Abstract
Salmonella infection is a major public health problem worldwide. Antibodies directed towards the O polysaccharide (OPS) of S. Typhimurium, a serogroup B non-typhoidal Salmonella serovar, have protected against fatal infection in animal models. The OPS is known to undergo O-acetylation, though the impact of these modifications on antibody binding is poorly understood. Using molecular simulations, we assessed the conformational properties and antigen-antibody interactions of de-acetylated and O-acetylated S. Typhimurium OPS when bound by monoclonal anti-OPS IgG Se155-4. Our findings indicate that (i) the α-D-abequose (8) monosaccharide makes important interactions with Se155-4, (ii) the de-acetylated form binds to the antibody in two conformations, (iii) the acetyl group at α-L-rhamnose (5) traps the acetylated O-antigenic saccharide in one of those two conformations when bound to the antibody; (iv) the dominant conformation sampled by both unbound saccharides only occurs in the deacetylated-antibody complex; and (v) both unbound saccharides sample the second bound conformation to a small extent (2 - 4%). These observations provide insights into the conformational preference of an antigenic saccharide when bound to a well-characterized specific monoclonal antibody, and suggest possible important properties of vaccine induced antibodies following immunization with live attenuated and OPS-based subunit vaccines.
Graphical abstract
Introduction
Non-typhoidal Salmonella (NTS) infections are a major problem worldwide. NTS infection in healthy adults generally results in gastroenteritis; however, infection in very young, elderly, and immunocompromised individuals can result in potentially fatal invasive disease.1-2 In sub-Saharan Africa, invasive NTS (iNTS) disease caused by S. Typhimurium is widespread in infants and toddlers, where fatality rates up to 30% occur.3-5 There are no available licensed NTS vaccines for humans, and treatment is complicated by increasing levels of antibiotic resistance. Development of an effective iNTS vaccine is thus a global health priority. 6
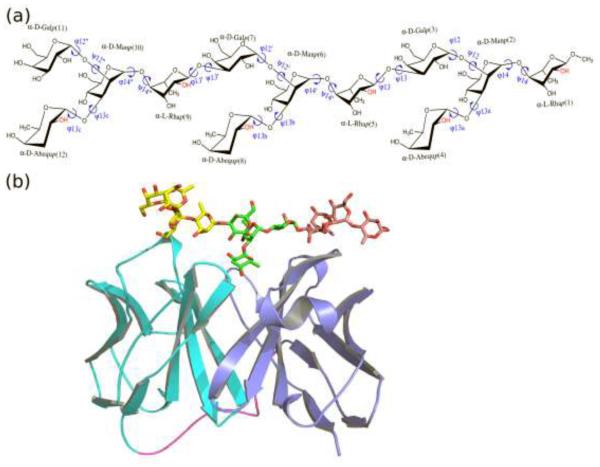
The surface carbohydrate in the majority of clinically important NTS serovars is the lipopolysaccharide (LPS) associated O polysaccharide (OPS). S. Typhimurium OPS is a virulence factor and protective antigen in animal models, and vaccines based on S. Typhimuirum OPS have shown promise in preclinical studies.7-11 Structural differences in Salmonella OPS are used to define serogroups. In S. Typhimurium, a serogroup B Salmonella, the backbone of the O-antigen repeat unit is a trisaccharide, →4)-L-rhamnose-α-(1→3)-D-galactose-α-(1→2)-D-mannose-α-(1→ (Figure 1a), which constitutes the O:12 antigen and is common to Salmonella serogroups A, B, and D.12 In each repeating unit, there is a serogroup defining immunodominant dideoxy hexose covalently connected to the α-D-mannose via a 1→3 glycosidic linkage. In serogroup B, a α-D-abequose at this position forms the O:4 antigen. The O-antigen from S. Typhimurium strains can undergo additional modification including variable glucosylation on the galactose and O-acetylation on the abequose (O:5 specificity) and rhamnose monosaccharides.13 These modifications are prevalent among invasive S. Typhimurium strains from sub-Saharan Africa.14
Figure 1.
Chemical structure of (a) the O-antigenic saccharide molecule and (b) a 3D structural model of the antibody-saccharide complex used in this study. (a) The depicted structure corresponds to the de-acetylated O-antigenic saccharide. The O-acetylated counterpart was constructed with the hydroxyl group substituted by an acetyl group at the C2 position of both α-D-Abequp and α-L-Rhap residues (red). The glycosidic torsional angles are defined as ϕ=O5-C1-On-Cn and ψ=C1-On-Cn-Cn+1, with the On, Cn and Cn+1 atoms in the reducing end monosaccharide. In the manuscript the first, second (central) and third tetrasaccharide units refer to residues 1-4, 5-8 and 9-12, respectively. (b) The single-chain variable fragments of the antibody is shown in cartoon with the light chain colored in cyan, heavy chain in blue, and the connecting loop in violet. The carbohydrate was represented as a stick model and colored brown, green, yellow for the 3 tetrasaccharide units, respectively. The hydrogen atoms in the carbohydrate were omitted for clarity and oxygen atoms are in red.
Serological evidence supports the notion that abequose acetylation generates unique epitopes, as monoclonal antibodies that reacted with the O-acetylated antigens (O: 4[5],12) did not bind robustly to the de-acetylated counterpart (O:4,12).15 The effect of acetylation on the conformational properties of the polysaccharide upon antibody binding is not well understood however, and could provide important insight into the host response to infection or vaccination with OPS based vaccines. Based on published experimental observations, two models could account for the effect of acetylation on antibody binding. One model proposes that the abequose residue in the repeating unit is the antigenic determinant and the majority of antibodies induced by serogroup B OPS prefer the respective acetylated or non-acetylated forms. NMR, crystallography, and thermodynamics studies have provided direct evidence of the abequose as the key structural determinant of the immunogenic epitope by interaction with amino acid side chains within the antibody binding pocket.16-20 The alternative model suggests that while direct interaction with abequose contributes to antigen recognition, the conformational properties of the O-antigen molecule upon acetylation confer additional antigenic preference.15, 21 Considering the conformational flexibility of the O-antigen along the glycosidic linkages, this second model is a reasonable hypothesis and is supported by the observation of monoclonal anti-group B OPS antibodies that bind complex epitopes that incorporate the abequose as well as other peripheral polysaccharide backbone determinants.15, 21-22
In a previous study the conformational properties of the de-acetylated O-antigenic saccharide and its acetylated counterpart in the unbound state were investigated using Hamiltonian replica exchange molecular dynamics (MD) simulations, from which a single dominant conformation for all assessed OPS forms was identified (manuscript submitted). In order to explore the role of S. Typhimurium OPS abequose O-acetylation on antibody binding, atomic-detail computer simulations were undertaken to assess the conformational properties of the O-antigen when unbound or bound to previously described Salmonella serogroup B OPS specific monoclonal antibody Se155-4 for which the X-ray crystal structure has been solved when bound to serogroup B OPS. 18, 23 These simulations were based on molecular dynamics (MD) simulations in conjunction with a recently developed Hamiltonian replica exchange enhanced sampling method24-25 along with the state-of-the-art CHARMM additive force field for proteins and carbohydrates.26-30 Two O-antigen models, corresponding to the respective acetylated and de-acetylated forms, were constructed and studied when bound in a complex with Se155-4 using previously published X-ray crystallography results. Results from the simulations suggest that binding of the antigen to the Se155-4 induces a conformational change in the antigen that is impacted by acetylation. In addition, it is observed that the binding pocket within the antibody is sufficiently large to accommodate the additional O-acetyl group at the abequose residue.
