wild-type parC genotype for predicting ciprofloxacin susceptibility compared to wild-type gyrA genotype (100% compared to 97.8%), with similar results reported by Shigemura et al. and Grad et al. Eleven other studies, however, found wild-type parC was not as sensitive or specific for predicting ciprofloxacin susceptibility as wild-type gyrA [30s, 31s, 44s, 45s, 46s, 47s, 51s, 53s, 54s, 56s, 57s].
Studies Using Real-Time Polymerase Chain Reaction Assays to Amplify Ser91 Region of the GyrA Gene
In total, eleven studies (N=4777 isolates) used RT-PCR techniques to amplify the Ser91 region of the gyrA gene. Samples were collected from ten countries between 2007 and 2016. All studies reported high sensitivity and specificity with ranges between 93.8-100% (mean of 98.8%) and 93.2-100% (mean of 99.3%), respectively (Figures 2a and 2b). Positive and negative predictive values ranged from 94.4-100% (mean of 99.4%) and 87.5-100% (mean of 98.2%), respectively. The pooled estimate of sensitivity and specificity based on the bivariate mixed-effect modeling approach were 98.2% (95% CI 96.5%-99.1%) and 98.6% (95% CI 97.0%-99.3%), respectively.
Siedner et al.were the first to use only real-time PCR techniques for determination of the gyrA genotype. They reported improvement of assay sensitivity after restricting the assay to amplification of only the Ser91 codon, compared to the entire amplicon of the gyrA gene as was reported previously [34s]. Furthermore, four studies reported 100% specificity of mutation in the Ser91 region of the gyrA gene for Neisseria gonorrhoeae compared with other subspecies [35s-37s, 41s].
Among 252 samples from Canada, Peterson et al. compared ciprofloxacin susceptibility results with the Asp95 region of the gyrA gene and the Asp86 region of the parC gene, both previously associated with ciprofloxacin resistance. They found comparable sensitivity for Asp86 parC in predicting ciprofloxacin resistance. Three other studies using real-time PCR techniques, however, report that amplification of the parC gene was less sensitive and specific than amplification of the gyrA gene, though additional alteration of the parC gene may confer a greater degree of antimicrobial resistance [32s, 38s, 39s].
Figure 3 shows the sROC for studies using non-real-time PCR and real-time PCR techniques.
Discussion
We systematically reviewed studies that have compared Neisseria gonorrheoae gyrA genotype results with conventional methods of ciprofloxacin susceptibility determination. Among studies using real-time PCR techniques there appears to be strong evidence that gyrA genotype results are both highly sensitive and specific for the prediction of Neisseria gonorrhoeae susceptibility to ciprofloxacin. Furthermore, studies have shown that gyrA genotype determined by real-time PCR is 100% specific to Neisseria gonorrhoeae compared to other Neisseria subspecies [35s-37s, 41s].
Studies using non-real-time PCR techniques such as PyrosequencingTM and probe hybridization reported wider ranges of sensitivities. That finding is likely in part due to the incorporation of multiple different mutations in the gyrA gene that may not be associated with resistance; the range of sensitivities improved when only analyzing known resistance mutations. Notably, the study by Gharizadeh et al. compared gyrA mutation with phenotypic susceptibility results; when looking at MIC assay results it was noted that samples with mutant gyrA genotypes had higher median MICs than wild-type samples.
We also compared the sROC between studies using real-time PCR and non-real-time PCR techniques. The summary estimates of sensitivity and specificity were well separated, though the confidence regions overlapped slightly. Based on that, we therefore conclude that the real-time PCR technique may be a more accurate technique in gyrase A genotype testing than other techniques.
Rapid molecular assays for determination of resistance are enabling targeted antimicrobial therapy that could curb the emergence of antimicrobial resistance. The FDA-approved molecular assay for Mycobacterium tuberculosis uses real-time PCR for amplification and molecular probes for the detection of mutations within the rifampin-resistance determining region. That assay has an overall sensitivity of 97.6% and specificity between 98.1-99.2% [15]. Real-time PCR amplification of known resistance genes has also been shown to be clinically effective in screening for Methicillin-resistant Staphylococcus aureus, with low false negative and false positive rates between 0.0% to 7.3% and 0.0% to 5.4% respectively [16]. Additionally, rapid molecular assays are available for carbapenem-resistant Enterobacteriaceae and have been shown to detect most resistance genes without yielding false positives [17].
