Abstract
Genetic circuits and reaction cascades are of great importance for synthetic biology, biochemistry, and bioengineering. An open question is how to maximize the modularity of their design to enable the integration of different reaction networks and to optimize their scalability and flexibility. One option is encapsulation within liposomes which enables chemical reactions to proceed in well-isolated environments. Here we adapt liposome encapsulation to enable the modular, controlled compartmentalization of genetic circuits and cascades. We demonstrate that it is possible to engineer genetic circuit-containing synthetic minimal cells (synells) to contain multiple-part genetic cascades, and that these cascades can be controlled by external signals as well as inter-liposomal communication without cross-talk. We also show that liposomes containing different cascades can be fused in a controlled way so that the products of incompatible reactions can be brought together. Synells thus enable more modular creation of synthetic biology cascades, an essential step towards their ultimate programmability.
Introduction
Chemical systems capable of performing biochemical reactions in the absence of live cells have been extensively used in research and industry to study and model biological processes1,2, to produce small molecules3,4, to engineer proteins5,6, to characterize RNAs7, as biosensors8,9 and molecular diagnostic tools10, and to extend the sensing abilities of natural cells11. Organisms from all three domains of life have been used to obtain transcription/translation (aka TX/TL) extracts for cell-free production of biochemical products from genetic codes12. Encapsulating cell-free TX/TL extracts into liposomes creates bioreactors often referred to as synthetic minimal cells (SMCs or synells)13–16. Although synells have been used to make functional proteins using encapsulated systems reconstituted from recombinant cell-free translation factors17–19, as well as cell-free extracts from bacterial6,20 and eukaryotic cells21, work on liposomal synells has so far focused on expression of single genes, with the goal of synthesizing a single gene product, and within a homogenous population of liposomes.
Here, we confront a key issue in synthetic biology: the modularity of multi-component genetic circuits and cascades. We show that by encapsulating genetic circuits and cascades within synells (Figs. 1a and 1b) and orchestrating the synells to either operate in parallel (Fig. 1c), communicate with one another (Fig. 1d), or fuse with one another in a controlled way (Fig. 1e). We can create genetic cascades that take advantage of the modularity enabled by liposomal compartmentalization. Thus, our strategy enables genetic cascades to proceed in well-isolated environments while permitting the desired degree of control and communication. We present design strategies for constructing and utilizing such synell networks, thus expanding the utility of liposome technology and improving the modularity of synthetic biology. Synell networks may support complex chemical reactions that would benefit from both the high-fidelity isolation of multiple reactions from one another, as well as controlled communication and regulatory signal exchange between those reactions. We show, for example, the controlled fusion of two populations of synells that contain mammalian transcriptional and mammalian translational machinery, respectively, which are normally incompatible when combined in the same compartment.
Fig. 1. An overview of genetic circuit interactions within and between synthetic minimal cells (synells).
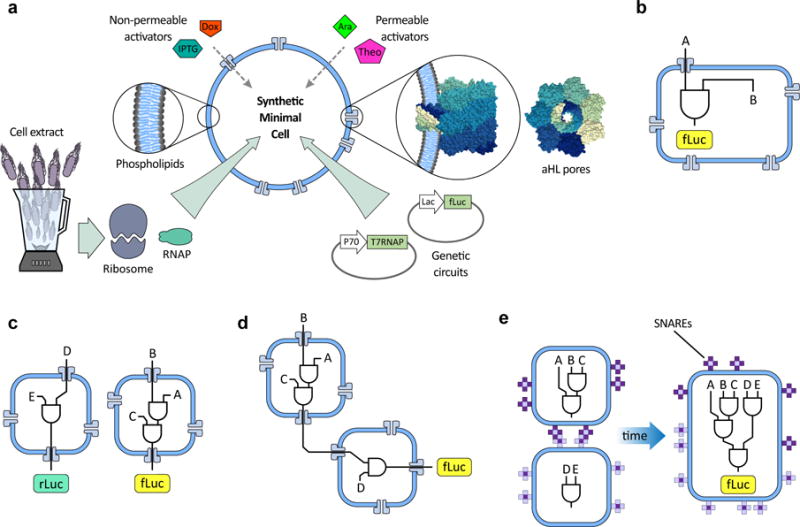
a. Synthetic minimal cells (synells) are semipermeable compartments made from a phospholipid bilayer membrane and various contents. The membrane can display a variety of proteins, including channel-forming proteins such as alpha-hemolysin (aHL). The phospholipid membranes of synells are permeable to molecules such as theophylline (Theo) and arabinose (Ara), and are permeable to others like β-D-1-thiogalactopyranoside (IPTG) and doxycycline (Dox) when aHL channels are present; these molecules can be used for triggering activity within synells. Synells can encapsulate cell lysates with transcriptional and/or translational activity, as well as DNA vectors encoding genes. In this paper, we demonstrate four novel competencies of synells that, together, can be used to create complex, modular genetic circuits. b. We show that synells can contain genetic circuits in which all the components and operations take place within the same liposome. c. We show that two genetic circuits can work independently in separate liposome populations. d. We show that genetic circuits within two different liposome populations can interact. e. We show that genetic circuits can run in parallel in separate compartmentalized reactions; if those reactions are encapsulated by liposomes carrying fusogenic peptides such as SNAREs, the reaction products can be joined together in a hierarchical fashion.
Results
Confinement of genetic circuits in liposomes
Before exploring the control of, and communication with, synells containing genetic cascades, we first characterized the basic structural and functional properties of individual synells. To characterize the size and functionality of our liposomes, we labeled liposome membranes with red dye (rhodamine functionalized with a lipid tail) and filled the liposomes with cell-free transcription/translation (TX/TL) extract derived from HeLa cells22–25, as well as DNA encoding either GFP or split GFP. Structured illumination microscopy (SIM) images showed that GFP liposomes had a diameter between 100 nm and 1 μm (Fig. 2a), a measurement that we confirmed with dynamic light scattering (Fig. S1). We used flow cytometry to quantify the functional expression of genes by synells; 68.4% of the GFP liposomes expressed fluorescence, along with 61.8% of those encapsulating split GFP (Figs. 2b – 2d; for control flow cytometry experiments, see Fig. S2). We characterized the enzymatic activity of several reporters in our liposomes (Fig. S3) and used a Western blot to provide an additional non-enzymatic characterization of luciferase expression (Fig. S4). We compared the performance of mammalian (HeLa) and bacterial (E. coli) TX/TL systems in our liposomes, finding the mammalian system to be slower and have a lower protein yield (Fig. S5).
Fig. 2. Molecular confinement of multicomponent genetic cascades.