Computational Details
MD simulations with the Hamiltonian replica exchange with concurrent solute scaling and biasing potential (HREST-BP) method
Four molecular models were compared to examine the conformational preference of the O-antigenic saccharides when bound to antibody. As shown in Figure 1, these models include the bound and unbound de-acetylated saccharide with 3 tetrasaccharide units and its O-acetylated counterpart. The amount and position of acetylated sites can differ between various S. Typhimurium isolates. Here we employed one representative model of the acetylation deduced from the analysis of Malawian invasive S. Typhimuirum isolate D23580, that maintains variable acetylation at the C2 position of both α-D-Abequp and α-L-Rhap residues.13 The simulations of the polysaccharides alone were from a previous study (manuscript submitted) while the simulations in the presence of the antibody were performed as part of the present study. Each system was immersed into a cubic box of the CHARMM-TIP3P water model with a minimum distance of 10 Å from any solute atom to the box boundary.31-33 In the complex models, the protein portion was derived from the Salmonella serogroup B OPS specific monoclonal antibody Se155-4 with only a single-chain variable domain (PDBID 1MFA and 1MFB)18, 23, which includes the antigen binding pocket. The missing loop, with a suggested sequence of PKSSPSVTLEPPSSNG, was constructed using FREAD to connect the variable domains of the light (Q111) and heavy (E251) chains, resulting in a total of 244 residues in the antibody model.34 In this study, we renumber the amino acids of the antibody model from 1 to 244 and, thus, residues 1-111 and 128-244 in the computational model correspond to the light chain (Q1-Q111) and heavy chain (E251-S367) in the crystal structure, respectively.16 The standard protonation states were used for the titratable amino acid residues predicted with PROPKA 35and crystal waters around the variable domains in 1MFB were included in the simulation system. The missing monosaccharides in the 3 tetrasaccharide units in the complex were manually constructed based on the bound fragment of the 1MFP crystal structure.18 The resulting complex models were equilibrated by initially subjecting them to energy minimization and 70-ns molecular dynamics simulations with gradually reduced positional harmonic restraints in NAMD in preparation for the subsequent enhanced sampling simulations.36
Enhanced sampling production simulations were performed with the program CHARMM37 or CHARMM-OpenMM38 under the CHARMM36 additive force field for proteins26-27 and carbohydrates28-30. In the production simulations the temperature was maintained at 298 K using the Hoover algorithm with a thermal piston mass of 1000 kcal/mol·ps239 and a constant pressure of 1 atm was realized using the Langevin piston algorithm with a collision frequency of 20 ps−1 and mass of 1630 amu.40 The covalent bonds involving hydrogens were constrained with SHAKE, which allows an integration step of 2 fs in the MD simulations.41 In the energy and force evaluations, the non-bonded Lennard-Jones interactions were computed within the cutoff of 12 Å with a switching function applied over the range from 10 to 12 Å. The electrostatic interactions were treated by the particle mesh Ewald method with a real space cutoff of 12 Å, a charge grid of 1 Å, a kappa of 0.34, and the 6-th order spline function for mesh interpolation.42
The Hamiltonian replica exchange with concurrent solute scaling and biasing potential (HREST-BP) was adopted to obtain adequate conformational sampling of each solvated system.24-25 HREST-BP only includes the subsystem of interest in the enhanced Hamiltonian and the effective biasing potentials and thus can minimize the number of replicas used in simulations. As the conformational propensity of the O-antigenic saccharides was the focus of the present study, only the carbohydrate molecule in each system was treated as the solute in the HREST-BP simulations. Enhanced sampling was achieved by subjecting the glycosidic linkages to potential biasing with concurrent effective temperature scaling of the intra-solute potential and solute-environment interactions. The biasing potentials were formulated with the 2-dimensional grid based correction map (bpCMAP) along the two dihedrals φ(O5-C1-On-Cn)/ψ(C1-On-Cn-Cn+1) for each glycosidic linkage and were constructed using the corresponding disaccharide model in the gas phase as described previously.43 For each simulation, 6 replicas were used in the HREST-BP and an exchange attempt was examined every 1000 MD steps according to the Metropolis criterion. The ground-state replicas were simulated unperturbed at 298 K and the other 5 perturbed replicas were assigned a scaling temperature of 316 K, 335 K, 356 K, 377 K and 400 K, respectively. The distribution of scaling factors was derived following the scheme described in the HREST-BP paper for the biasing potentials and a successful exchange ratio larger than 18% was observed for any neighboring replicas.25 For each of the four systems, a 100 ns production run was performed for every replica with HREST-BP to provide sufficient exploration of the conformational space of the O-antigenic saccharide. For the antibody/de-acetylated model, an independent 60 ns production run was carried out to examine the convergence of the sampling. Results from that simulation suggests that 60 ns of sampling can provide free energy profiles along the inter-monosaccharide distances very close to that of the 100 ns trajectory. The total 160 ns of the antibody/de-acetylated trajectory was used for data analysis in following discussions unless stated otherwise. To compare the flexibility of the protein scaffold in the absence and presence of the antigens, a standard MD simulation of 200 ns was performed for the apo antibody.
Glycosidic linkage based description of polysaccharide conformations
From the simulations, clustering analysis was performed on the basis of the glycosidic linkages (GL) to characterize the conformational heterogeneity or preference in each saccharide molecule.44 Each GL cluster or conformation can be represented based on torsion angles φ/ψ in the glycosidic linkages. Figure 2 shows the partitioning in φ/ψ space corresponding to different local minima on the free energy landscape of each GL. Each partitioned region is then assigned an integer identifier. All 1→3 and 1→4 linkages were included in the clustering analysis; 1→2 linkages were omitted as they only sampled one minima in the simulations. The definition of a GL conformation is based on the series of index numbers used to denote the state of each glycosidic linkage. This allows every recorded snapshot in the simulations to be assigned to a specific combination of the eight index numbers for clustering. For example, 11111111 indicates all the linkages are located in the region of φϵ(0°, 180°]/ψϵ(−180°, −120°) or (0°, 180°]. In this definition of a cluster, all the conformations in the cluster are in the same local free energy minima for all the linkages. We refer to these as glycosidic linkage, or GL, clusters or conformations. However, as shown below, conformations in the same cluster can vary in terms of Cartesian coordinates; however, as they occupy the same GL minima these different “Cartesian conformations” can readily interconvert without encountering any high free energy barriers associated with the glycosidic linkages. GL clusters, therefore provide a high dimensional representation of the carbohydrate conformation as compared to the 1-D or 2-D free energy or potential of mean force (PMF) profiles.
Figure 2.
Index of free energy minima for each glycosidic linkage used in the GL clustering analysis. One free energy map for 1→4 linkage in (a) unbound and (b) bound saccharides was used as example. In GL clustering, the indices in (a) were used for all glycosidic linkages except the ones defined in (b) were used for the ϕ14′/ψ14′ linkage of the bound saccharides.
Root-mean-square difference (RMSD) description of O-antigenic conformations
A convenient method to determine the overall extent of similarity between two conformations is based on the overall difference in the locations of the atoms of the saccharides. In the present study this was quantified based on the RMSD between the two conformations over the selected set of non-hydrogen atoms. Prior to RMSD determination the saccharides were aligned based on the non-hydrogen atoms in the pyranose rings in the central tetrasaccharide unit.
3D Cartesian coordinate volume description of O-antigenic saccharide conformations
The spatial distribution or range of conformational space as defined by Cartesian coordinates can be represented by the distribution or volume of the sampled conformations. To compute the sampled distribution or volume, a 3D grid with a voxel size of 1Å×1Å×1Å was constructed around the saccharide molecule. Then, each snapshot in the ground-state replica trajectories from the HREST-BP simulations was aligned over the non-hydrogen atoms of the pyranose rings of the central tetrasaccharide unit. For a given snapshot, each voxel was assigned a value of 1 if it was occupied by a non-hydrogen atom.45 The occupied volume is the total number of occupied voxels. This analysis was performed for all snapshots from the respective trajectories. Visualization of the sampled volume was performed by normalizing the voxel occupancies over all the snapshots. This approach allows for the volume sampled by selected portions of the saccharides as well as of the full saccharide to be quantified and visualized. To compare the similarity of the sampled conformational space between two simulations, e.g. A and B, the overlap coefficient (OC) was computed as follows,
| (1) |
where N is the number of voxels in the 3D grid map; and are the occupancies at the i-th voxel for simulations A and B, respectively.46 From the definition of OC, OC=0 and 1 suggest the two simulations sampled completely different and identical regions conformational space, respectively.