Therefore, given the robust performance of the real-time PCR gyrA assays, Neisseria gonorrhoeae genotyping of gyrA has the potential to enable reliable targeted therapy with ciprofloxacin. Targeted therapy with ciprofloxacin might reduce the ongoing selection pressure caused by the widespread empiric use of extended-spectrum cephalosporins and have implications for the enhanced control of ceftriaxone resistant Neisseria gonorrhoeae [8]. A recent study suggested that treatment might be a major driver of ceftriaxone resistance among Neisseria gonorrhoeae [18].
In November 2015, University California Los Angeles implemented routine gyrA testing on all Neisseria gonorrhoeae positive clinical specimens. Results from testing are used to promote targeted therapy with ciprofloxacin based on genotype susceptibility results [19, 20]. Further studies are underway to characterize the impact of the assay on patient clinical outcomes [21].
There were a few limitations to our study. Primarily this was an analysis of previous studies and therefore subject to the same limitations as the studies being reviewed. Additionally the prevalence of ciprofloxacin susceptible Neisseria gonorrhoeae differs in various regions of the world, [10, 22] and gyrA results may not be of substantial benefit in regions where the prevalence of ciprofloxacin resistance is very high. Furthermore, our study spanned twenty years, during which time technological advances likely impacted the sensitivity and specificity of test results. However, as most studies, including the earliest studies, report high sensitivity and specificity of gyrA testing for predicting ciprofloxacin susceptibility, the changes in technology likely did not impact the overall findings, but furthered the utility of gyrA testing, and therefore support, rather than negate our findings.
Conclusion
Gyrase A genotype testing is a novel approach to combating the emergence of multi drug-resistant Neisseria gonorrhoeae. Among studies comparing gyrA genotype results with conventional methods of susceptibility testing, there appeared to be strong evidence that gyrA genotype results are both sensitive and specific in predicting ciprofloxacin in vitro susceptibility, with real-time PCR techniques being a more accurate approach compared to non-real-time PCR techniques.
Supplementary Material
Summary.
A review of studies comparing gyrase A genotype of Neisseria gonorrhoeae to conventional susceptibility testing methods suggests strong evidence for use of rapid genotypic assays to predict in vitro susceptibility.
Acknowledgments
This research was supported by the United States National Institutes of Health grants R21AI117256 and R21AI109005.
References
- 1.Centers for Disease Control and Prevention . Sexually Transmitted Disease Surveillance 2013. U.S. Department of Health and Human Services; Atlanta: 2014. [Google Scholar]
- 2.Unemo M, Nicholas RA. Emergence of multidrug-resistant, extensively drug-resistant and untreatable gonorrhea. Future Microbiol. 2012;7(12):1401–22. doi: 10.2217/fmb.12.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolan GA, Sparling PF, Wasserheit JN. The emerging threat of untreatable gonococcal infection. N Engl J Med. 2012;366(6):485–7. doi: 10.1056/NEJMp1112456. [DOI] [PubMed] [Google Scholar]
- 4.Foerster S, Golparian D, Jacobsson S, et al. Genetic Resistance Determinants, In Vitro Time-Kill Curve Analysis and Pharmacodynamic Functions for the Novel Topoisomerase II Inhibitor ETX0914 (AZD0914) in Neisseria gonorrhoeae. Front Microbiol. 2015;6:1377. doi: 10.3389/fmicb.2015.01377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butler MS, Blaskovich MA, Cooper MA. Antibiotics in the clinical pipeline at the end of 2015. J Antibiot (Tokyo) 2016 doi: 10.1038/ja.2016.72. [DOI] [PubMed] [Google Scholar]
- 6.GlaxoSmithKline . A Dose-Ranging Study Evaluating the Efficacy, Safety, and Tolerability of GSK2140944 in the Treatment of Uncomplicated Urogenital Gonorrhea Caused by Neisseria Gonorrhoeae. National Library of Medicine (US); (Bethesda(MD): ClinicalTrials.Gov., 2014-[cited 2016 Aug 12] Available from: https://clinicaltrials.gov/ct2/show/NCT02294682 NLM Identifier: NCT02294682. [Google Scholar]