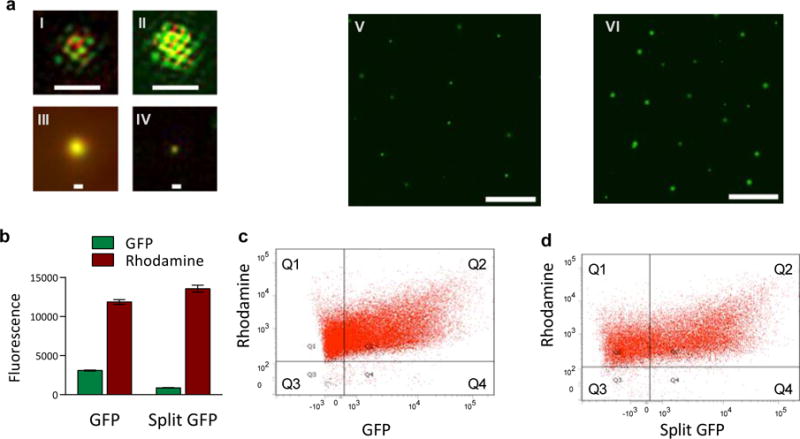
a. Images of liposomes expressing GFP. Sub-panels I–IV: structured illumination microscopy (SIM) images of representative liposomes expressing GFP and membrane-labeled with rhodamine. Every SIM image (panels labeled I, II, III, IV) represents a separate liposome; all liposomes were imaged on the same day and all liposomes came from the same sample, prepared 24h before imaging. All SIM images in this figure are at the same scale; the large scale bars in panels I and II are 1 μm, the small scale bars in panels III and IV are 200 nm. Sub-panels V–VI: widefield epifluorescent images of liposomes expressing GFP. The liposomes for this imaging sample were extruded through a 2 μm filter and dialyzed with a 1 μm membrane; panel V shows sample after 6 h incubation and panel VI shows an aliquot of the same sample after 24 h incubation. The scale bars on panels V and VI are 10 μm. b–d. Fraction of synells expressing GFP and split GFP, measured by flow cytometry (for control flow cytometry experiments, see Fig. S2). b. Bulk expression of GFP and fluorescence measured on the sample prior to the flow cytometry experiments. c. Analysis of samples expressing GFP; 68.4% of liposomes produced measurable green signal. d. Analysis of samples expressing split GFP; 61.8% of liposomes produced measurable green signal.
Having established that the liposomes were of proper size and functionality, we next sought to verify that a well-known advantage of liposomal compartmentalization—facilitated reaction efficacy due to molecular confinement (since encapsulating reactants within a liposome facilitates their interaction due to the small volume)26–29—can help support multi-component genetic circuits as well as chemical reactions of higher order. We compared cell-free transcription/translation (TX/TL) reactions that produce firefly luciferase (fLuc) from one, two, or three protein components, testing them in bulk solution vs. synells. In this experiment, we used HeLa cell extract constitutively expressing the Tet protein to mediate small-molecule induction of transcription of the one, two, or three fLuc components, as well as alpha-hemolysin (aHL), which serves as a pore to admit doxycycline (Dox) to trigger Tet function20,30,31. The one-component luciferase was simply conventional monolithic fLuc (Fig. 3a); the two-component system (i.e., to explore 2nd-order reactions) comprised the two halves of split firefly luciferase, each attached to a coiled coil and a split intein fragment to bring the halves together and covalently bridge them (Fig. 3b)32; and the three-component system involved the halves of split firefly luciferase bearing coiled coils and split inteins, with the coiled coils targeting a third protein, a scaffold (Fig. 3c)32.
Fig 3. Comparison of single- and multi-component genetic circuits.
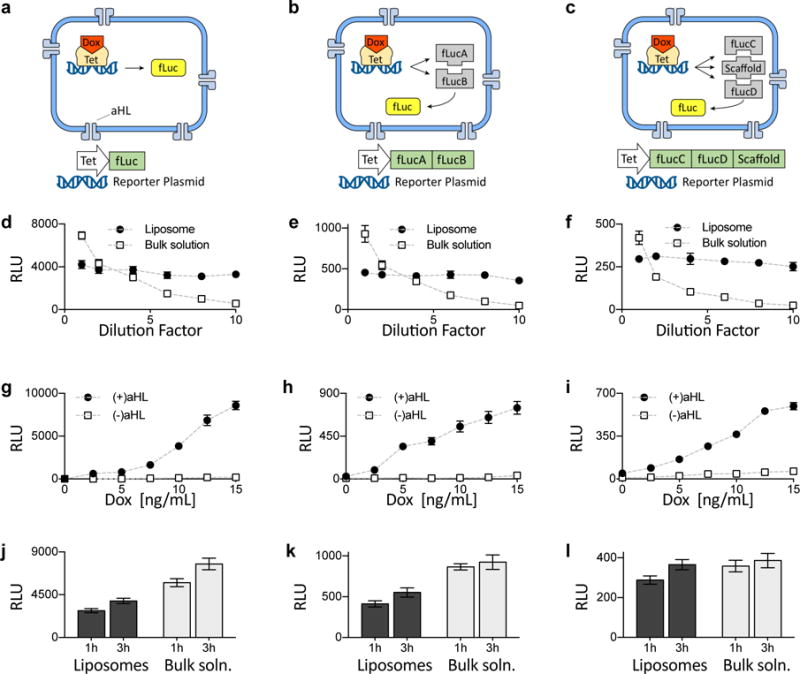
a–c. Genetic cascades involving one-, two-, or three-part luciferase protein assemblies. Expressed under doxycycline-inducible Tet promoters were whole firefly luciferase (fLuc) (a), the two halves (here denoted fLucA and fLucB) of split fLuc bearing split inteins and mutually binding coiled coils (b), and two halves (here denoted fLucC and fLucD) of split fLuc bearing split inteins and coiled coils that bind to a third common template (denoted “scaffold”) (c). d–f. Effects of dilution on fLuc expression in liposomes vs. bulk solution, for the fLuc assemblies described in a–c (see Fig. S6 for experiments under the control of a constitutive P70 promoter). Dotted lines throughout this figure are visual guides, not fits. RLU, relative light units. g–i. End-point expression of luciferase measured at the 3 h time point, for 7 different concentrations of doxycycline (Dox). See Fig. S7 for corresponding 1 h end-point expression data, and Figs. S8 – S10 for the same reactions in bulk solution. j–l. Comparison of liposomal vs. bulk solution expression of luciferase, at 2 different time points and for 10 ng/mL of Dox. The 2 plasmids in k and 3 plasmids in l were mixed at equimolar ratios, with total DNA concentration held constant. Error bars indicate S. E. M. n=4 replicates.