Interaction energy decomposition on per-residue basis
To examine the interactions between the model OPS and the surrounding protein residue, the molecular mechanics interaction energy, Emm, was calculated between the saccharides and the protein based on a cutoff value of 997 Å. Emm was then decomposed into the contribution of the electrostatic, Eele, and van der Waals, EvdW terms as well as being performed on the basis of individual monosaccharides or residues.
Free energy calculations
The free energy or potential of mean force (PMF) along one or two linkage dihedrals was computed from the unperturbed ground-state replica as,
| (2) |
where Ns is the number of snapshots recorded in the ground-state replica and if is within the bin [wi−Δw/2,wiΔw/2] and otherwise .
Results and Discussion
Presented is a study of the impact of acetylation on the conformational properties and antibody interactions of a specific OPS antigen. We note that in the biological context the antigen would be ultimately bound to the cell surface via the lipid A anchor of lipopolysaccharide (LPS). As the non-reducing end of the antigen that is being studied here is very distal from the lipid A membrane moiety it may be considered unlikely that interactions with the cell surface membrane will affect the conformational properties of the antigen, though such possible effects cannot be totally excluded.
Cluster analysis showing two distinct conformational states adopted by the antigenic saccharide in the antibody/de-acetylated saccharide system
GL clustering offers an approach to identify the sampling of individual clusters that is independent on the definition of alignment reference. For each linkage, one index number was assigned to represent the position of the free energy local minima as defined in Figure 2. Accordingly, one combination of 8 numbers, each corresponding to one of the 8 1→3 and 1→4 linkages, denotes the location of the respective 8 glycosidic linkages for a given conformation. Such a metric allows for the population of the different conformations to be readily calculated and, if necessary, subject to clustering analysis. Table 1 shows populations of all GL clusters sampled at greater than 0.1%. Two highly populated GL clusters are sampled in the bound de-acetylated saccharide with indices 11111133 (51.0%) and 12111133 (42.6%). These will subsequently be referred to as cluster 1 and cluster 2, respectively. The two clusters differ in the sampling of the 1→4 glycosidic linkage between α-L-rhamnose(5) and α-D-mannose(6) (Figure 1a). Interestingly, the bound O-acetylated saccharide only significantly samples GL cluster 12111133 (95.2% cluster 2) whereas the unbound saccharides predominately sample cluster 1, with populations of 91.0% in both cases, with the bound de-acetylated saccharide sampling both clusters (51.0 and 42.6%).
Table 1.
Probability distribution for each GL clustera
| Cluster | antibody/de- acetylated saccharide |
antibody/O- acetylated saccharide |
de-acetylated saccharide |
O-acetylated saccharide |
|---|---|---|---|---|
| 11111133 | 0.510 | 0.001 | 0.911 | 0.910 |
| 11111233 | 0.000 | 0.000 | 0.000 | 0.006 |
| 11112133 | 0.000 | 0.000 | 0.000 | 0.004 |
| 11121133 | 0.000 | 0.000 | 0.000 | 0.014 |
| 11111143 | 0.003 | 0.000 | 0.000 | 0.000 |
| 11211133 | 0.011 | 0.000 | 0.022 | 0.011 |
| 12111133 | 0.426 | 0.952 | 0.044 | 0.016 |
| 12111233 | 0.000 | 0.003 | 0.000 | 0.000 |
| 12121133 | 0.000 | 0.005 | 0.000 | 0.000 |
| 12111143 | 0.007 | 0.001 | 0.000 | 0.000 |
| 12211133 | 0.015 | 0.019 | 0.000 | 0.000 |
| 21111133 | 0.018 | 0.000 | 0.017 | 0.037 |
| 21111143 | 0.000 | 0.000 | 0.000 | 0.002 |
| 21211133 | 0.000 | 0.000 | 0.002 | 0.000 |
| 22111133 | 0.008 | 0.018 | 0.001 | 0.000 |
Only the clusters with a probability > 0.1% are listed. The index definition for φ/ψ was indicated in Figure 2. While a division of (0°, 180°] and (−180°, 0°) was used for ψ14' in the bound saccharides instead of (−180°, −120°) & (0°, 180°] and [−120°, 0°] for the unbound saccharides. The eight linkages correspond to φ14/ψ14, φ14'/ψ14', φ14"/ψ14", φ13a/ψ13a, φ13b/ψ13b, φ13c/ψ13c, φ13/ψ13, and φ13'/ψ13'. The population ratio is given for each GL cluster.
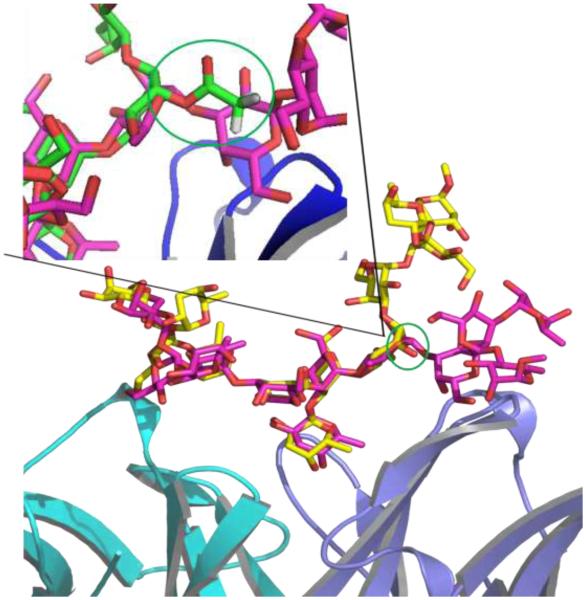
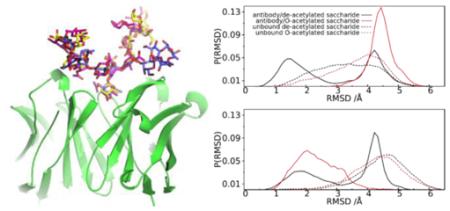
While defining conformations in terms of GL clusters is convenient due to the method being alignment independent, it does not allow for a detailed structural analysis. Indeed, while all the conformations in a given GL cluster sample the same minima in the 2D glycosidic linkage free energy surfaces, there is significant variability in the range of local conformations that can be sampled in the context of the Cartesian coordinates. To better understand the range of sampling in Cartesian coordinates, all conformations in cluster 1 or 2 were analyzed to identify the “central” conformation, as defined by that conformation that has the smallest overall RMSD with respect to all other conformations in the cluster. The two representative conformations for cluster 1 and 2 bound to the antibody are shown in Figure 3, which indicates distinct orientations of the first tetrasaccharide unit with respect to the antibody. RMSD probability distributions were then calculated relative to the individual central conformations for all the conformations sampled for the four systems, with the results presented in Figure 4. Results show the range of RMSD values to be broad. The bound O-acetylated and both the unbound de-acetylated and O-acetylated saccharides show a single distribution with a greater number of conformations sampled close to cluster 2 in the bound O-acetylated saccharide whereas for the unbound saccharides more conformations are close to the cluster 1 state than to the cluster 2 state. The de-acetylated bound saccharide demonstrates a bimodal distribution. These results are consistent with the GL cluster analysis showing the O-acetylated saccharide in the bound state to sample cluster 2, whereas the unbound de-acetylated and O-acetylated saccharides sample cluster 1 with the bound de-acetylated saccharide sampling both clusters. Notable is the broad nature of the RMSD distribution indicating that a range of local conformations are being sampled in the context of a single GL conformation, with the range of conformations being sampled being larger in the unbound versus the bound conformations.
Figure 3.
Structural model of two representative carbohydrate conformations identified from GL clustering analysis of the antibody/de-acetylated saccharide system. Cluster 1 conformation is colored in purple and cluster 2 in yellow with oxygen atoms in red. The antibody is shown in cartoon and the saccharides in stick representations. The circled group corresponds the O-acetylated site in rhamnose(5) in the antibody/O-acetylated system. The inset highlighted the position of the acetyl group in rhamnose(5) with respect to the Cluster 1 conformation.
Figure 4.