- 7.Buono SA, Watson TD, Borenstein LA, et al. Stemming the tide of drug-resistant Neisseria gonorrhoeae: the need for an individualized approach to treatment. J Antimicrob Chemother. 2015;70(2):374–81. doi: 10.1093/jac/dku396. [DOI] [PubMed] [Google Scholar]
- 8.Klausner JD, Kerndt P. Cephalosporin resistance in neisseria gonorrhoeae infections. JAMA. 2013;309(19):1989–1991. doi: 10.1001/jama.2013.4078. [DOI] [PubMed] [Google Scholar]
- 9.Low N, Unemo M. Molecular tests for the detection of antimicrobial resistant Neisseria gonorrhoeae: when, where, and how to use? Curr Opin Infect Dis. 2016;29(1):45–51. doi: 10.1097/QCO.0000000000000230. [DOI] [PubMed] [Google Scholar]
- 10.Kirkcaldy RD, Harvey A, Papp JR, et al. Neisseria gonorrhoeae Antimicrobial Susceptibility Surveillance - The Gonococcal Isolate Surveillance Project, 27 Sites, United States, 2014. MMWR Surveill Summ. 2016;65(7):1–19. doi: 10.15585/mmwr.ss6507a1. [DOI] [PubMed] [Google Scholar]
- 11.Cochran WG. The comparison of percentages in matched samples. Biometrika. 1950;37:3–4. 256–66. [PubMed] [Google Scholar]
- 12.Reitsma JB, Glas AS, Rutjes AW, et al. Bivariate analysis of sensitivity and specificity produces informative summary measures in diagnostic reviews. J Clin Epidemiol. 2005;58(10):982–90. doi: 10.1016/j.jclinepi.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 13.Irwig L, Macaskill P, Glasziou P, et al. Meta-analytic methods for diagnostic test accuracy. J Clin Epidemiol. 1995;48(1):119–30. doi: 10.1016/0895-4356(94)00099-c. discussion 131-2. [DOI] [PubMed] [Google Scholar]
- 14.R Development Core Team A Language and Environment for Statistical Computing, Vienna, Austria: R Foundation for Statistical Computing. 2016 [Google Scholar]
- 15.Boehme CC, Nabeta P, Hillemann D, et al. Rapid molecular detection of tuberculosis and rifampin resistance. N Engl J Med. 2010;363(11):1005–15. doi: 10.1056/NEJMoa0907847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim JU, Cha CH, An HK, et al. Multiplex real-time PCR assay for detection of methicillin-resistant Staphylococcus aureus (MRSA) strains suitable in regions of high MRSA endemicity. J Clin Microbiol. 2013;51(3):1008–13. doi: 10.1128/JCM.02495-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller S, Humphries RM. Clinical laboratory detection of carbapenem-resistant and carbapenemase-producing Enterobacteriaceae. Expert Rev Anti Infect Ther. 2016;14(8):705–17. doi: 10.1080/14787210.2016.1206815. [DOI] [PubMed] [Google Scholar]
- 18.Fingerhuth SM, Bonhoeffer S, Low N, et al. Antibiotic-Resistant Neisseria gonorrhoeae Spread Faster with More Treatment, Not More Sexual Partners. PLoS Pathog. 2016;12(5):e1005611. doi: 10.1371/journal.ppat.1005611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allan-Blitz L, Humphries R, Hemarajata P, Wang X, Klausner JD. Poster Presented at: ID Week. New Orleans: Oct, 2016. Implementation of a Rapid Molecular Assay for Determination of Neisseria gonorrhoeae Susceptibility in a Large Health System; pp. 26–30. [Google Scholar]
- 20.Allan-Blitz LT, Humphries RM, Hemarajata P, et al. Implementation of a Rapid Genotypic Assay to Promote Targeted Ciprofloxacin Therapy of Neisseria gonorrhoeae in a Large Health System. Clin Infect Dis. 2016 doi: 10.1093/cid/ciw864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klausner J. Clinical Validation of a Molecular Test for Ciprofloxacin-Susceptibility in Neisseria gonorrhoeae. National Library of Medicine (US); (Bethesda (MD): ClinicalTrials.Gov. 2000-[cited 2016 Nov 23]. Available from: https://www.clinicaltrials.gov/ct2/show/NCT02961751?term=NCT02961751&rank=1 NLM Identifier: NCT02961751) [Google Scholar]
- 22.Mlynarczyk-Bonikowska B, Kujawa M, Mlynarczyk G, et al. [Resistance to ciprofloxacin of Neisseria gonorrhoeae strains isolated in Poland in 2012-2013] Med Dosw Mikrobiol. 2014;66(2):99–104. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.