For all three orders of luciferase-producing reactions, the effect of dilution on fLuc expression was weaker for liposomes than for bulk solution (Figs. 3d – 3f; P < 0.0001 for interaction between factors of encapsulation and dilution factor; ANOVA with factors of encapsulation and dilution factor; see Tables S1 – S3 for full statistics and Fig. S6 for corresponding experiments under the control of a constitutive P70 promoter). As expected, fLuc expression was proportional to the concentration of Dox added to the external solution, and depended on aHL (Figs. 3g – 3i show end-point expression after 3 h; see Fig. S7 for the corresponding expression at a 1 h end-point, and Figs. S8 – S10 for the same reactions in bulk solution). Liposomes produced lower amounts of fLuc than the same volume of TX/TL extract in bulk solution—likely due to the well-known property of stochastic loading of reagents into liposomes27,28 (P < 0.0001 for factor of encapsulation in ANOVA with factors of time, encapsulation, and order; see Table S4 for full statistics). For the third-order reaction, we found that liposome encapsulation resulted in efficacy nearly equal to that of bulk solution (P = 0.1324 for factor of encapsulation in ANOVA with factors of time and encapsulation; Fig. 3l; see Table S7 for full statistics), whereas for the first-order and second-order reactions the liposomes resulted in lower efficacy (P < 0.0001 for factor of encapsulation in ANOVAs for both analyses, each with factors of time and encapsulation; Figs. 3j and 3k; see Tables S5 and S6 for full statistics). Molecular confinement in liposomes thus may help facilitate higher-order reactions that require multiple chemical building blocks to be brought together, since the restricted movement of reagents increases the probability of the requisite multi-way interactions.
Insulation of genetic circuits operating in parallel liposome populations
As a next step towards engineering sets of liposomes that can communicate with one another, we set out to determine whether liposomes could be used to insulate multiple and potentially incompatible genetic circuits from each other, so that they could operate in the same bulk environment. This insulation would enable modular design; each circuit could be optimized independently and deployed in the same environment as other circuits without interference. These circuits could reuse the same parts (proteins, DNA) for different purposes in different liposomes, thereby circumventing one limitation of genetic circuits designed to all operate within the same living cell (where one must assume that all circuit elements might encounter each other and must therefore be inherently orthogonal). Different liposome populations could also contain chemical micro-environments that are not mutually compatible (e.g., bacterial and mammalian extracts, or mammalian transcriptional and mammalian translational machinery)—there are numerous examples throughout chemistry of reactions being run under specialized, and thus often isolated, reaction conditions33.
We first assessed whether multiple liposomal circuits could operate in parallel without crosstalk. To do this, we created populations of liposomes that could respond differently to the same external activator. We built two populations of liposomes carrying mammalian TX/TL extract and the same amount of Dox-inducible luciferase DNA (either Renilla or firefly luciferase), but varied the amount of alpha-hemolysin DNA to result in high-aHL and low-aHL synell populations (Fig. 4a). High-aHL and low-aHL synells responded to the non-membrane-permeable Dox in the external solution, doing so proportionally to their own aHL concentration (Fig. 4b). We observed no evidence that doxycycline acting upon one liposome population affected expression of luciferase in the other population: specifically, there was no significant difference in fLuc expression in high-aHL fLuc liposomes when the rLuc liposomes were high-aHL vs. low-aHL, and the same held for the other combinations (Fig. 4b; Sidak’s multiple comparisons test after ANOVA with factors of luciferase type and alpha-hemolysin combination; see Table S8 for full statistics, and Figs. S11 and S12 for rLuc and fLuc expression data at different aHL plasmid concentrations, for two different time points). That is, luciferase expression from each liposome population depended only on the amount of aHL DNA present in that population, and not on that of the other population (Figs. 4c – 4e). This experiment thus not only verifies the independent operation of multiple non-interacting liposomes, but also verifies that multiple liposome populations can be programmed in advance to have varying response levels to a given trigger, and subsequently in the same internal solution, triggered to function simultaneously.
Fig. 4. Insulation of genetic circuits operating in parallel liposome populations.
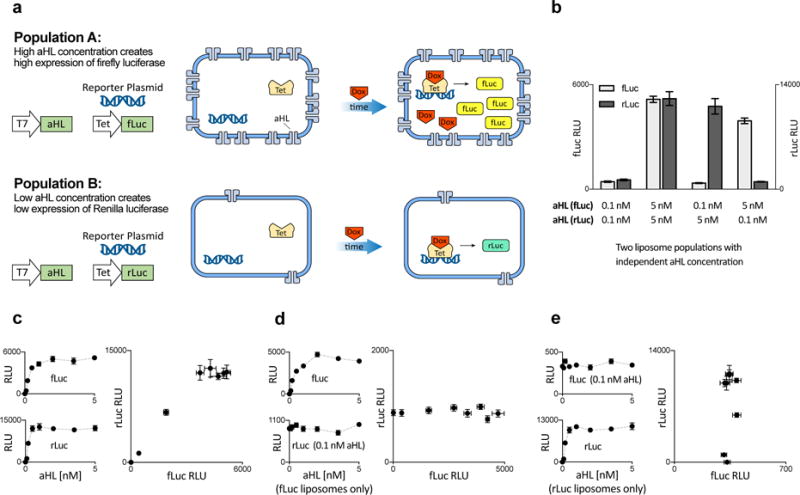
a. Schematic of liposome populations designed to contain similar genetic components but to respond differently to the same environmental concentration of the non-membrane-permeable small molecule activator doxycycline (Dox), by expressing different amounts of the alpha-hemolysin channel protein (aHL). These liposomes contain a measured amount of the plasmid for constitutively expressed aHL, and of a plasmid driving either firefly luciferase (fLuc) or Renilla luciferase (rLuc) from the Tet inducible promoter (the luciferase plasmids were always held at the same concentration). Throughout this figure, the two populations were incubated together in the solution containing Dox and harvested after 6 h (see Figs. S11 and S12 for rLuc and fLuc expression as a function of aHL plasmid concentration, after 2 h and 6 h, respectively). b. Each liposome contains either 0.1 nM or 5 nM of the aHL plasmid. c. Luciferase expression in symmetrical populations, where the amount of aHL DNA is the same across the two populations; the amount of fLuc and rLuc expression is graphed with respect to aHL plasmid concentration and to each other. d–e. Luciferase expression in asymmetrical populations. d. Luciferase expression when Renilla liposomes have a constant aHL plasmid concentration (0.1 nM) but the concentration of that plasmid is varied in the firefly liposomes. Expression of rLuc and fLuc are graphed against the plasmid concentration in firefly liposomes and against each other. e. Luciferase expression as in d, but with constant aHL plasmid concentration in firefly liposomes and variable concentration in Renilla liposomes. Error bars indicate S. E. M. n=4 replicates.