Probability distribution of the RMSD values with respect to the two representative conformations for cluster 1 (a) and cluster 2 (b), identified from cluster analysis of the antibody/de-acetylated saccharide system. The distributions were derived from simulations of the antibody/de-acetylated saccharide (solid black), antibody/O-acetylated saccharide (solid red), unbound de-acetylated (black dotted) and O-acetylated (red dotted) saccharides. The RMSD values were computed for all pyranose ring non-hydrogen atoms after alignment to the central tetrasaccharide unit.
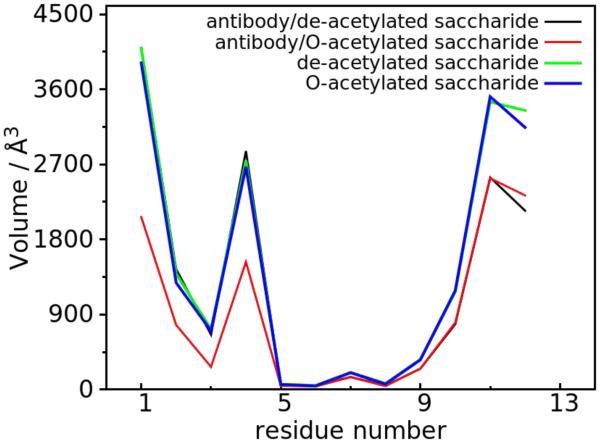
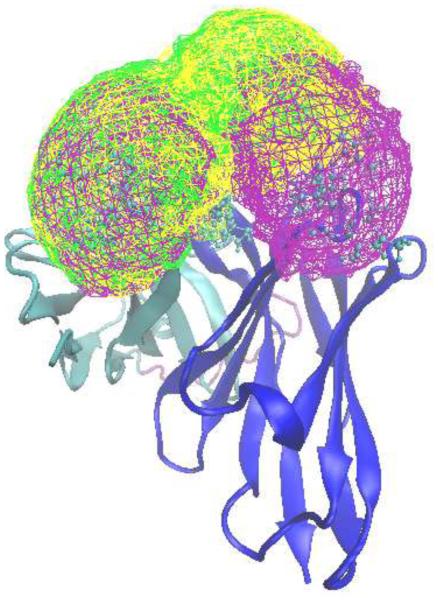
Volume analysis was undertaken to visually analyze the range of conformations being sampled as well as determine the similarity of the conformational space being sampled, following alignment of the central tetrasaccharide of the full saccharides. Results presented in Figure 5 show the total volumes sampled for each monosaccharide unit, and indicate a restricted motion of the O-acetylated saccharide in the bound state compared to its de-acetylated counterpart. This observation is consistent with the clustering and RMSD analyses, in which only one of the two representative states in antibody/de-acetylated saccharide was observed for the antibody/O-acetylated saccharide. Visual representation of the volume sampled for GL clusters 1 and 2 is shown in Figure 6. As expected, the bound O-acetylated and de-acetylated saccharides in cluster 2 sample similar regions of conformational space with cluster 1 for the antibody/de-acetylated complex sampling regions that overlap with cluster 2 as well as a range of new conformations. The primary difference in the region occupied by the bound O-acetylated and de-acetylated saccharides involves the first tetrasaccharide unit with respect to the antibody.
Figure 5.
The volume calculated from the 3D spatial distribution for the O-antigenic saccharide under different conditions. Each frame in the trajectory was aligned to the pyranose ring non-hydrogen atoms of the central tetrasaccharide unit.
Figure 6.
Spatial distribution sampled by the antigenic saccharides in the antibody/de-acetylated and antibody/acetylated systems. Shown are the GL clusters 11111133 (cluster 1, purple) and 12111133 (cluster 2, yellow) for antibody/de-acetylated complex and the GL cluster 12111133 (cluster 2, green) for the antibody/O-acetylated system. Distributions based on non-hydrogen atoms of the monosaccharide rings and the contour level is set to 10−6 for the full polysaccharide.
To understand the range of conformational sampling in Cartesian coordinates and the underlying reasons of the difference in bounded O-acetylated and de-acetylated saccharides, the 2D free energy profiles about the glycosidic linkages were analyzed for the bound saccharides (Figure 7) and the unbound saccharides (Figure S1, supporting information). The majority of linkages sample a single local GL minimum along the torsional angle ϕ. For all linkages with α-D configuration at the non-reducing end monosaccharide, the free energy landscape has one minima around 60° for ϕ and has multiple local minima along the ψ dihedrals except for the 1→2 glycosidic linkage. For linkages involving the α-L configuration, the only free energy minima along ϕ is located around −60°. This observation agrees with previous experimental and theoretical results on the conformational preference along the linkage dihedral ϕ.44 In the de-acetylated bound saccharide the presence of two deep minima for the 1→4 glycosidic linkage between α-L-rhamnose(5) and α-D-mannose(6), ϕ14′/ψ14′, corresponding to clusters 1 and 2, versus the single minimum in the O-acetylated bound saccharide free energy surface corresponding to cluster 2. The unbound saccharides also show a single low free energy minimum at the same region for this linkage, although sampling of a second GL conformation at ψ<0 region is evident with high free energy values. This corresponds to the small, but finite extent of sampling of cluster 2, as seen in Table 1 (cluster 12111133). For all the linkages the minima are generally broad, indicating that local conformational variations can take place without encountering significant free energy barriers. This leads to the wide range of RMSD values and broad volume distributions observed for the individual clusters (Figures 4, 5 and 6).
Figure 7.
2 dimensional free energy landscapes for each glycosidic linkage of the O-antigenic saccharide obtained from simulations of the (a) antibody/de-acetylated saccharide and (b) antibody/O-acetylated saccharide. The X- and Y-axis represent ϕ and ψ in degrees, respectively. The unit of free energy is kcal/mol.
The conformational analysis reveals a picture where two GL conformations, clusters 1 and 2, are sampled in the studied saccharides. Cluster 1 is sampled by the unbound saccharides (> 90%) as well as by the Se155-4 bound de-acetylated saccharide (~50%), which also samples cluster 2 (43%) with the antibody bound O-acetylated saccharide strongly favoring cluster 2 (95%). The difference in the two clusters is exemplified by differences in free energy surface for the 1→4 glycosidic linkage between α-L-rhamnose(5) and α-D-manose(6) in the bound de-acetylated and O-acetylated saccharides (Figure 7), This linkage determines the relative orientation of the first tetrasaccharide unit with respect to the central, second unit. Detailed examination of the complex conformation shows that the acetyl group at α-L-rhamnose(5) in cluster 2 occupies the region occupied by the linkage between α-D-galactose(3) and α-L-rhamnose(5) that is required for α-D-galactose(3) to sample the conformation present in cluster 1. This orientation of the acetyl group is associated with its interactions with the antibody, as discussed in the following section. Therefore, this particular packing of the acetyl group at α-L-rhamnose(5) with respect to the antibody locks the conformation of O-acetylated saccharide around cluster 2, whereas the de-acetylated saccharide can sample both clusters 1 and 2 when bound to the antibody.