Communication between genetic circuits operating in multiple liposome populations
Having established that genetic circuits in separate populations of liposomes could operate independently, we next sought to begin to create controlled communication pathways between populations of synells. In this way we could create a compartmentalized genetic circuit—which as noted above may need to be separated from others for reasons of control fidelity, toxicity, or reagent tunability—and connect it to other compartmentalized circuits. While previous works have emphasized the importance of modularity in genetic circuits34, to our knowledge nobody has approached the problem by physically separating circuit elements into different liposomes. We built two-component circuits by mixing together two populations of liposomes, a “sensor” that senses an external small molecule cue and a “reporter” that receives a message from the sensor population and produces an output; we could vary the occupancy of each population to achieve a different overall ratio of the two components (Fig. 5a; see Fig. S13 for additional characterization of the membrane-permeable small molecules used throughout this figure, and Tables S9 – S10 for the associated statistics). Our first version was built with bacterial TX/TL extract (Fig. 5b). The sensor liposomes contained IPTG (a small, non-membrane-permeable activator that induces the lac promoter) and the arabinose-inducible gene for aHL (arabinose, which unlike IPTG, is membrane-permeable); these liposomes thus sensed arabinose and released IPTG by expressing aHL channels. We combined these with reporter liposomes containing constitutively-expressed aHL, in which fLuc was under the control of the lac promoter—either directly (fLuc under lac promoter) or indirectly (T7RNAP under the lac promoter and fLuc under T7 promoter)—and found that multi-component compartmentalized genetic circuits thus constructed were able to operate as coherent wholes. We tested both systems with multiple dilutions of sensor and reporter liposomes, and found similar dose-response curves from titration of either species of liposome (Figs. 5c and 5d; bars in these panels represent final time points of 6 h; for the complete time series that includes the data in Fig. 5c, see Fig. S14; for the end-point expression of the circuit in Fig. 5c without arabinose triggering, see Fig. S15; for the complete time series that includes the data in Fig. 5d, see Fig. S16; for the end-point expression of the circuit in Fig. 5d without arabinose triggering, see Fig. S17). Using this modular architecture, we constructed a genetic circuit that combines both bacterial and mammalian components (Fig. 5e). The sensor liposome in this case responded to theophylline (membrane-permeable) to release doxycycline (non-membrane-permeable). Dox, in turn, activated fLuc expression in reporter liposomes built with mammalian components. As before, we showed that the multi-compartment genetic cascade could function as designed, with fLuc expression dose-response curves similar upon titrating either sensor or reporter liposome concentration (Fig. 5f; bars in this panel represent final time points of 6 h; for the complete time series that includes the data in Fig. 5f, see Fig. S18; for the end-point expression of the circuit in Fig. 5f without theophylline triggering, see Fig. S19). Thus, even multi-component genetic circuits with different chemical micro-environments (e.g., made from bacterial vs. mammalian cell extracts) can be assembled into coherent networks comprising multiple modules.
Fig. 5. Communication between genetic circuits operating in multiple liposome populations.
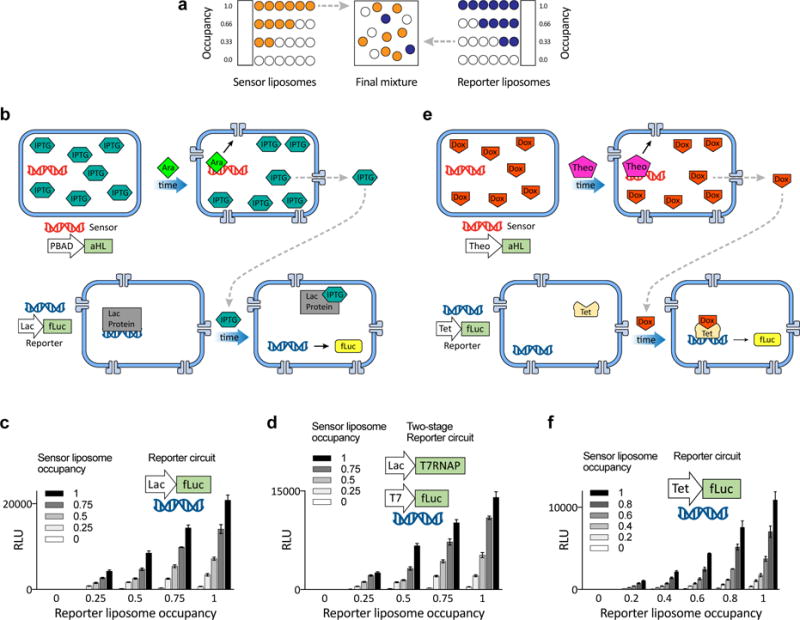
a. Scheme for mixing two populations of liposomes at different ratios of their components while maintaining a constant lipid concentration of 10 mM (the same scheme was used throughout this figure and Fig. 6). Each population contains the same amount of liposomes, but the liposome occupancy can vary between 0 (all liposomes are empty) and 1 (the maximum fraction of the liposomes contain reagents). b–d. Externally activated two-part circuits, with bacterial TX/TL. b. Scheme of interacting populations, denoted sensor and reporter. Sensor liposomes contain the alpha-hemolysin gene and are filled with IPTG; reporter liposomes contain machinery for firefly luciferase expression. During activation, arabinose (Ara) diffuses through the sensor liposome membrane and induces aHL expression, which releases IPTG, which induces fLuc expression in the reporter. c. Expression of fLuc for varying ratios of occupancy (as in a), for the sensor and reporter liposomes with indicated contents. This panel represents the time point 6h (for complete time series see Fig. S14). For this circuit without arabinose see Fig. S15. d. Expression of fLuc for a circuit in which the reporter liposomes contain DNA for a multicomponent genetic cascade, as indicated. This panel represents the 6h time point (for complete time series, see Fig. S16. For this circuit without arabinose, see Fig. S17). e–f. Externally activated two-part circuits, containing both bacterial and mammalian TX/TL components. e. Sensor vesicles contain the Theo-triggered aHL gene and Dox; reporter liposomes contain constitutively expressed aHL and Tet, and Dox/Tet-driven fLuc. During activation, Theo diffuses through the membrane of the activator liposomes and induces aHL expression, which creates pores that release Dox from the activator. Dox induces fLuc expression in the reporter liposomes. f. Expression of fLuc, for varying ratios of sensor and reporter liposomes (this panel represents 6h time point; for complete time series see Fig. S18. For this circuit without Theo, see Fig. S19). Error bars indicate S. E. M. n=4 replicates.