Interactions between the O-antigenic saccharide and the antibody
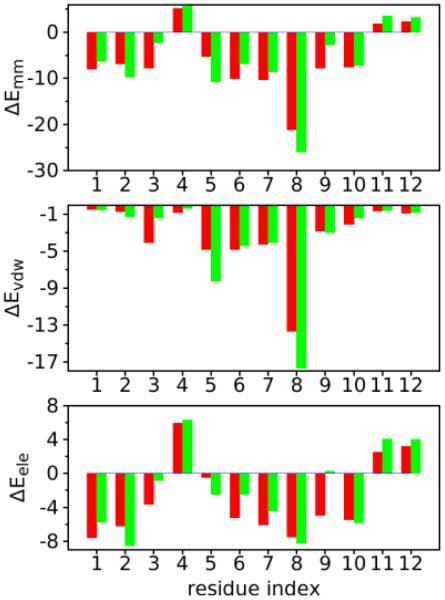
An unusual feature in the studied Salmonella Group B specific monoclonal antibody SE155-4 is the presence of multiple amino acid residues with aromatic side chains around the binding pocket.16, 20 This feature indicates an important contribution of nonpolar and hydrophobic interactions to binding between the antibody and the saccharide epitope. Accordingly, we compared the interaction energies of the de- and O-acetylated saccharides with the antibody using energy decomposition on a per residue basis (Figure 8). The result shows that 8 out of the 14 amino acid residues contributing significantly to the antibody-antigen binding energy have aromatic side chains (Figure 8). Antibody residues G32, H34, T185, and H228 have more favorable interaction energies with the de-acetylated saccharide than the O-acetylated form, arising mainly from the electrostatic interactions. This is due to these residues having more favorable hydrogen bonding with the saccharide in cluster 1 as compared to the O-acetylated saccharide (Table 2 and Figure 9). Specifically, stronger hydrogen bonding interactions in cluster 1 are formed between the backbone HN of G32 and the O4 and O6 atoms of α-D-mannose(10), the NE2 of H34 and HO2 of both α-D-galactose(7) and α-L-rhamnose(9) (see Figure S2 in supporting information), the O/HN of T185 and HO4/O4 of α-D-galactose(3), and the ND1 of H228 and HO4 of α-D-manose(6) (see Figure S3 and S4 in supporting information). On the other hand, residues W93, W96, N182, and F186 form more favorable interactions with the bound O-acetylated saccharide. This can be attributed to the hydrogen bonding interactions between the HE1 of W93 and O2 of both α-D-galactose(7) and α-L-rhamnose(9), the OD1/HD21 of N96 and HO4/O4 of α-D-galactose(7) (see Figure S5 in supporting information), HD21 of N182 and O5 of α-L-rhamnose(5), and the HZ of H186 and O5 of α-D-galactose(7) (see Figure S3 in supporting information) that can occur in cluster 2. These interaction differences are suggested to contribute to differential binding of the antibody to different modifications of the antigenic saccharide, especially the interaction involving HO2 atom of α-L-rhamnose(9) that is absent in the O-acetylated form. Of the residues contributing to antigen binding, experimental data is only available on the H34Q mutant, which has slightly decreased binding affinity. This result is consistent with the favorable interaction energy contribution of H34 to saccharide-antibody interactions (Figure 9). Other studied mutations show comparable binding affinities with respect to the wild-type antibody.20, 47-48
Figure 8.
Decomposition of molecular mechanics interaction energy (kcal/mol, ΔEmm=ΔEele+ΔEvdw with ΔEele and ΔEvdw the contributions from electrostatic and van der Waals interactions, respectively) between the saccharide and the protein for the antibody/de-acetylated saccharide (red) and antibody/O-acetylated saccharide (green) on a per-residue basis for the antibody. Residues with a contribution |ΔEMM|>2.0 kcal/mol in either system are presented in the histograms in the lower three panels.
Table 2.
Elctorstatic and van der Waals (vdW) interaction energies (kcal/mol) between each monosaccharide and the antibody for the two largest GL clusters
| Saccharide Residue |
Antibody/de-acetylated | Antibody/O-acetylated | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| all GL clusters | GL cluster 11111133 (1) |
GL cluster 12111133 (2) |
all GL clusters | GL cluster 11111133 (1) |
GL cluster 12111133 (2) |
|||||||
| Eele | EvdW | Eele | EvdW | Eele | EvdW | Eele | EvdW | Eele | EvdW | Eele | EvdW | |
| 1 | −8.1 | −0.5 | −10.0 | −0.6 | −6.1 | −0.3 | −5.8 (− 1.0)a |
−0.5 (− 0.1) |
−6.8 (− 1.0) |
−1.6 (− 0.3) |
−5.8 (− 1.0) |
−0.5 (− 0.1) |
| 2 | −7.2 | −0.8 | −6.6 | −0.6 | −8.0 | −1.0 | −8.4 | −1.3 | −10.7 | −4.9 | −8.5 | −1.3 |
| 3 | −3.4 | −3.7 | −5.1 | −5.6 | −1.4 | −1.6 | −0.8 | −1.4 | −2.8 | −2.9 | −0.8 | −1.4 |
| 4 | 6.5 | −0.7 | 7.1 | −1.0 | 5.9 | −0.3 | 6.3 (− 1.2) |
−0.4 (− 0.2) |
6.0 (−1.2) | −1.2 (− 0.5) |
6.3 (− 1.2) |
−0.4 (− 0.2) |
| 5 | −1.3 | −5.7 | 0.4 | −5.0 | −3.1 | −6.5 | −2.5 (− 2.0) |
−8.3 (− 1.3) |
−0.9 (− 1.6) |
−5.8 (− 0.8) |
−2.4 (− 2.0) |
−8.3 (− 1.3) |
| 6 | −5.0 | −5.5 | −6.2 | −5.4 | −3.6 | −5.7 | −2.4 | −4.4 | −1.5 | −3.0 | −2.4 | −4.4 |
| 7 | −5.7 | −4.4 | −7.0 | −4.5 | −4.2 | −4.2 | −4.5 | −4.1 | −8.8 | −2.2 | −4.5 | −4.1 |
| 8 | −7.7 | −15.5 | −8.2 | −15.1 | −7.1 | −16.1 | −8.4 (− 0.9) |
−17.7 (− 1.8) |
−4.3 (0.2) | −16.7 (− 1.2) |
−8.3 (− 0.9) |
−17.7 (− 1.8) |
| 9 | −5.9 | −3.1 | −5.5 | −3.1 | −6.5 | −3.1 | 0.3 (− 0.7) |
−3.0 (− 0.7) |
0.6 (−0.8) | −3.3 (− 0.7) |
0.3 (− 0.7) |
−3.0 (− 0.7) |
| 10 | −5.9 | −2.1 | −6.2 | −2.4 | −5.7 | −1.7 | −5.8 | −1.4 | −8.4 | −1.6 | −5.8 | −1.5 |
| 11 | 3.0 | −0.6 | 2.8 | −0.4 | 3.3 | −0.8 | 4.0 | −0.6 | 5.5 | −0.4 | 4.0 | −0.5 |
| 12 | 3.7 | −0.7 | 3.4 | −0.9 | 4.0 | −0.4 | 4.0 (− 1.2) |
−0.9 (− 0.2) |
5.7 (−1.3) | −0.2 (0.0) |
3.9 (− 1.2) |
−0.9 (− 0.2) |
| SUM | −24.0 | −44.0 | −26.4 | −43.8 | −24.0 | −44.0 | ||||||
| Total | −68.0 | −70.2 | −68.0 | |||||||||
The numbers in parenthesis are the contribution from the -CH3 atoms in acetyl group.
Figure 9.
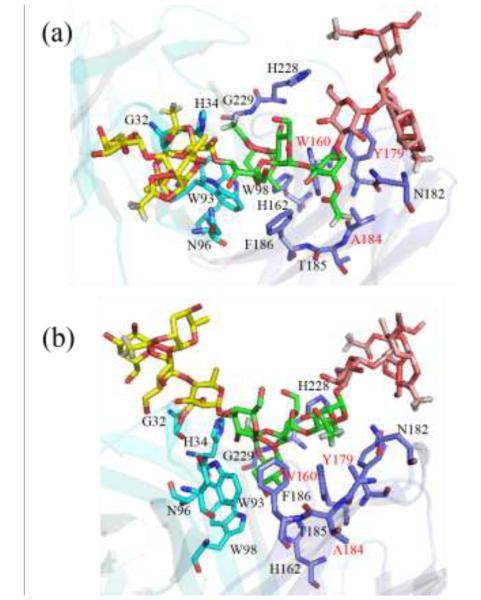
Structural model showing the interaction between the antigenic saccharide and antibody. The antibody is shown with cartoon representation with the light chain colored in cyan and heavy chain in blue, respectively. Stick representation is used to show the antigenic saccharide and the important antibody residues that contribute greatly to the binding. The antigenic saccharide was colored in brown, green, and yellow for the 1st, 2nd, and 3rd tetrasaccharide units. The hydrogen atoms except those for the acetyl groups were not depicted in the stick model for clarity. Three residues W160, Y179 and A184 form a hydrophobic local environment around the acetyl group of rhamnose(5) and their labels were colored red. (a) top view towards the binding pocket (b) side view towards the binding pocket.
From the view of the O-antigenic saccharides, monosaccharide α-D-abequose(8) has the most favorable interaction with the antibody for both species and monosaccharides α-D-galactose(3), α-L-rhamnose(5), α-D-abequose(8), and α-L-rhamnose(9) interact differently between the two saccharides. α-D-galactose(3) and α-L-rhamnose(9) interact more favorably in the de-acetylated saccharide and the other two more favorably in the O-acetylated saccharide (Figure 10). Upon acetylation at both α-D-abequose and α-D-rhamnose residues, a more favorable van der Waals interaction was observed for monosaccharide 5 and 8 in the O-acetylated saccharide. Both α-D-galactose(3) and α-L-rhamnose(9) have stronger electrostatic interaction with the antibody in the de-acetylated saccharide, which arise from the conformations around cluster 1 that are not sampled in the O-acetylated system (Table 2). These results suggest that the α-D-abequose(8) makes an important contribution to antibody binding and that the binding pocket of the SE155-4 can accommodate the addition of an extra methyl group of the α-D-abequose(8). However, it is evident that the additional monosaccharides, including those in tetrasaccharide units 1 and 3, contribute favorable interactions with the antibody, with those being impacted by O-acetylation.
Figure 10.
Decomposition of the molecular mechanics interaction energy (kcal/mol, ΔEmm=ΔEele+ΔEvdw with ΔEele and ΔEvdw the contributions from electrostatic and van der Waals interactions, respectively) between the saccharide and the protein for the antibody/de-acetylated saccharide (in red) and antibody/O-acetylated saccharide (in green) on a per-residue basis for the carbohydrate.
Table 2 shows the decomposition of the MM interaction energy contribution in the two largest GL clusters. It is interesting that more favorable electrostatic and van der Waals interactions were observed for GL cluster 2 than 1 in both bound systems for α-L-rhamnose(5). In addition, the O-acetylated form has a more favorable interaction than its de-acetylated counterpart. The stronger interaction is suggested to contribute to the acetyl group of α-L-rhamnose(5) locking the saccharide conformation in the cluster 2 state with W160, Y179 and A184 forming a favorable hydrophobic environment around the acetyl group. These results also indicate how the change in the position of tetrasaccharide unit 1 between clusters 1 and 2 significantly impact interactions with the antibody.
Conformational propensities of the unbound O-antigenic saccharides
As presented above the unbound saccharides primarily sample GL cluster 1 and show similar RMSD distributions, further suggesting similar sampling of conformational space (Figure S6 and S7 in supporting information), as reported in our previous study (manuscript submitted). The free energy landscape along each glycosydic linkage also shows very similar features between the two forms of the saccharide (Figure S1 supporting information). Detailed examination of the linkages shows the 1→3 linkages involving α-D-abequose to sample a slightly broader region in the O-acetylated saccharide. This is due to an additional hydrogen bond formed between the HO2 atom of the abequose residue in the de-acetylated saccharide, which is lost due to HO2 being replaced with the acetyl group (Figure 11). Thus, the loss of this extra hydrogen bonding interaction in O-acetylated saccharide makes the motion of abequose around the 1→3 linkage more flexible. Table 3 shows the overlap coefficients for individual saccharide residues between different systems. The result indicates a large coefficient of 0.79 between the two unbound saccharides, suggesting very similar sampling of conformational space. While subtle differences are present, the overall conformational propensities of the unbound de-acetylated and O-acetylated saccharides are very similar to each other. In addition, while cluster 1 is predominately sampled by the unbound saccharides, there is finite sampling of cluster 2 by 4 and 2 % for the de-acetylated and O-acetylated species, respectively, thereby facilitating binding of the O-antigens to the antibody in the cluster 2 conformation.
Figure 11.
The additional hydrogen bonding interactions formed in the unbound de-acetylated saccharide as compared to the unbounded O-acetylated saccharide.
Table 3.
Overlap coefficients of the spatial distributions for each monosaccharide between different systemsa
| Monosaccharide | 1-2 | 1-3 | 1-4 | 2-3 | 2-4 | 3-4 |
|---|---|---|---|---|---|---|
| 1 | 0.43 | 0.11 | 0.08 | 0.03 | 0.04 | 0.83 |
| 2 | 0.42 | 0.13 | 0.10 | 0.05 | 0.05 | 0.89 |
| 3 | 0.38 | 0.07 | 0.06 | 0.02 | 0.02 | 0.88 |
| 4 | 0.41 | 0.23 | 0.24 | 0.06 | 0.08 | 0.79 |
| 5 | 0.57 | 0.37 | 0.36 | 0.27 | 0.26 | 0.93 |
| 6 | 0.71 | 0.68 | 0.69 | 0.43 | 0.44 | 0.95 |
| 7 | 0.59 | 0.65 | 0.64 | 0.51 | 0.57 | 0.85 |
| 8 | 0.61 | 0.56 | 0.58 | 0.46 | 0.47 | 0.93 |
| 9 | 0.82 | 0.64 | 0.66 | 0.59 | 0.62 | 0.84 |
| 10 | 0.73 | 0.72 | 0.76 | 0.67 | 0.71 | 0.86 |
| 11 | 0.68 | 0.65 | 0.59 | 0.68 | 0.7 | 0.81 |
| 12 | 0.70 | 0.67 | 0.63 | 0.58 | 0.6 | 0.82 |
The index of the system is antibody/de-acetylated saccharide(1), antibody/O-acetylated saccharide (2), de-acetylated saccharide (3), and O-acetylated saccharide(4). The OC value for each pair of models as a function of the pyranose residue is given in the table.
Summary
The presented computational analysis yields a detailed molecular picture into the role of acetylation on the binding of the modeled Salmonella serogroup B O-antigens to monoclonal antibody Se155-4. We found that the α-D- abequose (8) monosaccharide contributes significant favorable interactions with the antibody regardless of O-acetylation. However, it is evident that additional monosaccharides make important contributions to antigen-antibody interactions, including those in tetrasaccharide units 1 and 3, with O-acetylation impacting all monosaccharide-antibody interactions.
The present results may be discussed in the context of the experimental investigations of saccharide binding to Se155-4. Microcalorimetry analysis using a Gal[Abe]Man trisaccharide yielded large changes in heat capacity that were suggested to be associated with hydrophobic contributions to binding.19 This is consistent with the presence of hydrophobic residues in the Se155-4 binding interface, with present results indicating selected hydrophobic residues to contribute to binding via favorable vdW interactions (Fig. 8). Binding studies of Se155-4 with various saccharides were reported by Bundle et al.16 While the polysaccharide sequences and lengths differed, abequose made the most favorable free energy contribution to binding in trisaccharides and the only favorable enthalpic contribution to binding consistent with the present energetic analysis (Fig. 10). That study also emphasized the role of the hydrophobic effect in binding and, interestingly, a significant enthalpy loss occurred upon removal of the methyl group in abequose, consistent with the favorable vdW interaction energy of that monosaccharide with the antibody. In addition, acetylation on the C2 position of abequose on a tetrasaccharide lead to only a 0.5 kcal/mol loss in inhibitory activity, consistent with the small difference in the interaction energy of the O-acetylated and de-acetylated species with the antibody in the present study.