Fusion of complementary genetic circuits
Finally, having established that it is possible to maintain liposomes in high-integrity states despite being mixed, we sought to engineer synells to fuse so that they could bring together two genetic cascades into the same environment in a programmable fashion. Two precursors might require synthesis in different milieus, but ultimately need to be reacted with one another. One prominent example is that of mammalian transcription and translation. Mixed mammalian transcription and translation cell-free extracts are not able to functionally result in transcription of DNA into RNA and then the translation of RNA into protein, perhaps because the micro-environments of the mammalian nucleus and cytoplasm are quite different, making their cell-free extracts incompatible (Fig. S20). Rather than mixing the two cell-free extracts into a single non-functioning mixture, it might be preferred to use synells to compartmentalize the reactions. Once nuclear-extract synells have completed transcription, it might be desirable to fuse them with cytoplasmic-extract synells for translation to take place.
Thus, we sought to make liposomes capable of controlled fusion (Fig. 6a). Fusing liposomes of opposite charge was previously demonstrated to activate gene expression in liposomes35. Our system uses only one kind of membrane composition (POPC cholesterol membranes, known to be a good environment for membrane channels like aHL), so to achieve fusion between liposomes we used SNARE/coiled-coil hybrid proteins (here called SNAREs for short), which can be generated in complementary pairs that are specific in their fusion properties36,37. We could thus fuse together complementary circuit elements by encapsulating them in separate populations of SNARE-fusable liposomes. We confirmed that SNAREs were mediating liposome fusion, through SIM imaging (Fig. 6a), by observing Fluorescence Resonance Energy Transfer (FRET) signals from lipid dyes added to the liposome membranes (FRET signals showed that the fusion process takes place within minutes; see Figs. S21 and S22) and by observing mixing of liposome content, reported as de-quenching of a molecular beacon encapsulated in one population of liposomes by a complementary target encapsulated in the other population (see Fig. S23). We observed large liposomes and also liposome aggregates (presumably in the process of fusing) of sizes on the order of 5–10 μm, and measured a minimal amount of leakage from the liposomes during the process of fusion (Fig. S24). We tried several combinations of complementary circuit elements: the gene for T7RNAP and a T7-driven fLuc (Fig. 6b); a non-membrane-permeable small molecule trigger (IPTG) and an IPTG-triggered (lac-promoter driven) fLuc (Fig. 6c); genes for a membrane pore (aHL) and a lac-promoter driven fLuc (Fig. 6d, in an IPTG-containing ambient); two different genes encoding for parts of split luciferase (Fig. 6e, using the same fLucA and fLucB as in Fig. 3b). For one final test, liposomes carrying mammalian nuclear (transcription) extract and the gene for fLuc, incubated for 12 hours, were then mixed with liposomes containing cytoplasmic (translation) extract, and further incubated for 12 hours (Fig. 6f). We were able to observe production of fLuc protein, even though a direct combination of transcriptional and translational machinery produced no fLuc above background levels (Fig. S20). Throughout all these cases, we observed production of the final output of the genetic cascade only when the two liposome populations were equipped with SNAREs, and only when they were a SNARE cognate pair (P < 0.0001 for factor of SNARE compatibility, ANOVA with factors of mechanism, occupancy, and SNARE compatibility; see Table S11 for full statistics; for systems in this figure, switching which liposome contained which SNARE had no effect on the results, as shown in Fig. S25).
Fig. 6. Fusion of complementary genetic circuits.
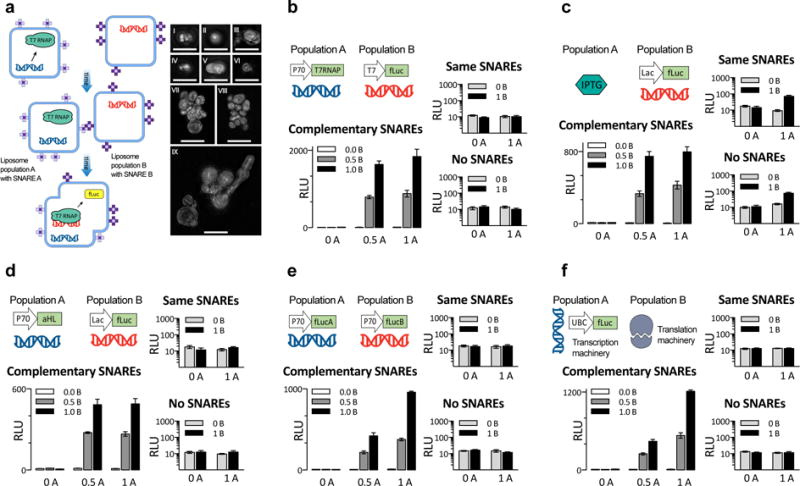
a. General scheme for SNARE-mediated liposome fusion. We created two populations of liposomes, A and B, decorated with complementary SNARE protein mimics in their outer leaflet. The images to the right, in sub-panels I through IX, are maximum-intensity projections of structured illumination microscopy (SIM) z-stacks of liposomes membrane-labeled with rhodamine, bearing complementary SNARE pairs, and fused for 4 hours. All images from panels I through IX represent separate fields of view. Scale bars, 5 μm. All liposomes in this figure, except f, contained bacterial TX/TL components. b–f. Five different instantiations of the liposome fusion concept, exploring several ways to distribute genetic circuits across fusable liposomes, with two different populations of liposomes at three occupancy levels for each case. b. Mixing of constitutively expressed T7 RNA polymerase with firefly Luciferase under T7 promoter. c. Mixing of a non-membrane-permeable small molecule activator IPTG with its inducible promoter, driving fLuc production. d. Mixing of a constitutively expressed membrane channel with an inducible promoter driving fLuc production, in the background of the small molecule that induces the promoter (IPTG). e. Mixing liposomes with genes encoding split protein. f. Mixing liposomes containing mammalian transcription (HeLa) and translation (HeLa) system, producing fLuc. For all five systems in b–f., experiments on the large graph are with one of a matching pair of SNARE on each population, the top of the two small panels is both liposomes with the same SNARE, and the bottom one neither population had any SNAREs. In both small graphs of b–f, the y-axis is in logarithmic scale to show the near-zero values for non-fusing liposomes. Switching which liposome contained which SNARE had no effect on the results (Fig. S25), whereas the absence of SNARE proteins or the presence of identical SNAREs on both populations hindered fusions (small graphs on b–f). Error bars indicate S. E. M. n=4 replicates.