We found that binding to Se155-4 produces differences in the conformations sampled by the O-antigens as a function of O-acetylation. The de-acetylated species bound to the antibody in two conformations whereas O-acetylation, specifically on the α-L-rhamnose (5) monosaccharide, lead to only a single conformation in the bound state. We previously found that both species of OPS (de- and O-acetylated) predominately sample the same conformation cluster 1 when unbound (manuscript submitted). This same cluster was sampled by the bound de-acetylated antigen, however it was not sampled significantly by the bound O-acetylated antigen. However, the conformational flexibility of the protein is similar in the apo form and in the presence of the different antigens as revealed from root mean square fluctuation analysis (see Figure S7 in supporting information). These results indicate that a conformational selection mechanism occurs for the de-acetylated saccharide binding to antibody, in which the antibody can identify and select the pre-organized primary conformation of the unbound saccharide from its equilibrium fluctuations. Kinetically, this mechanism will typically lower the energetic penalty for binding to the antibody. In contrast, the bound acetylated saccharide mainly samples the cluster 2 conformation; however the saccharide does sample the cluster 1 conformation (1.6%) in the unbound state (Table 1). Thus, it can be deduced that binding of the O-acetylated saccharide to the Se155-4 also follows a conformational selection mechanism such that when the cluster 1 conformation is sampled binding occurs. Moreover, as the conformational heterogeneity is more restricted for the bound acetylated saccharide than its bound de-acetylated counterpart the entropy change is more unfavorable for binding of the acetylated form. Thus, a larger entropy penalty is required for binding of the acetylated saccharide than the de-acetylated form. To our knowledge, there have been no published studies that assessed binding of acetylated saccharide to Se155-4. Taken together these results indicate that the abequose plays an central role in antigenicity, as previously discussed,16-20 while O-acetylation can impact the conformational properties of the O-antigen when bound to antibody as was previously hypothesized.15, 21 Interestingly, this change in conformational sampling occurs primarily when bound to Se155-4 versus in the unbound state.
Our finding that Se155-4 interacts with both conformations of the de-acetylated species versus a single conformation for the O-acetylated OPS suggests a possible new mechanism for antigen specificity. The analyses herein regarding the contribution of the peripheral monosaccharides in the OPS unit to antibody binding could offer additional insight into the previously published study that found that the specificity of individual anti- S. Typhimurium OPS monoclonal antibodies was formed by a combination of antigenic determinants.22 They may additionally provide a framework for analysis of vaccine induced anti-carbohydrate antibodies, wherein different monoclonal components of the polyclonal immune response may exhibit unique modes of saccharide epitope binding and specificity. Additional studies will be needed to determine whether conformational preferences are present for other monoclonal antibodies specific for Salmonella serogroup B OPS, that may vary as a function of the acetylation status of the immunizing antigen, and also to determine the possible influence of the lipid A moiety on the saccharide conformational preference.
Supplementary Material
Acknowledgements
This work was supported by grants from NIH (GM070855) to ADM Jr. and computational support from the University of Maryland Computer-Aided Drug Design Center is acknowledged.
Footnotes
Supporting Materials
The PMF maps of the unbound saccharides in Figure S1, distance distribution of the hydrogen bonding pairs between antibody and saccharide in Figures S2-S5, conformational space represented by the RMSD distribution between the unbound O-acetylated and de-acetylated saccharides in Figure S6 and the conformational fluctuation of the antibody residues in Figure S7.
References
- 1.Ao TT, Feasey NA, Gordon MA, Keddy KH, Angulo FJ, Crump JA. Global Burden of Invasive Nontyphoidal Salmonella Disease, 2010. Emerg. Infect. Dis. 2015;21:941–949. doi: 10.3201/eid2106.140999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hendriksen RS, Vieira AR, Karlsmose S, Wong DMALF, Jensen AB, Wegener HC, Aarestrup FM. Global Monitoring of Salmonella Serovar Distribution from the World Health Organization Global Foodborne Infections Network Country Data Bank: Results of Quality Assured Laboratories from 2001 to 2007. Foodborne Pathog. Dis. 2011;8:887–900. doi: 10.1089/fpd.2010.0787. [DOI] [PubMed] [Google Scholar]
- 3.MacLennan CA, Levine MM. Invasive Nontyphoidal Salmonella Disease in Africa: Current Status. Expert Rev. Anti. Infect. Ther. 2013;11:443–446. doi: 10.1586/eri.13.27. [DOI] [PubMed] [Google Scholar]
- 4.Tapia MD, Tennant SM, Bornstein K, Onwuchekwa U, Tamboura B, Maiga A, Sylla MB, Sissoko S, Kourouma N, Toure A, et al. Invasive Nontyphoidal Salmonella Infections among Children in Mali, 2002–2014: Microbiological and Epidemiologic Features Guide Vaccine Development. Clin. Infect. Dis. 2015;61:S332–S338. doi: 10.1093/cid/civ729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feasey NA, Dougan G, Kingsley RA, Heyderman RS, Gordon MA. Invasive Non-Typhoidal Salmonella Disease: An Emerging and Neglected Tropical Disease in Africa. The Lancet. 2012;379:2489–2499. doi: 10.1016/S0140-6736(11)61752-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kingsley RA, Msefula CL, Thomson NR, Kariuki S, Holt KE, Gordon MA, Harris D, Clarke L, Whitehead S, Sangal V, et al. Epidemic Multiple Drug Resistant Salmonella Typhimurium Causing Invasive Disease in Sub-Saharan Africa Have a Distinct Genotype. Genome Res. 2009;19:2279–2287. doi: 10.1101/gr.091017.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lanzilao L, Stefanetti G, Saul A, MacLennan CA, Micoli F, Rondini S. Strain Selection for Generation of O-Antigen-Based Glycoconjugate Vaccines against Invasive Nontyphoidal Salmonella Disease. PLoS ONE. 2015;10:e0139847. doi: 10.1371/journal.pone.0139847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simon R, Levine MM. Glycoconjugate Vaccine Strategies for Protection against Invasive Salmonella Infections. Hum. Vacc. Immunother. 2012;8:494–498. doi: 10.4161/hv.19158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rondini S, Micoli F, Lanzilao L, Gavini M, Alfini R, Brandt C, Clare S, Mastroeni P, Saul A, MacLennan CA. Design of Glycoconjugate Vaccines against Invasive African Salmonella Enterica Serovar Typhimurium. Infect. Immun. 2015;83:996–1007. doi: 10.1128/IAI.03079-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goh YS, Clare S, Micoli F, Saul A, Mastroeni P, MacLennan CA. Monoclonal Antibodies of a Diverse Isotype Induced by an O-Antigen Glycoconjugate Vaccine Mediate in Vitro and in Vivo Killing of African Invasive Nontyphoidal Salmonella. Infect. Immun. 2015;83:3722–3731. doi: 10.1128/IAI.00547-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raetz CRH, Whitfield C. Lipopolysaccharide Endotoxins. Annu. Rev. Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Popoff MY, Le ML. World Health Organization Collaborating Centre for Reference and Research on Salmonella. Pasteur Institute; Paris, France: 2001. Antigenic Formulas of the Salmonella Serovars. [Google Scholar]
- 13.Micoli F, Ravenscroft N, Cescutti P, Stefanetti G, Londero S, Rondini S, MacLennan CA. Structural Analysis of O-Polysaccharide Chains Extracted from Different Salmonella Typhimurium Strains. Carbohydr. Res. 2014;385:1–8. doi: 10.1016/j.carres.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 14.Onsare RS, Micoli F, Lanzilao L, Alfini R, Okoro CK, Muigai AW, Revathi G, Saul A, Kariuki S, MacLennan CA, et al. Relationship between Antibody Susceptibility and Lipopolysaccharide O-Antigen Characteristics of Invasive and Gastrointestinal Nontyphoidal Salmonellae Isolates from Kenya. PLoS Negl Trop Dis. 2015;9:e0003573. doi: 10.1371/journal.pntd.0003573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Slauch JM, Mahan MJ, Michetti P, Neutra MR, Mekalanos JJ. Acetylation (O-Factor 5) Affects the Structural and Immunological Properties of Salmonella Typhimurium Lipopolysaccharide O Antigen. Infect. Immun. 1995;63:437–41. doi: 10.1128/iai.63.2.437-441.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bundle DR, Eichler E, Gidney MAJ, Meldal M, Ragauskas A, Sigurskjold BW, Sinnott B, Watson DC, Yaguchi M, Young NM. Molecular Recognition of a Salmonella Trisaccharide Epitope by Monoclonal Antibody Se155-4. Biochemistry. 1994;33:5172–5182. doi: 10.1021/bi00183a022. [DOI] [PubMed] [Google Scholar]
- 17.Bundle DR, Baumann H, Brisson J-R, Gagne SM, Zdanov A, Cygler M. The Solution Structure of a Trisaccharide-Antibody Complex: Comparison of NMR Measurements with a Crystal Structure. Biochemistry. 1994;33:5183–5192. doi: 10.1021/bi00183a023. [DOI] [PubMed] [Google Scholar]
- 18.Cygler M, Wu S, Zdanov A, Bundle DR, Rose DR. Recognition of a Carbohydrate Antigenic Determinant of Salmonella by an Antibody. Biochem. Soc. Trans. 1993;21:437–441. doi: 10.1042/bst0210437. [DOI] [PubMed] [Google Scholar]
- 19.Sigurskjold BW, Bundle DR. Thermodynamics of Oligosaccharide Binding to a Monoclonal Antibody Specific for a Salmonella O-Antigen Point to Hydrophobic Interactions in the Binding Site. J. Biol. Chem. 1992;267:8371–6. [PubMed] [Google Scholar]
- 20.Cygler M, Rose D, Bundle D. Recognition of a Cell-Surface Oligosaccharide of Pathogenic Salmonella by an Antibody Fab Fragment. Science. 1991;253:442–445. doi: 10.1126/science.1713710. [DOI] [PubMed] [Google Scholar]
- 21.Ronholm J, Zhang Z, Cao X, Lin M. Monoclonal Antibodies to Lipopolysaccharide Antigens of Salmonella Enterica Serotype Typhimurium Dt104. Hybridoma. 2011;30:43–52. doi: 10.1089/hyb.2010.0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elkins K, Metcalf ES. Monoclonal Antibodies Demonstrate Multiple Epitopes on the O Antigens of Salmonella Typhimurium Lps. J. Immunol. 1984;133:2255–60. [PubMed] [Google Scholar]
- 23.Zdanov A, Li Y, Bundle DR, Deng SJ, MacKenzie CR, Narang SA, Young NM, Cygler M. Structure of a Single-Chain Antibody Variable Domain (Fv) Fragment Complexed with a Carbohydrate Antigen at 1.7-Å Resolution. Proc. Natl. Acad. Sci. U. S. A. 1994;91:6423–6427. doi: 10.1073/pnas.91.14.6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang M, MacKerell AD., Jr. Conformational Sampling of Oligosaccharides Using Hamiltonian Replica Exchange with Two-Dimensional Dihedral Biasing Potentials and the Weighted Histogram Analysis Method (WHAM) J. Chem. Theory Comput. 2015;11:788–799. doi: 10.1021/ct500993h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang M, Huang J, MacKerell AD. Enhanced Conformational Sampling Using Replica Exchange with Concurrent Solute Scaling and Hamiltonian Biasing Realized in One Dimension. J. Chem. Theory Comput. 2015;11:2855–2867. doi: 10.1021/acs.jctc.5b00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang J, MacKerell AD., Jr. CHARMM36 All-Atom Additive Protein Force Field: Validation Based on Comparison to NMR Data. J. Comput. Chem. 2013;34:2135–2145. doi: 10.1002/jcc.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Best RB, Zhu X, Shim J, Lopes PEM, Mittal J, Feig M, MacKerell AD. Optimization of the Additive CHARMM All-Atom Protein Force Field Targeting Improved Sampling of the Backbone Φ, Ψ and Side-Chain χ1 and χ2 Dihedral Angles. J. Chem. Theory Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guvench O, Mallajosyula SS, Raman EP, Hatcher E, Vanommeslaeghe K, Foster TJ, Jamison FW, MacKerell AD., Jr. CHARMM Additive All-Atom Force Field for Carbohydrate Derivatives and Its Utility in Polysaccharide and Carbohydrate–Protein Modeling. J. Chem. Theory Comput. 2011;7:3162–3180. doi: 10.1021/ct200328p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guvench O, Hatcher E, Venable RM, Pastor RW, MacKerell AD., Jr. CHARMM Additive All-Atom Force Field for Glycosidic Linkages between Hexopyranoses. J. Chem. Theory Comput. 2009;5:2353–2370. doi: 10.1021/ct900242e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guvench O, Greene SN, Kamath G, Brady JW, Venable RM, Pastor RW, Mackerell AD., Jr. Additive Empirical Force Field for Hexopyranose Monosaccharides. J. Comput. Chem. 2008;29:2543–2564. doi: 10.1002/jcc.21004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neria E, Fischer S, Karplus M. Simulation of Activation Free Energies in Molecular Systems. J. Chem. Phys. 1996;105:1902–1921. [Google Scholar]
- 32.Reiher WE. Theoretical Studies of Hydrogen Bonding. Harvard University; Cambridge, MA: 1985. Ph.D. Thesis. [Google Scholar]
- 33.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 34.Choi Y, Deane CM. Fread Revisited: Accurate Loop Structure Prediction Using a Database Search Algorithm. Proteins: Struct., Funct., Bioinf. 2010;78:1431–1440. doi: 10.1002/prot.22658. [DOI] [PubMed] [Google Scholar]
- 35.Olsson MHM, Søndergaard CR, Rostkowski M, Jensen JH. Propka3: Consistent Treatment of Internal and Surface Residues in Empirical Pka Predictions. J. Chem. Theory Comput. 2011;7:525–537. doi: 10.1021/ct100578z. [DOI] [PubMed] [Google Scholar]
- 36.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K. Scalable Molecular Dynamics with Namd. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brooks BR, Brooks CL, III, Mackerell AD, Jr., Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, et al. CHARMM: The Biomolecular Simulation Program. J. Comput. Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eastman P, Friedrichs MS, Chodera JD, Radmer RJ, Bruns CM, Ku JP, Beauchamp KA, Lane TJ, Wang L-P, Shukla D, et al. OpenMM 4: A Reusable, Extensible, Hardware Independent Library for High Performance Molecular Simulation. J. Chem. Theory Comput. 2013;9:461–469. doi: 10.1021/ct300857j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoover WG. Canonical Dynamics: Equilibrium Phase-Space Distributions. Phys. Rev. A. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 40.Feller SE, Zhang Y, Pastor RW, Brooks BR. Constant Pressure Molecular Dynamics Simulation: The Langevin Piston Method. J. Chem. Phys. 1995;103:4613–4621. [Google Scholar]
- 41.Ryckaert J-P, Ciccotti G, Berendsen HJC. Numerical Integration of the Cartesian Equations of Motion of a System with Constraints: Molecular Dynamics of N-Alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 42.Darden T, York D, Pedersen L. Particle Mesh Ewald - an N. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 43.Vanommeslaeghe K, Yang M, MacKerell AD. Robustness in the Fitting of Molecular Mechanics Parameters. J. Comput. Chem. 2015;36:1083–1101. doi: 10.1002/jcc.23897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang M, Angles d'Ortoli T, Sawen E, Jana M, Widmalm G, MacKerell AD. Delineating the Conformational Flexibility of Trisaccharides from NMR Spectroscopy Experiments and Computer Simulations. Phys. Chem. Chem. Phys. 2016;18:18776–18794. doi: 10.1039/c6cp02970a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guvench O, MacKerell AD., Jr. Computational Fragment-Based Binding Site Identification by Ligand Competitive Saturation. PLoS Comput. Biol. 2009;5:e1000435. doi: 10.1371/journal.pcbi.1000435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raman EP, Yu W, Lakkaraju SK, MacKerell AD. Inclusion of Multiple Fragment Types in the Site Identification by Ligand Competitive Saturation (SILCS) Approach. J. Chem. Inf. Model. 2013;53:3384–3398. doi: 10.1021/ci4005628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Young NM, Watson DC, Cunningham AM, MacKenzie CR. The Intrinsic Cysteine and Histidine Residues of the Anti-Salmonella Antibody Se155-4: A Model for the Introduction of New Functions into Antibody-Binding Sites. Protein Eng. Des. Sel. 2014;27:383–390. doi: 10.1093/protein/gzu018. [DOI] [PubMed] [Google Scholar]
- 48.Deng SJ, MacKenzie CR, Hirama T, Brousseau R, Lowary TL, Young NM, Bundle DR, Narang SA. Basis for Selection of Improved Carbohydrate-Binding Single-Chain Antibodies from Synthetic Gene Libraries. Proc. Natl. Acad. Sci. U. S. A. 1995;92:4992–4996. doi: 10.1073/pnas.92.11.4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.