Discussion
Liposomes are key in chemistry and chemical biology for compartmentalizing chemical reactions that require different environments or act on different samples. In this work, we show how synthetic minimal cells (synells)—liposomes containing genes as well as transcriptional and/or translational machinery—enable a great level of modularity for genetic circuit design and execution. We showed that circuits could be designed to run in synell populations in the same container, independent of each other due to the insulation provided by the liposomal membrane. Genetic circuits could also be connected to communicate with one another through small molecule messengers. This communication was possible even across liposomes containing incompatible micro-environments, as we showed by constructing the first genetic circuit containing bacterial and mammalian cell-free extracts and genetic elements. Finally, we explored the use of SNARE mimics to fuse synells together, enabling the direct union of separately synthesized reaction components. Using this strategy, we were able to produce RNA encoding for firefly luciferase (fLuc) in one population of liposomes containing mammalian transcriptional extract, which upon fusion with liposomes containing mammalian translational extract resulted in protein production—an outcome which does not occur if the gene is simply added to a mixture of the two extracts.
Synells thus enable a new level of modularity for synthetic biology. Modularity is key in engineering, since breaking a complex synthetic biology system into parts that can be independently controlled or regulated, without crosstalk, and that communicate only in well-defined ways, enables each part to be individually optimized while supporting their incorporation into an emergent whole. Our technologies will enable a large number of different synthetic biology problems to be made modular, even those that involve genetic cascades that might interfere with each other (or pose toxicity issues) if they were to all occur in one pot. Since our method of compartmentalization is liposomal, there is no need for specialized hardware to mediate the communication and control of multiple interacting reaction systems. Precise temporal control of synell networks could be even further enhanced by using light to trigger optogenetic signaling cascades, which in turn can trigger downstream effects38,39. We also show that the molecular confinement of liposomes can facilitate multicomponent protein-protein interactions.
Our synells, in addition to the power they offer to synthetic biology, may also enable the simulation of various complex behaviors that have been proposed as characteristics of early life forms. Controlled communication between cells, the fusion of genetic elements across cells, and the assembly of complex genetic cascades towards defined cellular behaviors, are all traits that arose in the course of early evolution. Synthetic minimal cells have been widely used as models for studying the origin and earliest evolution of life40–44. For example, one of us has previously shown that liposomes encapsulating a simple catalyst can be used to model early Darwinian competition mechanisms41. Interacting encapsulated genetic circuits will hopefully enable the study of the more complex characteristics that have been proposed for the last universal common ancestor (LUCA)45–47, perhaps helping reveal the dynamic and boundary conditions underlying the mechanisms of Darwinian evolution.
Materials and Methods
Cloning of expression constructs
The P70 (OR2-OR1-Pr50) and lac (Llac-0-151) promoter constructs were used in a modified pCI vector (Promega). The original promoter region of the vector was replaced by the appropriate promoter to make our constructs51. For bacterial expression, the previously described transcription terminator T500 was added at the end of each ORF. The original UTR was also removed and replaced with the previously described UTR150. The mammalian Tet constructs were built into Tet-On 3G bi-directional vector (Clontech) by cloning the genes into MCS1. The araBAD constructs were built using a PBAD vector52 (Thermo). We used PBAD-hisB, removing the His-tag and the enterokinase recognition site prior to inserting the genes used in this study.
Flow cytometry with GFP and split GFP
Fluorescence signal from these GFP liposomes was measured after 12 h of incubation for the experiments in Figs. 2b – 2d. Membranes (red fluorescence) were labeled with Lissamine Rhodamine B 1,2-Dihexadecanoyl-sn-Glycero-3-Phosphoethanolamine Triethylammonium Salt (rhodamine DHPE), used at 0.2 molar percentage of the POPC concentration. GFP was expressed from a plasmid with the T7 promoter. The halves of split GFP were fused to complementary coiled coils and expressed from two different plasmids (both with the T7 promoter). Flow cytometry analysis: events in two fluorescent channels were analyzed: GFP and red fluorescence. Each dataset consists of minimum 19,000 events. Fig. 2c shows an analysis of liposomes expressing GFP and Fig. 2d shows an analysis of liposomes expressing split GFP. The percentage of liposomes expressing protein was calculated as the percentage of events in the quadrant positive in both green and red channels (Q2 on both plots). The flow cytometer was not calibrated using size standards, therefore all information about the size of the particles in the experiment are approximate. For the detailed size measurements of the liposomes in this work see Fig. S1 data from the DLS experiments. The flow cytometry analysis was performed on FACSCanto II, and the data analysis was performed using FACSDiva 8.0.
Firefly luciferase assays
Firefly luciferase (fLuc) activity was assayed using the Steady-Glo Luciferase Assay System (Promega). The protein analysis was performed according to the manufacturer’s instructions. The cell lysis protocol was replaced with a modified procedure for lysing liposome-encapsulated expression reactions. The 50 μL liposome reactions were quenched by 10 μL of Quench Mix containing 0.3% v/v Triton-X100 (to disrupt vesicles), TURBO DNAse (Thermo; final concentration ~2U/60 μL; 1 μL used), TURBO DNAse buffer (final concentration ~0.5×, 2.5 μL 10× stock used), RNase Cocktail Enzyme Mix (Thermo, mixture of RNAse A and RNAse T1, 3 μL per 60 μL reaction). The samples were incubated with the Quench Mix for 15 min at 37°C. The resulting sample was used directly with the Steady-Glo luciferase assay, according to the manufacturer’s instructions.
The result is given in RLU—relative light units with 10 s integration time.
Enzyme activity assays
Renilla, NanoLuc luciferase, Beta-lactamase, Beta-galactosidase and Chloramphenicol acetyltransferase activity were assayed using commercially available kits, according to the manufacturer’s instructions. See Supplementary Information for detailed procedures.
E. coli cell-free TX/TL extract
Our E. coli cell-free extract was prepared according to the Noireaux Lab protocol, from Rosetta 2 BL21 cells (Novagen)50,53. The entire extract preparation was performed in a cold room (4°C).
HeLa cell-free extract
The HeLa cell-free translation (TL) extract was prepared according to a previously published protocol24. The entire extract preparation was performed in a cold room (4°C). For the mammalian in vitro transcription, we used the HeLa cell-free nuclear fraction transcription system HeLaScribe (Promega).
SNARE protein mimics
The SNARE protein mimics were chemically synthesized by solid phase protein synthesis (Genscript). SNARE-A was a fusion of the E3 coiled-coil motif and the trans-membrane region of the VAMP2 protein (residues 85–116). SNARE-B was a fusion of the K3 coiled-coil motif with a trans-membrane region from syntaxin-1A protein (residues 258–288), as described before36. The SNARE peptide-to-lipid molar ratio used in all experiments was 1:500.
It’s worth noting that liposomes undergoing SNARE-mediated fusion will form large aggregates made from multiple starter liposomes36,37; this does not affect the results in Fig. 6, but it would probably reduce the molecular confinement effects observed in Fig. 3.
Supplementary Material
Acknowledgments
We thank Eliza Vasile and Fei Chen for help with SIM microscopy, and Glenn Paradis and Kiryl Piatkevich for help with flow cytometry experiments. We thank Neha Kamat and Lin Jin for help troubleshooting the DLS machine. We thank Jack Szostak for sharing the liposome encapsulation formula. We thank Vincent Noireaux, Andreas Mershin, and Aaron Engelhart for helpful discussions about cell-free TX/TL systems. E.S.B. acknowledges, for funding, NIH 1U01MH106011, Jeremy and Joyce Wertheimer, NIH 1RM1HG008525, the Picower Institute Innovation Fund, NIH 1R01MH103910, NIH 1R01NS075421, NSF CBET 1053233, the New York Stem Cell Foundation-Robertson Award, and NIH Director’s Pioneer Award 1DP1NS087724. D.A.M.-A. acknowledges support from the Janet and Sheldon Razin (1959) Fellowship.
Footnotes
Author contributions
KPA and DAM-A contributed equally to this work. KPA, DAM-A and KRG-H performed experiments. KPA, DAM-A and ESB analyzed data and wrote the manuscript.
Literature
- 1.Carlson ED, Gan R, Hodgman CE, Jewett MC. Cell-free protein synthesis: Applications come of age. Biotechnol Adv. 2012;30:1185–1194. doi: 10.1016/j.biotechadv.2011.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith MT, Wilding KM, Hunt JM, Bennett AM, Bundy BC. The emerging age of cell-free synthetic biology. FEBS Lett. 2014;588:2755–2761. doi: 10.1016/j.febslet.2014.05.062. [DOI] [PubMed] [Google Scholar]
- 3.Hodgman CE, Jewett MC. Cell-free synthetic biology: Thinking outside the cell. Metab Eng. 2012;14:261–269. doi: 10.1016/j.ymben.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller D, Gulbis J. Engineering Protocells: Prospects for Self-Assembly and Nanoscale Production-Lines. Life. 2015;5:1019–1053. doi: 10.3390/life5021019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shimizu Y, Kuruma Y, Ying BW, Umekage S, Ueda T. Cell-free translation systems for protein engineering. FEBS J. 2006;273:4133–4140. doi: 10.1111/j.1742-4658.2006.05431.x. [DOI] [PubMed] [Google Scholar]
- 6.Shin J, Noireaux V. An E. coli cell-free expression toolbox: Application to synthetic gene circuits and artificial cells. ACS Synth Biol. 2012;1:29–41. doi: 10.1021/sb200016s. [DOI] [PubMed] [Google Scholar]
- 7.Takahashi MK, et al. Rapidly Characterizing the Fast Dynamics of RNA Genetic Circuitry with Cell-Free Transcription–Translation (TX-TL) Systems. ACS Synth Biol. 2015;4:503–515. doi: 10.1021/sb400206c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Michener JK, Thodey K, Liang JC, Smolke CD. Applications of genetically-encoded biosensors for the construction and control of biosynthetic pathways. Metab Eng. 2012;14:212–222. doi: 10.1016/j.ymben.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vamvakaki V, Chaniotakis Na. Pesticide detection with a liposome-based nano-biosensor. Biosens Bioelectron. 2007;22:2848–53. doi: 10.1016/j.bios.2006.11.024. [DOI] [PubMed] [Google Scholar]
- 10.Pardee K, et al. Paper-Based Synthetic Gene Networks. Cell. 2014;159:940–954. doi: 10.1016/j.cell.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lentini R, et al. Integrating artificial with natural cells to translate chemical messages that direct E. coli behaviour. Nat Commun. 2014;5:4012. doi: 10.1038/ncomms5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zemella A, Thoring L, Hoffmeister C, Kubick S. Cell-Free Protein Synthesis: Pros and Cons of Prokaryotic and Eukaryotic Systems. ChemBioChem. 2015 doi: 10.1002/cbic.201500340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forster AC, Church GM. Towards synthesis of a minimal cell. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brea RJ, Hardy MD, Devaraj NK. Towards Self-Assembled Hybrid Artificial Cells: Novel Bottom-Up Approaches to Functional Synthetic Membranes. Chem Pub Soc Euro. 2015 doi: 10.1002/chem.201501229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luisi PL, Ferri F, Stano P. Approaches to semi-synthetic minimal cells: a review. Naturwissenschaften. 2006;93:1–13. doi: 10.1007/s00114-005-0056-z. [DOI] [PubMed] [Google Scholar]
- 16.Stano P, Luisi PL. Semi-synthetic minimal cells: Origin and recent developments. Curr Opin Biotechnol. 2013;24:633–638. doi: 10.1016/j.copbio.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 17.Murtas G, Kuruma Y, Bianchini P, Diaspro A, Luisi PL. Protein synthesis in liposomes with a minimal set of enzymes. Biochem Biophys Res Commun. 2007;363:12–17. doi: 10.1016/j.bbrc.2007.07.201. [DOI] [PubMed] [Google Scholar]
- 18.Yu W, et al. Synthesis of functional protein in liposome. J Biosci Bioeng. 2001;92:590–593. doi: 10.1263/jbb.92.590. [DOI] [PubMed] [Google Scholar]
- 19.Oberholzer T, Nierhaus KH, Luisi PL. Protein expression in liposomes. Biochem Biophys Res Commun. 1999;261:238–241. doi: 10.1006/bbrc.1999.0404. [DOI] [PubMed] [Google Scholar]
- 20.Noireaux V, Libchaber A. A vesicle bioreactor as a step toward an artificial cell assembly. Proc Natl Acad Sci U S A. 2004;101:17669–74. doi: 10.1073/pnas.0408236101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stech M, et al. Production of functional antibody fragments in a vesicle-based eukaryotic cell-free translation system. J Biotechnol. 2012;164:220–231. doi: 10.1016/j.jbiotec.2012.08.020. [DOI] [PubMed] [Google Scholar]
- 22.Weber La, Feman ER, Baglioni C. A cell free system from HeLa cells active in initiation of protein synthesis. Biochemistry. 1975;14:5315–5321. doi: 10.1021/bi00695a015. [DOI] [PubMed] [Google Scholar]
- 23.Wimmer E. Cell-free, de novo synthesis of poliovirus. Science. 1991;254:1647–51. doi: 10.1126/science.1661029. [DOI] [PubMed] [Google Scholar]
- 24.Mikami S, Masutani M, Sonenberg N, Yokoyama S, Imataka H. An efficient mammalian cell-free translation system supplemented with translation factors. Protein Expr Purif. 2006;46:348–57. doi: 10.1016/j.pep.2005.09.021. [DOI] [PubMed] [Google Scholar]
- 25.Mikami S, Kobayashi T, Masutani M, Yokoyama S, Imataka H. A human cell-derived in vitro coupled transcription/translation system optimized for production of recombinant proteins. Protein Expr Purif. 2008;62:190–198. doi: 10.1016/j.pep.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 26.Tan C, Saurabh S, Bruchez MP, Schwartz R, Leduc P. Molecular crowding shapes gene expression in synthetic cellular nanosystems. Nat Nanotechnol. 2013;8:602–8. doi: 10.1038/nnano.2013.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Souza TP, et al. Encapsulation of Ferritin, Ribosomes, and Ribo-Peptidic Complexes Inside Liposomes: Insights Into the Origin of Metabolism. Orig Life Evol Biosph. 2012;42:421–428. doi: 10.1007/s11084-012-9303-4. [DOI] [PubMed] [Google Scholar]
- 28.de Souza TP, Fahr A, Luisi PL, Stano P. Spontaneous Encapsulation and Concentration of Biological Macromolecules in Liposomes: An Intriguing Phenomenon and Its Relevance in Origins of Life. J Mol Evol. 2014;79:179–192. doi: 10.1007/s00239-014-9655-7. [DOI] [PubMed] [Google Scholar]
- 29.Caschera F, Noireaux V. Integration of biological parts toward the synthesis of a minimal cell. Curr Opin Chem Biol. 2014;22:85–91. doi: 10.1016/j.cbpa.2014.09.028. [DOI] [PubMed] [Google Scholar]
- 30.Stefureac R, Long YT, Kraatz HB, Howard P, Lee JS. Transport of α-helical peptides through α-hemolysin and aerolysin pores. Biochemistry. 2006;45:9172–9179. doi: 10.1021/bi0604835. [DOI] [PubMed] [Google Scholar]
- 31.Gouaux E, Hobaugh M, Song L. Alpha-Hemolysin, Gamma-Hemolysin, and Leukocidin from Staphylococcus Aureus: Distant in Sequence but Similar in Structure. Protein Sci. 1997;6:2631–2635. doi: 10.1002/pro.5560061216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Selgrade DF, Lohmueller JJ, Lienert F, Silver PA. Protein Scaffold-Activated Protein Trans-Splicing in Mammalian Cells. 2013;135:7713–19. doi: 10.1021/ja401689b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tu Y, et al. Mimicking the Cell: Bio-Inspired Functions of Supramolecular Assemblies. Chem Rev. 116:2023–2078. doi: 10.1021/acs.chemrev.5b00344. [DOI] [PubMed] [Google Scholar]
- 34.Del Vecchio D, Ninfa AJ, Sontag ED. Modular cell biology: retroactivity and insulation. Mol Syst Biol. 2008;4 doi: 10.1038/msb4100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caschera F, et al. Programmed vesicle fusion triggers gene expression. Langmuir. 2011;27:13082–13090. doi: 10.1021/la202648h. [DOI] [PubMed] [Google Scholar]
- 36.Meyenberg K, Lygina AS, van den Bogaart G, Jahn R, Diederichsen U. SNARE derived peptide mimic inducing membrane fusion. Chem Commun. 2011;47:9405–9407. doi: 10.1039/c1cc12879e. [DOI] [PubMed] [Google Scholar]
- 37.Robson Marsden H, Korobko AV, Zheng T, Voskuhl J, Kros A. Controlled liposome fusion mediated by SNARE protein mimics. Biomater Sci. 2013;1:1046–1054. doi: 10.1039/c3bm60040h. [DOI] [PubMed] [Google Scholar]
- 38.Inglés-Prieto Á, et al. Light-assisted small-molecule screening against protein kinases. Nat Chem Biol. 2015;11:952–954. doi: 10.1038/nchembio.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boyden ES. A history of optogenetics: the development of tools for controlling brain circuits with light. F1000 Biol Rep. 2011;3:11. doi: 10.3410/B3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hanczyc MM, Fujikawa SM, Szostak JW. Experimental models of primitive cellular compartments: encapsulation, growth, and division. Science. 2003;302:618–622. doi: 10.1126/science.1089904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adamala K, Szostak JW. Competition between model protocells driven by an encapsulated catalyst. Nat Chem. 2013;5:495–501. doi: 10.1038/nchem.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Balaram P. Synthesizing life. Current Science. 2003;85:1509–1510. [Google Scholar]
- 43.Adamala K, et al. Open questions in origin of life: experimental studies on the origin of nucleic acids and proteins with specific and functional sequences by a chemical synthetic biology approach. Comput Struct Biotechnol J. 2014;9:e201402004. doi: 10.5936/csbj.201402004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ruiz-Mirazo K, Briones C, de la Escosura A. Prebiotic Systems Chemistry: New Perspectives for the Origins of Life. Chem Rev. 2014;114:285–366. doi: 10.1021/cr2004844. [DOI] [PubMed] [Google Scholar]
- 45.Glansdorff N, Xu Y, Labedan B. The Last Universal Common Ancestor: emergence, constitution and genetic legacy of an elusive forerunner. Biology Direct. 2008;3:29. doi: 10.1186/1745-6150-3-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Woese C. The universal ancestor. Proc Natl Acad Sci U S A. 1998;95:6854–6859. doi: 10.1073/pnas.95.12.6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Theobald DL. A formal test of the theory of universal common ancestry. Nature. 2010;465:219–222. doi: 10.1038/nature09014. [DOI] [PubMed] [Google Scholar]
- 48.Spencer AC, Torre P, Mansy SS. The Encapsulation of Cell-free Transcription and Translation Machinery in Vesicles for the Construction of Cellular Mimics. J Vis Exp. 2013;(80):e51304. doi: 10.3791/51304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adamala K, Engelhart AE, Kamat NP, Jin L, Szostak JW. Construction of a liposome dialyzer for the preparation of high-value, small-volume liposome formulations. Nat Protoc. 2015;10:927–938. doi: 10.1038/nprot.2015.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shin J, Noireaux V. Efficient cell-free expression with the endogenous E. Coli RNA polymerase and sigma factor 70. J Biol Eng. 2010;4:8. doi: 10.1186/1754-1611-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lutz R, Bujard H. Independent and tight regulation of transcriptional units in escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–30. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun ZZ, et al. Protocols for Implementing an Escherichia coli Based TX-TL Cell-Free Expression System for Synthetic Biology. J Vis Exp. 2013:1–15. doi: 10.3791/50762. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


